Abstract
Reduction in vegetative branching is commonplace when crops are domesticated from their wild progenitors. We have identified genetic loci responsible for these changes in foxtail millet (Setaria italica), a crop closely related to maize but whose genetics are little known. Quantitative trait locus (QTL) analysis and comparative genomics reveal that basal branching (tillering) and axillary branching are partially controlled by separate loci, and that the orthologue of teosinte branched1, the major gene controlling branching phenotype in maize, has only a minor and variable effect. We identify other candidate genes for control of branching, including a number of hormone biosynthesis pathway genes. These results suggest that similar phenotypic effects may not be produced by orthologous loci, even in closely related species, and that results from well characterized model systems such as maize must be reviewed critically before being applied to other species.
The study of maize domestication has become a paradigm for developmental evolution, with genetic control of vegetative branching being particularly well elucidated (1, 2). Foxtail millet is genetically less well known, but its presence in the same grass subfamily as maize [Panicoideae (3, 4)] suggests that genetic information from maize may be applicable in foxtail millet. The change in vegetative branching that occurred in the domestication of foxtail millet from its presumed progenitor green millet (Setaria viridis) closely parallels that of maize when compared with its ancestor, teosinte (1, 5). In both cases, the domesticated form has many fewer vegetative branches than its presumed wild progenitor (Fig. 1), presumably as a result of selection for increased allocation to seeds (6). Paterson et al. (7, 8) advanced the hypothesis that morphological convergence among domesticated cereals might reflect selection on corresponding loci and supported this hypothesis with data from genome maps of maize, rice, and sorghum. This hypothesis represents a testable formalization of the idea that humans have selected repeatedly for similar phenotypes in their crops, commonly referred to as the “domestication syndrome” (9), convincingly documented for many crops by Harlan (referred to by him as “adaptation syndrome”) (10). Accordingly, we have tested the hypothesis that similar architecture in maize and foxtail millet reflects human selection on orthologous genetic loci.
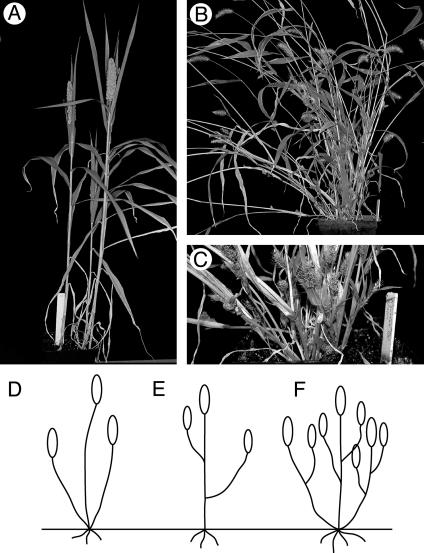
Fig. 1.
Vegetative branching traits. (A) Foxtail millet. (B) Green millet. (C) Green millet. Shown is close-up of base showing tillers and axillary branches. (D-F) Diagrams of various branching patterns, leaves not shown. (D) Tillers. (E) Axillary branches. (F) Tillers and axillary branches.
Materials and Methods
Mapping. A genetic map of a cross between foxtail millet (Setaria italica acc. B100) and green millet (S. viridis acc. A10) using restriction fragment length polymorphism (RFLP) markers was previously constructed at the John Innes Centre (11). The original molecular map used 160 RFLP probes, consisting of anonymous foxtail millet, pearl millet, and wheat genomic clones, and two known function clones identifying the waxy and carboxypeptidase loci (11). Additional rice probes were added to investigate the synteny of foxtail millet with rice, giving a map containing 257 loci and spanning 1,050 cM (12). For the QTL analysis, 119 of these markers were chosen to cover the genome at ≈10-cM intervals. F3 offspring selfed from 120 of the original 127 F2 plants were used to evaluate the number and location of QTL controlling the morphological characters distinguishing the two parents.
QTL Trials. Multiple replicates of the 120 F3 families were grown in two separate trials in a climate-controlled greenhouse at the University of Missouri (St. Louis). Fifteen plants (one per pot) of each family were used per trial, and position of pots was randomized to minimize the effect of differences in light intensity and other environmental variables within the greenhouse. Parental populations were grown in trial 2 but not in trial 1. Trial 1 was grown in May-June and trial 2 in July-August, with standardized soil, fertilizer, and water conditions, and a 16-h day length maintained by artificial lighting when necessary. Trial 2 had both a higher natural light intensity and higher average temperatures than trial 1 although temperatures in both trials were kept between 25°C and 35°C.
