Abstract
Mucosa-associated lymphoid tissue (MALT) lymphoma is the most common extranodal lymphoid cell neoplasia; it frequently follows chronic bacteria-induced inflammation in various tissues. MALT lymphomas are characterized genetically by the t(11;18)(q21;q21) translocation, which yields chimeric transcripts encoding structurally distinct API2/MALT1 fusion proteins. In this study, we provide functional evidence for the contribution of API2/MALT1 fusion proteins to transformation of cells in culture by activating the NF-κB pathway through a RelB/p50 dimer. Using microchip gene expression analysis, we demonstrate that different forms of the API2/MALT1 proteins activate both unique and overlapping gene programs in cells. In addition to this genome reprogramming, expression of distinct API2/MALT1 fusion products inhibits DNA damage-induced, p53-mediated apoptosis in an NF-κB-dependent manner. Collectively, these data reveal previously unknown functional diversity among API2/MALT1 fusion products and their function in NF-κB signaling as it connects to the apoptotic program, a pathway with strong relevance to cancer. Furthermore, they provide evidence underlying the emerging role of the NF-κB signaling pathway in the inhibition of apoptosis.
Helicobacter pylori infection is one of the main risk factors linked to gastric cancer, the second most common malignancy worldwide. In some individuals with chronic H. pylori infection, responding B lymphocytes acquire genetic changes during somatic hypermutation and become malignant, transforming into a non-Hodgkins lymphoma called mucosa-associated lymphoid tissue (MALT) lymphoma (1).
Two characteristic MALT lymphoma-specific chromosomal translocations have been observed: the t(11;18)(q21;q21) and the t(1;14)(p22;q32) (2, 3). The t(1;14)(p22;q32) is a rare genetic event that juxtaposes the entire coding region of the BCL10 gene to chromosome 14, wherein its expression is deregulated through the Ig enhancers (4). Experiments in BCL10-deficient mice demonstrated that this gene is a positive regulator of lymphocyte proliferation and a mediator of NF-κB activation in response to antigen receptor-mediated signaling in both B and T cells (5).
The most common genetic lesion associated with MALT Lymphoma is the t(11;18)(q21;q21) translocation. It occurs as the sole known genetic abnormality in ≈43% of the tumors, underscoring its significance in lymphomagenesis (2, 6). The genes involved in this translocation are API2 (also known as c-IAP2, HIAP1, and MIHC) on chromosome 11q21, and MALT1 on chromosome 18q21 (7-10). API2 belongs to the inhibitor of apoptosis (IAP) family of proteins and contains three tandem copies of the baculovirus IAP repeat (BIR) domain, a caspase recruitment domain (CARD) and a carboxyl-terminal RING domain (11, 12). API2 was initially identified in a complex with tumor necrosis factor (TNF) receptor 2 through direct interaction with TNF receptor-associated factors (TRAFs) 1 and 2, involving the BIR and TRAF domains of the respective proteins (13).
MALT1 is a human paracaspase and consists of a death domain, two Ig-like domains, and an interleukin-1-converting enzyme (ICE)-like protease (caspase) p20 domain. It does not seem to function as a traditional caspase, but recent in vitro and in vivo studies suggest that the MALT1 protein forms a specific complex with BCL10 and CARMA1 within the cell, and that this complex mediates NF-κB activation in both B and T cells (14-17).
Several variants of the API2/MALT1 fusion gene are present in patients with the t(11;18)(q21;q21) translocation (6, 8, 18). In all cases, the breakpoints within the API2 gene occur consistently between the third BIR domain and the caspase recruitment domain whereas the breakpoints within the MALT1 coding DNA are variable. All cases contain the caspase-like domain of the MALT1 carboxyl terminus. Recent evidence suggests that the API2/MALT1 fusion proteins can activate the NF-κB survival pathway, but the precise mechanism used to accomplish this activation, as well as the subsequent downstream events that lead to lymphomagenesis, is unknown (14, 15).
