Significance
The study of membrane proteins is often hampered by their tendency to misfold when extracted by detergent. Here, we explore a detergent-free approach to isolating membrane proteins while retaining their native lipid environment, making use of an amphipathic polymer that solubilizes intact membrane patches in the form of nanodiscs. Using a potassium channel as a model protein, we show that these “native nanodiscs” are highly thermostable particles that are suitable for spectroscopic studies, allowing structural characterization of the protein in its native environment and direct analysis of the lipids in its immediate surroundings. We also demonstrate that the channel can be reconstituted from nanodiscs into planar lipid bilayers for functional characterization, thus making native nanodiscs an excellent alternative to detergent solubilization.
Keywords: membrane–protein solubilization, styrene-maleic acid copolymer, lipid–protein interactions, nanodisc, ion channels
Abstract
A major obstacle in the study of membrane proteins is their solubilization in a stable and active conformation when using detergents. Here, we explored a detergent-free approach to isolating the tetrameric potassium channel KcsA directly from the membrane of Escherichia coli, using a styrene-maleic acid copolymer. This polymer self-inserts into membranes and is capable of extracting membrane patches in the form of nanosize discoidal proteolipid particles or “native nanodiscs.” Using circular dichroism and tryptophan fluorescence spectroscopy, we show that the conformation of KcsA in native nanodiscs is very similar to that in detergent micelles, but that the thermal stability of the protein is higher in the nanodiscs. Furthermore, as a promising new application, we show that quantitative analysis of the co-isolated lipids in purified KcsA-containing nanodiscs allows determination of preferential lipid–protein interactions. Thin-layer chromatography experiments revealed an enrichment of the anionic lipids cardiolipin and phosphatidylglycerol, indicating their close proximity to the channel in biological membranes and supporting their functional relevance. Finally, we demonstrate that KcsA can be reconstituted into planar lipid bilayers directly from native nanodiscs, which enables functional characterization of the channel by electrophysiology without first depriving the protein of its native environment. Together, these findings highlight the potential of the use of native nanodiscs as a tool in the study of ion channels, and of membrane proteins in general.
Integral membrane proteins (MPs) are an abundant class of proteins that play key roles in a wide range of essential cellular processes (1). To facilitate their study in vitro, detergent molecules are commonly used to extract MPs out of their native lipid–bilayer environment (2). However, the use of detergents has some inherent disadvantages. Most importantly, even though there are promising developments to improve their properties (3, 4), the insufficient mimicking of a lipid bilayer by detergent micelles often leads to destabilization and rapid loss of function of the incorporated protein (5). For many functional and structural studies, it is thus necessary to reconstitute the MP into a more stabilizing environment; for example, by replacing the detergent with amphipathic polymers (amphipols) (6) or incorporating the MP into lipid nanodiscs with a surrounding protein scaffold (7). Both approaches have proven to be valuable tools for the study of structural and functional properties of MPs (8, 9); however, a limitation remains, as transfer of MPs into any of these systems requires initial solubilization by detergent.
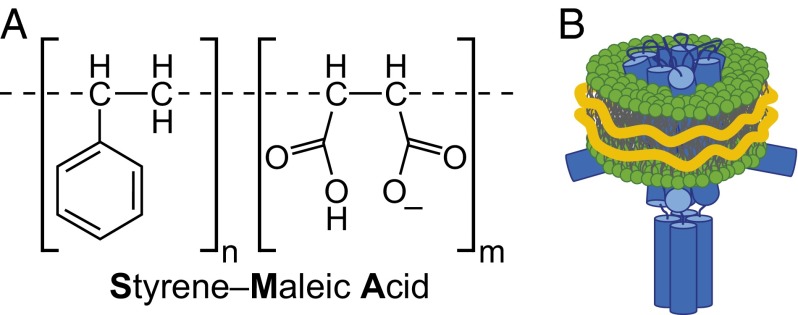
Recently, a detergent-free approach has been described using amphipathic styrene-maleic acid copolymers (SMAs) as an alternative to solubilize MPs directly from biological membranes in the form of nanodiscs, referred to as “Lipodisq” or SMA lipid particles (10–13) (Fig. 1). The mechanism of action of SMA differs fundamentally from that of detergents: instead of disrupting the lipid bilayer completely, SMA spontaneously self-inserts and extracts intact membrane patches in the form of discoidal particles that are stabilized by a SMA annulus (14, 15). Because these nanodiscs conserve a spatially delimited native biomembrane including MPs, we term them “native nanodiscs.” One of the main advantages of this system is the straightforward extraction protocol without the need for detergent. It has been shown that the SMA polymer is capable of directly extracting native nanodiscs containing large functional protein complexes from yeast (12), bacterial proteins involved in cell division (16) and photosynthesis (17), and several members of the ABC transporter family (13). The isolation of these proteins from a variety of different organisms suggests a general applicability of SMA solubilization for all MPs, irrespective of their expression host or native organism.
Fig. 1.
(A) Chemical structure of SMA polymers at neutral pH. For this study, a polymer with an average SMA ratio of n:m = 2:1 was used. (B) Schematic representation of a native nanodisc containing a KcsA tetramer (blue) and native lipids (green). The outer hydrophobic surface of the lipids is shielded by SMA (orange).
To further explore the potential of native nanodiscs, we used the SMA polymer to isolate an oligomeric bacterial membrane protein: the tetrameric potassium channel from Streptomyces lividans (KcsA) (18), expressed in Escherichia coli. KcsA is an ideal model protein for such studies because it is well-characterized and because reconstitution studies have shown that both its function and stability are strongly affected by lipid composition (19–21). In this work, we apply SMA to prepare and purify native nanodiscs with KcsA to compare the conformational properties and stability of the protein with those in detergent micelles. In addition, we use native nanodiscs to investigate preferential lipid–protein interactions by analyzing the composition of small patches of native membrane that are copurified with the protein. Finally, we study the functional properties of KcsA on reconstitution from native nanodiscs into a planar lipid bilayer system. Our results underscore the huge potential of SMA as a membrane-solubilizing agent, as well as the use of native nanodiscs as a membrane-mimetic system for biophysical studies on ion channels, and MPs in general.