Measurement of Phenotypic Traits. Plants were harvested after the seeds had ripened. During harvesting, measurements were made of the number of tillers and the number of axillary branches. Tillers were defined as those branches coming from the base of the plant, often forming adventitious roots, whereas axillary branches were found in the axils of leaves on each tiller (Fig. 1).
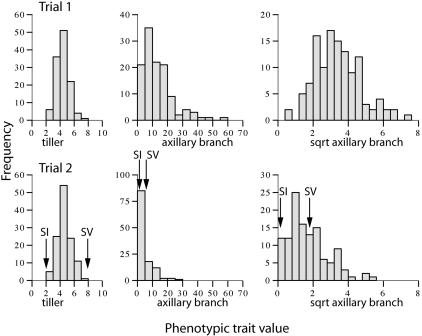
Data Transformation. Analyses were done on tiller number and number of axillary branches. Means were calculated for each trait for each of the families (13). Tiller number was normally distributed, but the distribution of the number of axillary branches was skewed [Fig. 2; Kolmogorov-Smirnov (K-S) test; P < 0.05] (13). This skew was corrected by means of a square root transformation of the data, after which a K-S test indicated that the distribution of the number of axillary branches was normalized in both trials (Fig. 2; K-S test; P > 0.05). Both raw and transformed values for axillary branch number were analyzed for presence and position of QTL, and most of the same QTL were detected (Fig. 3).
Fig. 2.
Histograms of phenotypic values for tiller, axillary branch, and transformed axillary branch number in trials 1 and 2. The mean values for the foxtail millet (SI) and green millet (SV) parents are marked for trial 2; these were not measured in trial 1.
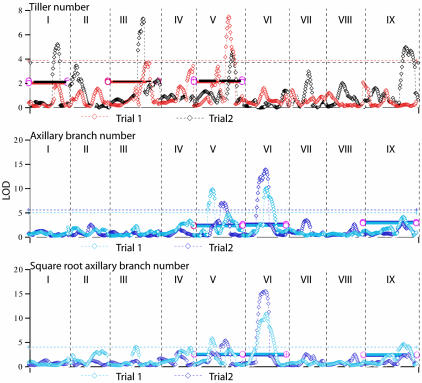
Fig. 3.
Likelihood profiles for tillering, axillary branch number, and square root transformed axillary branch number in the two trials. Each graph shows the likelihood in LOD scores of a QTL at 2-cM intervals along the nine linkage groups (here represented as lying end to end and numbered I to IX). The significance level at the genome-wide level of P < 0.05 level is shown for each trait in the appropriately colored dashed line; the significance level at the chromosome-wide level of P < 0.01 is shown by solid lines for the three chromosomes where shared QTL between trials are found.
QTL Detection. Each trait was analyzed in each trial by using composite interval mapping (CIM) as implemented in qtlcartographer (14). Significance thresholds for QTL were calculated by 1,000 permutations of the original data by using the same parameter settings as for the original analysis (15-17). To strike a balance between detection of true QTL and a high rate of acceptance of false positive results, a chromosome-wide significance level of P < 0.01 was used to declare the presence of QTL. A more stringent genome-wide significance level of P < 0.05 was also calculated. The identification of QTL based on multiple chromosome-wide significance levels will increase type I error compared with the genome-wide level, because nine different tests are being performed (one for each chromosome), but will also increase the probability of identifying more true QTL (18). Note was also taken of logarithm of odds (LOD) peaks in either trial that were significant at the chromosome level of P < 0.05. These peaks were declared as QTL when they were in the same position as significant QTL in the other trial. Where double peaks occurred, only one QTL was declared when the dip between the peaks was <1 LOD interval.
Epistasis. Epistatic interactions were identified by examining each pair of markers for each trait and testing for a significant interaction term, by using the program epistacy (19). Significant interactions were reported at P < 0.001, and the type of interaction (additive by additive, additive by dominant, dominant by additive, or dominant by dominant) was reported as significant at P < 0.01 (see Table 3, which is published as supporting information on the PNAS web site).
Mapping Additional Maize Clones. Additional RFLP markers from maize were added to the original map by using probes from the University of Missouri (Columbia), a teosinte branched1 (tb1) cDNA clone kindly provided by J. Doebley (University of Wisconsin, Madison), and probes amplified from foxtail millet by using primers for terminal ear1 (te1) (20), phytochrome B (phyb) (21), and knotted1 (kn1) (unpublished primers provided by Anthony Verboom, University of Missouri, St. Louis). Southern hybridizations were performed against restricted F2 DNA of the original S. italica acc. B100 × S. viridis acc. A10 population (11, 22). Marker data were scored by two people separately, and then results were cross-checked. Position of these markers on the genetic map was established by using the two-point linkage routine in mapmaker 3 (23).