In this study, the oncogenic properties of two distinct API2/MALT1 fusion products were investigated. In addition, the molecular basis of the API2/MALT1-mediated NF-κB activation and the ability of both fusion products to inhibit apoptosis were analyzed. Our results suggest that the API2/MALT1 fusion products promote tumorigenesis by the bilateral ability to accelerate cell proliferation and to inhibit programmed cell death. They also provide additional evidence for the inhibitory role of NF-κB signaling in apoptosis.
Materials and Methods
Transient, Stable Transfections and Luciferase Assays. pcDNA3-FUS1, pcDNA3-FUS2, pcDNA3-DEL, pcDNA3-API2, and empty vector constructs were transfected by lipofection by using the LipofectAMINE PLUS Reagents (Life Technologies) according to the manufacturer's protocol. Sixteen hours later, cell lysates were prepared and luciferase activities were measured with Dual-luciferase assay kits (Promega). NF-κB activities were determined by normalization of NF-κB-dependent luciferase to β-galactosidase. For stable transfection, FUS1 and FUS2 clones were isolated under G418 selection (1,000 μg/ml; Cellgro, Mediatech, Herndon, VA).
Cell Growth and Colony Formation Assays. Anchorage-independent proliferation was determined as follows. Supplemented agar (1%, 4 ml) in DMEM with 10% FCS was poured into a 60-mm dish to form bottom agar. The agar mix (0.7%, 1 ml) was layered with 2,000 cells on top and incubated at 37°C in a 5% CO2 atmosphere. Plates were stained with crystal violet (Sigma) for 1 h and destained, and colonies were counted under a dissecting microscope (19).
Flow Cytometry Analysis. pcDNA3-FUS1-3T3, pcDNA3-FUS2-3T3, and vector control 3T3 cells were collected by trypsinization and resuspended in PBS. The cells were then fixed in 70% ethanol, digested with RNase A (0.02 μg/μl), stained with 50 μg/ml ethidium bromide, and analyzed by using Becton Dickinson FAC-Scan and cellquest software. RNA extraction and semiquantitative PCR analysis were performed as described (20).
Analysis of Gene Expression Using Oligonucleotide Arrays. Gene expression profiling was performed on commercially available mouse gene probe arrays with the capacity to display >36,000 mRNA transcript levels of mouse genes and ESTs (Murine Genome U74 version 2, Affymetrix, Santa Clara, CA). Sample labeling and processing were performed according to the manufacturer's protocols (21). Data were collected by laser scanning, and pixel levels were analyzed with commercially available software (genechip, Affymetrix).
Quantification of mRNA Levels Using Real-Time PCR Analysis. Two micrograms of total RNA was reverse transcribed with MultiScribe Reverse Transcriptase (Applied Biosystems) according to the manufacturer's protocol. Conditions for all PCRs were optimized with regard to primers and various annealing temperatures (55-60°C). Specificity of the RT-PCR amplification products was documented with a 4% high resolution NuSieve agarose gel electrophoresis and resulted in a single product with the desired length. Optimized results were transferred on an iCycler Detection System (Bio-Rad) using SYBR green detection. The following iCycler run protocol was used: denaturation program (95°C for 10 min), amplification and quantification program repeated 30-40 cycles (95°C for 30 s, specified annealing temperature for 30 s, 72°C for 30 s, and specified acquisition temperature for 15 s). Melting curve analysis was performed after amplification. The relative quantification of a target gene in comparison with a reference gene (18S rRNA) was performed as described (22). Expression levels of five transcripts were reconfirmed by RT-PCR: necdin, CD24, HNF3, DCM, Apo-D, and CMOCR.