Results
KcsA Can Be Solubilized and Purified in Native Nanodiscs.
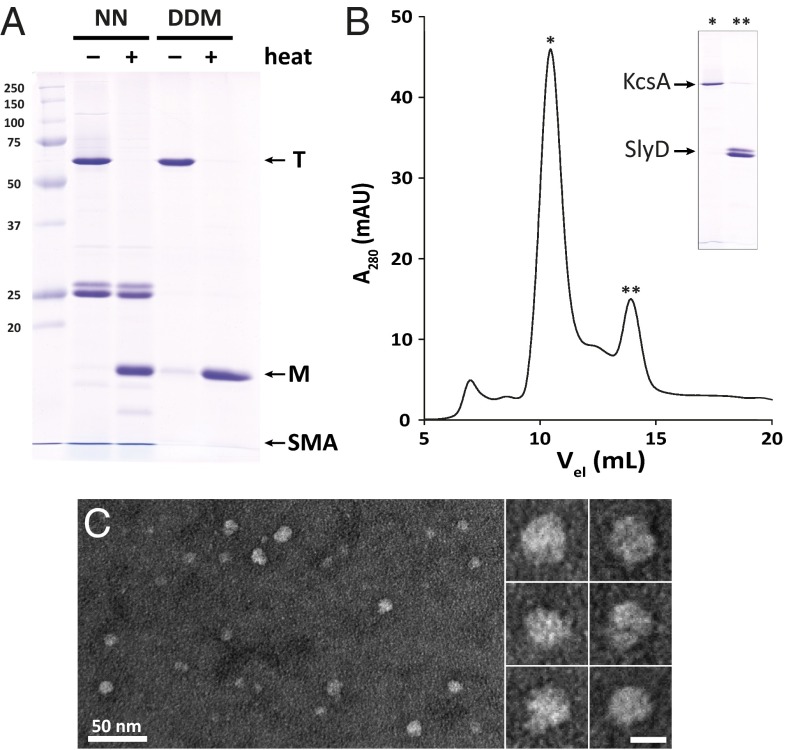
Preparations of KcsA in native nanodiscs were obtained by the addition of SMA polymer directly to lysed E. coli whole cells with overproduced His-tagged KcsA, followed by isolation of the KcsA-containing nanodiscs by Ni-affinity chromatography. SDS/PAGE analysis of purified KcsA in native nanodiscs shows a pronounced band at a molecular weight of ∼60 kDa (Fig. 2A, lane 2), similar to that observed with tetrameric KcsA purified in a standard protocol, using the nonionic detergent n-dodecyl-β-d-maltoside (DDM) (lane 4) (22). Exposing the samples to 95 °C in the presence of SDS results in a virtually complete transition to the monomeric form of KcsA, as evident from the appearance of a band at ∼17 kDa (Fig. 2A, lanes 3 and 5). Thus, KcsA can be isolated in native nanodiscs as a stable tetramer that is resistant to SDS at room temperature, similar to what is described for KcsA in detergent (23). In the isolations of KcsA in nanodiscs, we observed an additional pronounced double band of varying intensity at ∼25 kDa that was heat-stable. Mass spectrometric analysis on a tryptic-digested sample of this band identified it as SlyD, a stress-up-regulated, soluble 20.9-kDa protein endogenous to E. coli that contains multiple histidine residues in its C terminus (24). This common contaminant of Ni-affinity purifications could be removed completely by size-exclusion chromatography (Fig. 2B) (25). The SMA polymer itself migrates at a low apparent molecular weight because of its high intrinsic negative charge and is observed as a blue-stained front on the depicted gels (Fig. 2A, lanes 2 and 3).
Fig. 2.
Purification of KcsA in native nanodiscs. (A) SDS/PAGE analysis of purified KcsA in native nanodiscs (NN) and DDM micelles at room temperature (−) and after incubation at 95 °C (+). Complete transitions from tetrameric (T) to monomeric (M) state are visible. (B) Size exclusion chromatogram showing effective separation of KcsA (*) from the soluble contaminant SlyD (**). (C) Negative stain transmission electron micrograph of native nanodiscs with KcsA after size exclusion chromatography. Round particles of an average size of 10 ± 2 nm are visible. (Scale bar in the enlarged images, 10 nm.)
The size and shape of purified KcsA-containing nanodiscs were further investigated, using negative-stain transmission electron microscopy (Fig. 2C). The particles exhibit a round shape with a fairly homogeneous size distribution, with diameters of 10 ± 2 nm, which is in agreement with previous studies on native nanodiscs (10, 12) and on protein-free nanodiscs obtained by SMA solubilization of synthetic liposomes (14, 15).
Native Nanodiscs Surpass Detergent Micelles in Conserving Structural Stability of KcsA.