Identification of possible candidate genes from maize was accomplished by defining intervals on the maize map by using markers mapped on both maize and foxtail millet. In some cases, gaps between common markers were larger than the 1-LOD confidence intervals of the QTL, but estimates of QTL positions were done as precisely as possible. maizegdb (24) and gramene (25, 26) were used to identify genes that had mutant phenotypes in which vegetative branching was affected. These genes were then located on the foxtail millet map by means of the common markers.
Tests for Cosegregation of tb1 with Phenotype. The relationship between variation in tb1 alleles and the phenotypic data was tested by means of ANOVA and analysis of covariance (ANCOVA). The ANCOVA tested the relationship between tb1 and one of the traits in a trial while using the other trait in that trial as the covariate. In doing so, the variance associated with the covariate was removed from the analysis of tb1 vs. the trait of interest (13, 27).
Identification of Candidate Genes from the Rice Genome. Here, we used regions defined by markers common between foxtail millet and rice that contained the identified QTL. These regions were in most cases covered by a number of unassembled contigs (Oryza sativa var japonica, http://rgp.dna.affrc.go.jp/). Each of these contigs was scanned by using fgenesh (28), and identified ORFs were translated and compared with ORFs from other contigs from the same QTL region to reduce redundancies by using blast (29). The final data set of translated proteins for each region was used to query the NCBI database (29). Hits with eV values <10-7 were evaluated, and possible candidate genes were identified.
Results
Phenotypic Distribution of Traits. The phenotypic distribution of the two traits is not always predictable from the parent means (Fig. 2). Axillary branch number showed transgressive segregation (30) toward the upper end of the range of values where some F3 hybrids had many more branches than either parent (only measured in trial 2). Means for tiller number for trial 1 were slightly but significantly lower than for trial 2 (P < 0.01) whereas the opposite was the case for axillary branch number (P < 0.001). Correlations between tiller and axillary branch number within each trial were significant for trial 1 (R = 0.3, P < 0.01) but not for trial 2 (R = 0.15, P = 0.1).
QTL Detection. In the two trials, we found 11 and 14 QTL, respectively (Table 1). These we divide into (i) those that are reproducible between trials, (ii) those with a general effect on branching but not fully reproducible, and (iii) those that affect only one trait in one trial.
Table 1. Significant QTL for the two trials at the chromosome-wide level of P < 0.01 for tillers and numbers of axillary branches (square root transformed).
| Trial 1
|
Trial 2
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Chromosome | Position, cM | Additive | Dominance | R2, % | Candidate genes | Chromosome | Position, cM | Additive | Dominance | R2, % | Candidate genes |
| Tiller no. | I | 95 | +0.22 | −0.27 | 5.5 | ATSUR1 | I* | 89 | +0.40 | +0.22 | 11.5 | ATSUR1 |
| II | 20 | +0.02 | −0.43 | 5.5 | ||||||||
| † | III* | 114 | +0.51 | +0.12 | 14.9 | |||||||
| III | 140 | +0.23 | −0.46 | 10.6 | ||||||||
| † | III | 160 | +0.27 | +0.07 | 4.6 | |||||||
| IV | 109 | +0.51 | +0.32 | 11.3 | OSMOC1 | |||||||
| V | 80 | +0.42 | +0.21 | 9.1 | ||||||||
| V* | 115 | −0.70 | +0.24 | 28.1 | V* | 122 | −0.39 | +0.16 | 9.1 | |||
| VII | 65 | +0.31 | +0.02 | 5.4 | ||||||||
| IX* | 135 | +0.46 | +0.39 | 12.4 | ATSPS1 | |||||||
| IX* | 149 | +0.47 | +0.30 | 9.2 | ZMTB1, ATAXR1 | |||||||
| Axillary branch no. | II | 108 | −0.49 | +0.33 | 8.8 | |||||||
| III | 96 | +0.45 | −0.36 | 9.2 | ||||||||
| IV | 76 | +0.51 | −0.17 | 6.9 | IV | 69 | −0.39 | +0.14 | 5.2 | |||
| IV | 109 | +0.74 | +0.63 | 11.8 | OSMOC1 | |||||||
| V | 60 | +0.74 | +0.32 | 14.4 | V | 59 | +0.37 | +0.33 | 4.9 | |||
| V | 117 | +0.62 | +0.12 | 8.7 | V | 103 | +0.53 | +0.29 | 9.5 | |||
| VI | 73 | −1.02 | +0.32 | 24.8 | VI | 71 | −1.11 | +0.31 | 37.7 | |||
| VII | 62 | +0.32 | +0.12 | 3.8 | ||||||||
| IX | 131 | +0.75 | +0.42 | 14.8 | ATSPS1 | IX | 147 | +0.38 | +0.08 | 4.2 | ZMTB1, ATAXR1 | |
QTL significant at the genome level of P < 0.05 are marked with asterisks. Positive additive and dominance effects correspond to an increase in the phenotypic value for that trait (numbers represent deviation from the mean inter-parental value). R2 values are the percentage of phenotypic variation explained by a QTL. QTL in bold represent those that are shared between the two trials. Positions marked with † indicate QTL in one trial at the chromosome-wide level of P < 0.05 that correspond to a QTL in the other trial at the chromosome-wide level of P < 0.01. ATAXR1, Arabidopsis thaliana auxin resistant1; ATSUR1, Arabidopsis thaliana superroot1; ATSPS1, Arabidopsis thaliana supershoot1; OSMOC1, Oryza sativa monoculm1; ZMTB1, Zea mays teosinte branched1.
