Abstract
Glycoproteins containing Galα1-4Gal (galabiose) had been rarely found in vertebrates, except in a few species of birds and amphibians. We had previously reported that pigeon (Columba livia) egg white and serum glycoproteins are rich in N-glycans with Galα1-4Gal at nonreducing termini. To investigate the origin of Galα1-4Gal expression in avian evolution, we examined the presence of Galα1-4Gal glycoproteins in egg whites from 20 orders, 88 families, 163 genera, and 181 species of birds, as probed by Western blot with Griffonia simplicifolia-I lectin (terminal α-Gal/GalNAc-specific) and anti-P1 mAb (Galα1-4Galβ1-4GlcNAcβ1-specific). One of the significant observations is the total absence of Galα1-4Gal glycoproteins in Struthioniformes (four species), Tinamiformes (three species), Craciformes (two species), Galliformes (14 species), and Anseriformes (10 species), which are phylogenetically separated from other orders at earlier stage of modern bird diversification (100-65 million years ago). The presence or absence of Galα1-4Gal glycoproteins in other avian orders varied by the species (104 species positive, and 44 species negative), even though some of them belong to the same order or family. Our results revealed that the expression of Galα1-4Gal glycoproteins is not rare among avians, and is correlated with the phylogeny. The expression was most likely differentiated at earlier stage of diversification in modern birds, but some birds might have lost the facility for the expression relatively recently.
One of the characteristics of glycans attached to glycoproteins or glycolipids is their highly diversified structures with branches and different anomeric isomers and linkage positions of glycosidic bonds, in addition to variable chain lengths. For example, asparagine-linked glycans (N-glycans) on glycoproteins possess a common trimannosyl-core structure, from which variations can develop by branching with N-acetylglucosamine and by extension/repetition with galactosylation, sialylation, fucosylation, sulfation, and/or polylactosaminylation on the nonreducing ends. In many cases, glycans on proteins can be modulated into slightly different forms without significant changes of overall conformations of individual glycoproteins. However, even a subtlest change in oligosaccharide structures influences their interaction with carbohydrate-binding proteins expressed in self (endogenous receptors) and/or derived from nonself organisms (exogenous receptors).
Diversity of glycans is often found in species-specific manner, and certain carbohydrate chains become strong antigens in animals not expressing the same structure. The reason for existence of species-specific carbohydrates is unknown. However, several indirect investigations suggest that they may be for defense against infection of bacteria, virus, and parasites (1-3). This notion is because specific glycans expressed in animals are often targeted by carbohydrate-binding proteins on pathogens, for the first step of invasion. To reduce serious risks of pathogenic microbial infections, some hosts might have changed their oligosaccharide structures and made them highly heterogeneous. On the other hand, microbes might also have changed structures of their surface glycans to evade hosts' immune system. Thus, the glycan diversity in nature is considered to be compound consequences of survival race among hosts and microorganisms.
Investigation of species-specific oligosaccharides is important in understanding how glycan diversity was created during the course of evolution and diversification of animals. However, only limited information is available in this regard, especially with respect to the relationship between phylogeny and expression of species-specific carbohydrate antigens. Two of the better known examples are the expression of Galα1-3Gal and N-glycolylneuraminic acid (NeuGc). Galα1-3Gal is expressed in all mammals except human, apes, and Old World monkeys (catarrihins). Inactivated genes encoding α-1,3-galactosyltransferase (α-1,3-GalT), the enzyme to form Galα1-3Gal, had been found in catarrihins (4-6). The occurrence of this gene mutation in catarrihins was estimated to be between 40 and 25 millions years ago (mya) (5, 7), after the ancestors of Old World monkeys and New World monkeys diverged from the common ancestral primates (8, 9). In another example, deficiency of NeuGc in humans is unique among mammals (10). The human-specific inactivation of a gene encoding CMP N-acetylneuraminic acid hydroxylase, which is one of the critical enzymes to produce CMP-NeuGc, was estimated to have occurred 2.7-2.8 mya (11). The investigations of Galα1-3Gal and NeuGc-expression indicated that some glycan diversifications are inscribed in genes and maintained for a long period. However, these studies are mostly on mammals, and little systematic studies for nonmammalian vertebrates had been reported.
We recently showed that pigeon (Rock Dove, Columba livia) express Galα1-4Gal (galabiose) antigens attached on glycoproteins from egg white (12, 13) and serum (14), and analyzed their N-glycan structures (Fig. 1). The presence of Galα1-4Gal glycoproteins, however, had been rarely found in nature, except in a few birds (15-18) and amphibians (19-21). In mammals, the Galα1-4Gal sequence is only found in glycolipids expressed on cell surfaces. Whereas endogenous receptors for Galα1-4Gal had not been found in vertebrates, the Galα1-4Gal sequence is a minimum structure recognized by several bacterial adhesins and enterotoxins (see ref. 12 and references therein). It is demonstrated that pigeon egg white glycoproteins are recognized by P-fimbriae of uropathogenic Escherichia coli (12) and Shiga-like toxin type 1 (22), although whether Galα1-4Gal on glycoproteins in pigeon actually serves to protect the host as a decoy against such pathogens remains to be established.