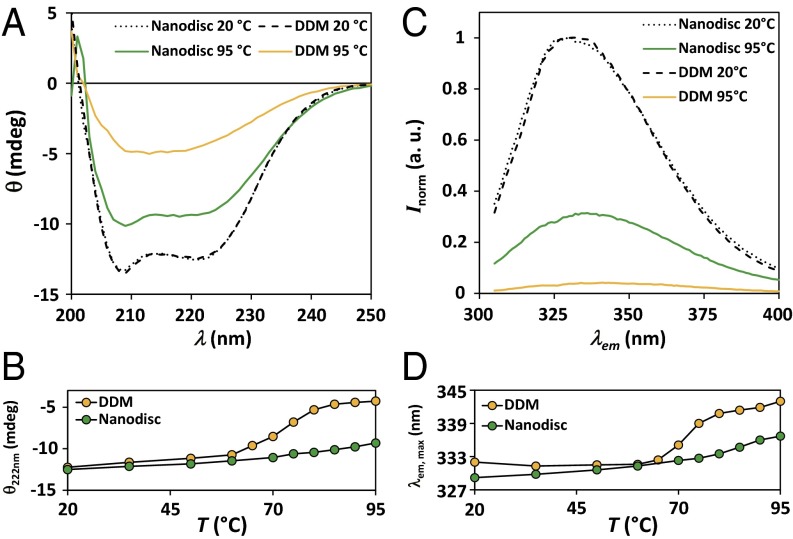
The purity of KcsA nanodiscs after size-exclusion chromatography renders them directly suitable for studies with circular dichroism (CD) and tryptophan fluorescence spectroscopy. Far-UV CD spectra of KcsA in native nanodiscs and DDM micelles at different temperatures are depicted in Fig. 3A. The shapes of the spectra acquired at room temperature are virtually indistinguishable and show the features of a predominantly α-helical protein with minima at around 208 and 222 nm, in good agreement with previous studies (20, 26) and available X-ray structures (27). Upon heat exposure, differences between the spectra of KcsA in the different systems become visible. At 95 °C, KcsA in DDM shows a loss of ∼50% in secondary structure, with a well-defined transition at 72 °C, as measured by the ellipticity at 222 nm (Fig. 3B). In contrast, KcsA in nanodiscs lacks a genuine transition as a function of temperature, and the amount of helicity decreases only slightly with increasing temperature, with a loss of ∼20% at 95 °C. In addition, the characteristic features of α-helical proteins are qualitatively conserved over the whole temperature range for KcsA in nanodiscs, whereas they are lost in DDM micelles above the transition.
Fig. 3.
Comparison of the thermal stability of KcsA in different environments. (A) Circular dichroism spectra of KcsA in native nanodiscs and DDM micelles at 20 °C and 95 °C. Data are offset corrected averages of 8–10 scans. (B) Corresponding thermal unfolding traces monitored by the ellipticity at 222 nm. Data are averages of two experiments with errors too small to be depicted. Solid lines are depicted to guide the eye. (C) Normalized fluorescence intensity of the intrinsic tryptophans of KcsA at 20 °C and 95 °C. Data are averages of three scans normalized to the intensity at 330 nm of the respective spectra at 20 °C. (D) Corresponding thermal unfolding traces monitored by the wavelength of maximum fluorescence emission. Solid lines are depicted to guide the eye.
Similar observations were made when the intrinsic fluorescence of KcsA in different environments was monitored. Increasing the temperature resulted in a strong decline in intensity of the recorded emission spectra of KcsA that was more pronounced in detergent than in nanodiscs (Fig. 3C). At 20 °C, the maximum emission wavelengths on excitation at 295 nm were ∼329 and ∼332 nm for KcsA in nanodiscs and micelles, respectively. The blue-shifted emission maximum in nanodiscs can be explained by the decreased solvent exposure of some tryptophans resulting from better shielding by conserved lipid molecules than is seen in detergent micelles. The thermal stability of the tertiary structure of KcsA was investigated by following the shift in the emission maximum as a function of temperature. Consistent with the CD analysis, KcsA in detergent micelles shows a genuine transition at ∼70 °C, whereas in nanodiscs, changes are less pronounced, without a clear transition for KcsA (Fig. 3D). The emission maximum of micellar KcsA at 95 °C is ∼343 nm, and is thus considerably more red-shifted than in nanodiscs (∼337 nm). Virtually the same results were obtained when the intensity of the fluorescence at 330 nm was monitored as function of temperature. Thus, the tryptophan residues of detergent-solubilized KcsA show larger conformational changes than those in nanodiscs, where the tertiary structure is more stable even at high temperatures.
The higher stability of KcsA in nanodiscs compared with DDM micelles is further supported by the observation of a faint monomer band of KcsA purified in DDM on SDS/PAGE at room temperature that is absent for nanodiscs (Fig. 2A, lanes 2 and 4). Upon storage at 4 °C, we observed an increase of this difference over time. Tetrameric KcsA in nanodiscs was stable over months, with a constant tetramer fraction above 95%, whereas the tetramer fraction of DDM-solubilized protein decreased to ∼60% after 6 weeks. In line with this, we found that neither freeze thawing nor lyophilization of KcsA in nanodiscs affects the integrity of the quaternary structure, whereas DDM-solubilized tetramers dissociate to some extent (Fig. S1). Thus, using a complementary set of techniques, it is shown that native nanodiscs serve as a good tool to conserve the structural stability of the oligomeric KcsA.
A possible explanation for the lower stability of detergent-solubilized KcsA is the dynamic equilibrium between monomeric and micellar DDM (2), which may promote the dissociation of the KcsA tetramer by segregation of monomers into separate micelles, thus favoring further unfolding. In contrast, native nanodiscs are stable particles, and no free SMA is needed in solution to maintain their stability. Thus, the more rigid structure of the particles could convey the higher stability of KcsA. However, it should also be noted that the mere presence of lipids can increase the stability of KcsA, as suggested by studies on reconstituted protein (20, 22). Conserving and stabilizing a lipid environment around MPs during their extraction by SMA polymers thus directly yields particles that maintain a higher stability of the trapped protein than detergent micelles.
Biochemical Analysis of the Local Lipid Environment of KcsA Reveals Enrichment of Anionic Lipids.
