Abstract
The orphan nuclear receptor (NR5A2), which belongs to the NR5A subfamily of nuclear receptors, is expressed in developing and adult tissues of endodermal origin, and can contribute to the development of several cancers through regulating cell proliferation. NR5A2 (rs3790843 and rs3790844) single nucleotide polymorphisms (SNPs) genotyping were examined in DNA samples, extracted from paraffin-embedded cancer tissue. Clinicopathologic and follow-up data were collected from 944 patients with gastric cancer (GC). Associations of the 2 SNPs with the progression and prognosis in gastric cancer patients were analyzed using the SPSS version 18.0. We found that NR5A2 rs3790843 polymorphism was significantly associated with the risk of GC which had regional lymph node metastasis (p = 0.044) or distant metastasis (p = 0.020). Our results also indicated that rs3790844 polymorphism was associated with the increased overall survival (OS) of GC patients in the dominant model (GG vs. GA/AA, HR (hazard ratio) = 0.823, 95% CI (confidence interval) = 0.679–0.997), suggesting a potential protective role of the variant A allele. Additionally, in the stratified analysis, both NR5A2 rs3790843 and rs3790844 polymorphism were associated with significantly lower risk of death in the groups of female, tumor size >5 cm in a dominant model. Our results represent the first demonstration that the NR5A2 rs3790844 polymorphism is associated with increased OS of GC patients in the dominant model, and similar results were found among the female group and tumor size >5 cm group for NR5A2 rs3790843 polymorphism. Further validation in other larger studies with different ethnic populations and functional evaluations are needed.
Keywords: NR5A2, rs3790843, rs3790844, single nucleotide polymorphism, gastric cancer
1. Introduction
Gastric cancer (GC), as the third leading cause of cancer death in men and the fifth in women, remains one of the major public health problems worldwide [1]. About two-thirds of the cases occur in developing countries and 42% in China alone, while the estimated high-risk areas in developed countries include Eastern Europe, and parts of Central and South America [2]. In recent decades, its incidence rate has declined, and remarkable progress has been achieved in comprehensive treatment strategies of combined therapy. However, the prognosis for patients still remains poor, with 5-year overall survival rates of 30% [3]. The development of gastric cancer is a multi-step, sequential process, which initiates from chronic gastritis, atrophy, intestinal metaplasia, dysplasia, and finally malignant transformation to invasive gastric cancer [4]. Moreover, multiple genetic and epigenetic alterations are implicated in the multi-step process of human stomach carcinogenesis and development [5]. Therefore, discovery of these biomarkers could be helpful in the improvement of early diagnosis, screening of high-risk individuals, as well as patient care [6,7]. In recent years, increased studies have focused on the detection of genetic variants that could play roles in the development, progression and prognoses of gastric cancer [8].
The orphan nuclear receptor (NR5A2), also known as liver receptor homologue-1 (LRH-1) and fetoprotein transcription factor (FTF), belongs to the NR5A or FTZ-F1 subfamily of nuclear receptors. NR5A2 is expressed in developing and adult tissues of endodermal origin, including liver, pancreas, intestine and the ovary [9,10]. Functionally, NR5A2 has been implicated in the regulation of bile acid and cholesterol homeostasis [9,10] and the regulation of inflammatory responses in the liver and gut [11]. Aromatase is an NR5A2 target gene, which catalyses the conversion of androgens (testosterone and primarily androstenedione) to oestrogens. NR5A2 may aid breast cancer progression in postmenopausal women by promoting local oestrogen biosynthesis [12,13,14]. The expression of NR5A2 is also elevated in pancreatic cancer and promotes pancreatic cancer cell growth through stimulation of cyclin D1, cyclin E1 and c-Myc [15]. A recent genome-wide association study (GWAS) identified rs3790844 and rs3790843, located in the first intron of NR5A2 in 1q32.11, as associated with pancreatic cancer susceptibility [16]. In the gastrointestinal tract, NR5A2 has been shown to participate in intestinal cell renewal [17] and is expressed in the stomach epithelium [18]. Therefore, we conducted this study to examine whether NR5A2 rs3790844 and rs3790843 polymorphisms are associated with clinical outcomes of gastric cancer. The two SNPs may serve as potential molecular prognostic markers for gastric cancer, which will promote further defined sub-populations at higher risk of the disease. Consequently, these sub-populations may require more rigorous treatment and postoperative follow up.
2. Results
2.1. Associations between Clinicopathological Variables and Overall Survival
The patients’ characteristics and clinical information are summarized in Table 1. In the follow-up period of 119 months, 442 patients died. There were 727 males (77.0%) and 217 females (23.0%), with a median age of 62 years ranging from 28 to 83 years. Clinicopathological characteristics including tumor size, depth of invasion, lymph node metastasis, distant metastasis, Tumor, Node and Metastasis (TNM) stage and Lauren classification were significantly associated with survival time (log-rank p < 0.05). Specifically, patients with tumor size >5 cm (median survival time (MST), 48 months) had a 42% significantly higher risk of death (HR (hazard ratio) = 1.42, 95% CI (confidence interval) = 1.178–1.716), compared with those with tumor size <5 cm (MST, 98 months), and the patients with lymph node metastasis or distant metastasis had significantly higher risk of death than those patients without lymph node metastasis or distant metastasis (log-rank p < 0.05). In addition, as the depth of invasion and TNM stage increased, the risk of death for gastric cancer showed a significant increase in a dose-response manner (log-rank p < 0.05).
Table 1.
