Abstract
Vibrio parahaemolyticus is the causative agent of food-borne gastroenteritis disease. Once consumed, human acid gastric fluid is perhaps one of the most important environmental stresses imposed on the bacterium. Herein, for the first time, we investigated Vibrio parahaemolyticus CHN25 response to artificial gastric fluid (AGF) stress by transcriptomic analysis. The bacterium at logarithmic growth phase (LGP) displayed lower survival rates than that at stationary growth phase (SGP) under a sub-lethal acid condition (pH 4.9). Transcriptome data revealed that 11.6% of the expressed genes in Vibrio parahaemolyticus CHN25 was up-regulated in LGP cells after exposed to AGF (pH 4.9) for 30 min, including those involved in sugar transport, nitrogen metabolism, energy production and protein biosynthesis, whereas 14.0% of the genes was down-regulated, such as ATP-binding cassette (ABC) transporter and flagellar biosynthesis genes. In contrast, the AGF stress only elicited 3.4% of the genes from SGP cells, the majority of which were attenuated in expression. Moreover, the number of expressed regulator genes was also substantially reduced in SGP cells. Comparison of transcriptome profiles further revealed forty-one growth-phase independent genes in the AGF stress, however, half of which displayed distinct expression features between the two growth phases. Vibrio parahaemolyticus seemed to have evolved a number of molecular strategies for coping with the acid stress. The data here will facilitate future studies for environmental stresses and pathogenicity of the leading seafood-borne pathogen worldwide.
Keywords: Vibrio parahaemolyticus, acid stress, transcriptome, gene expression, growth phase
1. Introduction
Vibrio parahaemolyticus, autochthonous to estuarine, marine, and coastal environments worldwide, is the causative agent of food-borne gastroenteritis disease and even death [1]. V. parahaemolyticus was first identified in 1950 in Osaka, Japan, where an outbreak of acute gastroenteritis following the consumption of semidried juvenile sardines sickened 272 and killed 20 individuals [2]. To date, more than eighty V. parahaemolyticus serotypes have been described on the basis of the somatic (O) and capsular (K) antigens [1]. Epidemic V. parahaemolyticus O3:K6 emerged in Calcutta, India in 1996 [3], was subsequently isolated in many Asian countries, and recently reported in Europe, Africa and America [1,4], arguing a pandemic of V. parahaemolyticus worldwide.
V. parahaemolyticus is a Gram-negative bacterium that is able to grow at pH 5–11, 1%–7% NaCl, 22–42 °C [5,6]. Once consumed with raw, undercooked or mishandled seafood, V. parahaemolyticus is challenged with the extremely low pH environment in the human stomach (pH of the human stomach normally ranges from 1–3 but can rise above 6.0 after food consumption) [7,8], before reaching the human gastrointestinal tract where it elicits gastroenteritis [9]. The molecular mechanisms of acid stress response in some Gram positive and Gram-negative bacteria (e.g., Escherichia coli, Salmonella enterica) have been reported, such as the pumping out of protons, production of ammonia and proton-consuming decarboxylation reactions, as well as modifications of the lipid content in the membrane (for a review, see [10]).
To date, the general stress response of Vibrionaceae-related bacteria under detrimental acid conditions remains largely unknown, despite their great significance in human health and economy in aquaculture industry. Some studies have revealed that Vibrios have a similar lysine-decarboxylation pathway in response to acid stress as E. coli, which consists of a lysine decarboxylase (CadA) and a lysine/cadaverine antiporter (CadB). The cadA and cadB genes were transcribed at low constitutive levels in an acid-independent manner and induced during infection and acid tolerance in Vibrio cholerae [11], and the genes were activated sequentially by two transcriptional regulators AphB and CadC of Vibrio vulnifus in acid stress [12]. Short preadaptation to a 6% salt concentration increased survival of the wild-type strain but not that of a cadA mutant of V. parahaemolyticus under lethal acid conditions [13]. Previous research on specific genes also revealed a few regulatory proteins (e.g., ToxRS and OmpU) involved in V. parahaemolyticus response to acid, bile salts, and sodium dodecyl sulfate stresses (e.g., [14]). In this study, for the first time, we investigated global-level gene expression profiles of V. parahaemolyticus CHN25 in response to artificial gastric fluid (AGF) stress by using full-genome microarray analysis. The information will facilitate our better understanding of molecular mechanisms underlying environmental stresses and pathogenicity of the leading seafood-borne pathogen worldwide.
2. Results and Discussion
2.1. Survival of V. parahaemolyticus CHN25 under Acid pH Conditions
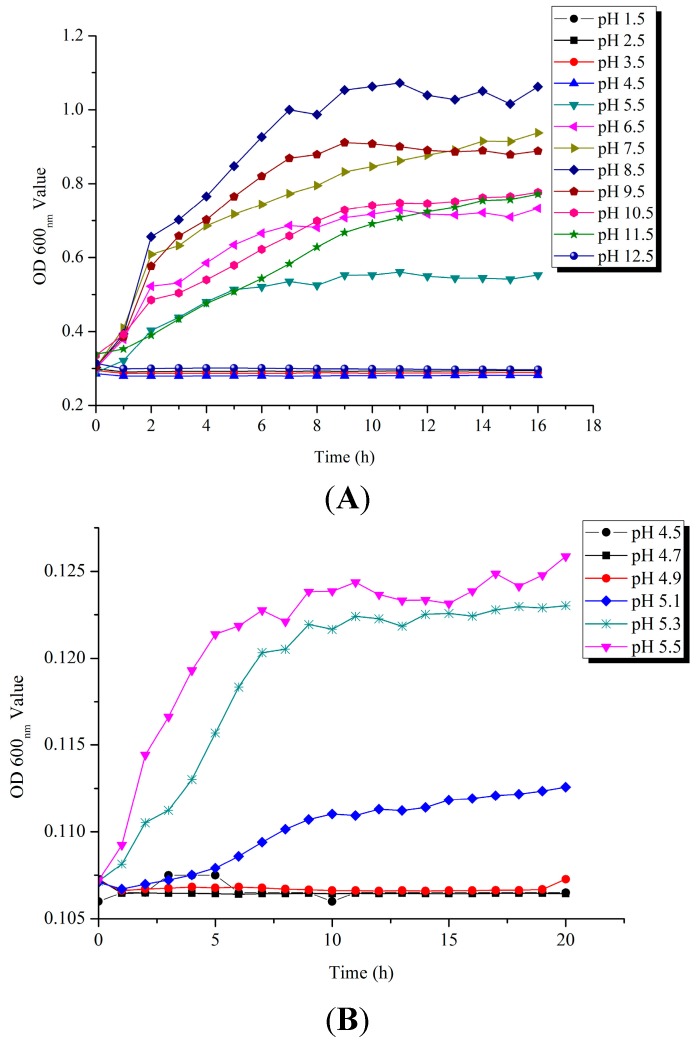
To gain an insight into the V. parahaemolyticus CHN25 tolerance to acid conditions, we determined growth curves of the bacterium, recently isolated and identified by Song et al. [15], in Tryptic Soy Broth (TSB) with the pH range of 1.5–12.5 at 37 °C. As illustrated in Figure 1A, V. parahaemolyticus CHN25 grows at pH 5.5–11.5, optimally at pH 8.5, demonstrating it is a moderately basophilic bacterium, consistent with previous studies (e.g., [5]). No cell growth was observed under more acidic conditions with pH values lower than 4.5. More detailed tests on the pH range between 4.5 and 5.5 revealed that V. parahaemolyticus CHN25 was able to grow at pH 5.0, but not at pH ≤ 4.9 (Figure 1B), suggesting the latter being a sub-lethal pH condition for V. parahaemolyticus CHN25.
Figure 1.
Survival of V. parahaemolyticus CHN25 under different pH conditions. The bacterium was grown in TSB liquid medium at pH 1.5–12.5 (A) and pH 4.5–5.5 (B), 37 °C, and growth curves were determined using a BioScreener.