Four QTL for tillering (one each on chromosomes I and V and two on chromosome III) and four for axillary branching (one each on chromosomes VI and IX and two on chromosome V) were significant in both trials and therefore reproducible. The additive effects of these QTL were consistent in sign, supporting the hypothesis that they refer to the same genetic locus or loci. We were surprised to find that none of the reproducible QTL affect both traits, despite the anatomical similarity of tillers and axillary branches. The possible exception is on chromosome V, where the LOD peaks of tillering and axillary branching QTL overlap (see Discussion).
Some QTL seem to have a general effect on branching although this effect is not always significant at the P < 0.01 level for both traits (Table 1 and Fig. 3). These putative loci occur on chromosomes II, III, IV, V, and VI in trial 1, and on V, VII, and IX in trial 2. The third set of QTL appear in only one trial or the other. Variation in number and position of detected QTL between trials is evidence for genotype by environment (G × E) interactions.
The amount of phenotypic variation explained was high in both cases although it varied from trial to trial. Significant QTL explained 66-73% of the variation for tillering, and 65-99% of the variation for axillary branches (Table 1). In general, individual QTL were only of moderate size although one QTL for axillary branching on chromosome VI explained between 25% and 38% of the variation (Table 1). LOD score peaks that just failed to reach significance were found in both trials, and these may represent further QTL (Fig. 3).
Each trait in each trial was controlled by QTL of both positive and negative effect, indicating that foxtail millet contains alleles that act to increase branching as well as the expected alleles that act to decrease branching (Table 1). The additive effects of these QTL, if all alleles were of the same sign, are sufficient to explain the range in variation seen in the F3 hybrids.
Epistasis. Significant digenic epistatic effects between marker loci were observed between markers associated with QTL and between unassociated markers (see Table 3). Only one epistatic effect was significant for tillering (in trial 1), but there were eight significant epistatic effects for axillary branching in trial 1, and nine in trial 2 (P < 0.001) (Table 3). The effects associated with these epistatic interactions explained between 13% and 27% of the phenotypic variance.
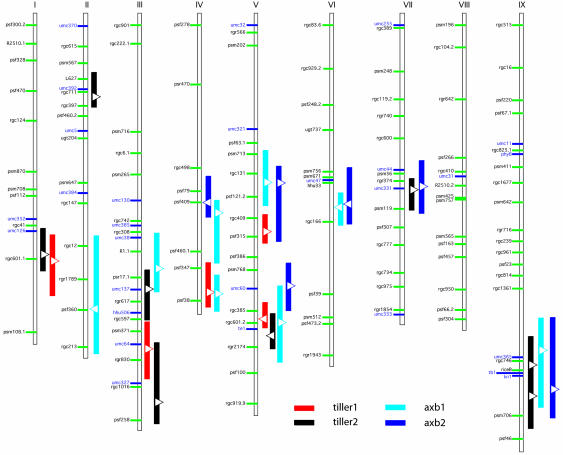
Comparative Mapping. We used comparative mapping to determine whether the loci controlling vegetative branching in foxtail millet were the same as identified in maize. We confirmed colinearity, predicted from rice and other cereal alignments, between the millet and maize genomes by placing 23 new RFLP markers and four genes from maize on the millet genome map (Fig. 4). All markers fell into regions that would be predicted on the basis of synteny with other markers. In particular, we verified that millet chromosomes V and IX, which contain QTL affecting both tillering and axillary branching, are indeed colinear with regions on maize chromosomes 3 and 1, respectively, that also contain QTL for vegetative branching (1, 31).
Fig. 4.