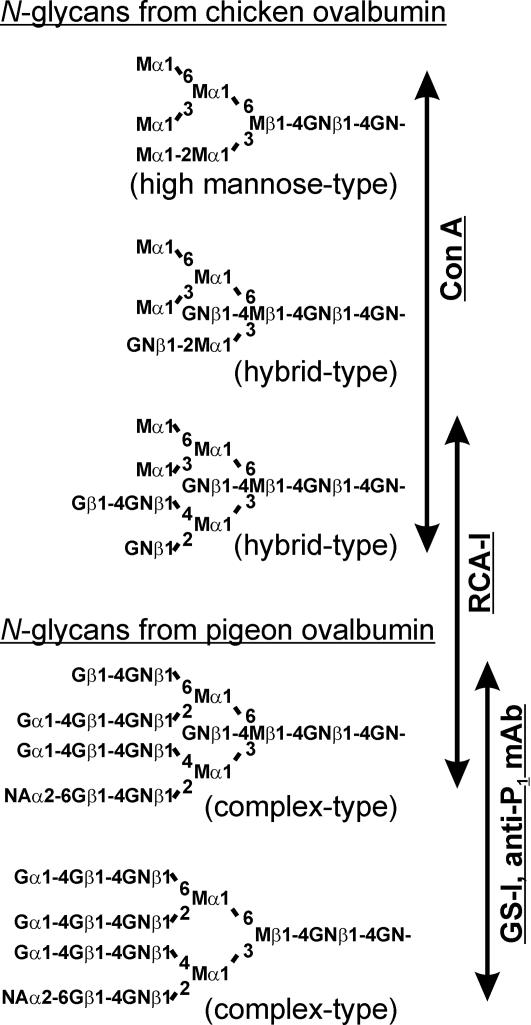
Fig. 1.
Representative N-glycan structures of ovalbumins from chicken (36-38) and pigeon (12, 13). Specificity of lectins (Con A, RCA-I, and GS-I) and anti-P1 mAb for the detection of oligosaccharides were also indicated. Monosaccharides were denoted as follows: M, mannose; GN, N-acetylglucosamine; G, galactose; NA, N-acetylneuraminic acid.
Based on the limited available information concerning the expression of Galα1-4Gal moiety in birds, we have assumed that Galα1-4Gal is absent in the avian orders Galliformes (chicken, turkey, quail, and pheasant) and Anseriformes (duck and gander; refs. 15 and 16), but is present in the orders Columbiformes (pigeon and turtle dove), Psittaciformes (budgerigar and cockatiel; refs. 15 and 17), and Apodiformes (swiftlet; ref. 18). Here, to trace the origin of Galα1-4Gal expression in birds, we examined the relationship between the expression of Galα1-4Gal antigen and phylogeny of birds by using avian egg whites from 181 species. We found that expression of Galα1-4Gal glycoproteins are actually distributed much wider among avians than what have ever been believed. Our investigation revealed that the ability to express Galα1-4Gal glycoproteins might have been possessed in common ancestors of modern birds, except Ratitae [Struthioniformes (e.g., ostrich) and Tinamiformes (e.g., tinamou)] and Galloanserae [Craciformes (e.g., curassow), Galliformes, and Anseriformes]. Galα1-4Gal glycoproteins are absent in egg whites from Ratitae and Galloanserae, which are phylogenetically separated from other orders at earlier stage of modern bird diversification (100-65 mya). Moreover, some species of birds belonging to non-Ratitae/Galloanserae might have lost ability to express Galα1-4Gal glycoproteins in egg white as recent as during or after diversification of avian families or genera.
Materials and Methods
Egg Whites. Egg whites from various species of birds (20 orders, 88 families, 163 genera, and 181 species) were collected by M.L. (23-25), and were maintained at -20°C. The Latin names of the birds were as given in Gruson and Forster (26) or in Sibley and Monroe (27). Three-letter abbreviations based on the common (English) name (Table 1, which is published as supporting information on the PNAS web site) were followed as described (23-25). Orders and families are listed according to Sibley and Monroe (27, 28). Phylogeny of birds based on DNA-DNA hybridization by Sibley et al. (27-29) was used for our basic reference because their classification of birds in the world are complementary to the phylogenetic analysis. Consequently, we used a term parvclass “Ratitae” designated by Sibley et al. (27, 29, 30), which is traditionally called “Palaeognathae.”
Materials. Alkaline phosphatase-conjugated lectins, concanavalin A (Con A), Ricinus communis agglutinin I (RCA-I), Griffonia simplicifolia I (GS-I), and peanut agglutinin were purchased from EY Laboratories (San Mateo, CA). Anti-P1 mAb (mouse IgM) was from Gamma Biologicals (Houston). Anti-Galα1-3Gal mAb (M86, mouse IgM) (31) was a generous gift from Dr. Galili (Rush University, Chicago). Alkaline phosphatase-conjugated anti-mouse IgM was from Sigma. Poly(vinylidene difluoride) membranes for blotting was from Millipore.
Methods. Procedures for SDS/PAGE and lectin/antibody blotting have been described (12, 32). In general, 2.5 μg of total egg white proteins from each species were loaded onto a lane of SDS/PAGE gels (10% acrylamide) under reducing condition. Egg white proteins were blotted on poly(vinylidene difluoride) membranes and detected by Coomassie brilliant blue staining. Oligosaccharides attached on egg white glycoproteins were probed by lectin/mAb staining. The results of lectin/mAb staining are recorded as follows: +, all major glycoproteins were stained; ±, some glycoproteins were stained; and -, no proteins were stained. This marking system does not take staining intensity into consideration because the protocol used are only qualitative and are not quantitative. Protein concentrations were measured by the BCA assay (33) by using BSA as a standard.
