Abstract
Different types of gross chromosomal rearrangements (GCRs), including translocations, interstitial deletions, terminal deletions with de novo telomere additions, and chromosome fusions, are observed in many cancers. Multiple pathways, such as S-phase checkpoints, DNA replication, recombination, chromatin remodeling, and telomere maintenance that suppress GCRs have been identified. To experimentally expand our knowledge of other pathway(s) that suppress GCRs, we developed a generally applicable genome-wide screening method. In this screen, we identified 10 genes (ALO1, CDC50, CSM2, ELG1, ESC1, MMS4, RAD5, RAD18, TSA1, and UFO1) that encode proteins functioning in the suppression of GCRs. Moreover, the breakpoint junctions of GCRs from these GCR mutator mutants were determined with modified breakpoint-mapping methods. We also identified nine genes (AKR1, BFR1, HTZ1, IES6, NPL6, RPL13B, RPL27A, RPL35A, and SHU2) whose mutations generated growth defects with the pif1Δ mutation. In addition, we found that some of these mutations changed the telomere size.
Maintaining genome stability is crucial for cell survival and normal cell growth. Different genetic disorders, including most human cancers, display different forms of genome instability (1-3). The mutator hypothesis suggests that cancer cells must acquire a mutator phenotype to account for the high rate of accumulating genetic changes observed (4, 5). It is best established in the case of the inherited (HNPCC) and sporadic mismatch repair-defective cancers, in which the mismatch-repair defect that underlies cancer development causes increased mutation rates (1). There is growing evidence supporting that the presence of genome rearrangements, known as gross chromosomal rearrangements (GCRs), reported in many different cancers could reflect the acquisition of a mutator phenotype (6-11).
Multiple pathways that suppress GCRs were characterized by using yeast, Saccharomyces cerevisiae, as a model system (12-17). At least five different pathways have been identified for the suppression of GCRs, including (i) cell-cycle checkpoints during DNA replication, (ii) a recombination pathway known as a break-induced replication, (iii) a pathway that suppresses de novo telomere addition, (iv) at least two pathways for the proper chromatin assembly, and (v) a possible mismatch-repair pathway that prevents recombination between divergent DNA sequences. The importance of these multiple mechanisms for suppression of genome instability in human cancer development has been uncovered by observations of mutations in genes of which encoded proteins function in the multiple mechanisms identified in studies using other model organisms. It includes ATM/ATR in ataxia telangiectasia (AT); MRE11 in AT-like disorder (ATLD); NBS1 in Nijmegen breakage syndrome (NBS); hCHK2 in Li-Fraumeni syndrome; and BLM/WRN/RTS in the Bloom, Werner, and Rothmund-Thomson syndrome (12, 18).
Although many pathways that function in the suppression of GCRs have been discovered, complete understanding of how GCRs arise and are suppressed has not been accomplished because of the difficulty in implementing forward genetic screens for mutations with elevated GCR rates. Here, we report the development of a screening method that allows the systematic analysis of the complete set of gene deletion mutants to identify genes whose mutations increased GCR formation. This screening method can be applicable to almost all known assays. To enhance the basal level of GCR rate, the pif1 deletion mutation was incorporated for this screen.
The PIF1 gene encodes two helicases: one helicase that functions as an inhibitor of telomerase (19, 20) and another helicase that functions in mitochondria (21). pif1 mutations are known to increase the GCR rate (19, 21), and inactivation of almost all known GCR suppression pathways in combination with the pif1-m2 mutation increases GCR rates synergistically compared with the strain carrying the pif1-m2 allele alone (19). Thus, the sensitivity of a GCR assay for genome-wide screening of GCR suppressors can be enhanced by an additional mutation in the PIF1 gene. We identified most of the known GCR mutator genes and 10 GCR mutator genes whose mutations increased GCR formation by using the screening method (ALO1, CDC50, CSM2, ELG1, ESC1, MMS4, RAD5, RAD18, TSA1, and UFO1). Mutator genes identified in this study strongly suggest that there are more pathways that prevent the formation of GCRs, such as transcriptional regulation, chromosome segregation, and postreplication repair, including ubiquitination and oxidative-stress response.
Redundant functions can be discovered by synthetic genetic interactions when a specific mutation suppresses or enhances the original mutant phenotype. A synthetic lethal phenotype may occur when two mutations inactivate a single pathway completely or cause defects in two redundant but dependent pathways (22-24). Because of the pif1Δ mutation in the screen, we were able to identify nine genes whose mutations generated growth defects in combination with the pif1Δ mutation (AKR1, BFR1, HTZ1, ISE6, NPL6, RPL13B, RPL27A, RPL35A, and SHU2), and we subsequently found that some of these mutations changed the telomere size.
The deletion mutation of the PIF1 gene also inactivates the mitochondrial PIF1 protein. The defect in mitochondrial function causes a slow rate of cell division, resulting in small colonies, known as petites, that have growth defects in media with a nonfermentable carbon source. We identified 22 hypothetical ORFs whose mutations induce the petite phenotype because of their putative role in mitochondrial function.