Genome map of the S. italica × S. viridis population. Shown is the location of major QTL and mapped genes for tillering and square root transformed axillary branch number. QTL are shown with 1-LOD support intervals. Triangles on QTL indicate position of maximum LOD score; direction of triangle indicates direction of effect (right, increase; left, decrease). Markers with green bars are from the original genome map and were used in the QTL analyses; those with blue bars represent maize markers or genes added for comparative analysis.
tb1. The strongest candidate gene for control of branching in the cereals is the maize gene tb1, which has been associated with QTL controlling vegetative branching in maize × teosinte crosses, and which has been hypothesized to suppress axillary meristem elongation (2, 32). We hybridized a cDNA clone of tb1 to the F2 mapping filters for millet and placed the gene at the bottom of linkage group IX, between markers riceB and psm706, as also predicted on the basis of synteny with knotted 1, phytochrome B, and various RFLP probes. The gene is in the same region as QTL for axillary branching in both trials and for tillering in trial 2 but not trial 1. However, the amount of variation explained by these QTL was appreciably less than for QTL from chromosomes V and VI.
We directly tested the role of tb1 in controlling vegetative branching by analyzing the relationship between tb1 and each of the traits in each of the trials. The ANOVA analyses showed a highly significant relationship between tb1 and tiller number in trial 2 (P = 0.001), a weakly significant relationship between tb1 and axillary branch number in trial 1 (P = 0.046), and nonsignificant relationships between tb1 and the other two trait/trial combinations (Table 2). When each of the traits was used in turn as a covariate for the other trait in trials 1 and 2, the weak relationship between tb1 and axillary branching in trial 2 disappears although the relationship between tb1 and tiller number in trial 2 remains highly significant (Table 2).
Table 2. ANOVA and ANCOVA for tiller and axillary branch number vs. tb 1.
| Trait | P value (ANOVA) | Trait (covariate) | P value (ANCOVA) |
|---|---|---|---|
| TILL1 | 0.438 | TILL1 (sqaxb1) | 0.801 |
| SQAXB1 | 0.046* | SQAXB1 (till1) | 0.086 |
| TILL2 | 0.001** | TILLL2 (sqaxb2) | 0.001** |
| SQAXB2 | 0.135 | SQAXB2 (till2) | 0.416 |
P value (ANOVA) is the significance for the trait without a covariate; P value (ANCOVA) is the significance for the trait with a covariate. Trait being tested is in uppercase; covariate is in lower case. TILL, till, tiller number; SQAXB, sqaxb, square root transformed axillary branch number. The suffix 1 or 2 refers to trials 1 or 2, respectively. *, P < 0.05; **, P < 0.01.
Identification of Candidate Genes by Using the Rice Genome. We also examined regions of the sequenced rice genome corresponding to the QTL regions on foxtail millet, using common markers mapped on both the rice and foxtail millet genomes to delimit appropriate rice genomic regions. Because many of these rice regions were not yet annotated, the genome sequence of these regions was analyzed by finding all ORFs, which were then translated and compared with the database to find similar genes in other organisms. We identified several hormone biosynthesis pathway genes and many transcription factors in the putatively colinear regions on rice. These genes included a number of auxin and gibberellin pathway genes in the rice region colinear to the QTL for tillering and axillary branching on millet chromosome IX, including a rice orthologue of dwarf plant8 (d8), a maize gene believed to be involved in a late stage of gibberellin synthesis (33). Auxin pathway genes in the rice region colinear to the QTL region on millet chromosome IX include a cytochrome P450 gene with high similarity to the lateral meristem proliferating gene supershoot1, from Arabidopsis, and a gene with high similarity to the Arabidopsis auxin resistant1 gene, all of which are involved in auxin regulation (34, 35). Rice genomic regions colinear to QTL regions on chromosomes I, V, VI, and VII also include a variety of auxin and gibberellin pathway mediators, including Arabidopsis superroot1 (36), and semidwarf1, the so-called “green revolution” gene in rice (37). We also identified a gene involved in regulation of tillering in rice, monoculm1,in the region of a QTL on chromosome IV correlated with both tillering and axillary branching in trial 1 (38).
Discussion
Phenotypic Differences and QTL Detected. Phenotypic traits, especially axillary branch number, differed markedly between the two trials. Plants in trial 2 had more tillers but fewer axillary branches, relative to plants in trial 1, resulting in a more elongate and lax appearance. Trial 2 experienced increased temperature and insolation, which may have caused the differences in plant architecture. Plants in both trials produced tillers predominantly during early growth and axillary branches only when the main stem had commenced production of an inflorescence.