Results
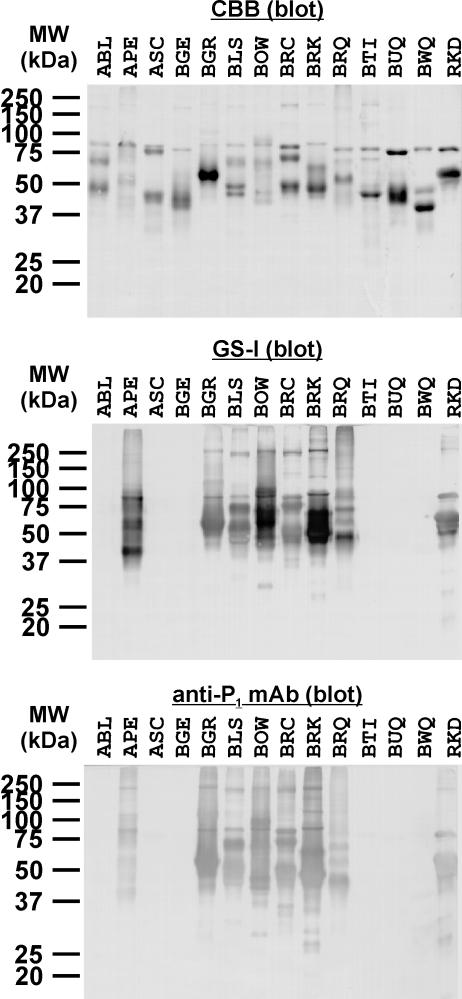
Lectin/Antibody Staining of Avian Egg White Glycoproteins. It is reported that major egg white glycoproteins in chicken are ovotransferrin (77 kDa, 12% of total protein), ovalbumin (45 kDa, 54% of total protein), and ovomucoid (28 kDa, 11% of total protein; ref. 34). All of them contain only N-glycans but not O-glycans. These glycoproteins are biosynthesized in tubular gland cells of oviduct (34). Pigeon egg white also contains these three major glycoproteins, and there are two variants of pigeon ovalbumin (12). The mobility of these major egg white glycoproteins on SDS/PAGE were different between chicken and pigeon, even though the corresponding glycoproteins share certain peptide sequence homology with each other. We have previously demonstrated (12) that the apparent mobility differences on SDS/PAGE are mostly due to the differences of number and size of N-glycans as well as amino acid sequences of individual glycoproteins. The presence of these three major glycoproteins (ovotransferrin, ovalbumin, and ovomucoid) are probably common in all avian egg whites, and indeed ovoumcoids had been isolated from approximately half of the egg white samples (23-25) used in our studies here. In this study, we first confirmed the presence of major egg white proteins from each bird, by SDS/PAGE, blotting onto poly(vinylidene difluoride) membrane, and staining with Coomassie brilliant blue. As expected, major proteins from all samples were visualized in the range of 30-80 kDa, although the mobility patterns varied from species to species. Different properties of avian egg white proteins, attributable to amino acid sequences and glycosylations in a species-specific manner, might have manifested in the differential mobilities. The examples of Coomassie brilliant blue-stained electrophoregrams are shown in Fig. 2.
Fig. 2.
Example of lectin/immunoblottings of avian egg white glycoproteins. Major egg white glycoproteins were visualized with Coomassie brilliant blue (CBB) staining. Galα1-4Gal was detected by GS-I and anti-P1 mAb. Abbreviations for samples based on the common (English) name of birds are shown in Table 1.
Glycoproteins blotted on poly(vinylidene difluoride) membranes were probed by staining with Con A (specific for high mannose-type, hybrid-type, and biantennary oligosaccharides), RCA-I (higher affinity for terminal β-galactoside), peanut agglutinin (higher affinity for Galβ1-3GalNAc, often found in O-glycans), GS-I (specific for terminal α-Gal/GalNAc), or anti-P1 mAb (specific for Galα1-4Galβ1-4GlcNAcβ1-; Fig. 1). The results were summarized in Table 1, and the examples of lectin staining are shown in Fig. 2. All of the major glycoproteins of avian egg whites from 100 of 181 species were stained by both GS-I and anti-P1 mAb. In four other species [Pennant-winged Nightjar, Willet, Australian Pratincole, and Wandering Albatross (WAL)], most major egg white glycoproteins were stained by both GS-I and anti-P1 mAb. The staining patterns of GS-I and anti-P1 mAb for all of the egg white glycoproteins tested were apparently the same. It is evident from the results of GS-I and anti-P1 mAb staining that α-Gal is always found with Galα1-4Galβ1-4GlcNAc. None of the GS-I-positive samples were stained with anti-Galα1-3Gal mAb (data not shown), indicating that the α-galactosyl residues of avian egg whites were not of Galα1-3Gal, which is expressed in mammals (except Old World monkeys, apes, and human). The GS-I-negative egg whites from 77 species were not stained with anti-P1 mAb either. All GS-I/anti-P1 mAb-positive samples, except WAL, were also stained with RCA-I, although it was not vice versa. It is highly likely that exposed β-galactosyl residues arose from incomplete α-galactosylation and/or α-sialylation, as found in pigeon egg white glycoproteins (Fig. 1 and ref. 13). WAL could not be stained with RCA-I distinctly, probably due to the nearly complete α-galactosylation, which masks terminal β-galactosyl residues. Indeed, after the treatment with α-galactosidase (from green coffee bean), major glycoproteins of WAL could be stained with RCA-I (data not shown).