Materials and Methods
Strains and Gene Disruption of S. cerevisiae. Media for propagation of strains have been described (25). All strains were propagated at 30°C. Yeast strains containing the hygromycin resistant cassette (hphMX) or the G418-resistant cassette (kanMX) were selected on plates with 300 μg/ml hygromycin B or 200 μg/ml G418, respectively. YKJM1347, the parental strain for the genome-wide screening of GCR suppressors, was generated as follows. The PIF1 gene in y2454 (mfa1Δ::MFA1pr-HIS3, can1Δ, his3Δ1, leu2Δ0, ura3Δ0, MET15+ lys2Δ0) (26) was disrupted with a hygromycin cassette, and the resulting strain was named YKJM1241. Then, YKJM1241 and RDKY3615 (ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8, YEL069::URA3) were mated and sporulated. Spores that were unable to grow without histidine but could grow in the absence of uracil were chosen. Strains carrying the MFA1pr-HIS3 gene were confirmed by PCR and by mating with a mating type A tester strain. The presence of the CAN1 gene in YKJM1347, along with three other isolates, was confirmed by testing the sensitivity of these strains to canavanine. The yeast deletion mutant library was purchased from the American Type Culture Collection (GSA-4). Most strains were constructed in the BY4741 background (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) with 11 exceptions that were constructed in the BY4730 background (MATa, leu2Δ0, met15Δ0, ura3Δ0). To confirm the GCR mutator mutations identified in the screening, identified genes were disrupted by standard PCR-based gene-disruption methods in YKJM1347, RDKY3615, and RDKY4343 (ura3::KAN, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8, HO::hisG, pif1-m2, YEL069::URA3), and correct gene disruptions were verified by PCR, as described (25). The sequences of primers used to generate disruption cassettes and confirm disruption of indicated genes are available on request.
Genome-Wide Screening of GCR Mutators. The strain YKJM1347 was crossed with the 4,644 individual MATa deletion mutant strains (Fig. 1). Mated diploid cells were selected by G418 and hygromycin on synthetic drop out (SD) plates with all amino acids except uracil [SD (-Ura)]. After sequentially replica plating twice onto SD (-Ura) plates with G418 and hygromycin, the selected diploid cells were replica plated onto sporulation plates. The resulting spores were replica plated onto SD (-His -Ura) plates containing hygromycin and G418 to select for haploids carrying the GCR assay and pif1 deletion mutation. Resulting cells were tested for CAN mutators that increase CAN1 gene-inactivation rate and GCR mutators that increase GCR rate. The number of colonies from each clone grown after 3-5 days on selection plates (CAN, containing canavanine, for CAN mutator screen and FC containing canavanine and 5FOA, for GCR screen, as described in ref. 25) was then compared with the colony number of YKJM1347 strain. Individual cells that gave an increased number of resistant colonies compared with YKJM1347 either in the CAN plates because of the inactivation mutation of CAN1 gene or in the FC plates because of the GCRs were collected from the original yeast extract/peptone/dextrose (YPD) plates and streaked onto fresh YPD plates. Three patches from individual colonies from candidates made on YPD plates were replica plated on to CAN or FC plates after 3 days of incubation for a secondary screening. There were 11 deletion mutant strains whose strain background is BY4730 (MATa, leu2Δ0, met15Δ0, ura3Δ0) in knock-out library. Because of the lack of the his3Δ200 mutation, ORFs deleted in these strains were excluded from the initial screen. Thus, these 11 strains were tested individually in RDKY3615 and RDKY4343.
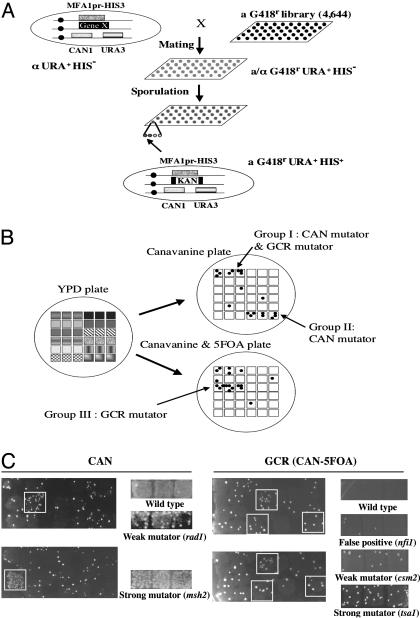
Fig. 1.
Genome-wide screening to find GCR mutators. (A) A library composed of strains carrying the GCR assay system, a pif1Δ mutation, and 1 of 4,644 gene deletion mutations was generated. (B) Secondary screening identified three types of GCR mutator mutations (see Table 1). (C) Screen results for CAN and GCR (CAN-5FOA) mutator screens. Primary screen is shown in Left, and secondary screen is shown in Right. In the CAN mutator assay, only one plate was used for the primary screen. Putative GCR mutators were selected only when two independent FC plates generated more than five colonies.