Associations between clinicopathological variables and overall survival.
| Variable | Patients, n = 944 | MST (Months) | Log-Rank p | HR (95% CI) d |
|---|---|---|---|---|
| Age (years) | ||||
| ≤60 | 442 | 88 | 0.239 | 1.000 |
| >60 | 502 | 59 | 1.118 (0.927–1.349) | |
| Sex | ||||
| Male | 727 | 70 | 0.536 | 1.000 |
| Female | 217 | 63 | 1.071 (0.860–1.335) | |
| Tumor Size | ||||
| ≤5 cm | 581 | 98 | <0.001 | 1.000 |
| >5 cm | 363 | 48 | 1.422 (1.178–1.716) | |
| Location | ||||
| Non-Cardia | 626 | 78 | 0.253 | 1.000 |
| Cardia | 318 | 63 | 0.891 (0.730–1.088) | |
| Histological Types a | ||||
| Intestinal | 370 | 74 | 0.436 | 1.000 |
| Diffuse | 497 | 60 | 1.069 (0.918–1.244) | |
| Differentiation a | ||||
| Well to Moderate | 305 | 80 | 0.518 | 1.000 |
| Poorly | 493 | 59 | 1.165 (0.942–1.443) | |
| Mucinous/Signet-Ring Cell | 69 | 62 | 1.199 (0.825–1.742) | |
| Lauren a | ||||
| 1 | 399 | 76 | <0.001 | 1.000 |
| 2 | 541 | 50 | 1.467 (1.211–1.776) | |
| Depth of Invasion b | ||||
| T1 | 182 | 84 | <0.001 | 1.000 |
| T2 | 138 | 78 | 0.540 (0.408–0.714) | |
| T3 | 8 | 70 | 0.800 (0.608–1.054) | |
| T4 | 594 | 51 | 1.008 (0.416–2.442) | |
| Lymph Node Metastasis c | ||||
| N0 | 378 | 81 | <0.001 | 1.000 |
| N1/N2/N3 | 566 | 43 | 1.814 (1.480–2.222) | |
| Distant Metastasis | ||||
| M0 | 886 | 74 | 0.004 | 1.000 |
| M1 | 58 | 26 | 1.646 (1.165–2.326) | |
| TNM Stage | ||||
| I | 250 | 83 | <0.001 | 1.000 |
| II | 203 | 88 | 1.231 (0.909–1.666) | |
| III | 458 | 41 | 1.949 (1.524–2.492) | |
| IV | 25 | 44 | 2.046 (1.163–3.600) | |
| Chemotherapy | ||||
| No | 638 | 61 | 0.684 | 1.000 |
| Yes | 306 | 69 | 1.043 (0.852–1.276) |
Mean survival time was presented when the median survival time could not be measured. TNM, Tumor, Node and Metastasis; MST, median survival time; HR, hazard ratio; CI, confidence interval; AJCC, American Joint Commission on Cancer. a: Partial data were not available, and statistics were based on available data; b: Invaded depth of tumor was classified according to the criteria of AJCC 7th; c: Lymph nodes were staged according to tumor node metastasis classification of the 7th edition of AJCC in which the number of lymph nodes with a metastasis of 1–2, 3–6, and ≥7 were classified as N1, N2, and N3, respectively; d: Adjusted by age and sex.
2.2. Association Analyses of NR5A2 rs3790843 and rs3790844 Genotypes with Clinicopathological Features
Among 944 specimens from patients with gastric cancer, NR5A2 rs3790843 was successfully genotyped in 907 specimens, and NR5A2 rs3790844 was successfully genotyped in 912 specimens. The frequency of each NR5A2 rs3790843 genotypes was 45.6% (414 specimens) for the TT variant, 45.2% (410 specimens) for the CT variant, and 9.2% (83 specimens) for the CC variant. Table 2 indicates that the risk of GC for the CT and CC variants compared with that for the TT variant in NR5A2 rs3790843 was associated significantly with regional lymph node metastasis (p = 0.044) and distant metastasis (p = 0.020) but not with age, sex, tumor size or location, histological type, differentiation, depth of invasion, chemotherapy history, American Joint Committee on Cancer staging, or Lauren classification. In reference to NR5A2 rs3790844, the GG, GA, and AA genotypes were identified in 399 specimens (43.8%), 416 specimens (45.6%), and 97 specimens (10.6%), respectively. Compared with the GG genotype, the GA/AA genotypes were not associated with any clinicopathologic data.
Table 2.
Association analyses of NR5A2 rs3790843 and rs3790844 genotypes with clinicopathological features.
| Variable | rs3790843 (n = 907) | rs3790844 (n = 912) | |||||
|---|---|---|---|---|---|---|---|
| TT | TC/CC | p | GG | GA/AA | p | ||
| Age (years) | ≤60 | 192 | 237 | 0.610 | 182 | 249 | 0.380 |
| >60 | 222 | 256 | 217 | 264 | |||
| Sex | Male | 321 | 375 | 0.601 | 311 | 391 | 0.539 |
| Female | 93 | 118 | 88 | 122 | |||
| Location | Non-Cardia Cancer | 279 | 320 | 0.432 | 264 | 338 | 0.930 |
| Cardia Cancer | 135 | 173 | 135 | 175 | |||
| Tumor Size | ≤5 cm | 261 | 302 | 0.581 | 243 | 324 | 0.486 |
| >5 cm | 153 | 191 | 156 | 189 | |||
| Lymph Node Metastasis a | N0 | 180 | 182 | 0.044 | 168 | 197 | 0.257 |
| N1/N2/N3 | 234 | 311 | 231 | 316 | |||
| Distant Metastasis | M0 | 398 | 456 | 0.020 | 382 | 478 | 0.098 |
| M1 | 16 | 37 | 9 | 35 | |||
| Histological Types b | Intestinal | 168 | 187 | 0.273 | 156 | 205 | 0.541 |
| Diffuse | 207 | 270 | 206 | 270 | |||
| Differentiation b | Well to Moderate | 137 | 155 | 0.556 | 127 | 170 | 0.687 |
| Poorly | 208 | 267 | 205 | 270 | |||
| Mucinous/Signet-Ring Cell | 30 | 35 | 30 | 35 | |||
| Lauren b | 1 | 182 | 199 | 0.253 | 169 | 218 | 0.450 |
| 2 | 229 | 293 | 227 | 294 | |||
| Chemotherapy | No | 275 | 333 | 0.721 | 266 | 348 | 0.709 |
| Yes | 139 | 160 | 133 | 165 | |||
| Depth of Invasion c | T1 | 86 | 93 | 0.854 | 76 | 101 | 0.929 |
| T2 | 60 | 69 | 59 | 71 | |||
| T3 | 3 | 3 | 2 | 4 | |||
| T4 | 257 | 320 | 254 | 326 | |||
| TNM Stage | I | 120 | 118 | 0.369 | 111 | 128 | 0.533 |
| II | 83 | 111 | 78 | 119 | |||
| III | 198 | 248 | 198 | 250 | |||
| IV | 10 | 13 | 9 | 13 | |||
HR, hazard ratio; CI, confidence interval; AJCC, American Joint Commission on Cancer; TNM, Tumor, Node and Metastasis. a: Lymph nodes were staged according to tumor node metastasis classification of the 7th edition of AJCC in which the number of lymph nodes with a metastasis of 1–2, 3–6, and ≥7 were classified as N1, N2, and N3, respectively; b: Partial data were not available, and statistics were based on available data; c: Invaded depth of tumor was classified according to the criteria of AJCC 7th.