2.2. Tolerance of V. parahaemolyticus CHN25 at Logarithmic Growth Phase (LGP) and Stationary Growth Phase (SGP) to the AGF (Artificial Gastric Fluid) Stress
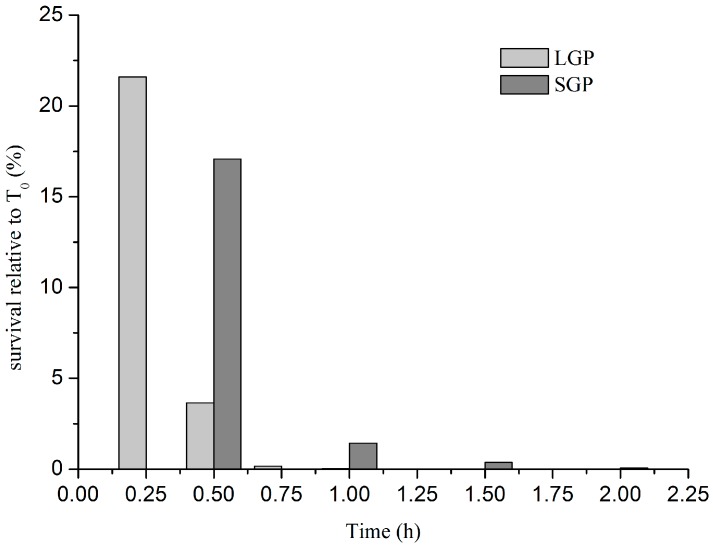
To investigate the possible effects of the human acidic stomach environment in vivo on V. parahaemolyticus CHN25 survival, we utilized the AGF (pH 4.9) to treat the bacterium in vitro grown to LGP and SGP in TSB (pH 8.5) at 37 °C, respectively. As shown in Figure 2, V. parahaemolyticus CHN25 cells at LGP displayed relatively lower survival rates when compared to the bacterial cells at SGP. Treating the LGP cells for 15 min resulted in a significantly decreased survival rate (21.6%), and further elevating exposure time (≥30 min) yielded a steep reduction in the survival (≤3.7%). For the SGP cells, the relative survival rate was 17.0% after exposed to AGF for 30 min, which was 4.6-fold higher than that for LGP cells. Nevertheless, the SGP cells also showed considerable loss in culturability after 30 min exposure to AGF. Thus, we extracted total RNA of the samples at both growth phases in TSB after treated for 30 min with AGF (pH 4.9) for the further transcriptomic analysis (see below). The samples cultured under the same condition without the AGF treatment were used as a control, respectively.
Figure 2.
Tolerance of V. parahaemolyticus CHN25 at LGP (logarithmic growth phase) and SGP (stationary growth phase) to the AGF (artificial gastric fluid) (pH 4.9) stress.
2.3. Transcriptome Profiles of V. parahaemolyticus CHN25 in the Response to the AGF Stress
We determined the global-level gene expression profiles of V. parahaemolyticus CHN25 after the AGF treatment by using full-genome microarray chips (see the Experimental Section). This analysis revealed a considerable number of differentially expressed genes involved in the response to acid stress in the bacterium. A total of 1210 genes were significantly changed when V. parahaemolyticus CHN25 grown to LGP in the AGF stress, which represented approximately 25.6% of the expressed genes in the bacterium. Of these, a total of 547 genes showed higher transcriptional levels (change ≥ 2.0-fold), whereas the expression of a total of 663 genes were down-regulated (change ≤ 0.5-fold). All the genes were grouped into ninety four gene functional catalogues identified in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (data not shown). When V. parahaemolyticus CHN25 grown to SGP, the AGF stress only elicited 160 differentially expressed genes, accounting for 3.4% of the expressed genes in the bacterium, which consisted of 52 up-regulated and 108 down-regulated genes falling into twenty two gene functional catalogues (data not shown). A complete list of the differentially expressed genes at both growth phases is available in NCBI Gene Expression Omnibus under the accession number GSE63167, of which 10.9% were annotated as hypothetical proteins with currently unknown functions in the public databases. To validate the transcriptome data, we chose eleven representative genes for quantitative real-time reverse transcription PCR (qRT-PCR) analysis. The resulting data were correlated with those yielded from the transcriptomic analysis (Table S1).
Consistent with previous studies (e.g., [16]), the majority of the expressed genes remained unaltered in SGP cells in the AGF stress, in which substrate metabolism, energy production, and cell division were turned down. Nevertheless, the transcriptome profile at this growth phase indeed provided a comparative mode to investigate growth-phase independent acid stress response in V. parahaemolyticus CHN25. Comparison of the transcriptome data revealed forty-one differentially expressed genes that were synchronously elicited from both LGP and SGP cells in the AGF stress, however, approximately half of which coded for hypothetical proteins. Of these, interestingly, almost half displayed different expression features between the two growth phases, e.g., the genes encoding the fructose-specific Enzyme IIABC subunits and prophage-associated proteins (see below). Overall, our data highlighted characteristic and distinct gene expression patterns of V. parahaemolyticus CHN25 with considerable variation over growth phases in the AGF stress.
2.4. Major Metabolic Pathways Involved in the Response of V. parahaemolyticus CHN25 to the AGF Stress
2.4.1. Major Metabolic Pathways Involved in V. parahaemolyticus CHN25 Cells at LGP in the AGF Stress
Based on gene set enrichment analysis (GSEA) of the transcriptome data against the KEGG database, nine significantly affected metabolic pathways with enrichment test p value below 0.05 were identified in LGP cells after exposed to the AGF stress (Table 1). They included the phosphotransferase system (PTS); galactose, nitrogen, fructose and mannose, and pyruvate metabolisms; ribosome and aminoacyl-tRNA biosynthesis; glycolysis/gluconeogenesis; and oxidative phosphorylation. The significantly changed metabolic pathways have also been reported in some other bacteria under acid conditions (e.g., [16,17,18,19,20]).
Table 1.
Major metabolic pathways and cellular functions involved in the response of V. parahaemolyticus CHN25 at LGP and SGP to the AGF (pH 4.9) stress for 30 min.
| Metabolic Pathway/Cellular Function | Locus/Gene in V. parahaemolyticus | Fold Change | Description of Encoded Protein | |
|---|---|---|---|---|
| RIMD2210633 | CHN25 | |||
| LGP cells | ||||
| Phosphotransferase system (PTS) | VPA1667 | Chn25A_1555 | 0.0384 | Glucose-specific IIBC component |
| VPA1424 | Chn25A_1312 | 27.9696 | Fructose-specific IIABC component | |
| VPA1422 | Chn25A_1310 | 2.2688 | Nitrogen regulatory IIA component | |
| VPA1421 | Chn25A_1309 | 2.3425 | Fructose-specific IIB component | |
| VPA1420 | Chn25A_1308 | 2.8724 | Fructose-specific IIABC component | |
| VPA0501 | Chn25A_1196 | 2.6299 | Mannitol-specific enzyme II component | |
| VPA0500 | Chn25A_1197 | 2.6362 | Mannitol-specific enzyme II component | |
| VPA0297 | Chn25A_0302 | 8.029 | fructose-specific IIBC component | |
| VPA0298 | Chn25A_0303 | 2.8846 | Fructose-specific IIA component | |
| VPA0231 | Chn25A_0232 | 2.0079 | Phosphotransferase enzyme II, A component | |
| VPA0230 | Chn25A_0231 | 5.501 | Putative sugar phosphotransferase component II B | |
| VP2637 | Chn25_2566 | 2.2518 | Cellobiose-specific IIB component | |
| VP2636 | Chn25_2565 | 2.6756 | Cellobiose-specific IIC component | |
| VP2046 | Chn25_1932 | 2.6196 | Glucose-specific IIBC components | |
| VP0831 | Chn25_0826 | 4.8477 | N-acetylglucosamine-specific IIABC component | |
| VP0370 | Chn25_0356 | 2.7113 | Mannitol-specific IIABC component | |
| VPA0229 (ulaA) | Chn25A_0230 | 6.4614 | Ascorbate-specific enzyme IIC | |
| VP0366 | Chn25_0352 | 3.2996 | Putative PTS, enzyme I | |
| Galactose metabolism | VPA0879 | Chn25A_0828 | 0.3848 | UDP-glucose 4-epimerase |
| VP2400 | Chn25_2264 | 4.517 | UDP-glucose 4-epimerase | |
| VP2400 | Chn25A_1071 | 3.5838 | UDP-glucose 4-epimerase | |
| VP2399 | Chn25A_1070 | 4.4699 | Galactose-1-phosphate uridylyltransferase | |
| VP2398 | Chn25_2262 | 3.8991 | Galactokinase | |
| VP2398 | Chn25A_1069 | 2.497 | Galactokinase | |
| VP2077 | Chn25_1963 | 2.0087 | Maltodextrin glucosidase | |
| Galactose metabolism | VP0839 | Chn25_0834 | 2.4836 | Phosphoglucomutase |
| VP2403 (ebgA) | Chn25_2266 | 2.4115 | Cryptic beta-d-galactosidase subunit alpha | |