The range of F3 mean phenotypic values for tillering in trial 2 matched that of the difference between the two parents whereas the range for axillary branch number greatly exceeded that of the two parents. Each parent has axillary branch QTL with both positive and negative effects, so that, under random segregation in the F2 generation, these alleles will be found in all combinations in the different F2 (and F3) hybrids. Some of the hybrids will have all alleles of positive or negative sign, producing the extremes of the trait distributions observed. This phenomenon has been termed “transgressive” segregation (30), because the hybrid values transgress the range of values found between the two parents, and has been observed in many crosses between species (39, 40). However, in maize, there are no reports of transgressive segregation for tillering, and QTL are of the same sign, all supporting a reduction in tillering between teosinte and maize (1, 5).
Summation of the additive effects in trial 2 accounts for variable amounts of the difference in phenotypic values between the parents. Additive effects account for ≈50% of the difference in tiller number, but almost none of the difference in axillary branch number. Several QTL have the expected positive effect on tiller and branch number, corresponding to the larger number of branches in green millet relative to foxtail. For axillary branching, however, the QTL with the largest additive effect is negative in sign, indicating that the green millet allele causes fewer branches and the foxtail millet allele more.
Significant digenic epistatic effects were found between some markers, whether associated with QTL or not (Table 3). Epistatic effects may account for some of the discrepancies between the range of parental values and the sum of the additive effects. Patterns of combinatorial additive effects and digenic epistasis have also been observed in sunflower hybrids (41).
Some QTL that affect tillering also affect axillary branching, suggesting a degree of common control between the two traits. This finding is not surprising, given that they are both branching phenotypes. The commonalities are more evident in trial 1 than in trial 2, a finding supported by the significant correlation between the two traits in trial 1 but not in trial 2. The QTL that affect both traits presumably have an effect on general plant robustness, increasing overall branching.
Eight QTL were found in both trials, four for tillering and four for axillary branching. We infer that the repeatable QTL represent loci that control differences in branching between the two species. Two of the QTL on chromosome V, one for tillering (115 and 122 cM in trials 1 and 2, respectively) and one for axillary branching (117 and 103 cM in trials 1 and 2, respectively), may reflect the same locus because they are close to each other. However, the signs of their effects are opposite, implying either that they are two separate loci affecting branching in different directions, or that the same locus has opposite effects on tillering and axillary branching. The sign of these QTL suggests that the allele for increased axillary branching is carried by the green millet parent whereas that for increased tillering is carried by the foxtail millet parent.
In our mapping population, some of the variation in tillering is controlled by loci separate from those controlling axillary branching. A similar result has been reported in pearl millet (P. glaucum) (42), a species more closely related to foxtail millet (43) than either is to maize. Grass taxonomists have also frequently noted a lack of correlation between the presence of tillers and axillary branches (44).
Correlation of QTL with tb1. QTL for tillering in trial 2 and for axillary branching in both trials are in the same region as tb1 but explain only ≈9% of the variance for tillering and 4-14% for axillary branching. ANOVAs found a significant relationship between tb1 and tillering in trial 2 and a weakly significant relationship with axillary branching in trial 1 (P = 0.001 and P = 0.046, respectively). However, the two traits were strongly correlated in trial 1, and, when the effect of tillering was removed statistically by means of analysis of covariance, the relationship with axillary branching was no longer significant. This lack of relationship between tb1 and axillary branching was unexpected. Thus, the tb1 region plays a role in branching, but it is not the only, or the most important, locus controlling vegetative branching in foxtail millet.
Other Candidate Genes for Vegetative Branching. We searched for candidate genes for vegetative branching other than tb1 by comparative mapping of QTL onto the maize genome using common markers. This method helped eliminate potential candidate genes with the appropriate mutant phenotype but that map outside the QTL region. For example, three QTL for branching on foxtail millet chromosome V are found between the common markers umc321 and umc60, and two between umc60 and terminal ear1 (te1) (Fig. 3). The markers appear in the same order on maize chromosome 3. The maize genes Corn-grass1 and -2, whose mutant phenotypes display profuse tillering, map to maize 3, but are placed by synteny between umc32 and umc321 and therefore apparently do not correspond to any of the foxtail millet QTL. Although we cannot rule out local genome rearrangements that might place such genes in the QTL in millet, such an approach allows us to prioritize our research effort on candidate genes that are found near detected QTL.
A number of genes were found through analysis of rice genome regions orthologous to QTL in foxtail millet. This list of genes is limited by the incomplete assembly and annotation of the rice genome, coupled with the large size of our QTL. Future fine-mapping will be necessary to delimit the regions more precisely.