All egg white glycoproteins tested in this study, except for Buttonquail (BUQ) and WAL, were stained with RCA-I, whereas none of the egg white glycoproteins examined were visibly stained with peanut agglutinin (data not shown). BUQ clearly stained only with Con A. This result indicates that BUQ is unable to produce galactosylated oligosaccharides, presumably due to lack of certain processing enzyme activities or deficient in some sugar-nucleotide donors, i.e., UDP-GlcNAc or UDPGal in the oviduct to form complex-type oligosaccharides (35). The fact that major egg white glycoproteins from almost all birds could be stained with RCA-I indicates that these birds are capable of producing β-galactosylated glycans, most likely as Galβ1-4GlcNAc, which are commonly found in the known N-glycan structures from chicken (Gallus Gallus; Fig. 1 and refs. 36-41), quail (Coturnix japonica; ref. 42), duck (Anas platyrhynchos hybrid; ref. 43), and pigeon (Columba livia; Fig. 1 and refs. 12 and 13) egg white glycoproteins. Because Galβ1-4GlcNAc sequence is a direct precursor of Galα1-4Galβ1-4GlcNAc, we regard that RCA-I-positive species possess potential substrates for putative α-1,4-GalT.
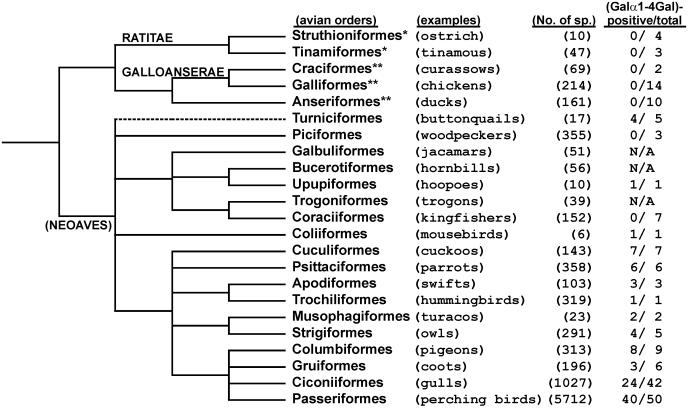
Relationship Between Expression of Galα1-4Gal in Egg White Glycoproteins and Phylogeny of Birds. Significantly, Galα1-4Gal glycoproteins are totally absent in Struthioniformes (four species), Tinamiformes (three species), Craciformes (two species), Galliformes (14 species), and Anseriformes (10 species), which are phylogenetically separated from other orders at earlier stage of modern bird diversification (Table 1 and Fig. 3). Some in the remaining orders express Galα1-4Gal glycoproteins in all species examined: Upupiformes (1/1, positive species/total species examined), Coliiformes (1/1), Cuculiformes (7/7), Psittaciformes (6/6), Apodiformes (3/3), Trochiliformes (1/1), Musophagiformes (2/2). However, Piciformes (0/3) and Coraciiformes (0/7) were negative in Galα1-4Gal expression in the species examined.
Fig. 3.
Phylogeny of modern birds and expression of Galα1-4Gal on avian egg white glycoproteins. The phylogenetic tree is based on DNA-DNA hybridization by Sibley et al. (29). The position of the Turniciformes in their classification is uncertain (p. 255 in ref. 29), and is indicated with dashed line. Avian orders belonging to Ratitae (*) and Galloanserae (**) are indicated with asterisks. Non-Ratitae/Galloanserae modern birds are sometimes called Neoaves. The examples of birds and recorded number of species in each order (27) are indicated in parentheses. Ciconiiformes in Sibley's classification (27, 28) are traditionally classified as Charadriiformes (e.g., gulls), Falconiformes (e.g., eagles), Podicipediformes (grebes), Pelecaniformes (e.g., tropicbirds), Ciconiiformes (e.g., storks), Sphenesciformes (penguins), Gaviiformes (loons), and Procellariformes (e.g., albatrosses). The number of Galα1-4Gal-positive species and the total species examined in this study are indicated in the right column. N/A, not available. [Modified from ref. 29 with permission from Yale Univ. Press (Copyright 1990, Yale Univ. Press).]
Interestingly, expression of Galα1-4Gal glycoproteins in birds in Turniciformes (4/5), Strigiformes (4/5), Columbiformes (8/9), Gruiformes (3/6), Ciconiiformes, (24/42), and Passeriformes (40/50) is not uniform, even within the same orders or families. For example, eight species of family Columbidae (pigeons and doves) differ in expression of Galα1-4Gal glycoproteins. The one negative species Common Crowned Pigeon is reported to be a more distant relative than other positive species (44). Similarly, among nine species in five genera of family Spheniscidae (penguins), three species in one genus (Pygoscelis) were positive, while other six species in four genera were negative for Galα1-4Gal expression. Thus, it is most likely that the facility for expression of Galα1-4Gal was lost in some species (or genera) after diversification of their families.