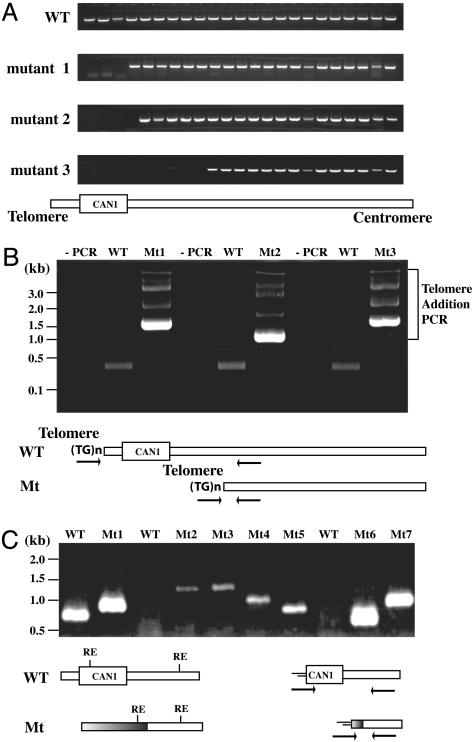
GCRs. All GCR rates were determined by fluctuation analysis at least two times by using either 5 or 11 cultures, and average values are reported. The sequences of the independent rearrangement breakpoints were determined by breakpoint-mapping methods (Fig. 2) and classified, as described (25). First, the chromosomal DNA from clones resistant to both canavanine and 5-FOA were analyzed by genome walking PCRs (Fig. 2 A). We used 23 different PCRs to narrow the breakpoint location to within 400 bp. Then, a PCR was carried out by using a primer complementary to the end of the rearranged chromosome V and a primer complementary to telomeric repeat sequence (CA16; 5′-CACCACACCCACACAC-3′). Genome rearrangements with de novo telomere-addition GCRs produced a ladder of PCR bands (Fig. 2B). To make sure that these PCR products were specific for de novo telomere addition, wild-type chromosomal DNA was used as a negative PCR control. If there was no difference in de novo telomere-addition PCR product between the wild-type chromosomal DNA and chromosomal DNA-carrying GCRs, the linker-mediated PCR, a modified version of mapping PCR of murine leukemia virus integration site (27) was performed. Chromosomal DNAs from wild type and clones carrying GCRs were digested by one of four restriction enzymes (BfaI, DpnII, HinPLI, and MspI) based on the restriction map near the breakpoint junction. Restriction enzymes were then inactivated, and linkers were ligated to the ends of the DNA. A sequence-specific primer and a linker-specific primer were used to amplify the breakpoint junctions by PCR. If PCR products of different sizes were generated in PCRs with chromosomal DNA from clones carrying GCRs compared with the wild-type control PCR product, these bands were purified and sequenced. The detailed sequences of primers and linkers are available on request.
Fig. 2.
Rearranged breakpoint junctions were determined with three PCR steps. (A) Genome walking PCR was performed to narrow the breakpoint junctions to within 400 bp. WT shows PCR products generated from an intact chromosome V. PCR patterns generated with three different chromosomes isolated from mutants carrying GCRs were visualized in 2% agarose gel. (B) De novo telomere-addition PCR identified GCRs carrying a terminal deletion, followed by telomere sequence addition. Chromosomal DNAs from wild type (WT) and Mt, representing intact chromosome V and clones carrying GCRs, respectively, were used as templates for PCR. (C) The linker-mediated PCR was performed with chromosomal DNA that did not generate ladder bands in the de novo telomere-addition PCR. Chromosomal DNAs from WT and GCR clone (Mt) were digested with different restriction enzyme (RE) and ligated with linkers. Then, a primer that can bind at the end of remaining chromosome V of GCR clone and a linker-specific primer were used for amplification of rearranged chromosomes.
Results
GCR Mutator Genes Were Identified with a GCR Mutator Screen. Multiple pathways suppressing GCRs have been characterized by using the yeast S. cerevisiae (12-17). However, it is not clear whether the hypothesis-driven approach that has identified the pathways has discovered all genes and pathways that function in this process. To find other pathways that suppress GCRs, we developed a method to screen a yeast deletion mutant library by modifying the method used for synthetic genetic array analysis (26). For this purpose, a pif1 mutant strain carrying the GCR assay and the HIS3 gene under the mating type A-specific promoter regulation was constructed (Fig. 1A and see Materials and Methods).
After sequential selections, we identified three groups of genes showing different mutator phenotypes (Table 1). Almost all previously identified GCR mutator genes that showed synergistic interaction with a pif1 mutation were identified in this screening. Genes that had not been implicated previously in suppression of GCRs, that are not just hypothetical ORFs, and whose mutations increased CAN-5FOA rates were redisrupted in the RDKY3615 (wild type), RDKY4343 (pif1-m2), and YKJM1347 (pif1Δ) strains, and GCR rates were determined. Mutations in the CDC50, CSM2, ELG1, ESC1, MMS4, RAD5, RAD18, or TSA1 genes increased the GCR rates compared with wild type and showed synergistic interactions when these mutations were combined with the pif1-m2 mutation (Table 2). However, mutations in EMP47, GIP2, GYP8, MGS1, MPH1, NFI1, PES4, STN7, or VPS1 did not increase the GCR rates compared with wild type (data not shown), and their mutations did not enhance the pif1 GCR rate (see Table 3, which is published as supporting information on the PNAS web site); they are likely to be false positives, or a second mutation in the deletion mutant library might have caused the increased GCR rates observed in the primary screen.
Table 1. Three different types of mutators identified in genome-wide screen of S. cerevisiae.
| Mutator | Group I (CAN and GCR) | Group II (CAN) | Group III (GCR) |
|---|---|---|---|
| Newly identified | CSM2, ELG1, MMS4, RAD5, RAD18, TSA1 | — | ALO1, CDC50, ESC1, UFO1 |
| Previously known | CAN1, MRE11, RAD52, RAD54 | MLH1, MMS2, MSH2, MSH3, MSH6, OGG1, RAD1, RAD27, SHU1 | ASF1, CAC1, RAD57, RDH54 |
| Not tested | YDL162c | APP1, ECM17, FYV8, RIB7, SRP72, SSP1, SWF5, TRF5, UNG1, YAP1801, YDC1, YRA2, YNL228w, YGL218w, YJR154w, YHR198c, YPR114w | YLR445w, YBR226c, YCR016w, YNL171c, YOR225w, YOR286w, YGL050w |
| False positive | EMP47, MGS1, MPH1 | — | GIP2, GYP8, NFI1, PES4, STN7, VPS1 |
Group I represents mutators that increased CAN1 inactivation mutation rate and GCR rate, Group II represents mutators that increased only CAN1 inactivation mutation rate, and Group III represents mutators that increased only GCR rates.