Electrophoretic Mobility-Shift Assay. Nuclear protein extracts (10 μg) prepared from NIH 3T3 cells expressing FUS1 and FUS2 were incubated with 32P-labeled NF-κB consensus oligonucleotide (sc-2505; Santa Cruz Biotechnology) for 25 min at room temperature. After incubation with antibodies against p65, p50, RelB, and c-Rel (sc-372 X, sc-114 X, sc-7386, and sc-226X; Santa Cruz Biotechnology), the mixtures were subjected to electrophoresis in 5% polyacrylamide gel and autoradiography. Competition assays were performed with unlabeled NF-κB consensus oligonucleotide and mutant oligonucleotide (sc-2511; Santa Cruz Biotechnology)
SDS/PAGE and Western Blot Analysis. Proteins were resolved on SDS-polyacrylamide gels under reducing conditions and blotted onto nylon membranes (Bio-Rad) according to standard procedures. Mouse monoclonal antibody against p53 (Ab-1, Oncogene), rabbit polyclonal against poly(ADP-ribose) polymerase (PARP) (sc-1152, Santa Cruz Biotechnology), and rabbit polyclonal against p21 (sc-397, Santa Cruz Biotechnology) were used according to the manufacturer's protocols.
Results
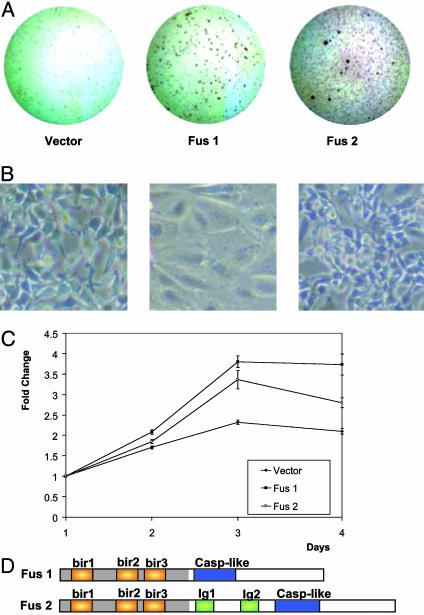
Oncogenic Potential of API2/MALT1 Fusion Products. The transforming potential of two distinct API2/MALT1 chimeric products (FUS1 and FUS2) was examined by performing in vitro transformation assays using stably and/or transiently transfected NIH 3T3 cells. FUS1 contains the 5′ portion of API2, consistently found in API2/MALT1 fusion proteins, fused to the MALT1 carboxyl terminus, including the caspase-like domain. FUS2 contains, in addition to the caspase-like domain, both Ig domains of the MALT1 protein. Increased growth rates and morphological changes characteristic of malignant transformation including loss of spindle cell morphology, contact growth inhibition, increased nuclear to cytoplasmic ratio, and increased nuclear size were identified in FUS1 and FUS2 stable NIH 3T3 clones (Fig. 1). The same stable FUS1 and FUS2 cell lines, as well as transiently transfected NIH 3T3 cells, were used in a soft agar transformation assay. Colony formation was rapid (≈1 week) in both cases, with a size of >100 μm and >200 μm in FUS1 and FUS2 transfected cells, respectively. Vector-transfected clones served as control for these experiments (Fig. 1).
Fig. 1.
Transformation of NIH 3T3 cells by Fus1 and Fus2. (A) pcDNA3, pcDNA3-Fus1, and pcDNA3-Fus2 constructs, transiently and stably transfected NIH 3T3 cells, were plated on soft agar. Rapid colony formation was observed in the Fus1- and Fus2-transfected cells as compared with vector control. (B) Transfection of Fus1 and Fus2 resulted in morphological changes associated with dedifferentiation, including loss of spindle cell morphology and contact growth inhibition compared with vector-transfected cells. (C) pcDNA3, pcDNA3-Fus1, and pcDNA3-Fus2 stably expressing cells were subjected to cell viability measurement with the MTT assay (±1 SD) showing significantly enhanced growth in FUS1- and FUS2-expressing cells compared with vector control. (D) Schematic representation of the structural features of Fus1 and Fus2 chimeric proteins.