The distribution of genotypes in the population was consistent with Hardy–Weinberg equilibrium (p = 0.20 for rs3790843 SNP, p = 0.46 for rs3790844 SNP), while strong linkage disequilibrium (LD) exists between alleles of the two loci with the following R2 values: rs3790843/rs3790844 = 0.798. Haplotype analyses were performed for all patients using the SHEsis software (http://analysis. bio-x.cn), and the six possible haplotype frequencies of the two SNPs are shown in Table 3. According to the results, the TTGG and CTGA haplotypes are the most common and represent 41.5% and 40.8%, respectively, in all groups. After haplotype analyses, however, no significant differences in these haplotype frequencies were found in any group.
Table 3.
Haplotype analyses of NR5A2 rs3790843 and rs3790844 genotypes.
| Haplotype | Frequencies | Patients/Deaths | HR (95% CI) a |
|---|---|---|---|
| TTGG | 0.415 | 379/184 | 1.00 |
| CTGA | 0.408 | 373/170 | 0.90 (0.73–1.10) |
| CCAA | 0.079 | 72/30 | 0.79 (0.54–1.17) |
| TTGA | 0.036 | 33//11 | 0.54 (0.30–1.00) |
| CTAA | 0.024 | 22//6 | 0.47 (0.21–1.07) |
| CTGG | 0.015 | 14//10 | 1.55 (0.81–2.94) |
a: Adjusted by age and sex.
2.3. Associations of NR5A2 rs3790843 and rs3790844 with Clinical Outcomes of OS
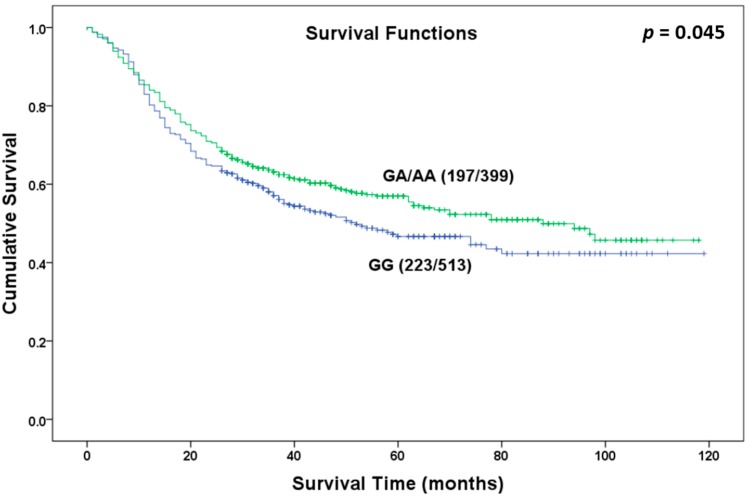
The Kaplan-Meier survival analysis and Cox proportional hazard models were used to assess the prognostic effect of NR5A2 rs3790843 and rs3790844 on GC patients in different genetic models (Table 4). We found rs3790844 polymorphism was associated with the OS of GC patients in the dominant model (GG vs. GA/AA, log-rank p = 0.045, Figure 1). Patients with the GA or AA genotype (MST 88 months) had a 17.7% lower risk of death than that of patients with the GG genotype (MST 52 months) (HR = 0.823, 95% CI = 0.679–0.997), suggesting a potential protective role of the variant A allele. No significant associations were observed between the rs3790843 genotypes and OS of GC patients in any genetic models. We further evaluated the associations by stratified analysis of variants of clinicopathological features (Table 5). In the dominant model, NR5A2 rs3790844 polymorphism was associated with significantly lower risk of death in the groups of patients with lymph node metastasis, no distant metastasis, diffuse type and no chemotherapy history (log-rank p < 0.05). Additionally, both NR5A2 rs3790843 and rs3790844 polymorphism were associated with significantly better prognosis among the female patients group and tumor size >5 cm group in a dominant model. Cox stepwise regression analysis was conducted to evaluate the independent effect of clinicopathological variables, rs3790843 and rs3790844 SNP on the OS of the patients with GC. As shown in Table 6, two variables (lymph node metastasis and NR5A2 rs3790844) were included in the regression model by stepwise selection of the covariant variables and rs3790844 SNP was shown to be an independent protective factor for GC with a 21.6% decreased risk (HR = 0.784, 95% CI = 0.646–0.951, p = 0.014).
Table 4.