Table 2. Effect of new GCR mutator gene defects on the rate of accumulating GCRs in wild-type and pif1-m2 backgrounds.
| Wild type
|
pif1-m2
|
|||
|---|---|---|---|---|
| Relevant genotype | Strain | GCR rate (CANr-5FOAr) | Strain | GCR rate (CANr-5FOAr) |
| Wild type | RDKY3615 | 3.5 × 10−10 (1) | RDKY4343 | 6.3 × 10−8 (180) |
| alo1 Δ | YKJM1921 | 4.7 × 10−8 (134) | YKJM1925 | 1.1 × 10−7 (314) |
| cdc50 Δ | YKJM1379 | 4.8 × 10−9 (14) | YKJM1401 | 2.6 × 10−7 (743) |
| csm2 Δ | YKJM1535 | 2.7 × 10−9 (8) | YKJM1529 | 1.6 × 10−7 (457) |
| elg1 Δ | YKJM1405 | 1.7 × 10−8 (49) | YKJM1403 | 3.0 × 10−7 (857) |
| esc1 Δ | YKJM1479 | 2.3 × 10−9 (7) | YKJM1838 | 1.1 × 10−7 (314) |
| mms4 Δ | YKJM1525 | 5.9 × 10−8 (169) | YKJM1527 | 2.3 × 10−7 (657) |
| rad5 Δ | YKJM1385 | 6.3 × 10−8 (181) | YKJM1387 | 2.2 × 10−7 (633) |
| rad18 Δ | YKJM1389 | 7.1 × 10−8 (202) | YKJM1391 | 2.5 × 10−7 (714) |
| tsa1 Δ | YKJM1467 | 2.6 × 10−9 (7) | YKJM1836 | 3.6 × 10−7 (1,029) |
| ufo1 Δ | YKJM1919 | 2.6 × 10−8 (74) | YKJM1923 | 1.4 × 10−7 (400) |
All strains are isogenic with the wild-type strain RDKY3615 (MATa, ura3-52, leu2Δ1, trp1Δ63, his3Δ200, lys2ΔBgl, hom3-10, ade2Δ1, ade8, hxt13::URA3), with the exception of the indicated mutations. The rate relative to wild type is given in parentheses.
Strain-background differences in 11 strains excluded them from the initial screen. Each ORF was deleted in RDKY3615 and RDKY4343 strains, and the GCR rates of these strains were determined. The deletion mutations in two ORFs that encode ALO1 and UFO1 increased the GCR rate 134- and 74-fold, respectively (Table 2). In the pif1-m2 mutation background, modest synergistic increases were observed by the deletion of these genes.
Mutations in New GCR Mutator Genes Increased the GCR Rates. ELG1 was first identified in a screen for mutations that elevate Ty1 cDNA-mediated mobility (28). Recently, an alternate replication factor complex (RFC) containing ELG1 that is different from RFC1, RAD24, and CTF18 RFCs and its role in DNA replication and checkpoints were reported (29-31). To determine which types of GCRs are suppressed by ELG1, 13 GCRs were isolated in an elg1Δ mutant background, and their breakpoint junctions were determined. The breakpoint junctions were mapped by de novo telomere-addition PCR and linker-mediated PCR (Fig. 2). Of 13 analyzed breakpoint junctions, 8 were de novo telomere addition, 4 were translocation, and 1 had independent point mutations in the CAN1 and URA3 genes. Thus, ELG1 mainly suppresses de novo telomere-addition GCRs. Consistent with this hypothesis, the deletion of EST2 in the elg1Δ mutant reduced the GCR rate observed in the elg1 strain significantly, indicating that telomerase is required for most elg1-induced GCR (data not shown). The RAD5 and RAD18 genes encode proteins functioning in postreplication DNA repair (32). Mutations in either RAD5 or RAD18 increased the GCR rate up to 200-fold, and breakpoint analysis showed that 9 of 10 were de novo telomere addition. The MMS4 gene was identified previously by means of a screen for sensitivity to methyl methane sulfonate (MMS) (33). Subsequent studies found that MMS4 and MUS81 form a heterodimer endonuclease complex that plays a role in processing lesions that block DNA replication, possibly by means of stalled replication forks during S phase (34, 35). Similar to the MMS4 mutation, mutation of the MUS81 gene increased the GCR rate almost 200-fold (V. Pennaneach, K.M., and R. D. Kolodner, unpublished data).