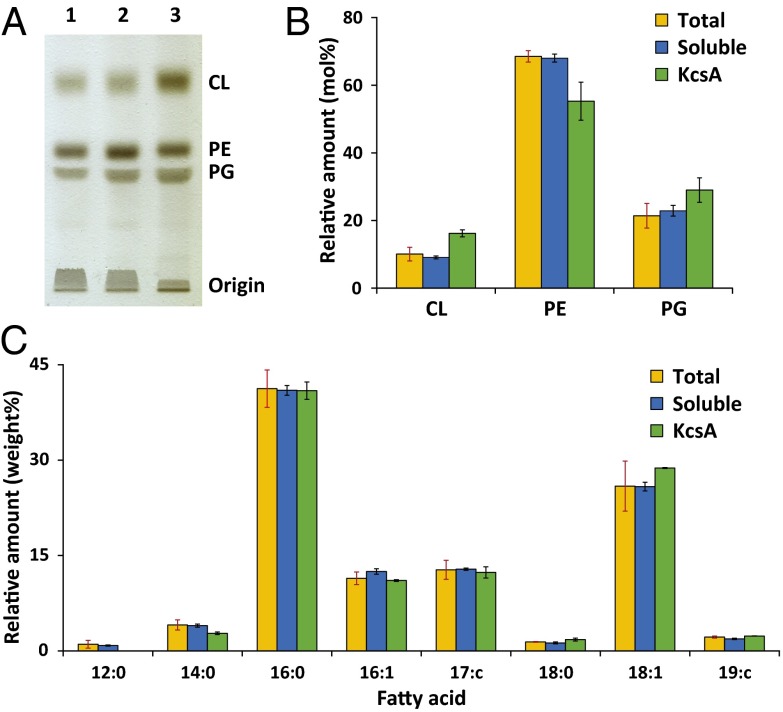
The isolation of intact nanopatches of biological membranes in native nanodiscs provides a unique opportunity to investigate the immediate local lipid environment of KcsA, and thus characterize preferential lipid–protein interactions. Using TLC, the isolated lipids from native nanodiscs can be separated according to headgroup. Fig. 4A shows a representative chromatogram with lipid samples from the total cell lysate, the complete SMA-solubilized fraction, and purified KcsA nanodiscs. In all samples, three pronounced bands are visible that can be identified by comparison with synthetic reference lipids as the zwitterionic phosphatidylethanolamine and the anionic lipids phosphatidylglycerol and cardiolipin. The lipid composition of independent bacterial cultures was found to be very sensitive to the growth conditions and time, resulting in a variation of absolute values in the range of 5–10 mol%. For comparison, data from each independent nanodisc preparation were therefore normalized to the corresponding total cell lysate. Quantitative analysis of the intensity of the chromatogram bands (Fig. 4B) shows that the lipid compositions of the total cell lysate and the soluble fraction are very similar and in good agreement with other studies on lipids of E. coli K 12 wild-type strains (28, 29). Thus, SMA does not preferentially solubilize any specific lipids in the E. coli membrane. Strikingly, KcsA nanodiscs show higher amounts of anionic lipids, with increases of 36% in phosphatidylglycerol and 61% in cardiolipin content compared with the total extract. This isolation of enriched lipid species can hence be attributed to the preferential interaction of anionic lipids with KcsA.
Fig. 4.
Analysis of coisolated lipids in native nanodiscs. (A) TLC on extracted lipids from lysed whole bacterial cells after addition of SMA (1), complete soluble fraction separated by ultracentrifugation (2), and native nanodiscs with KcsA purified by Ni-affinity chromatography (3). (B) Molar composition of the extracted lipid species according to headgroup of total lysate (orange), soluble fraction (blue), and KcsA nanodiscs (green). Data are shown as average mol percentages with SDs (red error bars) of three independent nanodisc isolations. Data for the soluble fraction and KcsA nanodiscs were normalized to the corresponding total lysate. Black error bars denote SDs of the change compared with total lysate. (C) Gas chromatography analysis of the fatty acid composition of all isolated lipids. Depicted data are averages of two independent experiments. Errors are given as in B.
Preferential lipid–protein interactions were further analyzed by investigating the composition of the acyl chains of all isolated lipids (Fig. 4C). Lipids from all samples predominantly contained palmitic (16:0) and cis-vaccenic acid (18:1) chains with a combined weight percentage of ∼70%, in agreement with other studies (28). In our analysis, no strong enrichment of specific acyl chains could be detected when comparing the lipids that were copurified with KcsA with the total extract or with the SMA-solubilized fraction. Only the amount of short lauric (12:0) and myristic acid (14:0) chains appeared to be slightly decreased in purified KcsA nanodiscs in favor of a slightly higher cis-vaccenic acid content. These results suggest a minor preference of KcsA for lipids with longer chains, possibly to promote hydrophobic matching (30, 31).
Reconstitution of KcsA into Planar Lipid Bilayers Enables Functional Characterization.
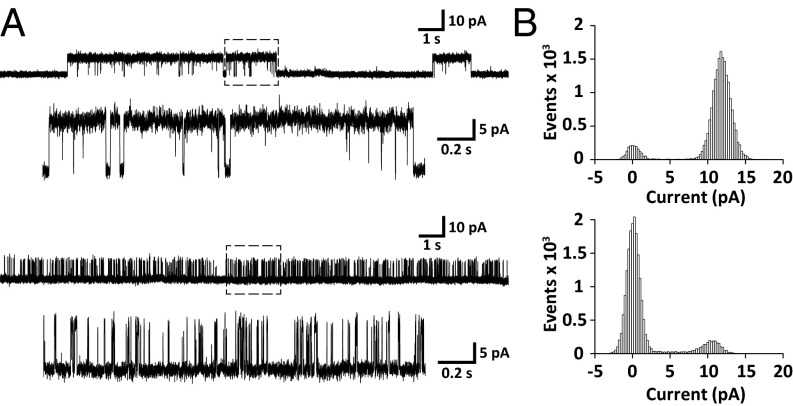
A final challenge remains in assessing the functionality of KcsA in native nanodiscs. Because analysis of ion transport is not possible in nanodiscs, we attempted to reconstitute the channel into a compartment-forming bilayer system that enables investigations of protein-facilitated potassium conductivity across membranes. To this end, we used a planar lipid bilayer setup that has been used successfully for functional studies on KcsA delivered in protein-stabilized nanodiscs (32). Seconds to minutes after the addition of native nanodiscs with KcsA, single-channel conductivity was observed as transient discrete current changes with an amplitude of 10–15 pA (Fig. 5), implying spontaneous fusion events of single nanodiscs with the bilayer. The recorded traces show both high and low open probability states that are characteristic for KcsA monitored under similar conditions by single-channel recordings of KcsA fused from liposomes (33) or incorporated via a cell-free expression protocol (34). The results are also comparable with those obtained by patch clamp studies of giant unilamellar vesicles with KcsA reconstituted from detergent (35). Together, this suggests identical functional properties of KcsA reconstituted from native nanodiscs compared with standard reconstitution protocols. Upon addition of the K+-channel blocker tetraethylammonium, we observed a reversible complete depletion of all opening events (Fig. S2), further confirming the presence of reconstituted KcsA in the bilayer. Native nanodiscs thus may serve as a powerful alternative to conventional approaches for the functional reconstitution of ion channels.