Different species of the same genus examined here are as follows: Tragopan (two species), Turnix (five species), Amazona (two species), Apus (two species), Columba (two species), Zenaida (two species), Grus (two species), Falco (two species), Egretta (two species), Aptenodytes (two species), Pygoscelis (three species), Spheniscus (two species), Turdus (two species), and Passer (two species). The species belonging to the same genera, except Turnix, revealed the same pattern in terms of expression of Galα1-4Gal glycoproteins (Table 1). In the exception, Turnix, only one of the five species (BUQ) showed different expression pattern from the other species in the same genus. Unfortunately, phylogenetic analysis for Turniciformes is very incomplete, and we could not survey the differences of these species in this genus. BUQ is, however, the only species that could not be stained with both RCA-I and GS-I, indicating that BUQ cannot express β-galactosyl residues. Therefore, we speculate that this species lost its ability to express Galα1-4Gal on egg white glycoproteins not by inactivation of α-1,4-GalT, but by some other mechanism.
Discussion
Birds are one of the branches of higher vertebrates believed to have evolved from a lineage of reptiles. Ancestors of birds and of mammals survived in the Cretaceous-Tertiary boundary (65 mya) when dinosaurs became extinct, and then radiated in Tertiary (65-1.6 mya). The number of species of living birds (9,672 recognized species, by the year 1990; ref. 27) is largest among vertebrates. Phylogenetic classification of birds has been an especially difficult task. They are underrepresented in the fossil records, due to their brittle bones. After the plumage is removed, the birds are taxonomically quite similar. Yet the study based on morphology (45, 46), DNA-DNA hybridization (27, 29), nuclear/mitochondrial DNA sequences (47-51), and protein sequences (52) mostly agree that modern birds (Neornithes) are monophylic and had been divided into three large taxa, (i) Ratitae (traditionally called Palaeognathae), (ii) Galloanserae, and (iii) non-Ratitae/Galloanserae (sometimes called Neoaves§), at the earlier stage of modern birds history. The divergence dates are supposed to be in the Cretaceous (≈100 mya) according to molecular clock interpretations (53-55), or soon after the Cretaceous-Tertiary extinction event (65 mya) estimated by limited fossil records (56, 57). The root placement of Galloanserae, i.e., whether it is related to Ratitae or to other avian orders, remains to be settled, because available results are varied and controversial, depending on the used methods and data sets (55). DNA-DNA hybridization method by Sibley et al. (29, 30) placed Galloanserae as a closer relative to Ratitae as shown in Fig. 3; however, Sibley et al. (p. 255 in ref. 29) considered that the branch was probably misplaced.
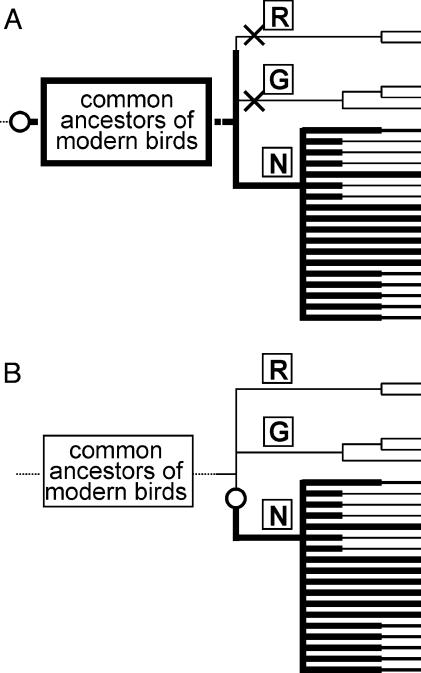
We showed that the expression of Galα1-4Gal on egg white glycoproteins is totally absent in Ratitae and Galloanserae, whereas majority of other groups express Galα1-4Gal. Because the expression of Galα1-4Gal is widely distributed among non-Ratitae/Galloanserae birds, it is most likely that the ancestor of non-Ratitae/Galloanserae birds might be able to express Galα1-4Gal before further diversification. Although the root placement of Galloanserae is not clear, the major branching point in expression of Galα1-4Gal on egg white glycoproteins seems to be between Ratitae/Galloanserae and non-Ratitae/Galloanserae. For this event, we can consider two hypothetical models for Galα1-4Gal expression (Fig. 4). In the first model (Fig. 4A), the common ancestor of all modern bird lineages could express Galα1-4Gal, but this facility was lost in Ratitae/Galloanserae after they diversified. In the second model (Fig. 4B), the common ancestor of the modern birds did not have ability to express Galα1-4Gal, but it was acquired somehow only in ancestors of non-Ratitae/Galloanserae birds. The relationship of expression of Galα1-4Gal glycoproteins in birds and amphibians is unclear, but they might have occurred independently, in the case of the second model. Fig. 4A involves a possibility that Galα1-4Gal expression in avians was inherited from amphibians. However, the conclusion can only be tentative unless gene structures of α-1,4-GalT in avians and amphibians are carefully compared. Some non-Ratitae/Galloanserae birds might have lost Galα1-4Gal expression capability during or after diversification at the level of families or genera, so that the differential expression of Galα1-4Gal was also observed within the same orders or families. None of the Piciformes and Coraciiformes species examined were Galα1-4Gal-positive in egg white glycoproteins. However, the negligible expression in these orders might have occurred independently from those in Ratitae/Galloanserae, because both Piciformes and Coraciiformes are phylogenetically distinct from Ratitae/Galloanserae (29, 46, 49, 50). In the case of second model (Fig. 4B), there is a possibility that after Piciformes was divided from others, the Galα1-4Gal expression capability was acquired in ancestors of the rest of non-Ratitae/Galloanserae birds. However, this speculation can only be tentative, because there is no consensus on the phylogenetic position of Piciformes within non-Ratitae/Galloanserae, as determined by DNA-DNA hybridization and by several other methods (47, 50, 55).