Compared with ELG1, RAD5, RAD18, and MMS4, there are few studies implicating ALO1, CDC50, CSM2, ESC1, TSA1, and UFO1 in DNA metabolism. ALO1 encodes a D-arabinono-1,4-lactone oxidase functioning in D-erythroascorbic acid biosynthesis. The higher sensitivity of the alo1Δ mutant to oxidative stress suggests that ALO1 also functions in the oxidative stress response (36). Interestingly, TSA1, another GCR mutator gene identified in this study, encodes a protein functioning in oxidative response to protect cells from free-radical damage (37). Mutations of ALO1 and TSA1 genes increased the GCR rate 134- and 7-fold compared with wild type, respectively (Table 2). Combining an alo1Δ or a tsa1Δ mutation with the pif1-m2 mutation caused a synergistic increase in the GCR rate. Recent studies (38) have shown also that a tsa1Δ mutation increases both the GCR rate and the rate of accumulating point mutations in CAN1. CDC50 is a cell-cycle protein that is localized mainly in the cytoplasm (39) and involved in protein maturation and transport. MMS and hydroxyurea sensitivities conferred on yeast cells by a cdc50Δ mutation suggest a possible role for CDC50 in DNA metabolism linked to cell-cycle regulation. CSM2 functions during meiosis and is required for proper segregation of chromosomes (40). Inactivation of the CSM2 gene increased the GCR rate 8-fold in the wild-type background and 457-fold in the pif1-m2 background (Table 2). ESC1 encodes a nuclear protein that makes a complex with the SIR4 protein to silence chromatin (41). Although there was only a 7-fold increase in the GCR rate in the esc1Δ strain, a 314-fold increase in the GCR rate was observed when the esc1Δ mutation was combined with a pif1-m2 mutation. UFO1 encodes a protein required for the degradation of Ho endonuclease by recruiting it to the cytoplasm for ubiquitination (42). The mutation of the UFO1 gene increased the GCR rate 74-fold in wild type and 400-fold in the pif1-m2 strain background (Table 2).
Genes Showing Synergistic Growth Defects with a pif1Δ Mutation and Genes Whose Mutations Cause Petite Phenotypes Were Identified. Because our screen included the pif1Δ mutation to enhance the basal level of GCR formation, strains that caused growth defects with the pif1Δ mutation were excluded (see Table 4, which is published as supporting information on the PNAS web site). Nine genes that generated growth defects with the pif1Δ mutation were identified (AKR1, BFR1, HTZ1, IES6, NPL6, RPL13B, RPL27A, RPL35A, and SHU2). The identified genes encode proteins that have been implicated in nuclear functions (HTZ1 and SHU2), as ribosome constituents (RPL13B, RPL27A, and RPL35A), and in protein trafficking (AKR1, BFR1, and NPL6). There was one gene whose function has not yet been determined (IES6). Genes whose mutations caused defects in mating, meiosis, and sporulation when the pif1Δ mutation was combined were also excluded from our GCR suppressor screening (see Table 4). Many hypothetical ORFs whose functions are not yet known were identified in this procedure. It is possible that these hypothetical ORFs function in the mitochondria for energy metabolism based on their inability to grow in a nonfermentable carbon source. From the genes mentioned above, several mutations that caused growth defects with the pif1Δ mutation (AKR1 and BFR1) or that were defective in mating, meiosis, or sporulation (HTZ1, SHU2, and SUV3) were chosen based on their possible role in DNA metabolism. Mutations in these genes in wild type or the pif1-m2 mutant background did not change the GCR rate significantly (data not shown). Also, the telomeres of strains defective in the NPL6, SHU2, and IES6 genes showed larger sizes, and the telomeres of strains defective in the RPL13B and SUV3 genes showed smaller sizes compared with wild type (data not shown).
Discussion
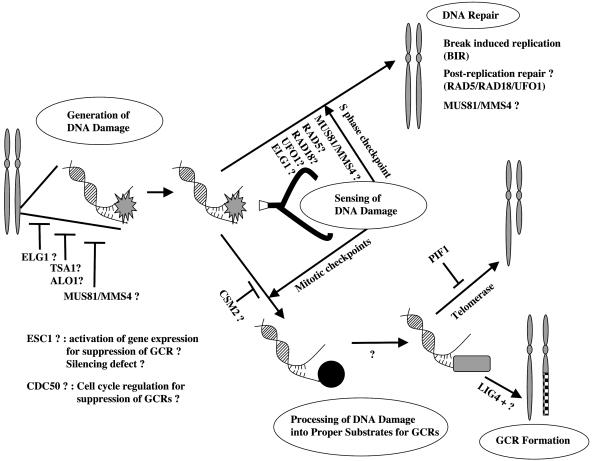
Studies (12, 14-17) have identified GCR suppression pathways based on gene groups known to encode proteins functioning in cell-cycle checkpoints, DNA repair, DNA recombination, and telomere maintenance. In this study, a genome-wide screening method identified 10 genes whose encoded proteins might function differently from proteins implicated previously in the suppression of GCRs. There are at least the following five steps for the formation and suppression of GCRs: generation of DNA damage, sensing the DNA damage, DNA repair that corrects the damage, processing of DNA damage in a manner that generates substrates for GCRs, and GCR-producing machinery (Fig. 3). In each step, different groups of proteins normally act to suppress GCRs. Defects in DNA replication or in the suppression of toxic cellular products, such as hydrogen peroxide, can induce DNA damage. DNA damage by diverse insults, MMS, γ-ray, camptothecin, and bleomycin, as well as a double strand break (DSB) generated by an Ho endonuclease, increased GCR frequency. Therefore, the DSB seems to be an intermediate for GCR formation (13). ALO1, ELG1, TSA1, and MMS4/MUS81 might function to suppress the generation of DNA damages. The S-phase cell-cycle checkpoints function in sensing DNA damage to suppress GCRs (25, 43), whereas the mitotic checkpoint senses the same DNA damage and allows the formation of GCRs (K.M., S.S., and R. D. Kolodner, unpublished data). RAD5/RAD18 and MMS4/MUS81 could function in sensing DNA damage along with the S-phase checkpoint to suppress GCRs based on the observation of no synergistic increase of GCR rates in strains carrying rad5Δ, rad18Δ, or mms4Δ, with a mec1Δ mutation (data not shown). On the other hand, the E3 ligase function for ubiquitination of target proteins, such as proliferating cellular nuclear antigen (PCNA) by RAD5 and RAD18, might be important for the suppression of GCR. The identification of another E3 ligase, UFO1, during this screen supports the possible role of ubiquitination for the suppression of GCR. The lack of ubiquitination, which increases the half-life of proteins because of defects in these E3 ligases, might