| VP2404 (ebgC) | Chn25_2267 | 2.9965 | Cryptic beta-d-galactosidase subunit beta | |
| VP2397 (galM) | Chn25_2261 | 2.1978 | Aldose 1-epimerase | |
| VP2855 (pfkA) | Chn25_2776 | 0.3195 | 6-phosphofructokinase | |
| Ribosome biosynthesis | VP2772 | Chn25_2698 | 3.0673 | 30S ribosomal protein S7 |
| VP1210 | Chn25_1217 | 2.6329 | 50S ribosomal protein L25 | |
| VP2925 (rplA) | Chn25_2832 | 2.3329 | 50S ribosomal protein L1 | |
| VP2926 (rplK) | Chn25_2833 | 2.1866 | 50S ribosomal protein L11 | |
| VP2923 (rplL) | Chn25_2830 | 2.7378 | 50S ribosomal protein L7/L12 | |
| VP0264 (rplP) | Chn25_0255 | 2.2242 | 50S ribosomal protein L16 | |
| VP1282 (rplT) | Chn25_1289 | 6.5113 | 50S ribosomal protein L20 | |
| VP0262 (rplV) | Chn25_0253 | 2.5181 | 50S ribosomal protein L22 | |
| VP0259 (rplW) | Chn25_0250 | 2.2719 | 50S ribosomal protein L23 | |
| VP0329 (rpmA) | Chn25_0317 | 5.5738 | 50S ribosomal protein L27 | |
| VP0185 (rpmB) | Chn25_0181 | 2.7392 | 50S ribosomal protein L28 | |
| VP0265 (rpmC) | Chn25_0256 | 2.5323 | 50S ribosomal protein L29 | |
| VP0255 (rpmE) | Chn25_0246 | 5.0193 | 50S ribosomal protein L31 | |
| VP0186 (rpmG) | Chn25_0182 | 7.2192 | 50S ribosomal protein L33 | |
| VP2030 (rpsA) | Chn25_1918 | 3.9866 | 30S ribosomal protein S1 | |
| VP0263 (rpsC) | Chn25_0254 | 2.6074 | 30S ribosomal protein S3 | |
| VP2740 (rpsF) | Chn25_2668 | 2.818 | 30S ribosomal protein S6 | |
| VP0439 (rpsI) | Chn25_0398 | 2.1455 | 30S ribosomal protein S9 | |
| VP2453 (rpsO) | Chn25_2391 | 3.7221 | 30S ribosomal protein S15 | |
| VP0266 | Chn25_0257 | 3.3225 | Ribosomal protein S17 | |
| VP0531 (rpsT) | Chn25_0481 | 6.127 | 30S ribosomal protein S20 | |
| Fructose and mannose metabolism | VPA1425 | Chn25A_1313 | 23.9303 | Mannose-6-phosphate isomerase |
| VP2599 | Chn25_2528 | 0.2922 | Fructose-bisphosphate aldolase | |
| VP2488 | Chn25_2426 | 4.6724 | Putative phosphoglucomutase/phosphomannomutase | |
| Amino sugar and nucleotide sugar metabolism | VP0543 | Chn25_0493 | 2.2962 | N-acetylmuramic acid-6-phosphate etherase |
| VPA0833 (glgC) | Chn25A_0780 | 0.4431 | Glucose-1-phosphate adenylyltransferase | |
| VP1023 (glgC) | Chn25_1047 | 2.336 | Glucose-1-phosphate adenylyltransferase | |
| VP0829 (nagA) | Chn25_0825 | 2.027 | N-acetylglucosamine-6-phosphate deacetylase | |
| VPA0038 (nagB) | Chn25A_0033 | 2.5647 | Glucosamine-6-phosphate deaminase | |
| Glycolysis/gluconeogenesis | VPA0566 | Chn25A_1136 | 5.8196 | Alcohol dehydrogenase |
| VPA0180 | Chn25A_0182 | 2.2002 | Phospho-beta-glucosidase B | |
| VP2157 | Chn25_2026 | 4.6439 | Glyceraldehyde-3-phosphate dehydrogenase | |
| Aminoacyl-tRNA biosynthesis | VP2470 | Chn25_2407 | 2.0793 | Tyrosyl-tRNA synthetase |
| VP1280 | Chn25_1286 | 0.4891 | Threonyl-tRNA synthetase | |
| VP2548 (alaS) | Chn25_2482 | 2.6177 | Alanyl-tRNA synthetase | |
| VP0861 (argS) | Chn25_0854 | 2.7778 | Arginyl-tRNA synthetase | |
| VP1150 (cysS) | Chn25_1159 | 2.0297 | Cysteinyl-tRNA synthetase | |
| VP0021 (glyS) | Chn25_0010 | 3.0031 | Glycyl-tRNA synthetase subunit beta | |
| VP0534 (ileS) | Chn25_0484 | 2.3183 | Isoleucyl-tRNA synthetase | |
| VP0727 (leuS) | Chn25_0685 | 2.1816 | Leucyl-tRNA synthetase | |
| VP2069 (metG) | Chn25_1955 | 2.2087 | Methionyl-tRNA synthetase | |
| VP1291 (pheT) | Chn25_1298 | 5.4809 | Phenylalanyl-tRNA synthetase subunit beta | |
| VP2646 (valS) | Chn25_2575 | 2.077 | Valyl-tRNA synthetase | |
| Pyruvate metabolism | VPA1567 | Chn25A_1456 | 2.1699 | Putative pyruvate formate lyase |
| VPA1123 | Chn25A_0560 | 0.4528 | Putative acyl-CoA thiolase | |
| VPA0823 | Chn25A_0771 | 2.0537 | Pyruvate kinase | |
| VPA0646 | Chn25A_1010 | 0.3383 | Putative pyruvate dehydrogenase E1 component, beta subunit | |
| Pyruvate metabolism | VPA0620 | Chn25A_1034 | 0.4695 | Putative acyl-CoA thiolase |
| VPA0611 | Chn25A_1091 | 0.3706 | Acetate kinase | |
| VPA0372 | Chn25A_0367 | 3.9537 | Phosphoenolpyruvate synthase | |
| VPA0144 | Chn25A_0145 | 0.4364 | d-lactate dehydrogenase | |
| VP2881 | Chn25_2801 | 3.0293 | Acetyl-CoA carboxylase, biotin carboxylase subunit | |
| VP2878 | Chn25_2798 | 2.514 | Acetyl-CoA synthetase | |
| VP2545 | Chn25_2479 | 2.1607 | Oxaloacetate decarboxylase subunit gamma | |
| VP2517 | Chn25_2451 | 3.4256 | Dihydrolipoamide dehydrogenase | |
| VP2039 | Chn25_1927 | 2.6156 | Pyruvate kinase II | |
| VP1627 | Chn25_1620 | 3.6175 | Acylphosphatase | |
| VP1258 | Chn25_1264 | 2.6731 | Malate dehydrogenase | |
| VP0325 | Chn25_0313 | 3.5504 | Malate dehydrogenase | |
| VP2519 (aceE) | Chn25_2453 | 2.132 | Pyruvate dehydrogenase subunit E1 | |
| VPA1499 (lldD) | Chn25A_1389 | 8.5267 | l-lactate dehydrogenase | |
| Oxidative phosphorylation | VPA0631 | Chn25A_1023 | 0.464 | Putative protoheme IX farnesyltransferase |
| VPA0544 | Chn25A_1156 | 0.4221 | Protoheme IX farnesyltransferase | |
| VP2841 | Chn25_2763 | 2.102 | Fumarate reductase iron-sulfur subunit | |
| VPA0539 | Chn25A_1161 | 0.2437 | Cytochrome c oxidase, subunit III | |
| VP1543 | Chn25_1521 | 2.2303 | Cytochrome c oxidase, subunit CcoO | |
| VP1541 | Chn25_1519 | 2.2683 | Cytochrome c oxidase, subunit CcoP | |
| VP1054 | Chn25_1074 | 2.0957 | Cytochrome d ubiquinol oxidase, subunit II | |
| VP1053 | Chn25_1073 | 2.3358 | Cytochrome d ubiquinol oxidase, subunit I | |
| VPA0628 | Chn25A_1026 | 2.0087 | Cytochrome o ubiquinol oxidase, subunit I | |
| VP1165 | Chn25_1174 | 2.4205 | Putative manganese-dependent inorganic pyrophosphatase | |
| VP0443 | Chn25_0401 | 2.4995 | Ubiquinol-cytochrome c reductase, cytochrome c1 | |
| VP0442 | Chn25_0400 | 2.0404 | Ubiquinol-cytochrome c reductase, cytochrome b | |
| Oxidative phosphorylation | VP3076 | Chn25_2976 | 0.4698 | F0F1 ATP synthase subunit I |
| VP3068 (atpC) | Chn25_2968 | 3.0803 | F0F1 ATP synthase subunit epsilon | |
| VP0844 | Chn25_0839 | 2.0288 | Succinate dehydrogenase, hydrophobic membrane anchor protein | |
| VP0843 (sdhC) | Chn25_0838 | 2.1718 | Succinate dehydrogenase cytochrome b556 large membrane subunit | |
| Ciliary or bacterial-type flagellar motility | VP0772 (flgA) | Chn25_0767 | 0.2303 | Flagellar basal body P-ring biosynthesis protein FlgA |
| VP2235 (flhA) | Chn25_2102 | 0.4555 | Flagellar biosynthesis protein FlhA | |
| VP2236 (flhB) | Chn25_2103 | 0.463 | Flagellar biosynthesis protein FlhB | |
| VP2255 | Chn25_2122 | 0.436 | Polar flagellar rod protein FlaI | |
| VP2256 (fliD) | Chn25_2123 | 0.4776 | Flagellar capping protein | |
| VP2257 | Chn25_2124 | 2.0357 | Flagellar protein FlaG | |
| VP2261 | Chn25_2127 | 0.2235 | Flagellin | |
| VPA0263 | Chn25A_0264 | 0.3335 | Flagellar basal body P-ring biosynthesis protein | |
| Polyamine transport | VP1332 | Chn25_1334 | 0.4055 | Binding protein component of ABC transporter |
| VP1336 | Chn25_1337 | 0.3839 | ABC transporter ATP-binding protein | |
| VP1337 | Chn25_1338 | 0.419 | Putative permease of ABC transporter | |
| VP1338 | Chn25_1339 | 0.371 | ABC transporter permease | |
| d-ribose transport | VPA1087 | Chn25A_0593 | 12.5376 | d-ribose pyranase |
| VPA1086 | Chn25A_0594 | 14.963 | d-ribose transporter ATP binding protein | |
| VPA1086 (rbsC) | Chn25A_0595 | 8.678 | Ribose ABC transporter permease protein | |
| VPA1084 | Chn25A_0596 | 7.7049 | d-ribose transporter subunit RbsB | |
| Maltose transport | VPA1399 (malG) | Chn25A_1076 | 2.0352 | Maltose transporter permease |