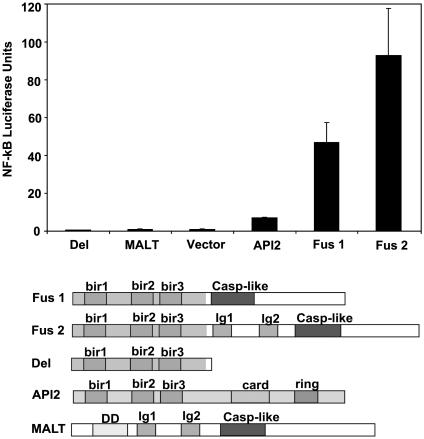
FUS1 and FUS2 Differentially Activate the NF-κB Signaling Pathway in 293 Cells, and the Ig-Like Domains of MALT1 Are Not Required for NF-κB Activation. Two studies recently suggested that the API2/MALT1 chimeric proteins (FUS1 and FUS2) activate the NF-κB pathway (14, 15). To assess how the structural differences of the chimeric proteins may affect NF-κB activation, pcDNA3-FUS1, -FUS2, -API2, and -MALT1 and a deletion construct (pcDNA3-DEL1) that represented the API2 portion of the fusion products were tested in an NF-κB luciferase promoter assay in 293 cells. 293 cells represent an appropriate system to examine activation of the NF-κB signaling because the signaling components are intact and can be activated upon stimulation. In contrast, most existing B cell lines are Epstein-Barr virus transformed and therefore have a deregulated NF-κB pathway. Moreover, the available B cell lines exhibit an array of chromosomal translocations that do not permit any specific conclusions about API2/MALT1-mediated effects. Expression of the chimeric proteins FUS1 and FUS2 resulted in a robust ≈45- and ≈ 95-fold NF-κB activation, respectively. In contrast, transfection of DEL1-, MALT1- and vector- expression plasmids had no effect on NF-κB activation. Finally, as expected, the API2 expression construct slightly activated NF-κB (5-fold) (Fig. 2) (23).
Fig. 2.
The fusion constructs Fus1 and Fus2 activate NF-κB in a luciferase promoter assay. 293 cells were transfected in triplicate with pcDNA3, pcDNA3-Fus1, pcDNA3-Fus2, pcDNA3-Del, pcDNA3-API2, or pcDNA3-MALT1 in the presence of pTK-Luc (an NF-κB responsive luciferase reporter) and pEF1-BOS-β-gal (used as a control for transfection efficiency). Strong activation of NF-κB levels was observed with the Fus 1 and Fus 2 constructs. However, the MALT1 full-length cDNA and the Del 1 construct failed to activate NF-κB. As expected, the pcDNA3-API2 full-length construct slightly activated NF-κB.
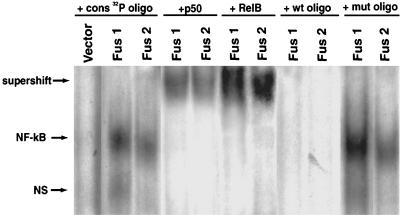
API2/MALT1-Mediated NF-κB Activation Involves a RelB/p50 Dimer. To further elucidate the influence of the API2/MALT1 fusion products on NF-κB signaling and to define which members of the NF-κB family are activated through the fusion products, nuclear extracts of FUS1- and FUS2-expressing cells were prepared and analyzed by electrophoretic mobility-shift assay and supershift experiments. NF-κB member-specific antibodies against p50, p65, RelB, c-Rel, and p52 were used (24-27). Supershifted complexes were detectable only with antibodies against p50 and RelB in both FUS1 and FUS2 stably expressing cell lines compared with vector control (Fig. 3). These results demonstrate that FUS1 and FUS2 can activate NF-κB in stable transfected cell lines.
Fig. 3.
Fus1 and Fus2 activate NF-κB through a RelB/p50 transcriptional dimer. Induction of NF-κB DNA activity was observed in nuclear extracts of Fus1 and Fus2 stably expressing cell lines compared with the vector. The identity of protein components of the NF-κB DNA-binding activity was determined by electrophoretic mobility-shift assay with antibodies to p50, p52 (not shown), RelA (not shown), RelB, and c-Rel (not shown). RelB and p50 generated shifted signal with both fusions. Competition experiments were carried out with mutant and wild-type NF-κB oligonucleotides in 20× excess. The data shown are representative of three independent experiments.