cause problems in cell-cycle progression. It has been established that the ubiquitination of the FANCD2 protein upon DNA damage is an important step for the suppression of the chromosome fragility syndrome Fanconi anemia (44). Thus, it is possible that there is a yeast FANCD2-like substrate that suppresses the GCR formation by means of regulation by RAD5, RAD18, and/or UFO1. The formation of the alternate replication factor complex and a role in the cell-cycle checkpoint by ELG1 (29-31) suggests that ELG1 functions in the S-phase checkpoint. Based on the synergistic increase of GCR rate by an additional mutation in the DNA damage checkpoint genes but not the DNA replication checkpoint genes along with the elg1 mutation, it is possible that ELG1 functions in the DNA replication checkpoint (data not shown). Although the role of CSM2 in mitosis is not clear, CSM2 could suppress GCR by means of the inhibition of the mitotic checkpoint function for GCR formation. DNA repair mechanisms, including the break-induced DNA replication pathway (19), correct DNA damage to suppress GCRs. Although there was no increase of GCR rate in strains defective in other postreplication repair proteins (data not shown), it is still possible that there are many redundancies that are under RAD5 and RAD18 regulation in the postreplication repair pathways to suppress GCR. MMS4/MUS81 might also be important in DNA repair for the suppression of GCRs. When DNA damage is not repaired, it can be processed by unknown endonucleases or exonucleases to generate substrates for GCR formation. Finally, de novo telomere-addition GCR by the telomerase complex (which is suppressed by PIF1), LIG4-dependent or -independent translocation, or chromosome fusion GCRs are generated (19, 45). Although it is unclear how CDC50, which is localized in the cytoplasm, suppresses GCRs, it is possible that the cdc50Δ mutation increased the GCR rate as a consequence of abnormal cell-cycle progression. ESC1 might function to suppress GCRs by means of its interaction with a chromatin-silencing factor, SIR4, that changes the expression of genes required for GCRs (41). The synergistic increase of the GCR rates when mutations in CDC50, CSM2, ESC1, and TSA1 were combined with a mec1Δ mutation suggests that at least these proteins seem to function independently from the cell-cycle checkpoint function (data not shown).
Fig. 3.
There are at least five different steps for GCR suppression and formation. Although genomes are highly protected, spontaneous DNA damages due to cellular errors can be generated. Inactivation of proteins such as ALO1, ELG1, TSA1, MMS4, and MUS81 may cause an increase in spontaneous DNA damage. The DNA damage that is generated is detected first by the DNA damage-sensing machinery. The S-phase checkpoints sense this damage and transfer the signal to the DNA repair machinery to facilitate repair. RAD5, RAD18, MUS81, and MMS4 seem to function in DNA repair as well as in the sensing of DNA damage. RAD5 and RAD18, along with UFO1, might function in the transfer of signal by means of the ubiquitination of substrates. There are many different DNA repair pathways that are used to fix broken DNA. Break-induced replication, postreplication repair, and MUS81/MMS4-mediated recombination repair seem to play major roles in the suppression of GCRs. DNA damage that is not corrected by DNA repair pathways is sensed by the mitotic checkpoint, and the GCR formation machinery becomes engaged. First, damaged DNA is processed by unknown endonucleases or exonucleases to generate appropriate substrates. Then, telomerase adds telomere sequences at the end of broken chromosomes to produce the de novo telomere addition that is inhibited by PIF1 or translocations with other chromosome sequences, or they are generated by LIG4-dependent or -independent pathways.
CAN1 inactivation mutators were screened recently (38) with a knock-out library. Comparison of identified CAN1 mutator genes showed that 28 of 33 genes were found in both studies. Four of the five remaining genes were selected in the primary screen of this study; however, they were not selected after secondary screening because of less severe CAN1 mutator phenotypes. The shu2Δ mutant was also not selected because of a growth defect caused by the addition of the pif1Δ mutation. Huang et al. (38) also tested the putative GCR mutator phenotype in some of the strong CAN1 mutators and found that TSA1 and ELG1 are strong GCR mutator genes as well. This study identified TSA1, ELG1, and eight other GCR muator genes.
The identification of new genes that encode proteins functioning in specific biological pathways is indispensable to understanding the mechanism completely. The screening method described in this study using the yeast deletion library can be applicable to the identification of new proteins in almost all of the different biological pathways that can be detected with specific assays.
Genome instability is characteristic of cancer cells (2, 3, 12). The present study has identified at least 10 genes whose mutations increased GCR rates and the human homologs of which might be found in GCR-prone human cancers. Indeed, there have been many recent reports (46-51) supporting the concept that genes identified in our screen have importance in cancer development. Abnormal expression of human TSA1 in many different cancers and severe hemolytic anemia and several malignant cancers in mice lacking murine homolog of TSA1 gene, Prdx1, were observed. The deletion of the human ELG1 gene locus is detected frequently in neurofibromatosis, several mutations in the human RAD5 gene were discovered in some melanomas, and the RAD18-defective mouse embryonic stem cell increased sister chromatid exchanges. Studies to verify the function of each protein found in this study by using yeast and mammalian systems and searching for more mutations in each gene in cancer patients may generate clues for understanding cancer development and finding possible targets to cure cancer.