Fig. 5.
Functional characterization of KcsA reconstituted from native nanodiscs into a planar lipid bilayer of E. coli polar lipid extract. (A) Typical single-channel current traces on addition of KcsA nanodiscs to a setup with a symmetric 150-mM KCl solution at +100 mV. The top part represents a channel in the high open probability mode that is characterized by long opening dwell times, as shown in the enlarged section indicated by a box of dashed lines. Channels in the low open probability mode exhibit a distinctly different pattern, with short opening times resulting in sequences of bursts (Bottom). (B) All-point histograms for open/closed distribution of channels with high (Top) and low (Bottom) open probability calculated from the enlarged 2-s single-channel current traces from A.
Discussion
We have shown that native nanodiscs with embedded KcsA are formed spontaneously from bacterial cells upon addition of SMA polymer and that, using Ni-affinity and size-exclusion chromatography, sufficient protein can be purified in nanodiscs for extensive biophysical characterization. The isolation of KcsA in this manner is less cumbersome than standard detergent protocols and has the additional advantage of conserving the native conformation of the protein in a stabilizing environment comprising native lipids. Our data suggest that native nanodiscs in general convey a higher stability of the incorporated protein than detergent micelles, in agreement with previous findings on the thermostability of an ABC transporter (13) and photosynthetic reaction centers (17). In addition, as shown in this study, the coisolation of MPs with a patch of biological membrane allows for the analysis of preferential interactions and further facilitates structural and functional studies on purified protein in a (near) native state, as discussed next.
Insights into preferential lipid–protein interactions are important to understand structural and functional properties of MPs in a membrane environment. However, detailed information is not easily obtained with standard methods. Here, we exploited the extraction of intact nanopatches of biological membranes by the SMA polymer to directly identify preferential lipid–protein interactions that allow approximations of the situation in vivo. Analysis of the lipid composition revealed an enrichment of anionic lipids in close proximity to KcsA. This finding is unlikely to be an artifact of the extraction protocol because we found that none of the three main E. coli lipid species is preferentially incorporated into nanodiscs. This is intriguing, as the SMA polymer has a high negative charge density at pH 8.0 because of its many carboxylic acid moieties, which could be expected to lead to electrostatic repulsion of anionic lipids. The absence of a measurable effect of this repulsion hence emphasizes the promiscuity of the polymer with respect to lipid species in biological membranes. Similarly, SMA did not preferentially solubilize lipids containing specific fatty acids. This indicates that lipid solubilization by SMA is applicable to all phospholipids in any membrane, irrespective of headgroup or acyl chains, as further supported by the successful extraction of all major phospholipids of the mitochondrial membrane in yeast (12), as well as the membrane of the photosynthetic bacterium Rhodobacter sphaeroides (17).
The importance of anionic lipids for KcsA functionality and their close association with the channel has been well established by a wealth of studies using fluorescence quenching by brominated lipids (33), mass spectrometry (36), electrophysiology (33, 37, 38), and molecular dynamic simulations (39, 40). In addition, the diacylglycerol fragment found in crystal structures of KcsA was attributed to phosphatidylglycerol (19), and it has been shown that anionic lipids strongly stabilize the KcsA tetramer against dissociation by both heat (20, 22) and small fluorinated alcohols (21). In these studies, KcsA was purified in detergent, and preferential interactions were either deduced from the copurification of single tightly bound lipids or were observed indirectly after reconstitution into synthetic lipid bilayer systems. In contrast, the method described in this work facilitates direct biochemical analysis of preferential lipid–protein interactions that involve the native annular lipid environment. This paves the way for a new application to study specific lipid–protein interactions by using the SMA polymer to isolate nanodiscs with protein that has been conventionally reconstituted into bilayers of well-defined composition. By systematically varying the composition of these bilayers, one can then obtain very detailed information on the specificity of KcsA or other proteins for both lipid headgroups and acyl chains. Aside from assessing lipid–protein interactions, native nanodiscs offer the important additional advantage of copurifying proteins or other molecules that weakly interact with the target MP, providing unique insights into the composition of its immediate membrane environment. This feature has been used very recently for the isolation of a detergent-labile complex of penicillin-binding proteins in Staphylococcus aureus (16). Native nanodiscs thus are a powerful tool to study preferential interactions of MPs both in vitro and in vivo.