Fig. 4.
Schematic models for acquisition/loss of Galα1-4Gal expression capability in the modern birds during their diversification. (A) The common ancestors of modern birds already possess the capability to express Galα1-4Gal glycoproteins (open circle, followed by thick lines) before the differentiation into Ratitae (R), Galloanserae (G), and non-Ratitae/Galloanserae (N), but this capability might have been lost (cross, followed by thin lines) in Ratitae and Galloanserae after they are separated from others. (B) The common ancestors of modern birds did not possess the capability, but it was acquired by non-Ratitae/Galloanserae after they are separated from others. In either models, some species of birds belonging to non-Ratitae/Galloanserae might have lost (thin lines) the capability to express Galα1-4Gal glycoproteins in egg whites at the levels of families and/or genera. The root position of Galloanserae was undefined.
Our results showed that the expression of Galα1-4Gal glycoproteins is actually not rare, and is in more than half of the avian species. Passeriformes (perching birds) are regarded as the most successful modern birds based on the number of species (5,712 species, 59% of total avian species), and are often compared with rodents, which constitute ≈40% of mammal species (56). Although we have analyzed only 50 species in Passeriformes, 40 of them were Galα1-4Gal-positive. The recorded number of species of non-Ratitae/Galloanserae group is 9,171 (95% of total avian species). In our survey, 104 of 148 species in this group were Galα1-4Gal-positive. Assuming that the ratio of Galα1-4Gal-positive versus total species in non-Ratitae/Galloanserae is also 104:148, 6,444 species (67% of total avian species) would be expected to express Galα1-4Gal-glycoproteins. If this is the case, Galα1-4Gal glycoproteins are far from “rare” in birds. Although the expression of Galα1-4Gal in birds is apparently not mandatory, we can speculate that the expression of Galα1-4Gal influences the fate of birds by glycan-based interaction with microbes or xenobiotics, either advantageously or disadvantageously. The loss of Galα1-4Gal in birds may also involve the potential ability to produce natural antibodies against Galα1-4Gal, when the epitope is absent in both glycoproteins and glycolipids. It is proposed that natural anticarbohydrate antibodies may act as barriers to retrovirus transmission between positive and negative taxa (1).
Formation of Galα1-4Gal on glycoproteins is presumably catalyzed by a putative α-1,4-GalT, which transfers Gal residue (most likely from UDP-Gal) to lactosamine (Galβ1-4GlcNAc) moieties on glycoproteins. When the substrates are adequately provided, the expression of Galα1-4Gal are mainly regulated by expression of the gene encoding α-1,4-GalT. Additionally, coenzymes or specific chaperons are sometimes also critical for the enzyme activity (58). Whereas pigeon expresses Galα1-4Gal glycoproteins not only in egg whites but also in other tissues and fluids (14), no Galα1-4Gal has been detected so far in chicken. The apparent absence of Galα1-4Gal glycoproteins in entire body of chicken probably reflects inactivation or absence of the α-1,4-GalT gene. In some species, lack of Galα1-4Gal in egg white glycoproteins might be due to tissue-specific inactivation of the α-1,4-GalT gene expression, or polymorphism. These speculations will be substantiated when sequence(s) of putative avian α-1,4-GalT gene(s) becomes available.
Species-specific glycan expression, such as Galα1-3Gal and NeuGc in mammals, showed that structural analysis of (pseudo)genes can provide useful information about the past events on the genes for glycan expression. However, the main emphasis of differentiation of both Galα1-3Gal and NeuGc is focused on glycan differentiation in human versus other mammals. Consequently, the timing of divergence, i.e., loss of Galα1-3Gal and NeuGc expression capability, could not be much earlier than history of human or primates. The origins of Galα1-3Gal and NeuGc expression are supposed to be in early stage of mammalian evolution, but it has not been determined precisely. Although we have not determined the actual origin of Galα1-4Gal expression in avians, our data clearly indicate that the glycan diversification occurred in the early history of modern birds (100-65 mya). Moreover, we found that the loss of Galα1-4Gal expression facility also occurred in some birds relatively recently (mostly 65 mya to the present). Therefore, comparison of the activated/inactivated α-1,4-GalT gene(s) among birds will be important in understanding the genetic mechanism of species-specific glycan differentiation at different evolutional stages, including more ancient events than those so far studied for mammals.
Supplementary Material
Acknowledgments
We thank Dr. Charles Sibley for the kind gift of his egg white collection (1,036 different species) to M.L., of which 116 species were used in this work; Dr. Uri Galili for his gift of anti-Galα1-3Gal mAb; and Yale University Press for the use the figure of a phylogenic tree (Fig. 3). This work was supported by National Institutes of Health Research Grant DK09970 (to N.S. and Y.C.L.). The acquisition and maintenance of M.L.'s egg white collection were supported by National Institutes of Health Grants GM10831 and GM63539.
Abbreviations: Con A, concanavalin A; GalT, galactosyltransferase; GS-I; Griffonia simplicifolia I; mya, million years ago; NeuGc, N-glycolylneuraminic acid; RCA-I, Ricinus communis agglutinin I; WAL, Wandering Albatross; BUQ, Buttonquail.