Associations of NR5A2 rs3790843 and rs3790844 with clinical outcomes of overall survival.
| Genetic Model | Genotypes | Patients | Deaths | MST (Months) | Log-Rank p | HR (95%CI) a |
|---|---|---|---|---|---|---|
| rs3790843 | ||||||
| Codominant Model | TT | 414 | 196 | 59 | 0.668 | 1.000 |
| CT | 410 | 187 | 70 | 0.934 (0.765–1.142) | ||
| CC | 83 | 36 | 88 | 0.872 (0.611–1.244) | ||
| Dominant Model | TT | 414 | 196 | 59 | 0.415 | 1.000 |
| CT or CC | 493 | 223 | 70 | 0.924 (0.762–1.119) | ||
| Recessive Model | TT or CT | 824 | 383 | 70 | 0.551 | 1.000 |
| CC | 83 | 36 | 88 | 0.902 (0.641–1.269) | ||
| rs3790844 | ||||||
| Codominant Model | GG | 399 | 197 | 52 | 0.075 | 1.000 |
| GA | 416 | 186 | 78 | 0.853 (0.698–1.043) | ||
| AA | 97 | 37 | 98 | 0.699 (0.492–0.993) | ||
| Dominant Model | GG | 399 | 197 | 52 | 0.045 | 1.000 |
| GA or AA | 513 | 223 | 88 | 0.823 (0.679–0.997) | ||
| Recessive Model | GG or GA | 815 | 383 | 65 | 0.103 | 1.000 |
| AA | 97 | 37 | 98 | 0.757 (0.540–1.061) | ||
MST, median survival time; HR, hazard ratio; CI, confidence interval. Mean survival time was presented when the median survival time could not be measured. a: Adjusted by age and sex.
Figure 1.
Overall survival curve in relation to NR5A2 rs3790844 polymorphism in patients with gastric cancer in dominant model.
Table 5.
Stratified analysis of rs3790843 and rs3790844 polymorphism among GC patients.
| rs3790843 | rs3790844 | |||||||
|---|---|---|---|---|---|---|---|---|
| TT | TC/CC | HR (95% CI) e | p | GG | GA/AA | HR (95% CI) e | p | |
| Age (years) | ||||||||
| ≤60 | 88/192 | 108/237 | 0.977 (0.737–1.294) | 0.869 | 90/182 | 107/249 | 0.806 (0.609–1.067) | 0.132 |
| >60 | 108/222 | 115/256 | 0.873 (0.672–1.136) | 0.313 | 107/217 | 116/264 | 0.837 (0.644–1.089) | 0.185 |
| Sex | ||||||||
| Male | 144/321 | 175/375 | 1.044 (0.838–1.302) | 0.700 | 149/311 | 172/391 | 0.877 (0.705–1.093) | 0.243 |
| Female | 52/93 | 48/118 | 0.619 (0.417–0.916) | 0.017 | 48/88 | 51/122 | 0.660 (0.445–0.981) | 0.040 |
| Location | ||||||||
| Non–CardiaCancer | 133/279 | 148/320 | 0.948 (0.750–1.198) | 0.652 | 133/264 | 148/338 | 0.810 (0.641–1.024) | 0.077 |
| CardiaCancer | 63/135 | 75/173 | 0.861 (0.615–1.204) | 0.381 | 64/135 | 75/175 | 0.840 (0.601–1.174) | 0.308 |
| Tumor Size | ||||||||
| ≤5 cm | 108/261 | 127/302 | 1.044 (0.807–1.349) | 0.745 | 106/243 | 132/324 | 0.927 (0.718–1.197) | 0.559 |
| >5 cm | 88/153 | 96/191 | 0.747 (0.558–0.999) | 0.049 | 91/156 | 91/189 | 0.693 (0.517–0.928) | 0.014 |
| Lymph Node Metastasis a | ||||||||
| N0 | 64/180 | 60/182 | 0.947 (0.666–1.347) | 0.763 | 63/168 | 63/197 | 0.865 (0.610–1.226) | 0.414 |
| N1/N2/N3 | 132/234 | 163/311 | 0.829 (0.659–1.043) | 0.109 | 134/231 | 160/316 | 0.744 (0.591–0.937) | 0.012 |
| Distant Metastasis | ||||||||
| M0 | 188/398 | 200/456 | 0.886 (0.726–1.081) | 0.232 | 188/382 | 202/478 | 0.793 (0.650–0.968) | 0.022 |
| M1 | 8/16 | 23/37 | 1.340 (0.599–2.998) | 0.476 | 7/9 | 21/35 | 1.189 (0.544–2.599) | 0.664 |
| Histological Types b | ||||||||
| Intestinal | 71/168 | 80/187 | 1.041 (0.756–1.434) | 0.805 | 69/156 | 84/205 | 0.952 (0.692–1.309) | 0.762 |
| Diffuse | 108/207 | 127/270 | 0.826 (0.639–1.068) | 0.145 | 111/206 | 123/270 | 0.735 (0.568–0.950) | 0.019 |
| Differentiation b | ||||||||
| Well to Moderate | 57/137 | 67/155 | 1.099 (0.772–1.565) | 0.601 | 56/127 | 69/170 | 0.962 (0.676–1.368) | 0.828 |
| Poorly | 104/208 | 126/267 | 0.882 (0.680–1.144) | 0.344 | 105/205 | 125/270 | 0.822 (0.634–1.067) | 0.140 |
| Others c | 18/30 | 14/35 | 0.522 (0.259–1.056) | 0.07 | 19/30 | 13/35 | 0.397 (0.195–0.808) | 0.011 |
| Lauren b | ||||||||
| 1 | 69/182 | 78/199 | 1.049 (0.759–1.451) | 0.771 | 69/169 | 80/218 | 0.899 (0.652–1.241) | 0.655 |
| 2 | 125/229 | 144/293 | 0.824 (0.648–1.047) | 0.113 | 126/227 | 142/294 | 0.763 (0.600–0.970) | 0.027 |
| Chemotherapy | ||||||||
| No | 132/275 | 151/333 | 0.908 (0.719–1.147) | 0.416 | 136/266 | 149/348 | 0.767 (0.608–0.968) | 0.025 |
| Yes | 64/139 | 72/160 | 0.957 (0.683–1.340) | 0.799 | 61/133 | 74/165 | 0.953 (0.679–1.338) | 0.783 |
| Depth of Invasion d | ||||||||
| T1 | 25/86 | 34/93 | 1.297 (0.774–2.173) | 0.324 | 24/76 | 33/101 | 1.051 (0.621–1.779) | 0.852 |
| T2 | 31/60 | 24/69 | 0.669 (0.393–1.141) | 0.140 | 31/59 | 25/71 | 0.648 (0.382–1.099) | 0.107 |
| T3 | 1/3 | 2/3 | 1.405 (0.125–15.838) | 0.782 | 1/2 | 2/4 | 0.809 (0.071–9.157) | 0.864 |
| T4 | 134/257 | 159/320 | 0.880 (0.699–1.108) | 0.276 | 135/254 | 157/326 | 0.809 (0.642–1.019) | 0.072 |
| TNM Stage | ||||||||
| I | 40/120 | 39/118 | 0.992 (0.638–1.542) | 0.972 | 40/111 | 40/128 | 0.862 (0.556–1.336) | 0.506 |
| II | 38/83 | 38/111 | 0.743 (0.474–1.166) | 0.196 | 37/78 | 41/119 | 0.711 (0.456–1.109) | 0.132 |
| III | 111/198 | 135/248 | 0.861 (0.670–1.108) | 0.245 | 113/198 | 132/250 | 0.800 (0.622–1.029) | 0.082 |
| IV | 4/10 | 8/13 | 1.855 (0.557–6.171) | 0.314 | 4/9 | 7/13 | 1.257 (0.368–4.302) | 0.715 |
HR, hazard ratio; CI, confidence interval; AJCC, American Joint Commission on Cancer; TNM, Tumor, Node and Metastasis. a: Lymph nodes were staged according to tumor node metastasis classification of the 7th edition of AJCC in which the number of lymph nodes with a metastasis of 1–2, 3–6, and ≥7 were classified as N1, N2, and N3, respectively; b: Partial data were not available, and statistics were based on available data; c: Others: mucinous carcinoma and Signet-ring cell carcinoma; d: Invaded depth of tumor was classified according to the criteria of AJCC 7th; e: Adjusted by age and sex.