In addition to purification of MPs and analysis of their preferential interactions, native nanodiscs also allow structural and functional characterization. Here, we used CD and fluorescence spectroscopy to study the thermostability of KcsA in a straightforward way. Moreover, their small size makes native nanodiscs, in principle, suitable for characterization by the full range of biophysical techniques, including NMR spectroscopy, that have been successfully applied to MPs incorporated into similar lipid nanodiscs that are stabilized by scaffold proteins instead of SMA (9). Native nanodiscs are also promising tools for studies on MPs in the rapidly developing field of single-particle cryo-electron microscopy. The use of amphipols to stabilize MPs has recently led to a high-resolution de novo structure determination of a cation channel at a resolution of 3.4 Å (41), breaking the side-chain resolution barrier. Together with promising studies using scaffold-protein nanodiscs (42, 43), as well as initial studies with native nanodiscs (44), these new developments suggest that native nanodiscs may become a powerful alternative to enable high-resolution structural characterization of MPs in their native environment.
Several proteins have so far been functionally characterized in native nanodiscs by investigating their ligand-binding (13) and optical (10–12, 17) properties, as well as their enzymatic activity (10, 12). Our data demonstrate that native nanodiscs can be also used to directly reconstitute KcsA into planar lipid bilayers, allowing electrophysiologic characterization of single channels in membranes with well-defined composition. To the best of our knowledge, this represents the first evidence of purification and transfer of an MP from the membrane of living cells to synthetic lipid bilayers without being deprived of its native lipid environment during any stage in the procedure. The ability of native nanodiscs to fuse with bilayers can be considered a promising first step toward enabling crystallization trials of MPs in a near-native environment. Other prospects of successful reconstitution include a plethora of assays available for MPs in liposomes and supported bilayers in applications that require a well-defined lipid environment. With the inherent advantage of conserving the native lipid environment of MPs, native nanodiscs constitute a highly promising and convenient alternative to detergent solubilization and may lead the way toward an in situ approach for structural and functional studies on MPs, including pharmacologic assays.
Materials and Methods
Materials and SMA Preparation.
All chemicals and enzymes were purchased from Sigma-Aldrich unless otherwise indicated. DDM was from Affymetrix, and all reference lipids used were from Avanti Polar Lipids. The used polymer was SMA2000, a styrene-maleic anhydride copolymer with a molar styrene maleic anhydride ratio of 2:1, a weight average molecular weight of 7.5 kDa, and a molar mass dispersity (polydispersity index) of 2.5 (Cray Valley). Conversion into SMA was achieved by hydrolysis in 1 M KOH and reflux for 2 h while heating the suspension to 100 °C. Subsequently, the polymer was precipitated by the addition of HCl and washed 6 times with 100 mM HCl to remove K+ ions. The sample was lyophilized, and hydrolysis was confirmed by Fourier transform infrared spectroscopy (17). SMA stock solutions were prepared by dissolving 6% (wt/vol) SMA powder in 50 mM unadjusted Tris buffer with gradual addition of NaOH solution until the pH reached the neutral range. The solution was then stored at −20 °C and adjusted to pH 8.0 and to the desired volume after thawing.
Gene Expression and Protein Purification.
KcsA was produced as described earlier (22). Cell pellets were resuspended in 50 mM Tris buffer at pH 8.0 containing 600 mM NaCl and 30 mM KCl (10 mL per 2 g wet cell weight) and incubated with 10 μg/mL lysozyme and 5 μg/mL DNase for 45 min on ice. The suspension was aliquoted and mixed 1:1 with 6% (wt/vol) SMA in 50 mM Tris at pH 8 for the preparation of native nanodiscs or 1:1 with 50 mM Tris buffer at pH 8 for detergent solubilization, respectively. The cell suspension with SMA was pushed through a disruptor at a pressure of 215 MPa and incubated overnight with gentle agitation at room temperature, followed by 45 min of centrifugation at 100,000 × g. SDS/PAGE analysis of supernatant and pellet after centrifugation revealed that 70–80% of the total amount of tetrameric KcsA was solubilized by SMA. The supernatant was then incubated for at least 4 h at 4 °C with 2.5 mL HisPur Ni-NTA agarose beads (Thermo Scientific) per liter of culture medium. The beads were transferred to a gravity-flow column and washed three times each with buffers containing 10 and 50 mM imidazole. Protein in native nanodiscs was then eluted with buffer containing 300 mM imidazole, yielding 1–2 mg protein per liter of culture medium on average, estimated by SDS/PAGE analysis and densitometric comparison of the protein bands after Coomassie staining with a gradient of BSA of known concentration, using the Quantity One software (Biorad). For biophysical studies, the pooled eluted fractions were loaded on a Superdex 200 10/300 GL size exclusion column (GE Healthcare) connected to an Äkta Prime Plus chromatography system (GE Healthcare) to separate the protein-containing nanodiscs from contaminating soluble proteins and free polymer. Thereby, the buffer was exchanged to 10 mM Tris at pH 8, 100 mM NaCl, and 5 mM KCl, which was used in all subsequent experiments.
Detergent purification was performed on aliquots of lysed cells, as described earlier (22), with the difference of using Tris buffer at pH 8.0 instead of Hepes. Eluted protein was extensively dialyzed against 1 mM DDM in the same buffer that was used for nanodiscs, using a dialysis membrane with a 50-kDa molecular weight cutoff. Protein concentration was determined by the absorption at 280 nm, using a calculated extinction coefficient of 34,950 M−1 ⋅ cm−1 (45). For protein in native nanodiscs, this can only be considered an estimate, as the phenyl groups of SMA contribute to the absorption in this wavelength range.
Spectroscopy.
Far-UV CD experiments were performed on a J-810 spectropolarimeter (Jasco) with a Peltier thermo control element (Jasco), using Teflon-sealed, polarimetrically checked quartz glass cuvettes with an optical pathlength of 1 mm and a volume of 350 μL (Hellma Analytics). Experimental parameters included a wavelength increment of 1 nm, a scan speed of 20 nm/min, a response time of 4 s, and a KcsA concentration of 0.06 mg/mL, as determined for DDM-solubilized protein. KcsA in native nanodiscs was diluted such that samples had the same signal intensity as protein in DDM, correcting for inaccuracies in concentration determination. Samples were allowed to equilibrate for 30 min at the desired temperature before measurement. All resulting spectra are buffer- and offset-corrected averages of 8–10 scans in the range of 200–250 nm.