Footnotes
References
- 1.Gagneux, P. & Varki, A. (1999) Glycobiology 9, 747-755. [DOI] [PubMed] [Google Scholar]
- 2.Dennis, J. W., Granovsky, M. & Warren, C. E. (1999) BioEssays 21, 412-421. [DOI] [PubMed] [Google Scholar]
- 3.Hooper, L. V. & Gordon, J. I. (2001) Glycobiology 11, 1R-10R. [DOI] [PubMed] [Google Scholar]
- 4.Larsen, R. D., Rivera-Marrero, C. A., Ernst, L. K., Cummings, R. D. & Lowe, J. B. (1990) J. Biol. Chem. 265, 7055-7061. [PubMed] [Google Scholar]
- 5.Joziasse, D. H., Shaper, J. H., Jabs, E. W. & Shaper, N. L. (1991) J. Biol. Chem. 266, 6991-6998. [PubMed] [Google Scholar]
- 6.Galili, U. & Swanson, K. (1991) Proc. Natl. Acad. Sci. USA 88, 7401-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koike, C., Fung, J. J., Geller, D. A., Kannagi, R., Libert, T., Luppi, P., Nakashima, I., Profozich, J., Rudert, W., Sharma, S. B., Starzl, T. E. & Trucco, M. (2002) J. Biol. Chem. 277, 10114-10120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galili, U. & LaTemple, D. C. (1997) Immunol. Today 18, 281-285. [DOI] [PubMed] [Google Scholar]
- 9.Galili, U., Shohet, S. B., Kobrin, E., Stults, C. L. & Macher, B. A. (1988) J. Biol. Chem. 263, 17755-17762. [PubMed] [Google Scholar]
- 10.Chou, H. H., Takematsu, H., Diaz, S., Iber, J., Nickerson, E., Wright, K. L., Muchmore, E. A., Nelson, D. L., Warren, S. T. & Varki, A. (1998) Proc. Natl. Acad. Sci. USA 95, 11751-11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, H. H., Hayakawa, T., Diaz, S., Krings, M., Indriati, E., Leakey, M., Pääbo, S., Satta, Y., Takahata, N. & Varki, A. (2002) Proc. Natl. Acad. Sci. USA 99, 11736-11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki, N., Khoo, K. H., Chen, H. C., Johnson, J. R. & Lee, Y. C. (2001) J. Biol. Chem. 276, 23221-23229. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi, N., Khoo, K. H., Suzuki, N., Johnson, J. R. & Lee, Y. C. (2001) J. Biol. Chem. 276, 23230-23239. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki, N., Khoo, K. H., Chen, C. M., Chen, H. C. & Lee, Y. C. (2003) J. Biol. Chem. 278, 46293-46306. [DOI] [PubMed] [Google Scholar]
- 15.François-Gérard, C. & Brocteur, J. (1980) Blood Transfus. Immunohematol. 23, 95-102. [DOI] [PubMed] [Google Scholar]
- 16.Brocteur, J., François-Gérard, C., André, A., Radermecker, M., Bruwier, M. & Salmon, J. (1975) Haematologia 9, 43-47. [PubMed] [Google Scholar]
- 17.Johnson, J. R., Swanson, J. L. & Neill, M. A. (1992) Infect. Immun. 60, 578-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wieruszeski, J. M., Michalski, J. C., Montreuil, J., Strecker, G., Peter-Katalinic, J., Egge, H., van Halbeek, H., Mutsaers, J. H. & Vliegenthart, J. F. (1987) J. Biol. Chem. 262, 6650-6657. [PubMed] [Google Scholar]
- 19.Guerardel, Y., Kol, O., Maes, E., Lefebvre, T., Boilly, B., Davril, M. & Strecker, G. (2000) Biochem. J. 352, 449-463. [PMC free article] [PubMed] [Google Scholar]
- 20.Delplace, F., Maes, E., Lemoine, J. & Strecker, G. (2002) Biochem. J. 363, 457-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strecker, G., Wieruszeski, J. M., Michalski, J. C., Alonso, C., Leroy, Y., Boilly, B. & Montreuil, J. (1992) Eur. J. Biochem. 207, 995-1002. [DOI] [PubMed] [Google Scholar]
- 22.Tomoda, H., Arai, M., Koyama, N., Matsui, H., Ōmura, S., Obata, R. & Lee, Y. C. (2002) Anal. Biochem. 311, 50-56. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski, M., Jr., Kato, I., Ardelt, W., Cook, J., Denton, A., Empie, M. W., Kohr, W. J., Park, S. J., Parks, K., Schatzley, B. L., et al. (1987) Biochemistry 26, 202-221. [DOI] [PubMed] [Google Scholar]
- 24.Laskowski, M., Jr., Apostol, I., Ardelt, W., Cook, J., Giletto, A., Kelly, C. A., Lu, W. Y., Park, S. J., Qasim, M. A., Whatley, H. E., et al. (1990) J. Protein Chem. 9, 715-725. [DOI] [PubMed] [Google Scholar]
- 25.Apostol, I., Giletto, A., Komiyama, T., Zhang, W. & Laskowski, M., Jr. (1993) J. Protein Chem. 12, 419-433. [DOI] [PubMed] [Google Scholar]
- 26.Gruson, E. S. & Forster, R. A. (1976) Checklist of the World's Birds: A Complete List of the Species, With Names, Authorities, and Areas of Distribution (Quadrangle/New York Times Book Co., New York).
- 27.Sibley, C. G. & Monroe, B. L. (1990) Distribution and Taxonomy of Birds of the World (Yale Univ. Press, New Haven, CT).