| VPA1400 (malF) | Chn25A_1077 | 2.8009 | Maltose transporter membrane protein | |
| VPA1401(malE) | Chn25A_1078 | 3.0226 | Maltose ABC transporter periplasmic protein | |
| VPA1402 | Chn25A_1079 | 2.561 | Maltose/maltodextrin transporter ATP-binding protein | |
| VPA1644 (lamB) | Chn25A_1532 | 3.35 | Maltoporin | |
| SGP cells | ||||
| Pyrimidine and purine metabolism | VPA1243 | Chn25A_0442 | 0.491 | Cytosine deaminase |
| VP0524 (thyA) | Chn25_0476 | 2.1909 | Thymidylate synthase | |
| VP1760 | Chn25_1387 | 2.1851 | Putative adenylate cyclase | |
| VPA1159 | Chn25A_0527 | 0.4775 | Guanosine 5'-monophosphate oxidoreductase | |
| VPA0855 | Chn25A_0801 | 0.3912 | Putative 5'-nucleotidase | |
| VPA0074 | Chn25A_0069 | 2.3652 | Putative DNA polymerase III, epsilon subunit | |
| VP2303 (dnaE) | Chn25_2169 | 0.4586 | DNA polymerase III subunit alpha | |
| Iron ion transport | VP2491 | Chn25_2429 | 0.4159 | Iron (III) ABC transporter, periplasmic Iron-compound-binding protein |
| VPA0310 | Chn25A_0316 | 2.1424 | Hypothetical protein | |
| PTS | VP2674 | Chn25_2602 | 2.2237 | Phosphocarrier protein NPr |
| VPA0297 | Chn25A_0302 | 0.4786 | PTS system, fructose-specific IIBC component | |
| VPA0298 | Chn25A_0303 | 0.4004 | PTS system, fructose-specific IIA component | |
| VPA1424 | Chn25A_1312 | 0.3946 | PTS system, fructose-specific IIABC component | |
| Quaternary ammonium group transport | VPA1111 | Chn25A_0571 | 0.453 | Putative glycine betaine-binding ABC transporter |
| Aromatic compound catabolic process | VP0240 | Chn25_0231 | 2.2949 | Putative 5-carboxymethyl-2-hydroxymuconate delta isomerase |
| Glycine betaine biosynthetic process from choline | VPA1114 | Chn25A_0568 | 0.4435 | Transcriptional regulator BetI |
| VPA1112 | Chn25A_0570 | 0.3776 | Choline dehydrogenase | |
| Pilus | VPA0725 | Chn25A_0670 | 3.0408 | Putative TadB |
| ATP binding | VPA0380 | Chn25A_0375 | 0.481 | Hypothetical protein |
| VPA1302 | Chn25A_0400 | 0.2424 | Hypothetical protein | |
| Outer membrane-bounded periplasmic space and nitrite reductase activity | VP1928 | Chn25_1816 | 2.0205 | Cytochrome c nitrite reductase pentaheme subunit |
| Cytolysis | VP3048 | Chn25_2947 | 2.0825 | Putative hemolysin III |
| Betaine-aldehyde dehydrogenase activity | VPA1113 | Chn25A_0569 | 0.3189 | Betaine aldehyde dehydrogenase |
| Phosphoglycerate transport | VPA0825 | Chn25A_0773 | 0.3614 | Putative phosphoglycerate transport regulatory protein PgtC |
| NADPH dehydrogenase activity | VPA0465 | Chn25A_1233 | 2.0487 | Putative NAD(P)H oxidoreductase |
| Regulation of DNA repair | VP2945 | Chn25_2852 | 2.205 | LexA repressor |
| 3-isopropylmalate dehydratase complex and 3-isopropylmalate dehydratase activity | VP0343 | Chn25_0331 | 2.0142 | Isopropylmalate isomerase large subunit |
| Triglyceride lipase activity | VP1181 | Chn25_1190 | 2.2305 | lactonizing lipase |
| Lactoylglutathione lyase activity | VP2166 | Chn25_2034 | 0.3883 | Putative lactoylglutathione lyase |
| Anaerobic electron transport chain and nitrogen compound metabolic process | VP1928 | Chn25_1816 | 2.0205 | Cytochrome c nitrite reductase pentaheme subunit |
| Alkaline phosphatase activity | VP2163 | Chn25_2032 | 0.4035 | Alkaline phosphatase |
| Zinc ion transmembrane transporter activity | VPA1287 | Chn25A_0416 | 0.2585 | Putative transporter |
| Transmembrane transport | VP1359 | Chn25_1745 | 2.0727 | Hypothetical protein |
The phosphoenolpyruvate-dependent PTS is known as a major sugar transport multicomponent system in bacteria, by which many sugars are transported into bacteria, concomitantly phosphorylated, and then fed into glycolysis [21]. In this study, thirteen genes in the PTS were significantly up-regulated in LGP cells in the AGF stress, which coded for glucose-, fructose-, mannitol-, cellobiose-, ascorbate-, and N-acetylglucosamine-specific enzyme II components. One of these genes, VPA1424 encoding fructose-specific enzyme II ABC subunits, was strongly up-regulated for 27.97-fold in mRNA level, suggesting extremely active transport and utilization of the fructose in the AGF stress. In addition, interestingly, two genes encoding glucose-specific enzyme II BC subunits were identified in V. parahaemolyticus CHN25, one of which (VP2046) was induced with a minor increase of 2.62-fold, whereas the other (VPA1667) strikingly displayed a 25-fold decrease in expression, implying possible unknown regulation mechanisms underlying the transport of the key sugar in central carbohydrate metabolism in the AGF stress.
The Leloir pathway for the catabolism of d-galactose was positively affected in the AGF stress, which produces UDP-glucose, an important building block for glycogen biosynthesis. Consistent with previous research (e.g., [16]), the genes encoding the pathway components were significantly up-regulated in the AGF stress (Table 1).
Approximately half of the genes linked to nitrogen metabolism were significantly changed by the AGF stress, the majority of which showed higher transcriptional levels. Interestingly, four enzymes: 2-nitropropane dioxygenase (VPA0296) catalyzing nitroalkane to nitrite; NrfBD (VP0987) and NrfAH (VP1989) involved in the conversion of nitrite to ammonia in dissimilar nitrate reduction; and nitrite reductase large subunit (VPA0987) involved in the catalysis of nitrite to ammonia, were significantly up-regulated. Moreover, the gene encoding a glutamate dehydrogenase (VP1602) that catalyzes l-glutamate to ammonia was up-regulated as well, suggesting possibly increased amount of ammonia in LGP cells that likely combined with intracellular protons to yield the ammonium ion and alkalized intracellular environment in the AGF stress [10]. On the other hand, the gene (VP0483) involved in the conversions of ammonia to l-glutamine and to l-glutamate was significantly down-regulated, implying perhaps attenuated ammonia utilization in the AGF stress. In addition, a carbonic anhydrase (VP2514) that converts carbon dioxide to HCO3− was notably down-regulated (5.27-fold), suggesting the repressed production of electrically negative acid ions in LGP cells, which may facilitate to maintain intracellular pH homeostasis in the AGF stress. To our knowledge, no linking to acid stress of the latter two genes has been described previously.
Bacterial ribosome consists of two major subunits, each of which is composed of a variety of proteins. Inconsistent with some previous studies showing down-regulated ribosomal genes under acidic conditions (e.g., [20]), in this study, twelve genes encoding the large 50S ribosomal subunit component were up-regulated (2.19–7.22-fold) in LGP cells after exposed to the AGF stress. Similarly, seven components of the small 30S subunit were also up-regulated in expression (2.15–6.13-fold) (Table 1). Despite a highly conserved translational machinery with invariable rRNA and protein components, the formation of distinct ribosomal subpopulations has been reported in bacteria when encountered adverse conditions, e.g., the S21, L2 and L20 subpopulations at pH 4.5 urea condition in E. coli [22,23]. In this study, L20 and some other components (S20, L27, L31 and L33) were highly up-regulated for more than 5.0-fold in the AGF stress. It will be interesting to elucidate biological significance of the enhanced ribosome synthesis and possible ribosomal subpopulations in V. parahaemolyticus CHN25 to the AGF stress in future research.