Gene Expression Analysis of NIH 3T3 Cells Stably Expressing FUS1 and FUS2. To further explore the molecular basis underlying the oncogenic activity of the API2/MALT1 fusion products, gene expression was studied by using Affymetrix oligonucleotide microarrays. Transcriptional profiles were measured in cells expressing FUS1 and FUS2 or vector control. To ensure that the obtained profiles are not due to overexpressed phenotypes, expression levels of the FUS1- and FUS2-expressing stable clones were compared with endogenous levels of FUS1 and FUS2 in primary tumors. We found that FUS1 and FUS2 expression levels in the stable clones were similar to the levels of chimeric transcripts in the examined tumor samples (data not shown). Several distinguishable clusters of genes (including immunoreceptors and cell surface receptors, stress responsive genes, transcription factors, enzymes, genes that regulate the extracellular matrix architecture, and genes involved in cell growth) were regulated by the API2/MALT1 fusions (Tables 1 and 2, which are published as supporting information on the PNAS web site). In fact, 86% of the genes deregulated by the FUS1 and 80% of the transcriptional targets of FUS2 are known inflammatory and immune system-responsive genes (28). Expression profiling of FUS1 and FUS2 stably expressing cells revealed up-regulation of several genes with antiapoptotic features (necdin, XMR, Kruppel-like factor, and cardiac-responsive adriamycin protein) and down-regulation of the proapoptotic prostate apoptosis response transcript (Tables 1 and 2).
The oligonucleotide arrays were performed twice in independent experiments. Real-time PCR reconfirmed the induction of representative genes in the clones used in the microarray analysis and in two additional stable cell lines of each fusion product.
FUS1 and FUS2 Inhibit p53-Mediated Apoptosis After DNA Damage. In consideration of the fact that antiapoptotic genes and genes involved in cell cycle regulation were transcriptionally activated by the fusion products, we further determined whether this activation could influence apoptotic events in the FUS1- and FUS2-expressing cells. Of specific interest was the high up-regulation of necdin by both fusion products, a nuclear protein that was shown to bind and inhibit p53-induced apoptosis in SAOS2 osteosarcoma cells (29).
It has been well documented that p53 triggers apoptosis or causes cell-cycle arrest after DNA damage (30, 31). To examine whether FUS1 and FUS2 can inhibit p53-mediated apoptosis, we treated FUS1, FUS2, and vector clones with 20 J/m2 of UV irradiation. Both floating and attached cells were collected 12 h, 24 h, and 36 h after irradiation, fixed, and subsequently stained with propidium iodide for fluorescence-activated cell sorter analysis. Treatment with UV induced a significant level of cell death in vector-expressing cells (6.5%/12 h, 13.4%/24 h, and 34.7%/36 h). Conversely, expression of the fusion proteins FUS1 or FUS2 was able to protect cells from UV-induced cell death with, respectively, 2.3%/12 h, 7.0%/24 h, and 14.6%/36 h, and 2.2%/12 h, 6.6%/24 h, and 13.6%/36 h (Fig. 4A). In addition, whole cell lysates were prepared from the FUS1-, FUS2-, and vector-expressing cell lines at 12 h and 24 h post UV irradiation, and cell pellets were analyzed by Western blotting. PARP cleavage was increased in vector clones and reduced in FUS1-and FUS2 clones with an ≈3.1-fold and ≈2.7-fold reduction of cleaved PARP product, respectively (Fig. 4B). p53 protein levels increased by 12 h and were equal in all three cell lines by 12 h and 24 h, suggesting that both fusion products do not block the activation of p53 after UV irradiation.
Fig. 4.
Fus1 and Fus2 inhibit p53-mediated apoptosis after UV treatment. (A) NIH 3T3 cells expressing Fus1, Fus2, and empty vector were treated with 20 J/m2 UVC. Twelve hours, 24 h, and 36 h after treatment, cells were stained with propidium iodide for fluorescence-activated cell sorter (FACS) analysis. Apoptotic percentage was calculated by scoring sub-G1 populations. The FACS data shown represent the mean ± SD obtained from three independent experiments. ANOVA gave P < 0.01 for all time points. (B) Western blot analysis on whole-cell extracts after 12 h and 24 h of UV treatment using a polyclonal α-PARP antibody showed ≈3.1-fold and ≈2.71-fold reduction in levels of cleaved PARP product in Fus1- and Fus2-expressing cells compared with the vector control by 24 h. p53 protein levels significantly increased 12 h after UV treatment in vector, Fus1, and Fus2 clones and remained high by 24 h post UV damage. Increased p21 protein levels were detected in all three cell lines 24 h after UV irradiation. Loading was controlled by anti-Ran monoclonal antibody.