- 28.Sibley, C. G. & Monroe, B. L. (1993) A Supplement to Distribution and Taxonomy of Birds of the World (Yale Univ. Press, New Haven, CT).
- 29.Sibley, C. G. & Ahlquist, J. E. (1990) Phylogeny and classification of birds: A Study in Molecular Evolution (Yale Univ. Press, New Haven, CT).
- 30.Sibley, C. G., Ahlquist, J. E. & Monroe, B. L. (1988) Auk 105, 409-423. [Google Scholar]
- 31.Galili, U., LaTemple, D. C. & Radic, M. Z. (1998) Transplantation 65, 1129-1132. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, N. & Lee, Y. C. (2004) Glycobiology 14, 275-292. [DOI] [PubMed] [Google Scholar]
- 33.Smith, P. K., Krohn, R. I., Hermanson, G. T., Mallia, A. K., Gartner, F. H., Provenzano, M. D., Fujimoto, E. K., Goeke, N. M., Olson, B. J. & Klenk, D. C. (1985) Anal. Biochem. 150, 76-85. [DOI] [PubMed] [Google Scholar]
- 34.Burley, R. W. & Vadehra, D. V. (1989) The Avian Egg: Chemistry and Biology (Wiley, New York).
- 35.Kornfeld, R. & Kornfeld, S. (1985) Annu. Rev. Biochem. 54, 631-664. [DOI] [PubMed] [Google Scholar]
- 36.Tai, T., Yamashita, K., Ogata-Arakawa, M., Koide, N., Muramatsu, T., Iwashita, S., Inoue, Y. & Kobata, A. (1975) J. Biol. Chem. 250, 8569-8575. [PubMed] [Google Scholar]
- 37.Tai, T., Yamashita, K., Ito, S. & Kobata, A. (1977) J. Biol. Chem. 252, 6687-6694. [PubMed] [Google Scholar]
- 38.Yamashita, K., Tachibana, Y. & Kobata, A. (1978) J. Biol. Chem. 253, 3862-3869. [PubMed] [Google Scholar]
- 39.Yamashita, K., Kamerling, J. P. & Kobata, A. (1982) J. Biol. Chem. 257, 12809-12814. [PubMed] [Google Scholar]
- 40.Yamashita, K., Kamerling, J. P. & Kobata, A. (1983) J. Biol. Chem. 258, 3099-3106. [PubMed] [Google Scholar]
- 41.Yamashita, K., Tachibana, Y., Hitoi, A. & Kobata, A. (1984) Carbohydr. Res. 130, 271-288. [DOI] [PubMed] [Google Scholar]
- 42.Hase, S., Sugimoto, T., Takemoto, H., Ikenaka, T. & Schmid, K. (1986) J. Biochem. 99, 1725-1733. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, N., Matsuda, T., Shikami, K., Shimada, I., Arata, Y. & Nakamura, R. (1993) Glycoconj. J. 10, 425-434. [DOI] [PubMed] [Google Scholar]
- 44.Johnson, K. P. & Clayton, D. H. (2000) Mol. Phylogenet. Evol. 14, 141-151. [DOI] [PubMed] [Google Scholar]
- 45.Cracraft, J. (1986) Paleobiology 12, 383-399. [Google Scholar]
- 46.Cracraft, J. (1988) in The Phylogeny and Classification of the Tetrapods, ed. Benton, M. J. (Oxford Univ. Press, Oxford), pp. 339-361.
- 47.Mindell, D. P., Sorenson, M. D., Huddleston, C. J., Miranda, H. C., Knight, A., Sawchuk, S. J. & Yuri, T. (1997) in Avian Molecular Evolution and Systematics, ed. Mindell, D. P. (Academic, San Diego), pp. 213-247.
- 48.Groth, J. G. & Barrowclough, G. F. (1999) Mol. Phylogenet. Evol. 12, 115-123. [DOI] [PubMed] [Google Scholar]
- 49.van Tuinen, M., Sibley, C. G. & Hedges, S. B. (2000) Mol. Biol. Evol. 17, 451-457. [DOI] [PubMed] [Google Scholar]
- 50.Prychitko, T. M. & Moore, W. S. (2003) Mol. Biol. Evol. 20, 762-771. [DOI] [PubMed] [Google Scholar]
- 51.Slack, K. E., Janke, A., Penny, D. & Arnason, U. (2003) Gene 302, 43-52. [DOI] [PubMed] [Google Scholar]
- 52.Stapel, S. O., Leunissen, J. A., Versteeg, M., Wattel, J. & de Jong, W. W. (1984) Nature 311, 257-259. [DOI] [PubMed] [Google Scholar]
- 53.Hedges, S. B., Parker, P. H., Sibley, C. G. & Kumar, S. (1996) Nature 381, 226-229. [DOI] [PubMed] [Google Scholar]
- 54.Cooper, A. & Penny, D. (1997) Science 275, 1109-1113. [DOI] [PubMed] [Google Scholar]
- 55.Cracraft, J. (2001) Proc. R. Soc. London Ser. B 268, 459-469. [Google Scholar]
- 56.Feduccia, A. (1995) Science 267, 637-638. [DOI] [PubMed] [Google Scholar]
- 57.Feduccia, A. (1996) The Origin and Evolution of Birds (Yale Univ. Press, New Haven, CT).
- 58.Ju, T. & Cummings, R. D. (2002) Proc. Natl. Acad. Sci. USA 99, 16613-16618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.