Table 6.
Stepwise Cox regression analysis on the survival of GC.
| Variables | β | SE | HR b | 95% CI | p Value |
|---|---|---|---|---|---|
| Age a | 0.067 | 0.100 | 1.070 | (0.878–1.302) | 0.503 |
| Sex | 0.04 | 0.118 | 1.041 | (0.825–1.313) | 0.743 |
| Lymph Node Metastasis | 0.567 | 0.112 | 1.763 | (1.414–2.198) | <0.001 |
| rs3790844 (GG vs. GA/AA) | −0.243 | 0.099 | 0.784 | (0.646–0.951) | 0.014 |
| rs3790843 (TT vs. CT/CC) | −0.152 | 0.101 | 0.859 | (0.705–1.046) | 0.131 |
β, regression coefficient; SE, standard error; HR, hazard ratio; CI, confidence interval. a: Age was included as a continuous variable in the Cox stepwise regression analysis; b: Adjusted by age and sex.
3. Discussion
In the present study, we investigated the effects of two SNPs (rs3790843 and rs3790844) of the NR5A2 gene on the progression and survival of GC in Chinese populations. We found that NR5A2 rs3790843 polymorphism was significantly associated with the risk of GC when compared with regional lymph node metastasis and distant metastasis. Our results also indicated that rs3790844 polymorphism was associated with the increased OS of GC patients in the dominant model, suggesting a potential protective role of the variant A allele. No significant associations were observed between the rs3790843 genotypes and OS of GC patients in any genetic models. Additionally, in the stratified analysis, NR5A2 rs3790844 polymorphism was associated with significantly lower risk of death in the groups of female, tumor size >5 cm, lymph node metastasis, no distant metastasis, diffuse type and no chemotherapy history in the dominant model. Similar results were found among the female patients group and tumor size >5 cm group for the NR5A2 rs3790843 polymorphism in a dominant model.
Nuclear receptor 5 subtype A2 (NR5A2), also known as Liver receptor homolog-1 (LRH-1), is a member of the orphan family of nuclear receptors that participates in a wide range of developmental processes [9,10]. Functionally, NR5A2 has been implicated in the regulation of bile acid and cholesterol homeostasis, and is expressed in developing and adult tissues of endodermal origin, including liver, pancreas, intestine and the ovary [9,10]. NR5A2 was first isolated as a transcriptional activator of the alpha-fetoprotein (AFP) gene, which encodes a factor that plays a crucial rolein hepatic specification [19]. Further, NR5A2 expression is elevated in pancreatic cancer andpromotes pancreatic cancer cell growth through stimulation of cyclin D1, cyclin E1 and c-Myc [15], while genome-wide association studies implicate mutations in the NR5A2 gene in pancreatic ductal adenocarcinoma [16]. In the gastrointestinal tract, NR5A2 has been shown to participate in intestinal cell renewal [17] and is expressed in the stomach epithelium [18]. However, it seemed no study focused on the association between NR5A2 and the progression and prognoses of gastric cancer. Thus, this study was conducted, and it indicates that the NR5A2 polymorphism is significantly associated with the development and overall survival of gastric cancer.
In the gastrointestinal tract, NR5A2 regulates proliferation through at least two different mechanisms: direct activation of the cyclin E1 promoter and, in cooperation with β-catenin, induction of cyclin D1 and c-Myc transcription [20]. A previous study showed that NR5A2, which interacts with β-catenin, induces cell proliferation through the concomitant induction of cyclin D1 and E1, which are downstream NR5A2 effectors whose induction may account for the diminished threshold for G/S transition [20]. Whereas β-catenin coactivates NR5A2 on the cyclin E1 promoter, NR5A2 acts as a potent tissue-restricted coactivator of β-catenin on the cyclin D1 promoter.