To determine whether the activated p53 is transcriptionally functional, we examined the protein levels of p21, a downstream target of p53, which is known to be involved in cell cycle arrest. Increased p21 protein levels were detected in FUS1-, FUS2-, and vector-expressing cells by 24 h.
Collectively, our data suggest that FUS1 and FUS2 inhibit cell death after DNA damage, but not the induction of p53 protein, and interestingly that induced p53 is transcriptionally functional. The experiments were reconfirmed with two additional clones for both fusions to exclude the possibility of clonal artifacts (data not shown).
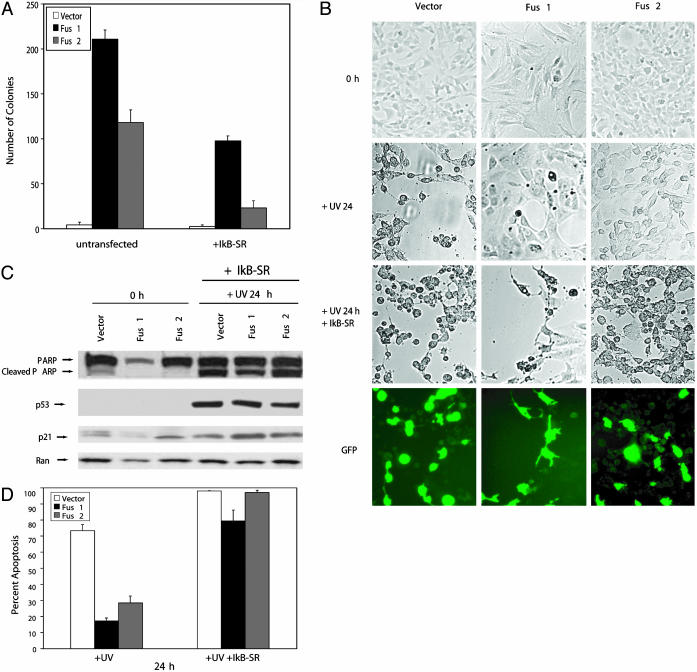
Transforming and Antiapoptotic Features of FUS1 and FUS2 Are NF-κB-Dependent. Having demonstrated that the fusion proteins activate the NF-κB survival signal and also inhibit p53-mediated apoptosis, we then tested whether the FUS1- and FUS2-mediated activation of NF-κB accounts for the observed ability to transform cultured cells. For this purpose, FUS1-, FUS2-, and vector-expressing cells were transiently transfected with the IκB superrepressor (IκB-SR). The IκB-SR blocks NF-κB translocation into the nucleus by inhibiting IκB phosphorylation/degradation, resulting in abrogation of NF-κB activity. Thirty-six hours after IκB-SR transfection, FUS1, FUS2 and vector-transfected cells were plated in soft agar to detect possible changes in the ability to form colonies. IκB-SR transfection resulted in 56% reduction of colonies in FUS1 cells and 80% reduction in FUS2 cells. Untransfected cells were used as a control (Fig. 5A).
Fig. 5.