In the fructose and mannose metabolisms, two enzymes, bifunctional phosphomannomutase/phosphoglucomutase (VPA2488) and mannose-6-phosphate isomerase (VPA1425) that functioned in the conversions of d-mannose-1 phosphate to β-d-fructose-6 phosphate, were up-regulated in LGP cells. Interestingly, the latter exhibited a 23.93-fold enhanced expression, which reinforced the extremely active fructose metabolism in the AGF stress. All the differentially expressed genes in aminoacyl-tRNA biosynthesis were also up-regulated (2.01–5.48-fold), except the gene (VP1289) with a minor decrease (Table 1).
In the glycolysis/gluconeogenesis, the pfkA (VP2855) gene encoding a 6-phosphofructokinase that catalyzes the second rate-limiting reaction in glycolysis was down-regulated. The following reaction catalyzed by a fructose-bisphosphate aldolase (VP2599) was repressed as well. In contrast, three genes (VP2157, VPA0823, VP2039) in the pathway were up-regulated, the latter two of which coded for pyruvate kinases catalyzing the last rate-limiting reaction in glycolysis, suggesting possibly active pyruvate metabolism in LGP cells in the AGF stress.
Approximately 20 genes linked to the pyruvate metabolism were significantly elicited from LGP cells by the AGF stress. Of these, five genes were down-regulated, and the others were up-regulated. Interestingly, the lldD (VPA1499) gene encoding an l-lactate dehydrogenenase displayed an increase of 8.53-fold in expression, which degrades l-lactate to pyruvate. Moreover, the conversion of malate to pyruvate catalyzed by a malate oxidoreductase (VP1258) was also enhanced. The increased amount of pyruvate was actively metabolized by a phosphoenolpyruvate synthase (VPA0372) to produce phosphoenolpyruvic acid that enters into glycolysis. Similarly, the acetyl-CoA synthetase (VP2878) that catalyzes acetate to acetyl-CoA, an efficient substrate for tricarboxylic acid (TCA) cycle, was also up-regulated. These data suggested active l-lactate, malate and acetate metabolisms in LGP cells in the acid stress, consistent with previous research (e.g., [17]).
In oxidative phosphorylation, strikingly, the most enhanced was the atpC gene (VP3068), encoding ε subunit of a multisubunit F0F1-ATPase, which synthesizes ATP aerobically, as a result of protons passing into the cell, or hydrolyzes ATP for the expulsion of protons from cytoplasm anaerobically [24]. The up-regulated F0F1-ATPase gene operon has been reported in some other bacteria in bile and acid stresses (e.g., [20]). In this study, the enhanced expression of ε subunit of the F0F1-ATPase, which is located in a common central stalk linking the F0 and F1 rotary motors [25], suggested perhaps active pumping of excessive protons from LGP cells after exposed to a sub-lethal acid condition (pH 4.9).
2.4.2. Major Metabolic Pathways Involved in V. parahaemolyticus CHN25 Cells at SGP in the AGF Stress
Based on the GESA-KEEG analysis, only four metabolic pathways, including pyrimidine, purine, as well as fructose and mannose metabolisms and the PTS, were identified to be significantly changed in SGP cells after exposed to the AGF stress (p < 0.05) (Table 1). Distinct from LGP cells, the genes encoding fructose-specific Enzyme IIA subunit (VPA0298) and fructose-specific enzyme II ABC subunits (VPA1424) involved in the PTS and fructose and mannose metabolisms were significantly down-regulated, indicating possibly reduced fructose transport in SGP cells in the AGF stress. In addition, consistent with some previous studies (e.g., [17]), the majority of the differentially expressed genes involved in pyrimidine and purine metabolisms were also down-regulated, e.g., DNA polymerase III α and ε subunits (VP2303, VPA0074), suggesting likely reduced DNA synthesis in SGP cells in the AGF stress.
2.5. Other Altered Biological Functions in V. parahaemolyticus CHN25 in the Response to the AGF Stress
The GSEA of the differentially expressed genes against the Go Ontology (GO) database revealed several significantly affected biological functions (p < 0.05) in LGP cells in the AGF stress (Table 1). Of these, the d-ribose and maltose/maltodextrin transport systems were significantly enhanced. The ATP-binding cassette (ABC) transporters are known as molecular pumps that harness the chemical energy of ATP hydrolysis to translocate solutes across the membrane [26]. Significantly changed ABC transporters have been reported in some other bacteria after acid shock (e.g., [17]). In this study, expression of the rbsABCD operon encoding d-ribose transporter components was strongly enhanced for 7.70–14.96-fold. Enhanced expression of several genes involved in sugar transport and utilization (e.g., ribose) has also been observed in Lactobacillus plantarum in the gastrointestinal tract of mice [27]. Similarly, five genes in the maltose/maltodextrin transport system were also up-regulated (Table 1). These data suggested active d-ribose and maltose/maltodextrin ABC transport systems in the AGF stress.
In contrast, two biological functions were significantly repressed in LGP cells in the AGF stress. One of these was the flagellar biosynthesis and motility, in which all the eight differently expressed genes were notably down-regulated (2.08–4.55-fold), except the flaG gene encoding a distal rod protein with a minor increase in mRNA level. They included the flhA, flhB, flicC, flicD, flgA, VPA0263 and VP2255. Flagellum motility is generally thought to be extremely energy consumptive under detrimental conditions. Albeit previous studies gave different expression characteristics of the genes involved in flagellar biosynthesis and motility under acidic conditions (e.g., [18,28,29,30]), our data strongly suggested the reduced biosynthesis of the flagellum structure and or flagellum motibity in V. parahaemolyticus CHN25 cells at LGP in the AGF stress. In addition, the polyamine transporter system was repressed as well, in which ATP-binding protein (VP1332), ABC transporter binding protein and permeases (VP1336–VP1338) were significantly down-regulated.
Based on the GSEA-GO analysis, a number of significantly changed biological functions were identified in SGP cells (p < 0.05) (Table 1), the majority of which were repressed in the AGF stress. Of these, the most down-regulated was a putative transporter (VPA1287) involved in Zn2+ transmembrane transport system. Likewise, the gene (VP2491) encoding a periplasmic iron-compound-binding protein in Fe3+ transport system was also significantly down-regulated. Low pH is thought to increase metal ion toxicity in bacteria, and an excess of metal ions causes oxidative damage [17]. Our data suggested perhaps decreased transport of the metal ions (Zn2+, Fe3+) into SGP cells after exposed to the AGF stress. In addition, three genes in the glycine betaine (GB) biosynthesis were down-regulated, which are involved in the conversions of choline to betaine aldehyde and betain aldehyde to GB. Among the most up-regulated biological functions in SGP cells was the pilus biosynthesis. The gene (VPA0725), encoding a putative TadB involved in Flp pili biogenesis [31], was up-regulated (3.04-fold) in the AGF stress, suggesting possibly enhanced biofilm formation to protect the bacterium from the detrimental acid stress.
Activation of phage-associated genes at low pH stress has been reported in Lactobacillus reuteri [20]. Strikingly, in this study, three genes (VPA1173–1175) encoding phage major capsid protein, phage capsid scaffolding protein and putative bacteriophage protein showed unusual expression features between the two growth-phases cells of V. parahaemolyticus CHN25 in the AGF stress. They were induced with a minor increase of 2.06–2.19-fold in LGP cells, but highly down-regulated in SGP cells, particularly the capsid-related genes showing strongly 40-fold reduced expression. In addition, expression of the bacteriophage Mu tail sheath protein (GpL, VP2725) was slightly repressed at both growth phases. It will be interesting to elucidate biological significance of the differently expressed phage-associated genes in the AGF stress in future studies.
2.6. Regulators Involved in the Response of V. parahaemolyticus CHN25 to the AGF Stress
The genome-wide transcriptome data also revealed a total of sixty-nine and nine changed regulators in V. parahaemolyticus CHN25 cells at LGP and SGP in the AGF stress, respectively (Table S2). They globally or specifically regulate a wide variety of cellular processes including environmental stresses in bacteria, such as DNA-binding transcriptional or response regulators; LysR-type transcriptional regulators (LTTRs); AraC/XylS-, AsnC-, LacI-, LuxR-, MarR- and TetR-family of regulators; and some other regulators involved in multiple metabolism pathways. Of these, the majority were down-regulated in LGP cells, whereas opposite expression characteristics were observed in SGP cells in the AGF stress.
Interestingly, several differentially expressed regulators were identified from the cells at the two growth phases, suggesting the growth phase-independent and AGF-dependent regulation in V. parahaemolyticus CHN25. Of these, the genes encoding a LacI-family transcriptional regulator (VP2393), repressing a lac operon in E. coli [32], and a putative transcriptional regulator (VPA0593) showed higher expression levels in the AGF stress. In contrast, expression of a regulator BetI (VPA1114) was repressed, which negatively regulated the betT and betIBA genes that govern GB biosynthesis from choline in response to choline in E. coli [33]. The repressed BetI perhaps in turn acivated the target genes in the AGF stress, which perhaps led to increased amount of GB to maintain the integrity of cell membranes against the damaging effects of the AGF stress, as in other stress responses to excessive salt, cold, heat and freezing in bacteria [34]. The possible link between the acid stress and GB, an osmoprotectant in osmotic stress, has also been reported in Streptococcus pneumoniae [16]. The molecular responses of bacteria to external environment signals are complex, but in which the two-component transduction systems have been known to play an important role [35]. Consistent with previous studies, a response regulator (VPA0737) belonging to the two-component signaling systems, which enable bacteria to sense, respond, and adapt to a wide range of environments, stressors, and growth conditions [35], was elicited by the AGF stress in V. parahaemolyticus CHN25. However, distinct responses of the regulator were detected, which was up- and down-regulated in expression in LGP and SGP cells, respectively, implying different regulatory strategies adopted by the bacterium for dealing with the same stressor between the two growth phases. In addition, interestingly, two AsnC-family transcriptional regulators (VPA1717, VPA0091), known as feast/famine regulatory proteins specifically involved in multiple cellular metabolisms in bacteria, displayed 5.0- and 3.5-fold increased expression in LGP and SGP cells, respectively, suggesting possible regulation functions in the AGF stress as those in the feast/famine stress [36].