Conclusions
Our results clearly show that human selection in domestication of foxtail millet had its main effect on loci different from those involved in domestication of maize. In foxtail millet, tb1 is not a major candidate for the control of vegetative branching and does not always have a significant effect. Prompted by these results, we dissected a number of foxtail millet plants and could not find visible axillary meristems, raising the possibility that axillary meristem initiation as well as elongation varies between green and foxtail millet. tb1 affects only branch elongation (2), which may explain why it is only a minor player in foxtail millet domestication. As well, the molecular signature of selection observed in tb1 in maize may reflect strong selection for traits other than vegetative branching because tb1 also affects inflorescence architecture and sex expression (1, 2, 5). Data on foxtail millet point to other candidate genes, many in hormone biosynthesis and response pathways, that have more important and consistent effects on both tillering and axillary branching between the domesticated species and its presumed progenitor.
Our data show that the domestication syndrome caused by human selection for a particular phenotype (10) may not always reflect corresponding loci in disparate species. Simple extrapolation from maize (the most closely related model system) to foxtail millet would have overestimated the role of tb1 in tillering and axillary branching. Extrapolation would also have failed to identify other more important loci. In contrast, the combination of QTL and comparative genome analysis has identified a number of possible candidate genes responsible for control of these traits. Although many of the loci underlying domestication QTL in foxtail millet thus seem to be different from those in maize, it will be intriguing to discover whether they correspond to important loci in other cereals. It will be of interest to know whether the correspondence of genomic regions identified by Patterson et al. (7, 8) is particularly evident for traits of the inflorescence, which are presumably the direct targets of human selection, rather than vegetative traits, which are presumably selected indirectly.
Supplementary Material
Acknowledgments
We thank Torbert Rocheford, Jeff Wong, James Beales, Chris Basten, Krista Nichols, Rebecca Doerge, Joe Williams, Simon Malcomber, Michael Zanis, and members of the Kellogg laboratory for helpful discussions, John Doebley for donating a clone of tb1, and Andrew Paterson and Loren Rieseberg for helpful comments on the manuscript. This work was supported by a National Science Foundation grant (to E.A.K.).
Abbreviations: QTL, quantitative trait locus; RFLP, restriction fragment length polymorphism; LOD, logarithm of odds; tb1, teosinte branched1.
References
- 1.Doebley, J. & Stec, A. (1991) Genetics 129, 285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hubbard, L., McSteen, P., Doebley, J. & Hake, S. (2002) Genetics 162, 1927-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grass Phylogeny Working Group (2000) in Grasses: Systematics and Evolution, eds. Jacobs, S. W. L. & Everett, J. E. (CSIRO, Collingwood, Victoria), pp. 3-7.
- 4.Grass Phylogeny Working Group (2001) Ann. Mo. Bot. Gard. 88, 373-457. [Google Scholar]
- 5.Doebley, J. & Stec, A. (1993) Genetics 134, 559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harlan, J. R. (1973) Evolution 27, 311-325. [DOI] [PubMed] [Google Scholar]
- 7.Paterson, A. H., Lin, Y.-R., Li, Z., Schertz, K. F., Doebley, J. F., Pinson, S. R. M., Liu, S.-C., Stansel, J. W. & Irvine, J. E. (1995) Science 269, 1714-1718. [DOI] [PubMed] [Google Scholar]
- 8.Paterson, A. H. (2002) New Phytol. 154, 591-608. [DOI] [PubMed] [Google Scholar]
- 9.Poncet, V., Lamy, F., Enjalbert, J., Joly, H., Sarr, A. & Robert, T. (1998) Heredity 81, 648-658. [Google Scholar]
- 10.Harlan, J. R. (1992) Crops and Man (Am. Soc. of Agronomy and Crop Science Soc. of America, Madison, WI).
- 11.Wang, Z. M., Devos, K. M., Liu, C. J., Wang, R. Q. & Gale, M. D. (1998) Theor. Appl. Genet. 96, 31-36. [Google Scholar]
- 12.Devos, K. M., Wang, Z. M., Beales, J., Sasaki, T. & Gale, M. D. (1998) Theor. Appl. Genet. 96, 63-68. [Google Scholar]
- 13.Anonymous (2001) spss (SPSS, Chicago).
- 14.Basten, C. J., Weir, B. S. & Zeng, Z.-B. (1994) in Proceedings of the 5th World Congress on Genetics Applied to Livestock Production: Computing Strategies and Software, eds. Smith, C., Gavora, J. S., Chesnais, B. B. J., Fairfull, W., Gibson, J. P., Kennedy, B. W. & Burnside, E. B. (Organizing Committee, 5th World Conference on Genetics Applied to Livestock Production, Guelph, ON, Canada), Vol. 22, pp. 65-66. [Google Scholar]
- 15.Churchill, G. A. & Doerge, R. W. (1994) Genetics 138, 963-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doerge, R. W. & Churchill, G. A. (1996) Genetics 142, 285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basten, C. J., Weir, B. S. & Zeng, Z.-B. (1999 1996) qtlcartographer (Department of Statistics, North Carolina State University, Raleigh).