Transformation of NIH 3T3 cells and inhibition of p53-mediated apoptosis by Fus1 and Fus2 depend on NF-κB activation. (A) IκB-SR-transfected and untrasfected Fus1, Fus2, and vector cells were subjected to an anchorage-dependent growth assay in soft agar. Colony formation was reduced by 56% and 80% in Fus1 and Fus2 cells transfected with the IκB-SR compared with vector control. The data shown represent the mean ± SD obtained from three independent experiments performed in duplicate. (B) Treatment of Fus1, Fus2, and vector control cells with UVC after IκB-SR transient transfection resulted in similar amounts of PARP cleaved protein in Fus1-, Fus2-, and vector-expressing cells. p53 and p21 protein levels increased 24 h post UV damage in the transfected cells. This result is similar to the protein changes detected in untransfected Fus1-, Fus2-, and vector-expressing cells (Fig. 4B). Loading was controlled by anti-Ran monoclonal antibody. (C and D) Similar morphological changes indicative of cell death were detected in IκB-SR-transfected Fus1-, Fus2-, and vector-expressing cells 24 h post UV irradiation. In contrast, in untransfected cells, cell death in vector-expressing cells was significantly increased compared with FUS1- and FUS2-expressing cells. Apoptotic cells were visualized by DAPI (4′,6-diamidino-2-phenylindole) staining, and IκB-SR transfection efficiency was determined by GFP cotransfection. The data shown represent the mean ± SD obtained from three independent experiments performed in duplicate. Loading was controlled by anti-Ran monoclonal antibody.
To further elucidate whether NF-κB activation is required for the antiapoptotic features of the fusion products, FUS1-, FUS2-, and vector-expressing cells that had been transfected with the IκB-SR were exposed to DNA damage as described above. Western blot analysis of PARP cleavage levels in FUS1 and FUS2 clones revealed similar levels of cell death in FUS1 and FUS2 cells compared with the vector (Fig. 5B). Levels of PARP cleavage dramatically increased in FUS1 and FUS2 cells transfected with the IκB-SR compared with untransfected cells 24 h post UV treatment. However, no significant changes in PARP cleavage product were observed in vector-transfected cells with IκB-SR compared with untransfected vector-expressing cells (Figs. 4B and 5B). p53 and p21 protein levels increased in IκB-SR-transfected cells 24 h after exposure to UV damage, but no significant differences were observed in the levels of p53 and p21 proteins in IκB-SR-transfected cells compared with untransfected cells. (Figs. 4B and 5B).
In addition to the similar PARP cleavage levels in IκB-SRFUS1-, IκB-SR-FUS2-, and vector-expressing cells, similar morphological changes indicative of apoptosis were observed in FUS1- and FUS2-expressing cells as compared with the vector. Changes in cell shape, detachment from the culture plate and nuclear fragmentation, occurred 24 h after UV damage in all three FUS1, FUS2, and vector control clones (Fig. 5C). DAPI (4′,6-diamidino-2-phenylindole) staining revealed comparable changes in cell death as detected by Western blot analysis (Fig. 5D). Three independent experiments were performed with IκB-SR-transfected and untransfected FUS1-, FUS2-, and vector-expressing cells. IκB-SR transfection efficiency was controlled by GFP cotransfection in the described experiments.
Collectively, our results support the notion that FUS1- and FUS2-mediated NF-κB activation is required for the transformation potential of the fusion products and the ability to inhibit p53-induced cell death.
Discussion
In this study, we demonstrated that two distinct API2/MALT1 fusion products (FUS1 and FUS2) exhibit in vitro oncogenic potential by transforming NIH 3T3 cells with distinct morphological changes, increased proliferative growth, and rapid development of colonies in soft agar. The structural composition of these fusion products differs only in their carboxyl terminal portion, with FUS1 containing only the caspase-like domain of the MALT1 protein and FUS2 containing the two Ig-like MALT1 domains in addition to the caspase-like domain.
We also showed that both fusion products (FUS1 and FUS2) strongly activate the NF-κB pathway in a transient luciferase assay in 293 cells. A deletion construct (DEL1) that represents the consistent API2 portion of the fusion products, as well as the full-length MALT1 cDNA construct, failed to activate NF-κB in the same assay. These results indicate that the BIR domains (API2) and/or the Ig and caspase domains (MALT1) alone are not sufficient for the observed NF-κB activation, but structural and/or conformational changes attributed to the fusion of the API2-BIR domains to the caspase-like domain of MALT1 may lead to the observed NF-κB up-regulation. These results also strongly suggest that the Ig domains are not required for this activation, but their presence may play a synergistic effect in enhancing NF-κB activity.