Tumor growth is the result of uncontrolled cell proliferation or a defective cell death program. For the proliferation of cells, the phosphorylation of pRb by cyclin–CDK complexes to release the transcription factor E2F is an essential step. The cyclin D1, cyclin E and c-Myc are important parts of expression peaks at the G1–S transition, and then decreases as cells proceed through the S phase [21,22,23]. The G1–S transition is the major regulation point of the cell cycle, and during the G1–S transition, alterations in cell cycle regulators lead to the deregulation of the cell cycle, which can cause unbridled cell division, contributing to cancer development. Previously, overexpression of cyclin E was demonstrated in many tumors and correlated with prognosis [24,25,26,27] including gastric cancer [28]. Wang et al. [29] found that the mRNA expression of NR5A2 was significantly upregulated in gastric cancer, as compared with self-paired normal control. In addition, overexpression of NR5A2 was shown to promote the proliferation of gastric adenocarcinoma SGC-7901 cells via induction of cyclin E1, which may lead to the tumorigenesis of gastric cancer [29]. Botrugno et al. [20] found that cyclin E was a direct NR5A2 target gene, which could partly explain the mechanism of NR5A2 polymorphism in the progression and prognoses of gastric cancer. On the other hand, β-catenin has been proposed to act as a docking protein that assembles both general and specific factors required for the activation of target genes. Botrugno et al. also found that NR5A2 could act as a coactivator for β-Catenin/Tcf4 to drive the expression of cyclin D1 and other β-catenin/Tcf target genes, such as c-Myc [30,31,32]. As reported, cyclin D1 is a proto-oncogene that belongs to the family of G1 cyclins, and plays an important role in cell cycle G1 to S transition by binding its partners cyclin dependent kinase 4 and 6 to phosphorylate and inactivate the Rb protein [33]. In addition, cyclin D1 over-expression is usually an early event in carcinogenesis and a prognostic indicator associated with poor survival in cancers [34]. Given above-mentioned evidences, NR5A2 polymorphism could also affect the carcinogenesis and prognoses of cancer through acting as a coactivator for β-catenin on the cyclin D1 promoter.
Interestingly, in the stratified analyses by sex, our results indicated that both the NR5A2 rs3790843 and rs3790844 polymorphisms were associated with significantly better prognosis in the female patient group. NR5A2 is a direct estrogen receptor α (ERα) target gene [35,36], its expression correlates with ERα in breast tumours [37] and it promotes breast cancer proliferation and invasion [38]. Previous study showed that NR5A2 is an important regulator of ERα target genes and it shares many binding sites with ERα. Importantly, at shared sites, NR5A2 promotes ERα recruitment and vice versa, ERα stimulates NR5A2 recruitment [39]. As a result, in the female patients with more ERα, more NR5A2 polymorphisms, which act as a protective factor, contribute to a better prognosis.
Taken together, the presented findings of the potential involvement of the NR5A2 gene in anti-tumorigenesis prompts us to further characterize its structure, biological function and interaction with other partners by in vitro or in vivo studies. However, some limitations of the present study should be addressed. First, Helicobacter pylorus, a known crucial factor in gastric carcinogenesis, was not considered due to the lack of related follow-up information. Second, only two SNPs in NR5A2 are evaluated, and it is possible that some other important SNPs are neglected or the observed associations may be due to other polymorphisms in linkage disequilibrium with the rs3790843 and rs3790844 polymorphisms. Finally, for validation of the genotype–phenotype relationship, further investigation is underway to clarify the association between rs3790843 and rs3790844 polymorphisms and expression levels of NR5A2 protein in gastric cancer tissues, and will be reported separately.
4. Experimental Section
4.1. Study Subjects
A retrospective cohort of 944 patients with gastric cancer who underwent a surgical resection at the Yixing People’s Hospital (Yixing, Jiangsu Province, China) between 1999 and 2006, who had a median follow-up of 35 months (range, 0–119 months) were recruited for this study [40]. The demographic features and clinicopathologic data were summarized in Table 1. All patients were diagnosed with gastric carcinoma histopathologically. None had received neoadjuvant radiochemotherapy or postoperative radiotherapy, and some had received adjuvant chemotherapy. Formalin-fixed and paraffin-embedded blocks of these patients were collected from the department of pathology of this hospital. The samples used for genotyping were reviewed and classified by two independent pathologists. Death dates were confirmed via review of death certificates of inpatient and outpatient records or obtained from patients’ families through follow-up telephone calls. The study protocol was approved by the Institutional Review Board of Nanjing Medical University (Nanjing, China), and all patients provided written informed consent on the use of paraffin specimens for gene polymorphism analyses.
4.2. Genotyping
Genomic DNA was extracted from tumor specimens using proteinase K digestion, followed by isopropanol extraction and ethanol precipitation. The NR5A2 (rs3790844 and rs3790843) SNPs were examined by multiplex snapshot technology using an ABI fluorescence-based assay allelic discrimination method (Applied Biosystems, Foster City, CA, USA), which has been described indetail in a previous study [41]. The primers were designed to anneal immediately adjacent to the nucleotideat the mutation site: rs3790844: forward, 5'-TTCCGTGTGGAAACACAGGTCA-3'; reverse, 5'-TCGACTGGAGCCCAAGATCA-3'; rs3790843: forward, 5'-TCTTTGCCCCGATGAGTTCG-3'; reverse, 5'-TCCCAGATGCTCTGGTGCAG-3'. The primers for extension were as follows: rs3790844, 5'-TTTTTTTTTTTTT…TTTTTTTTTTTTTTTTTGGAAACACAGGTCACTAAAACTGG-3'; and rs3790843, 5'-TCTGGTGCAGCCGAAGTAG-3'. The snapshot products were analyzed by using an ABI 3130 genetic analyzer (Applied Biosystems) and the genotypes were determined by GeneMapper Analysis Software version 4 (Applied Biosystems). Genotype analysis was performed by two investigators blinded to the survival end points. Genotyping was validated by sequencing a randomly selected 10% of samples, and the results were 100% concordant. However, 37 cases of rs3790843 and 32 cases of rs3790844 failed in genotyping because of poor DNA quality, which were excluded in further analysis. Finally, 907 cases of rs3790843 and 912 cases of rs3790844 were included in the analysis.