Tryptophan fluorescence measurements were performed on a Cary Eclipse spectrofluorometer (Varian) equipped with a thermo control unit (Varian) in sealed quartz glass cuvettes with optical path lengths of 4 × 10 mm and an inner volume of 1.4 mL (Hellma). The excitation light beam had a wavelength of 295 nm at a slit size of 5 nm, and the fluorescence emission was recorded in the range of 300–400 nm, using a slit size of 5 nm, a wavelength increment of 1 nm, a scan speed of 60 nm/min, and an integration time of 1 s. The KcsA concentration was 0.01 mg/mL. All samples were allowed to equilibrate for 30 min at the desired temperature. Spectra were recorded in triplicate and corrected for buffer contribution before analysis by nonlinear least squares fitting to a bimodal log-normal distribution, using the Excel add-in Solver (Frontline Systems) (46).
Lipid Analysis.
Before lipid isolation, KcsA-containing native nanodiscs that were eluted from Ni-NTA beads were washed with buffer on spin columns with a molecular weight cutoff of 30 kDa to remove imidazole. Lipids were then extracted according to a modified version of the method of Bligh and Dyer (47) and analyzed by quantitative TLC, as well as gas chromatography (for details, see SI Materials and Methods).
Electrophysiology.
Single-channel recordings of KcsA were performed on a Compact setup for planar lipid bilayer electrophysiology (Ionovation) connected to an EPC 10 amplifier (HEKA). Lipid bilayer formation was achieved by painting E. coli polar lipid extract dissolved in n-decane (50 mg/mL) over a 200-μm hole in a Teflon-septum separating two compartments. Both compartments contained 150 mM KCl solution that was buffered with 10 mM Hepes to pH 7.0 in the cis compartment and 10 mM succinic acid to pH 4.0 in the trans compartment, respectively. After channel insertion, the conductivity at a constant voltage of +100 mV was recorded for several minutes, using the PatchMaster software (HEKA). Data were sampled at 10 kHz and digitally filtered at 1 kHz. All measurements were performed at 22 °C.
For a more detailed description of the method used for incorporation of KcsA channels into planar bilayers as well as electron microscopy, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mohammed Jamshad and Rosemary Parslow for help with the initial trials of solubilization of KcsA in native nanodiscs. We are indebted to Mirjam Damen for performing the mass spectrometric analysis on the SlyD protein and Hans Meeldijk for help with the transmission electron microscopy measurements. We further thank Ruud Cox for help with the gas chromatography analysis, Hugo van den Hoek for assistance with size-exclusion chromatography, and Cray Valley for their kind gift of SMA2000 polymer. Financial support received from the seventh framework program of the European Union (Initial Training Network “ManiFold,” Grant 317371 to J.M.D.) and from the Netherlands Organisation for Scientific Research (Grants 700.11.334 and 700.58.102 to E.A.W.v.d.C. and M.B.), as well as via the research program of the Foundation for Fundamental Research on Matter (to S.S.), is gratefully acknowledged. T.R.D. acknowledges support from the Biotechnology and Biological Sciences Research Council (Grants BB/J017310/1, BB/I020349/1 and BB/G010412/1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416205112/-/DCSupplemental.
References
- 1.von Heijne G. The membrane protein universe: What’s out there and why bother? J Intern Med. 2007;261(6):543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 2.Garavito RM, Ferguson-Miller S. Detergents as tools in membrane biochemistry. J Biol Chem. 2001;276(35):32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 3.McQuade DT, et al. Rigid amphiphiles for membrane protein manipulation. Angew Chem Int Ed Engl. 2000;39(4):758–761. [PubMed] [Google Scholar]
- 4.Chae PS, et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7(12):1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowie JU. Stabilizing membrane proteins. Curr Opin Struct Biol. 2001;11(4):397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]
- 6.Tribet C, Audebert R, Popot JL. Amphipols: Polymers that keep membrane proteins soluble in aqueous solutions. Proc Natl Acad Sci USA. 1996;93(26):15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Civjan NR, Bayburt TH, Schuler MA, Sligar SG. Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques. 2003;35(3):556–560, 562–563. doi: 10.2144/03353rr02. [DOI] [PubMed] [Google Scholar]
- 8.Popot JL. Amphipols, nanodiscs, and fluorinated surfactants: Three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 9.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584(9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knowles TJ, et al. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J Am Chem Soc. 2009;131(22):7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 11.Orwick-Rydmark M, et al. Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano Lett. 2012;12(9):4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 12.Long AR, et al. A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC Biotechnol. 2013;13:41. doi: 10.1186/1472-6750-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gulati S, et al. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem J. 2014;461(2):269–278. doi: 10.1042/BJ20131477. [DOI] [PubMed] [Google Scholar]
- 14.Orwick MC, et al. Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angew Chem Int Ed Engl. 2012;51(19):4653–4657. doi: 10.1002/anie.201201355. [DOI] [PubMed] [Google Scholar]
- 15.Jamshad M, et al. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. NanoResearch. 2014 doi: 10.1007/s12274-014-0560-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulin S, et al. Surfactant-free purification of membrane protein complexes from bacteria: Application to the staphylococcal penicillin-binding protein complex PBP2/PBP2a. Nanotechnology. 2014;25(28):285101. doi: 10.1088/0957-4484/25/28/285101. [DOI] [PubMed] [Google Scholar]