Supplementary Material
Acknowledgments
We thank S. Burgess (National Institutes of Health), P. Hieter (University of British Columbia, Vancouver), S. Lee (University of Texas, San Antonio), and M. Lichten (National Institutes of Health) for helpful discussions; R. D. Kolodner (University of California at San Diego, La Jolla) for helpful discussions, sharing unpublished data, strains, and comments on the manuscript; C. Boone (University of Toronto) and J. Haber (Brandeis University, Waltham, MA) for strains; and D. Bodine (National Institutes of Health), E. Hendrickson (University of Minnesota), P. Liu (National Institutes of Health), R. Nussbaum (National Institutes of Health), and J. Puck (National Institutes of Health) for comments on the manuscript. K.M. thanks God. A.G. was supported by a Howard Hughes Medical Institute grant to Colgate University.
Abbreviations: GCR, gross chromosomal rearrangement; MMS, methyl methane sulfonate.
References
- 1.Kolodner, R. D. (1996) Genes Dev. 10, 1433-1442. [DOI] [PubMed] [Google Scholar]
- 2.Lengauer, C., Kinzler, K. W. & Vogelstein, B. (1998) Nature 396, 643-649. [DOI] [PubMed] [Google Scholar]
- 3.Vessey, C. J., Norbury, C. J. & Hickson, I. D. (1999) Prog. Nucleic Acid Res. Mol. Biol. 63, 189-221. [DOI] [PubMed] [Google Scholar]
- 4.Hartwell, L. (1992) Cell 71, 543-546. [DOI] [PubMed] [Google Scholar]
- 5.Loeb, L. A., Loeb, K. R. & Anderson, J. P. (2003) Proc. Natl. Acad. Sci. USA 100, 776-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albertson, D. G., Ylstra, B., Segraves, R., Collins, C., Dairkee, S. H., Kowbel, D., Kuo, W. L., Gray, J. W. & Pinkel, D. (2000) Nat. Genet. 25, 144-146. [DOI] [PubMed] [Google Scholar]
- 7.Padilla-Nash, H. M., Nash, W. G., Padilla, G. M., Roberson, K. M., Robertson, C. N., Macville, M., Schrock, E. & Ried, T. (1999) Genes Chromosomes Cancer 25, 53-59. [DOI] [PubMed] [Google Scholar]
- 8.Pinkel, D., Segraves, R., Sudar, D., Clark, S., Poole, I., Kowbel, D., Collins, C., Kuo, W. L., Chen, C., Zhai, Y., et al. (1998) Nat. Genet. 20, 207-211. [DOI] [PubMed] [Google Scholar]
- 9.Piper, J., Rutovitz, D., Sudar, D., Kallioniemi, A., Kallioniemi, O. P., Waldman, F. M., Gray, J. W. & Pinkel, D. (1995) Cytometry 19, 10-26. [DOI] [PubMed] [Google Scholar]
- 10.Schrock, E., Veldman, T., Padilla-Nash, H., Ning, Y., Spurbeck, J., Jalal, S., Shaffer, L. G., Papenhausen, P., Kozma, C., Phelan, M. C., et al. (1997) Hum. Genet. 101, 255-262. [DOI] [PubMed] [Google Scholar]
- 11.Sprung, C. N., Afshar, G., Chavez, E. A., Lansdorp, P., Sabatier, L. & Murnane, J. P. (1999) Mutat. Res. 429, 209-223. [DOI] [PubMed] [Google Scholar]
- 12.Kolodner, R. D., Putnam, C. D. & Myung, K. (2002) Science 297, 552-557. [DOI] [PubMed] [Google Scholar]
- 13.Myung, K. & Kolodner, R. D. (2003) DNA Repair (Amst.) 2, 243-258. [DOI] [PubMed] [Google Scholar]
- 14.Myung, K., Pennaneach, V., Kats, E. S. & Kolodner, R. D. (2003) Proc. Natl. Acad. Sci. USA 100, 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka, S. & Diffley, J. F. (2002) Genes Dev. 16, 2639-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lengronne, A. & Schwob, E. (2002) Mol. Cell 9, 1067-1078. [DOI] [PubMed] [Google Scholar]
- 17.Huang, D. & Koshland, D. (2003) Genes Dev. 17, 1741-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna, K. K. & Jackson, S. P. (2001) Nat. Genet. 27, 247-254. [DOI] [PubMed] [Google Scholar]
- 19.Myung, K., Chen, C. & Kolodner, R. D. (2001) Nature 411, 1073-1076. [DOI] [PubMed] [Google Scholar]
- 20.Zhou, J., Monson, E. K., Teng, S., Schulz, V. P. & Zakian, V. A. (2000) Science 289, 771-774. [DOI] [PubMed] [Google Scholar]
- 21.Schulz, V. P. & Zakian, V. A. (1994) Cell 76, 145-155. [DOI] [PubMed] [Google Scholar]
- 22.Novick, P., Osmond, B. C. & Botstein, D. (1989) Genetics 121, 659-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guarente, L. (1993) Trends Genet. 9, 362-366. [DOI] [PubMed] [Google Scholar]
- 24.Hartman, J. L. t., Garvik, B. & Hartwell, L. (2001) Science 291, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 25.Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104, 397-408. [DOI] [PubMed] [Google Scholar]