4.3. Statistical Methods
The associations of each genotype or combinations of genotypes with clinicopathologic features were compared using the Pearson chi-square test for categorical variables and the Student’s t-test for continuous data. Associations of genetic variants and clinicopathological features with the overall survival (OS) were estimated using Kaplan-Meier method and comparisons between different groups of patients were performed with the log-rank test. Univariate or multivariate Cox proportional hazard models was used to estimate the adjusted HR and their 95% CI. Moreover, Cox stepwise regression analysis was conducted to assess the independent impacts of SNP or clinicopathological features on the overall survival after adjusting for other covariates, with a significance level of p < 0.05 for entering and p > 0.10 for removing the respective explanatory variables. All tests were two-sided and p < 0.05 was considered statistically significant. All the statistical analyses were carried out using SPSS version 16.0 for Windows (SPSS Inc., Chicago, IL, USA).
5. Conclusions
In conclusion, our results represent the first demonstration that NR5A2 rs3790844 polymorphism was associated with the increased OS of GC patients in the dominant model, and similar results were found among the female patients group and tumor size >5 cm group for the NR5A2 rs3790843 polymorphism, suggesting that the mutant alleles may serve as suitable markers for predicting the survival of gastric cancer patients, especially in a Chinese population. Large population-based prospective studies with ethnically diverse populations are warranted to verify these findings.
Acknowledgments
The work was partly supported by grants from the National Natural Science Foundation of China (Grant No. 81272469), the National 973 Basic Research Program of China (Grant No. 2013CB911300) and the clinical special project for Natural Science Foundation of Jiangsu Province (Grant No. BL2012016), and a grant from The Project of Plans for the Development of Science and Technology of Nanjing, China (Grant No. 201208020), and the grant from Nanjing 12th Five-Year Key Scientific Project of Medicine.
Author Contributions
Xunlei Zhang, Mulong Du and Meilin Wang designed the study and applied for Research Ethics Board Approval; Chunxiang Cao, Lili Shen, Meng Kuang, Yongfei Tan and Xinying Huo recruited the patients and collected the data; Yongfei Tan, Weida Gong and Zhi Xu analyzed the data and prepared draft figures and tables. Zhi Xu and Dongying Gu prepared the manuscript draft. All authors approved the final article. Zhengdong Zhang, Cuiju Tang and Jinfei Chen had complete access to the study data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Santoro R., Carboni F., Lepiane P., Ettorre G.M., Santoro E. Clinicopathological features and prognosis of gastric cancer in young european adults. Br. J. Surg. 2007;94:737–742. doi: 10.1002/bjs.5600. [DOI] [PubMed] [Google Scholar]
- 4.Correa P. Human gastric carcinogenesis: A multistep and multifactorial process—First american cancer society award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 5.Resende C., Ristimaki A., Machado J.C. Genetic and epigenetic alteration in gastric carcinogenesis. Helicobacter. 2010;15(S1):34–39. doi: 10.1111/j.1523-5378.2010.00782.x. [DOI] [PubMed] [Google Scholar]
- 6.Ludwig J.A., Weinstein J.N. Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 7.Panani A.D. Cytogenetic and molecular aspects of gastric cancer: Clinical implications. Cancer Lett. 2008;266:99–115. doi: 10.1016/j.canlet.2008.02.053. [DOI] [PubMed] [Google Scholar]
- 8.Becker K.F., Keller G., Hoefler H. The use of molecular biology in diagnosis and prognosis of gastric cancer. Surg. Oncol. 2000;9:5–11. doi: 10.1016/S0960-7404(00)00016-5. [DOI] [PubMed] [Google Scholar]
- 9.Fayard E., Auwerx J., Schoonjans K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez-Marcos P.J., Auwerx J., Schoonjans K. Emerging actions of the nuclear receptor LRH-1 in the gut. Biochim. Biophys. Acta. 2011;1812:947–955. doi: 10.1016/j.bbadis.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venteclef N., Jakobsson T., Ehrlund A., Damdimopoulos A., Mikkonen L., Ellis E., Nilsson L.M., Parini P., Janne O.A., Gustafsson J.A., et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRβ in the hepatic acute phase response. Genes Dev. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clyne C.D., Kovacic A., Speed C.J., Zhou J., Pezzi V., Simpson E.R. Regulation of aromatase expression by the nuclear receptor LRH-1 in adipose tissue. Mol. Cell. Endocrinol. 2004;215:39–44. doi: 10.1016/j.mce.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Clyne C.D., Speed C.J., Zhou J., Simpson E.R. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J. Biol. Chem. 2002;277:20591–20597. doi: 10.1074/jbc.M201117200. [DOI] [PubMed] [Google Scholar]
- 14.Lazarus K.A., Wijayakumara D., Chand A.L., Simpson E.R., Clyne C.D. Therapeutic potential of liver receptor homolog-1 modulators. J. Steroid Biochem. Mol. Biol. 2012;130:138–146. doi: 10.1016/j.jsbmb.2011.12.017. [DOI] [PubMed] [Google Scholar]
- 15.Benod C., Vinogradova M.V., Jouravel N., Kim G.E., Fletterick R.J., Sablin E.P. Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc. Natl. Acad. Sci. USA. 2011;108:16927–16931. doi: 10.1073/pnas.1112047108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petersen G.M., Amundadottir L., Fuchs C.S., Kraft P., Stolzenberg-Solomon R.Z., Jacobs K.B., Arslan A.A., Bueno-de-Mesquita H.B., Gallinger S., Gross M., et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 2010;42:224–228. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoonjans K., Dubuquoy L., Mebis J., Fayard E., Wendling O., Haby C., Geboes K., Auwerx J. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc. Natl. Acad. Sci. USA. 2005;102:2058–2062. doi: 10.1073/pnas.0409756102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rausa F.M., Galarneau L., Belanger L., Costa R.H. The nuclear receptor fetoprotein transcription factor is coexpressed with its target gene HNF-3β in the developing murine liver, intestine and pancreas. Mech. Dev. 1999;89:185–188. doi: 10.1016/S0925-4773(99)00209-9. [DOI] [PubMed] [Google Scholar]