Among the differentially expressed regulators in LGP cells, the LTTRs were the most abundant in the AGF stress, except putative regulators with currently unknown regulatory functions in the public databases. The LTTRs represent the most abundant type of globally transcriptional regulators in bacteria, which are involved in a wide range of cellular processes, e.g., cell division, quorum sensing, oxidative stress, virulence, motility, attachment and secretion [37]. In this study, a total of ten LTTRs were identified in LGP cells in the AGF stress, however, all of which were significantly repressed in expression, except the one (VP0067) with an opposite minor increase. Similarly, expression of several regulators were suppressed as well, all of which have been reported to directly regulate gene expression in response to environmental stimuli or coordinately regulate in a complex network in bacteria [38,39,40]. For example, an AraC/XylS-family regulator (VPA0531), one of the most common positive regulators in bacteria, showed a decrease of 5.26-fold in the AGF stress. Regulators belonging to this family have three major regulatory functions including stress responses to alkylating agents, antibiotics, organic solvents and heavy metals, as well as the transition from LGP to SGP [38]. In addition, approximately a dozen regulators controlling multiple metabolic pathways were also repressed in the AGF stress, e.g., the DNA-binding transcriptional regulators AraC, HexR and YidZ (VPA1678, VP1236 and VPA1575), the key components in bacterial gene regulatory networks that can sense fluctuations under internal and external conditions [41,42,43]. In contrast, 26.1% of the differentially expressed regulators in LGP cells displayed significantly enhanced expression in the AGF stress. Of these, the regulator (VP2866) belonging to the LuxR-family transcriptional regulators, which are key players in quorum sensing and coordinate gene expression in a variety of cellular functions [44], showed a higher expression level. Similarly, expression of an osmolarity response regulator OmpR (VP0154) involved in the EnvZ/OmpR signal transduction system was also enhanced in the AGF stress, which positively or negatively modulates multiple gene expression implicated in the control of Y. enterocolitica adaptation to high osmolarity, oxidative and low pH stresses [45].
For the SGP cells, expression of a regulator belonging to the LTTRs (VP1316) and a putative transcriptional regulator (VPA1689) were significantly increased, whereas expression of a phospoglycerate transport regulatry protein PgtC (VPA0825) and a putative transcriptional regulator (VP1154) were decreased in the AGF stress.
Taken together, the transcriptome data figured out a complex molecular regulatory network in V. parahaemolyticus CHN25 after exposed to the AGF stress, which lead to three major molecular snapshots. A number of regulators, acting as activators and or repressors of single or operonic genes or a series of regulatory cascades under different environmental stresses in bacteria, were elicited from LGP cells, which perhaps globally or specifically triggered cell responses to the AGF stress and controlled intracellular processes. In contrast, a considerable number of regulators remained unchanged in SGP cells under the same stress condition, which was consistent with the turndown feature at this growth phase. In addition, some growth-phase independent regulators were identified, which likely played crucial roles specifically in the AGF stress response in V. parahaemolyticus CHN25. Finally, the AGF stress appeared to mediate cross-talk regulation with some other environmental stimuli, e.g., osmotic and feast/famine stresses. An in-depth regulatory network in future studies will allow for better understanding of acid stress mechanisms in V. parahaemolyticus.
2.7. Possible Acid Stress Mechanisms in V. parahaemolyticus CHN25
In this study, expression of the genes directly or indirectly associated with the pumping out of protons (e.g., F1F0-ATPase) was significantly enhanced in LGP cells after exposed to a sub-lethal acid condition (pH 4.9). Moreover, two genes (VP2125 and VP2718) encoding Na+/H+ antiporters were also up-regulated, which are important not only for energy transduction, but also for intracellular pH regulation, extrusion of toxic Li+ (and Na+) and cell volume regulation in bacteria [46].
Production of ammonia has been known to be one of the major mechanisms in acid stress response in bacteria. In this study, a number of up-regulated genes involved in nitrogen metabolism (e.g., NrfBD, NrfAH, a glutamate dehydrogenase) were identified, which likely increased intracellular ammonia in LGP cells in the AGF stress. Moreover, expression of the aspA (VP2863) and hutH (VP1273) genes encoding aspartate ammonia-lyase and histidine ammonia-lyase, as well as the gene (VPA0254) encoding l-serine dehydratase 1 that converts serine into ammonia and pyruvate were also significantly increased. In contrast, alanine dehydrogenase (VP1103) and d-amino acid dehydrogenase small subunit (VP0623) showed down-regulated expression. In addition, the nagE (VPA0038) gene encoding a glucosamine-6-phosphate deaminase that converts fructose-6-phpsphate to glucN-phosphate were up-regulated in the AGF stress. It has been reported that urease located on bacterial cell surface may create a neutral microenvironment by hydrolysis of urea to carbon dioxide and ammonia [47]. Unexpectedly, no urease-related genes were identified in V. parahaemolyticus CHN25 in the AGF stress. Overall, these data may have supposed a strong link between the enhanced ammonia production via multiple metabolic pathways in LGP cells and the acid stress imposed on the bacterium.
One interesting observation from the transcriptome data was that the regulator AphB (VP2184) belonging to the LTTRs was not significantly elicited by the AGF stress. Moreover, unexpectedly, expression of the cadAB operon (VP2890–VP2891) involved in the proton-consuming lysine-decarboxylation pathway was strikingly down-regulated (20–25-fold) in LGP cells. This finding was inconsistent with previous studies (e.g., [12,48,49]). We questioned whether the saline concentration of the AGF resulted in the distinct observation, since it has been reported that V. parahaemolyticus RIMD2210633 grown in Luria-Bertani (LB) supplemented with 3% NaCl induced a stronger cadA response after acidification than cells grown in LB with 1% NaCl [50]. To address the interesting result, we treated V. parahaemolyticus CHN25 grown to LGP with the AGF supplemented with 3% NaCl instead of 0.21% NaCl, and then determined the cadAB gene expression by qRT-PCR analysis. The resulting data revealed that both cadA and cadB genes were highly up-regulated in mRNA levels (data not shown). Our data, coupled with the previous results, demonstrated that environmental saline concentration likely mediated an important cross-regulation in acid stress response in V. parahaemolyticus.
In addition, consistent with previous research, the toxS (VP0819) gene was significantly up-regulated in the AGF stress, which belongs to the ToxR-ToxS signal transduction system required for the acid stress response in V. cholerae [11]. In addition, our transcriptome data also revealed some other possible mechanisms in V. parahaemolyticus CHN25, such as the attenuated consumption of ammonia (e.g., VP0483) and enhanced production of HCO3− (e.g., VP2514), to maintain intracellular pH homeostasis in the AGF stress.
3. Experimental Section
3.1. Bacterial Growth Conditions
V. parahaemolyticus CHN25 bearing a SXT/R391-like integrative and conjugative element has recently been characterized by Song et al. [15]. The bacterium was detected positive for the tlh gene, but featured no toxic tdh and trh genes. V. parahaemolyticus CHN25 was streaked from a frozen stock at −80 °C in our laboratory onto LB solid medium [51] adjusted to pH 8.5, 3% NaCl, and incubated at 37 °C overnight. One colony was then inoculated into 5 mL TSB liquid medium (pH 8.5, 3% NaCl) (Beijing Land Bridge Technology Co., Ltd., Beijing, China), and aerobically cultured at 37 °C with shaking at 175 rpm. The overnight culture was diluted 1:100 (v/v) into fresh TSB liquid medium adjusted to the pH range of 1.5–12.5 with 1 M HCl or 6 M NaOH, respectively, and incubated at 37 °C for 16–20 h. The growth curves were determined using a BioScreener (BioScreen, Helsinki, Finland).
3.2. AGF Survival Assay
The AGF survival assay was performed according to the method described previously [19] with slight modifications. Briefly, V. parahaemolyticus CHN25 was incubated in TSB liquid medium at 37 °C to LGP and SGP, defined as an optical density at 600 nm (OD600 nm) of 0.7 and 1.3, respectively. An aliquot of the bacterial culture (1 mL) was centrifuged at 3500 rpm for 2 min, and the cell pellet was resuspended with 1 mL of 1× AGF, containing 8.3 g proteose peptone, 3.5 g d-glucose, 2.05 g NaCl, 0.6 g KH2PO4, 0.147 g CaCl2, and 0.37 g KCl per/L [19]. The cell suspension was added into 4 mL of 1× AGF, and the acid-exposed cells were incubated at 37 °C for 0–60 min or 0–2 h for LGP and SGP cells, respectively. Culturable bacterial cells were enumerated at different time points via plating appropriate dilutions of cell culture onto LB solid medium.