- 18.Cheverud, J. M. (2001) Heredity 87, 52-58. [DOI] [PubMed] [Google Scholar]
- 19.Holland, J. B. (1998) J. Hered. 89, 374-375. [Google Scholar]
- 20.White, S. E. & Doebley, J. F. (1999) Genetics 153, 1455-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathews, S., Tsai, R. C. & Kellogg, E. A. (2000) Am. J. Bot. 87, 96-107. [PubMed] [Google Scholar]
- 22.Laurie, D. A., Pratchett, N., Devos, K. M., Leitch, I. J. & Gale, M. D. (1983) Theor. Appl. Genet. 87, 177-183. [DOI] [PubMed] [Google Scholar]
- 23.Lander, E. S., Green, P., Abrahamson, Barlow, A., Daly, M. J., Lincoln, S. E. & Newburg, L. (1987) Genomics 1, 174-181. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence, C. J., Dong, Q., Polacco, M. L., Seigfried, T. E. & Brendel, V. (2004) Nucleic Acids Res. 32, D393-D397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ware, D., Jaiswal, P., Ni, J., Pan, X., Chang, K., Clark, K., Teytelman, L., Schmidt, S., Zhao, W., Cartinhour, S., et al. (2002) Nucleic Acids Res. 30, 103-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ware, D. H., Jaiswal, P., Ni, J., Yap, I. V., Pan, X., Clark, K. Y., Teytelman, L., Schmidt, S. C., Zhao, W., Chang, K., et al. (2002) Plant Physiol. 130, 1606-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinn, G. P. & Keough, M. J. (2002) Experimental Design and Data Analysis for Biologists (Cambridge Univ. Press, Cambridge, U.K.).
- 28.Salamov, A. A. & Solovyev, V. V. (2000) Genome Res. 10, 516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lynch, M. & Walsh, B. (1998) Genetics and Analysis of Quantitative Traits (Sinauer, Sunderland, MA).
- 31.Lukens, L. N. & Doebley, J. (1999) Genet. Res. 74, 291-302. [Google Scholar]
- 32.Doebley, J., Stec, A. & Hubbard, L. (1997) Nature 386, 485-488. [DOI] [PubMed] [Google Scholar]
- 33.Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuok, M. & Yamaguchi, J. (2001) Plant Cell 13, 999-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tantikanjana, T., Yong, J. W. H., Letham, D. S., Griffith, M., Hussain, M., Ljung, K., Sandberg, G. & Sundaresan, V. (2001) Genes Dev. 15, 1577-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stirnberg, P., Chatfield, S. P. & Ottoline Leyser, H. M. (1999) Plant Physiol. 121, 839-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King, J. J., Stimart, D. P., Fisher, R. H. & Bleecker, A. B. (1995) Plant Cell 7, 2023-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monna, L., Kitazawa, N., Yoshino, R., Suzuki, J., Masuda, H., Maehara, Y., Tanji, M., Sato, M., Nasu, S. & Minobe, Y. (2002) DNA Res. 9, 11-17. [DOI] [PubMed] [Google Scholar]
- 38.Li, X., Qian, Q., Fu, Z., Wang, Y., Xiong, G., Zeng, D., Wang, X., Liu, X., Teng, S., Hiroshi, F., et al. (2003) Nature 422, 618-621. [DOI] [PubMed] [Google Scholar]
- 39.Burke, J. M., Tang, S., Knapp, S. J. & Rieseburg, L. H. (2002) Genetics 161, 1257-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieseberg, L. H., Widmer, A., Arntz, A. M. & Burke, J. M. (2003) Philos. Trans. R. Soc. London Ser. B 358, 1141-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieseberg, L. H., Raymond, O., Rosenthal, D. M., Lai, Z., Livingstone, K., Nakazato, T., Durphy, J. L., Schwarzbach, A. E., Donovan, L. A. & Lexer, C. (2003) Science 301, 1211-1216. [DOI] [PubMed] [Google Scholar]
- 42.Poncet, V., Lamy, F., Devos, K. M., Gale, M. D., Sarr, A. & Robert, T. (2000) Theor. Appl. Genet. 100, 147-159. [Google Scholar]
- 43.Doust, A. N. & Kellogg, E. A. (2002) Am. J. Bot. 89, 1203-1222. [DOI] [PubMed] [Google Scholar]
- 44.Clark, L. G. & Pohl, R. W. (1996) Agnes Chase's First Book of Grasses (Smithsonian Institution Press, Washington, DC).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.