- 17.Swainsbury DJK, Scheidelaar S, van Grondelle R, Killian JA, Jones MR. Bacterial reaction centers purified with styrene maleic Acid copolymer retain native membrane functional properties and display enhanced stability. Angew Chem Int Ed Engl. 2014;53(44):11803–11807. doi: 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schrempf H, et al. A prokaryotic potassium ion channel with two predicted transmembrane segments from Streptomyces lividans. EMBO J. 1995;14(21):5170–5178. doi: 10.1002/j.1460-2075.1995.tb00201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valiyaveetil FI, Zhou Y, MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41(35):10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 20.Triano I, et al. Occupancy of nonannular lipid binding sites on KcsA greatly increases the stability of the tetrameric protein. Biochemistry. 2010;49(25):5397–5404. doi: 10.1021/bi1003712. [DOI] [PubMed] [Google Scholar]
- 21.Raja M, Spelbrink REJ, de Kruijff B, Killian JA. Phosphatidic acid plays a special role in stabilizing and folding of the tetrameric potassium channel KcsA. FEBS Lett. 2007;581(29):5715–5722. doi: 10.1016/j.febslet.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 22.van Dalen A, Hegger S, Killian JA, de Kruijff B. Influence of lipids on membrane assembly and stability of the potassium channel KcsA. FEBS Lett. 2002;525(1-3):33–38. doi: 10.1016/s0014-5793(02)03061-2. [DOI] [PubMed] [Google Scholar]
- 23.Cortes DM, Perozo E. Structural dynamics of the Streptomyces lividans K+ channel (SKC1): Oligomeric stoichiometry and stability. Biochemistry. 1997;36(33):10343–10352. doi: 10.1021/bi971018y. [DOI] [PubMed] [Google Scholar]
- 24.Wülfing C, Lombardero J, Plückthun A. An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J Biol Chem. 1994;269(4):2895–2901. [PubMed] [Google Scholar]
- 25.Parsy CB, Chapman CJ, Barnes AC, Robertson JF, Murray A. Two-step method to isolate target recombinant protein from co-purified bacterial contaminant SlyD after immobilised metal affinity chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;853(1-2):314–319. doi: 10.1016/j.jchromb.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 26.van den Brink-van der Laan E, Chupin V, Killian JA, de Kruijff B. Stability of KcsA tetramer depends on membrane lateral pressure. Biochemistry. 2004;43(14):4240–4250. doi: 10.1021/bi036129d. [DOI] [PubMed] [Google Scholar]
- 27.Uysal S, et al. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci USA. 2009;106(16):6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raetz CR. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol Rev. 1978;42(3):614–659. doi: 10.1128/mr.42.3.614-659.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morein S, Henricson D, Rilfors L. Separation of inner and outer membrane vesicles from Escherichia coli in self-generating Percoll gradients. Anal Biochem. 1994;216(1):47–51. doi: 10.1006/abio.1994.1006. [DOI] [PubMed] [Google Scholar]
- 30.Williamson IM, Alvis SJ, East JM, Lee AG. Interactions of phospholipids with the potassium channel KcsA. Biophys J. 2002;83(4):2026–2038. doi: 10.1016/S0006-3495(02)73964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Killian JA. Hydrophobic mismatch between proteins and lipids in membranes. Biochim Biophys Acta. 1998;1376(3):401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee S, Nimigean CM. Non-vesicular transfer of membrane proteins from nanoparticles to lipid bilayers. J Gen Physiol. 2011;137(2):217–223. doi: 10.1085/jgp.201010558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marius P, et al. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys J. 2008;94(5):1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friddin MS, et al. Single-channel electrophysiology of cell-free expressed ion channels by direct incorporation in lipid bilayers. Analyst (Lond) 2013;138(24):7294–7298. doi: 10.1039/c3an01540h. [DOI] [PubMed] [Google Scholar]
- 35.Chakrapani S, Cordero-Morales JF, Perozo E. A quantitative description of KcsA gating II: Single-channel currents. J Gen Physiol. 2007;130(5):479–496. doi: 10.1085/jgp.200709844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demmers JAA, van Dalen A, de Kruijff B, Heck AJR, Killian JA. Interaction of the K+ channel KcsA with membrane phospholipids as studied by ESI mass spectrometry. FEBS Lett. 2003;541(1-3):28–32. doi: 10.1016/s0014-5793(03)00282-5. [DOI] [PubMed] [Google Scholar]
- 37.Cuello LG, Romero JG, Cortes DM, Perozo E. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry. 1998;37(10):3229–3236. doi: 10.1021/bi972997x. [DOI] [PubMed] [Google Scholar]
- 38.Iwamoto M, Oiki S. Amphipathic antenna of an inward rectifier K+ channel responds to changes in the inner membrane leaflet. Proc Natl Acad Sci USA. 2013;110(2):749–754. doi: 10.1073/pnas.1217323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deol SS, Domene C, Bond PJ, Sansom MSP. Anionic phospholipid interactions with the potassium channel KcsA: Simulation studies. Biophys J. 2006;90(3):822–830. doi: 10.1529/biophysj.105.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weingarth M, et al. Structural determinants of specific lipid binding to potassium channels. J Am Chem Soc. 2013;135(10):3983–3988. doi: 10.1021/ja3119114. [DOI] [PubMed] [Google Scholar]
- 41.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504(7478):107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gogol EP, et al. Three dimensional structure of the anthrax toxin translocon-lethal factor complex by cryo-electron microscopy. Protein Sci. 2013;22(5):586–594. doi: 10.1002/pro.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy JG, et al. A KcsA/MloK1 chimeric ion channel has lipid-dependent ligand-binding energetics. J Biol Chem. 2014;289(14):9535–9546. doi: 10.1074/jbc.M113.543389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Postis V, et al. The use of smalps as a novel membrane protein scaffold for structure study by negative stain electron microscopy. Biochim Biophys Acta – Biomembranes. 2014 doi: 10.1016/j.bbamem.2014.10.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 46.Kemmer G, Keller S. Nonlinear least-squares data fitting in Excel spreadsheets. Nat Protoc. 2010;5(2):267–281. doi: 10.1038/nprot.2009.182. [DOI] [PubMed] [Google Scholar]
- 47.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.