- 26.Tong, A. H., Evangelista, M., Parsons, A. B., Xu, H., Bader, G. D., Page, N., Robinson, M., Raghibizadeh, S., Hogue, C. W., Bussey, H., et al. (2001) Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- 27.Wu, X., Li, Y., Crise, B. & Burgess, S. M. (2003) Science 300, 1749-1751. [DOI] [PubMed] [Google Scholar]
- 28.Scholes, D. T., Banerjee, M., Bowen, B. & Curcio, M. J. (2001) Genetics 159, 1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Aroya, S., Koren, A., Liefshitz, B., Steinlauf, R. & Kupiec, M. (2003) Proc. Natl. Acad. Sci. USA 100, 9906-9911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellaoui, M., Chang, M., Ou, J., Xu, H., Boone, C. & Brown, G. W. (2003) EMBO J. 22, 4304-4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanellis, P., Agyei, R. & Durocher, D. (2003) Curr. Biol. 13, 1583-1595. [DOI] [PubMed] [Google Scholar]
- 32.Broomfield, S., Hryciw, T. & Xiao, W. (2001) Mutat. Res. 486, 167-184. [DOI] [PubMed] [Google Scholar]
- 33.Xiao, W., Chow, B. L. & Milo, C. N. (1998) Mol. Gen. Genet. 257, 614-623. [DOI] [PubMed] [Google Scholar]
- 34.Mullen, J. R., Kaliraman, V., Ibrahim, S. S. & Brill, S. J. (2001) Genetics 157, 103-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boddy, M. N., Gaillard, P. H., McDonald, W. H., Shanahan, P., Yates, J. R., 3rd, & Russell, P. (2001) Cell 107, 537-548. [DOI] [PubMed] [Google Scholar]
- 36.Huh, W. K., Lee, B. H., Kim, S. T., Kim, Y. R., Rhie, G. E., Baek, Y. W., Hwang, C. S., Lee, J. S. & Kang, S. O. (1998) Mol. Microbiol. 30, 895-903. [DOI] [PubMed] [Google Scholar]
- 37.Wong, C. M., Zhou, Y., Ng, R. W., Kung, H. F. & Jin, D. Y. (2002) J. Biol. Chem. 277, 5385-5394. [DOI] [PubMed] [Google Scholar]
- 38.Huang, M. E., Rio, A. G., Nicolas, A. & Kolodner, R. D. (2003) Proc. Natl. Acad. Sci. USA 100, 11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radji, M., Kim, J. M., Togan, T., Yoshikawa, H. & Shirahige, K. (2001) Yeast 18, 195-205. [DOI] [PubMed] [Google Scholar]
- 40.Rabitsch, K. P., Toth, A., Galova, M., Schleiffer, A., Schaffner, G., Aigner, E., Rupp, C., Penkner, A. M., Moreno-Borchart, A. C., Primig, M., et al. (2001) Curr. Biol. 11, 1001-1009. [DOI] [PubMed] [Google Scholar]
- 41.Andrulis, E. D., Zappulla, D. C., Ansari, A., Perrod, S., Laiosa, C. V., Gartenberg, M. R. & Sternglanz, R. (2002) Mol. Cell. Biol. 22, 8292-8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaplun, L., Ivantsiv, Y., Bakhrat, A. & Raveh, D. (2003) J. Biol. Chem. 278, 48727-48734. [DOI] [PubMed] [Google Scholar]
- 43.Myung, K. & Kolodner, R. D. (2002) Proc. Natl. Acad. Sci. USA 99, 4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D'Andrea, A. D. & Grompe, M. (2003) Nat. Rev. Cancer 3, 23-34. [DOI] [PubMed] [Google Scholar]
- 45.Mieczkowski, P. A., Mieczkowska, J. O., Dominska, M. & Petes, T. D. (2003) Proc. Natl. Acad. Sci. USA 100, 10854-10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenne, D. E., Tinschert, S., Stegmann, E., Reimann, H., Nurnberg, P., Horn, D., Naumann, I., Buske, A. & Thiel, G. (2000) Genomics 66, 93-97. [DOI] [PubMed] [Google Scholar]
- 47.Yanagawa, T., Iwasa, S., Ishii, T., Tabuchi, K., Yusa, H., Onizawa, K., Omura, K., Harada, H., Suzuki, H. & Yoshida, H. (2000) Cancer Lett. 156, 27-35. [DOI] [PubMed] [Google Scholar]
- 48.Noh, D. Y., Ahn, S. J., Lee, R. A., Kim, S. W., Park, I. A. & Chae, H. Z. (2001) Anticancer Res. 21, 2085-2090. [PubMed] [Google Scholar]
- 49.Tateishi, S., Niwa, H., Miyazaki, J., Fujimoto, S., Inoue, H. & Yamaizumi, M. (2003) Mol. Cell. Biol. 23, 474-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sood, R., Makalowska, I., Galdzicki, M., Hu, P., Eddings, E., Robbins, C. M., Moses, T., Namkoong, J., Chen, S. & Trent, J. M. (2003) Genomics 82, 153-161. [DOI] [PubMed] [Google Scholar]
- 51.Neumann, C. A., Krause, D. S., Carman, C. V., Das, S., Dubey, D. P., Abraham, J. L., Bronson, R. T., Fujiwara, Y., Orkin, S. H. & Van Etten, R. A. (2003) Nature 424, 561-565. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.