3.3. RNA Extraction and Microarray Analysis
Total RNA preparation was performed using RNeasy Protect Bacterial Mini Kit (QIAGEN Biotech Co., Ltd., Hilden, Germany) according to the manufacturer’s instructions. The DNA was removed from the samples using RNase-Free DNase Set (QIAGEN, Hilden, Germany), and its quality and quantity was assessed using the Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Two independently prepared RNA samples were used in each microarray experiment [52].
Microarray chip design, cRNA labeling, hybridization, scanning and analyses were conducted at Shanghai Biotechnology Co., Ltd. (Shanghai, China). An array of 15,000 specific 60-m oligonucleotides was designed based on predicted coding sequences from V. parahaemolyticus RIMD2210633 and V. parahaemolyticus CHN25, respectively. It contained 4711 probes and covered 99.72% of the genes in V. parahaemolyticus CHN25. A sample of 2 μg RNA was used to synthesize cDNA, which was further transcribed into cRNA using a transcription mix containing aa-UTP and T7 RNA polymerase. Cyanine-3 (Cy3) labeled cRNA was performed by Low Input Quick Amp Labeling Kit, One-Color (Agilent), followed by purification using RNeasy mini kit (QIAGEN), according to the manufacturer’s instructions. Each slide was hybridized with 600 ng Cy3-labeled cRNA using Gene Expression Hybridization Kit (Agilent) in an Agilent Microarray Hybridization Chamber (Agilent) at 65 °C. After 17 h hybridization, slides were washed with Gene Expression Wash Buffer Kit (Agilent) following the manufacturer’s instructions. Microarrays were scanned using Agilent Microarray Scanner (Agilent) and the data were extracted with Feature Extraction software version 10.7 (Agilent). Raw data were normalized by Quantile algorithm, Gene Spring software version 11.0 (Agilent). The average coefficient of variation (CV) was <0.15 as recommended by Agilent for the quality control. Normalized expression ratios were calculated for each gene and tested for significance with the criteria |fold change| > 2.0 and p < 0.05. The GSEA of differently expressed genes was supported by the eBioservice (http://sas.ebioservice.com/portal/root/molnet_shbh/index.jsp) (Shanghai Biotechnology Co., Ltd., Shanghai, China) against the GO (http://geneontology.org/) and KEGG (http://www.genome.jp/kegg/) database, respectively.
3.4. qRT-PCR Analysis
Selected differentially expressed genes and/or significantly enriched genes in microarray chip analysis were validated by qRT-PCR. Oligonucleotide primers were synthesized by Shanghai Sangon Biological Engineering Technology Services Co., Ltd. (Shanghai, China). The reverse transcription reaction was performed using PrimeScript RT reagent Kit With gDNA Eraser (Perfect Real Time) (Japan TaKaRa BIO, Dalian Company, Dalian, China) according to the manfacturer’s protocol. A 20 μL reaction volume contained 10 μL FastStart Universal SYBR Green Master (ROX), 5 μM each of the oligonucleotide primers, 2 μL template cDNA and appropriate volume of sterile ddH2O (Roche, Basel, Switzerland). All qRT-PCR reactions were performed in a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) under the following conditions: initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, and primer annealing at 60 °C for 60 s, according to the manufacturer’s instructions. The pvuA gene was used as the qRT-PCR reference gene as previously described [53,54]. Expression level of the pvuA gene in V. parahemolyticus CHN25 grown in TSB to LGP and SGP was used as a reference/baseline, respectively. The data were analyzed using the Applied Biosystems 7500 software, and the relative expression ratio was calculated for each gene by using the delta-delta threshold cycle (Ct) method [55].
3.5. Microarray Data Accession Number
The microarray data have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE63167.
4. Conclusions
This study constitutes the first investigation of Vibrio parahaemolyticus CHN25 response to the AGF under a sub-lethal acid condition using genome-wide microarray analysis. The transcriptome data revealed global-level distinct gene expression profiles of the bacterium with considerable variation over growth phases after exposed to the AGF (pH 4.9) for 30 min. Vibrio parahaemolyticus seemed to have evolved a number of molecular strategies for coping with the acid stress in a complex gene regulation network. Our data in this study will highly facilitate the in-depth research of environmental stresses and pathogenicity of the leading seafood-borne pathogen worldwide.
Acknowledgments
The work was supported by the Grants No. B-9500-10-0004, No. 13YZ098 and No. ZZhy12028 from Shanghai Municipal Education Commission, and a Grant No. 31271830 from National Natural Science Foundation of China.
Supplementary Materials
Supplementary tables can be found at http://www.mdpi.com/1422-0067/15/12/22539/s1.
Author Contributions
Xuejiao Sun, Taigang Liu, Xu Peng and Lanming Chen participated in the design and or discussion of the study. Xuejiao Sun carried out the major experiments. Xuejiao Sun, Taigang Liu and Lanming Chen analyzed the data. Lanming Chen wrote the manuscript, and Xu Peng revised it for important improvement. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ceccarelli D., Hasan N.A., Huq A., Colwell R.R. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front. Cell. Infect. Microbiol. 2013;3 doi: 10.3389/fcimb.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujino T., Okuno Y., Nakada D., Aoyama A., Fukai K., Mukai T., Ueho T. On the bacteriological examination of Shirasu food poisoning. Med. J. Osaka Univ. 1953;4:299–304. [Google Scholar]
- 3.Okuda J., Ishibashi M., Hayakawa E., Nishino T., Takeda Y., Mukhopadhyay A.K., Garg S., Bhattacharya S., Nair G.B., Nishibuchi M. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 1997;35:3150–3155. doi: 10.1128/jcm.35.12.3150-3155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Velazquez-Roman J., León-Sicairos N., de Jesus Hernández-Díaz L., Canizalez-Roman A. Pandemic Vibrio parahaemolyticus O3:K6 on the American continent. Front. Cell. Infect. Microbiol. 2013;3 doi: 10.3389/fcimb.2013.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Twedt R.M., Spaulding P.L., Hall H.E. Morphological, cultural, biochemical, and serological comparison of Japanese strains of Vibrio parahemolyticus with related cultures isolated in the United States. J. Bacteriol. 1969;98:511–518. doi: 10.1128/jb.98.2.511-518.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joseph S.W., Colwell R.R., Kaper J.B. Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 1982;10:77–124. doi: 10.3109/10408418209113506. [DOI] [PubMed] [Google Scholar]
- 7.Schubert M.L. Gastric exocrine and endocrine secretion. Curr. Opin. Gastroenterol. 2009;25:529–536. doi: 10.1097/MOG.0b013e328331b62a. [DOI] [PubMed] [Google Scholar]
- 8.Russell T.L., Berardi R.R., Barnett J.L., Dermentzoglou L.C., Jarvenpaa K.M., Schmaltz S.P., Dressman J.B. Upper gastrointestinal pH in seventy-nine healthy, elderly, North American men and women. Pharm. Res. 1993;10:187–196. doi: 10.1023/A:1018970323716. [DOI] [PubMed] [Google Scholar]
- 9.O’Boyle N., Boyd A. Manipulation of intestinal epithelial cell function by the cell contact-dependent type III secretion systems of Vibrio parahaemolyticus. Front. Cell. Infect. Microbiol. 2013;3 doi: 10.3389/fcimb.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lund P., Tramonti A., Biase D. Coping with low pH: Molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 2014;38 doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- 11.Merrell D.S., Camilli A. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J. Bacteriol. 2000;182:5342–5350. doi: 10.1128/JB.182.19.5342-5350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee J.E., Jeong H.G., Lee J.H., Choi S.H. AphB influences acid tolerance of Vibrio vulnificus by activating expression of the positive regulator CadC. J. Bacteriol. 2006;188:6490–6497. doi: 10.1128/JB.00533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalburge S.S., Whitaker W.B., Boyd E.F. High-salt preadaptation of Vibrio parahaemolyticus enhances survival in response to lethal environmental stresses. J. Food Prot. 2014;77:246–253. doi: 10.4315/0362-028X.JFP-13-241. [DOI] [PubMed] [Google Scholar]
- 14.Whitaker W.B., Parent M.A., Boyd A., Richards G.P., Boyd E.F. The Vibrio parahaemolyticus ToxRS regulator is required for stress tolerance and colonization in a novel orogastric streptomycin-induced adult murine model. Infect Immun. 2012;80:1834–1845. doi: 10.1128/IAI.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song Y., Yu P., Li B., Pan Y., Zhang X., Cong J., Zhao Y., Wang H., Chen L. The mosaic accessory gene structures of the SXT/R391-like integrative and conjugative elements derived from Vibrio spp. isolated from aquatic products and environment in the Yangtze River estuary, China. BMC Microbiol. 2013;13 doi: 10.1186/1471-2180-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martín-Galiano A.J., Overweg K., Ferrándiz M.J., Reuter M., Wells J.M., Adela G. Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology. 2005;151:3935–3946. doi: 10.1099/mic.0.28238-0. [DOI] [PubMed] [Google Scholar]