- 19.Galarneau L., Pare J.F., Allard D., Hamel D., Levesque L., Tugwood J.D., Green S., Belanger L. The α1-fetoprotein locus is activated by a nuclear receptor of the drosophila FTZ-F1 family. Mol. Cell. Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Botrugno O.A., Fayard E., Annicotte J.S., Haby C., Brennan T., Wendling O., Tanaka T., Kodama T., Thomas W., Auwerx J., et al. Synergy between LRH-1 and β-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell. 2004;15:499–509. doi: 10.1016/j.molcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Lew D.J., Dulic V., Reed S.I. Isolation of three novel human cyclins by rescue of G1 cyclin (cln) function in yeast. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-W. [DOI] [PubMed] [Google Scholar]
- 22.Sherr C.J. Mammalian G1 cyclins. Cell. 1993;73:1059–1065. doi: 10.1016/0092-8674(93)90636-5. [DOI] [PubMed] [Google Scholar]
- 23.Wimmel A., Lucibello F.C., Sewing A., Adolph S., Muller R. Inducible acceleration of G1 progression through tetracycline-regulated expression of human cyclin E. Oncogene. 1994;9:995–997. [PubMed] [Google Scholar]
- 24.Florenes V.A., Faye R.S., Maelandsmo G.M., Nesland J.M., Holm R. Levels of cyclin D1 and D3 in malignant melanoma: Deregulated cyclin D3 expression is associated with poor clinical outcome in superficial melanoma. Clin. Cancer Res. 2000;6:3614–3620. [PubMed] [Google Scholar]
- 25.Richter J., Wagner U., Kononen J., Fijan A., Bruderer J., Schmid U., Ackermann D., Maurer R., Alund G., Knonagel H., et al. High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am. J. Pathol. 2000;157:787–794. doi: 10.1016/S0002-9440(10)64592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamai T., Takagi K., Asami H., Ito Y., Oshima H., Yoshida K.I. Decreasing of p27kip1 and cyclin E protein levels is associated with progression from superficial into invasive bladder cancer. Br. J. Cancer. 2001;84:1242–1251. doi: 10.1054/bjoc.2000.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heah K.G., Hassan M.I., Huat S.C. p53 Expression as a marker of microinvasion in oral squamous cell carcinoma. Asian Pac. J. Cancer Prev. 2011;12:1017–1022. [PubMed] [Google Scholar]
- 28.Ahn M.J., Kim B.H., Jang S.J., Hong E.K., Lee W.M., Baik H.K., Park H.K., Lee C.B., Ki M. Expression of cyclin D1 and cyclin E in human gastric carcinoma and its clinicopathologic significance. J. Korean Med. Sci. 1998;13:513–518. doi: 10.3346/jkms.1998.13.5.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S.L., Zheng D.Z., Lan F.H., Deng X.J., Zeng J., Li C.J., Wang R., Zhu Z.Y. Increased expression of hLRH-1 in human gastric cancer and its implication in tumorigenesis. Mol. Cell. Biochem. 2008;308:93–100. doi: 10.1007/s11010-007-9616-1. [DOI] [PubMed] [Google Scholar]
- 30.He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-myc as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 31.Shtutman M., Zhurinsky J., Simcha I., Albanese C., D’Amico M., Pestell R., Ben-Ze’ev A. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tetsu O., McCormick F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 33.Kato J., Matsushime H., Hiebert S.W., Ewen M.E., Sherr C.J. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CKD4. Genes Dev. 1993;7:331–342. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- 34.Kim J.K., Diehl J.A. Nuclear cyclin D1: An oncogenic driver in human cancer. J. Cell. Physiol. 2009;220:292–296. doi: 10.1002/jcp.21791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Annicotte J.S., Chavey C., Servant N., Teyssier J., Bardin A., Licznar A., Badia E., Pujol P., Vignon F., Maudelonde T., et al. The nuclear receptor liver receptor homolog-1 is an estrogen receptor target gene. Oncogene. 2005;24:8167–8175. doi: 10.1038/sj.onc.1208950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiruchelvam P.T., Lai C.F., Hua H., Thomas R.S., Hurtado A., Hudson W., Bayly A.R., Kyle F.J., Periyasamy M., Photiou A., et al. The liver receptor homolog-1 regulates estrogen receptor expression in breast cancer cells. Breast Cancer Res. Treat. 2011;127:385–396. doi: 10.1007/s10549-010-0994-9. [DOI] [PubMed] [Google Scholar]
- 37.Miki Y., Clyne C.D., Suzuki T., Moriya T., Shibuya R., Nakamura Y., Ishida T., Yabuki N., Kitada K., Hayashi S., et al. Immunolocalization of liver receptor homologue-1 (LRH-1) in human breast carcinoma: Possible regulator of insitu steroidogenesis. Cancer Lett. 2006;244:24–33. doi: 10.1016/j.canlet.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 38.Chand A.L., Herridge K.A., Thompson E.W., Clyne C.D. The orphan nuclear receptor LRH-1 promotes breast cancer motility and invasion. Endocr. Relat. Cancer. 2010;17:965–975. doi: 10.1677/ERC-10-0179. [DOI] [PubMed] [Google Scholar]
- 39.Lai C.F., Flach K.D., Alexi X., Fox S.P., Ottaviani S., Thiruchelvam P.T., Kyle F.J., Thomas R.S., Launchbury R., Hua H., et al. Co-regulated gene expression by oestrogen receptor alpha and liver receptor homolog-1 is a feature of the oestrogen response in breast cancer cells. Nucleic Acids Res. 2013;41:10228–10240. doi: 10.1093/nar/gkt827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M., Bai J., Tan Y., Wang S., Tian Y., Gong W., Zhou Y., Gao Y., Zhou J., Zhang Z. Genetic variant in PSCA predicts survival of diffuse-type gastric cancer in a Chinese population. Int. J. Cancer. 2011;129:1207–1213. doi: 10.1002/ijc.25740. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y., Wang M., Gu D., Wu D., Zhang X., Gong W., Tan Y., Zhou J., Wu X., Tang C., et al. Association of XRCC1 gene polymorphisms with the survival and clinicopathological characteristics of gastric cancer. DNA Cell Biol. 2013;32:111–118. doi: 10.1089/dna.2012.1840. [DOI] [PubMed] [Google Scholar]