- 17.Bore E., Langsrud S., Langsrud Ø., Rode T.M., Holck A. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology. 2007;153:2289–2303. doi: 10.1099/mic.0.2007/005942-0. [DOI] [PubMed] [Google Scholar]
- 18.Hellweg C., Pühler A., Weidner S. The time course of the transcriptome response of Sinorhizobium meliloti 1021 following a shift to acidic pH. BMC Microbiol. 2009;9 doi: 10.1186/1471-2180-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivy R.A., Wiedmann M., Boor K.J. Listeria monocytogenes grown at 7 °C shows reduced acid survival and an altered transcriptional response to acid shock compared to L. monocytogenes grown at 37 °C. Appl. Environ. Microbiol. 2012;78:3824–3836. doi: 10.1128/AEM.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall T., Båth K., Britton R.A., Jonsson H., Versalovic J., Roos S. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 2007;73:3924–3935. doi: 10.1128/AEM.01502-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Postma P., Lengeler J., Jacobson G. Phosphoenolpyruvate: Carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bickle T.A., Howard G.A., Traut R.R. Ribosome heterogeneity the nonuniform distribution of specific ribosomal proteins among different functional classes of ribosomes. J. Biol. Chem. 1973;248:4862–4864. [PubMed] [Google Scholar]
- 23.Moll I., Engelberg-Kulka H. Selective translation during stress in Escherichia coli. Trends Biochem. Sci. 2012;37:493–498. doi: 10.1016/j.tibs.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okuno D., Iino R., Noji H. Rotation and structure of FoF1-ATP synthase. J. Biochem. 2011;149:655–664. doi: 10.1093/jb/mvr049. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J., Li Z., Zhang H., Wang J., Liu Y., Chen G. Rapid detection of several foodborne pathogens by F0F1-ATPase molecular motor biosensor. J. Microbiol. Methods. 2013;93:37–41. doi: 10.1016/j.mimet.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Oldham M.L., Chen S., Chen J. Structural basis for substrate specificity in the Escherichia coli maltose transport system. Proc. Natl. Acad. Sci. USA. 2013;110:18132–18137. doi: 10.1073/pnas.1311407110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bron P.A., Grangette C., Mercenier A., de Vos W.M., Kleerebezem M. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 2004;186:5721–5729. doi: 10.1128/JB.186.17.5721-5729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Jonge R., Ritmeester W.S., van Leusden F.M. Adaptive responses of Salmonella enterica serovar Typhimurium DT104 and other S. Typhimurium strains and Escherichia coli O157 to low pH environments. J. Appl. Microbiol. 2003;94:625–632. doi: 10.1046/j.1365-2672.2003.01875.x. [DOI] [PubMed] [Google Scholar]
- 29.Hickey E.W., Hirshfield I.N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl. Environ. Microbiol. 1990;56:1038–1045. doi: 10.1128/aem.56.4.1038-1045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maurer L.M., Yohannes E., Bondurant S.S., Radmacher M., Slonczewski J.L. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J. Bacteriol. 2005;187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomich M., Fine D.H., Figurski D.H. The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J. Bacteriol. 2006;188:6899–6914. doi: 10.1128/JB.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swint-Kruse L., Matthews K.S. Allostery in the LacI/GalR family: Variations on a theme. Curr. Opin. Microbiol. 2009;12:129–137. doi: 10.1016/j.mib.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rkenes T., Lamark T., Strøm A.R. DNA-binding properties of the BetI repressor protein of Escherichia coli: The inducer choline stimulates BetI-DNA complex formation. J. Bacteriol. 1996;178:1663–1670. doi: 10.1128/jb.178.6.1663-1670.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakamoto A., Murata N. The role of glycine betaine in the protection of plants from stress: Clues from transgenic plants. Plant. Cell Environ. 2002;25:163–171. doi: 10.1046/j.0016-8025.2001.00790.x. [DOI] [PubMed] [Google Scholar]
- 35.Laub M.T., Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 36.Deng W., Wang H., Xie J. Regulatory and pathogenesis roles of Mycobacterium Lrp/AsnC family transcriptional factors. J. Cell. Biochem. 2011;112:2655–2662. doi: 10.1002/jcb.23193. [DOI] [PubMed] [Google Scholar]
- 37.Maddocks S.E., Oyston P.C. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154:3609–3623. doi: 10.1099/mic.0.2008/022772-0. [DOI] [PubMed] [Google Scholar]
- 38.Gallegos M.T., Schleif R., Bairoch A., Hofmann K., Ramos J.L. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos J.L., Martínez-Bueno M., Molina-Henares A.J., Terán W., Watanabe K., Zhang X., Gallegos M.T., Brennan R., Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakhlanova I., Dudkina A., Baĭtin D. Enzymatic control of homologous recombination in Escherichia coli cells and hyper-recombination. Mol. Biol. 2012;47:205–217. doi: 10.7868/s0026898413020031. [DOI] [PubMed] [Google Scholar]
- 41.Brinkrolf K., Brune I., Tauch A. The transcriptional regulatory network of the amino acid producer Corynebacterium glutamicum. J. Biotechnol. 2007;129:191–211. doi: 10.1016/j.jbiotec.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 42.Justino G.C., Santos M.R., Canário S., Borges C., Florêncio M.H., Mira L. Plasma quercetin metabolites: Structure-antioxidant activity relationships. Arch. Biochem. Biophys. 2004;432:109–121. doi: 10.1016/j.abb.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 43.Leyn S.A., Li X., Zheng Q., Novichkov P.S., Reed S., Romine M.F., Fredrickson J.K., Yang C., Osterman A.L., Rodionov D.A. Control of proteobacterial central carbon metabolism by the HexR transcriptional regulator a case study in shewanella oneidensis. J. Biol. Chem. 2011;286:35782–35794. doi: 10.1074/jbc.M111.267963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Xie J. Role and regulation of bacterial LuxR-like regulators. J. Cell. Biochem. 2011;112:2694–2702. doi: 10.1002/jcb.23219. [DOI] [PubMed] [Google Scholar]
- 45.Brzostek K., Skorek K., Raczkowska A. Advances in Yersinia Research. Springer-Verlag; New York, NY, USA: 2012. OmpR, a central integrator of several cellular responses in Yersinia enterocolitica; pp. 325–334. [DOI] [PubMed] [Google Scholar]
- 46.Padan E., Schuldiner S. Molecular physiology of the Na+/H+ antiporter in Escherichia coli. J. Exp. Biol. 1994;196:443–456. doi: 10.1242/jeb.196.1.443. [DOI] [PubMed] [Google Scholar]
- 47.Merrell D.S., Camilli A. Acid tolerance of gastrointestinal pathogens. Curr. Opin. Microbiol. 2002;5:51–55. doi: 10.1016/S1369-5274(02)00285-0. [DOI] [PubMed] [Google Scholar]
- 48.Kovacikova G., Lin W., Skorupski K. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J. Bacteriol. 2010;192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka Y., Kimura B., Takahashi H., Watanabe T., Obata H., Kai A., Morozumi S., Fujii T. Lysine decarboxylase of Vibrio parahaemolyticus: Kinetics of transcription and role in acid resistance. J. Appl. Microbiol. 2008;104:1283–1293. doi: 10.1111/j.1365-2672.2007.03652.x. [DOI] [PubMed] [Google Scholar]
- 50.Whitaker W.B., Parent M.A., Naughton L.M., Richards G.P., Blumerman S.L., Boyd E.F. Modulation of responses of Vibrio parahaemolyticus O3:K6 to pH and temperature stresses by growth at different salt concentrations. Appl. Environ. Microbiol. 2010;76:4720–4729. doi: 10.1128/AEM.00474-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sambrook J., Russell D.W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press; New York, NY, USA: 2001. [Google Scholar]
- 52.Tessema G.T., Moretro T., Snipen L., Heir E., Holck A., Naterstad K., Axelsson L. Microarray-based transcriptome of Listeria monocytogenes adapted to sublethal concentrations of acetic acid, lactic acid, and hydrochloric acid. Can. J. Microbiol. 2012;58:1112–1123. doi: 10.1139/w2012-091. [DOI] [PubMed] [Google Scholar]
- 53.Coutard F., Lozach S., Pommepuy M., Hervio-Heath D. Real-time reverse transcription-PCR for transcriptional expression analysis of virulence and housekeeping genes in viable but nonculturable Vibrio parahaemolyticus after recovery of culture ability. Appl. Environ. Microbiol. 2007;73:5183–5189. doi: 10.1128/AEM.02776-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Funahashi T., Moriya K., Uemura S., Miyoshi S.I., Shinoda S., Narimatsu S., Yamamoto S. Identification and characterization of pvuA, a gene encoding the ferric vibrioferrin receptor protein in Vibrio parahaemolyticus. J. Bacteriol. 2002;184:936–946. doi: 10.1128/jb.184.4.936-946.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]