To decipher the molecular mechanism of the API2/MALT1- mediated NF-κB activation, we then examined, in FUS1- and FUS2-expressing cells, which NF-κB transcriptional dimer is activated by the API2/MALT1 chimeric proteins. We provide evidence that both fusion products activate an NF-κB transcriptional dimer consisting of the RelB and p50 polypeptides. It is well established that RelB is an essential component for germinal center (GC) and marginal zone (MZ) formation and dimerizes only with either p50 or p52 (32). Taking into consideration that MALT lymphomas originate in marginal-zone B lymphocytes and are of GC/post-GC origin, the observed activation of the RelB/p50 dimer reaffirms that our experimental system may mimic the cellular context of MZ/GC B cells from which MALT lymphomas are derived (33).
A plethora of genes transcriptionally regulated by NF-κB have been reported (30). Our study identifies the sets of genes that are deregulated by the FUS1 and FUS2 chimeric products. Whereas 86% of the FUS1 and 80% of the FUS2 transcriptional targets are associated with inflammatory and immune responses, several previously uncharacterized genes are described here that are involved in cell proliferation and also genes of yet unknown function. None of the detected genes belong to the normal expression profile of NIH 3T3 cells, an observation that underlines the physiological relevance of our results. Necdin was the most highly up-regulated gene in FUS1 cells. It belongs to the MAGE family of proteins and was shown to interact with p53 and inhibit p53-mediated apoptosis (29, 34). It was also markedly overexpressed in FUS2-expressing cells. This finding prompted us to determine whether activation of API2/MALT1 downstream targets could result in inhibition of apoptosis. The results presented here demonstrate that both fusion products provide efficient protection from UV-induced, p53-dependent cell death. p53 and p21 protein levels increased upon UV treatment in FUS1, FUS2, and the vector control, strongly suggesting that both fusions inhibit p53-mediated cell death but they do not prohibit the induction of p53 protein or its transcriptional activity. It has been established that regulation of p53 function after exposure to DNA-damaging agents not only is based on p53 protein concentration, but mainly occurs by means of posttranslational modifications and alterations in p53-binding proteins (35). Although p53 protein is induced after UV treatment, inability to induce apoptosis may be the result of an inactive or altered protein form, or protein complex.
The question was then asked: Is the observed oncogenic potential and the ability to protect from apoptosis in FUS1 and FUS2 cells an autonomous effect or a result of the activation of the NF-κB signaling pathway? Using the IκB-SR inhibitor of NF-κB, we demonstrated that persistent activation of the NF-κB signaling, mediated by the API2/MALT1 chimeric products, is required for the observed transformation potential of both chimeric products as well as the ability to resist apoptotic stimuli.
In summary, the data provided in this study show that structurally different API2/MALT1 fusion products exhibit common functional features, but also have distinct mechanistic characteristics. Although expression of both fusion proteins leads to anchorage-independent growth in soft agar, colony size in FUS2-expressing cells is larger. Activation levels of NF-κB in transient transfected cells are higher in FUS2 as compared with FUS1, and FUS2 cells become more sensitive to cell death after reduction of the NF-κB proliferative signal compared with FUS1. In addition, FUS1- expressing cells are more resistant to apoptotic stimuli compared with FUS2-expressing cells, a phenomenon that could be attributed to the higher number of cell cycle regulators that are up-regulated in FUS1 cells compared with FUS2-expressing cells. These genes include, in addition to necdin, the Kruppel-like factor, which is a mediator of TNF receptor-associated factor 2 (TRAF2) antiapoptotic effects; the inhibitor of DNA binding 1 (ID 1), a protein required for neurogenesis, angiogenesis, vascularization of tumor xenografts, and G1 progression; and four and a half LIM domain (FHL), which enhances β-catenin activity in cancer cells (36-38). Further studies would be required to investigate whether MALT lymphoma patients with the FUS1 chimeric product have a more aggressive disease or an earlier disease onset. In contrast, the observed FUS1- and FUS2-mediated NF-κB activation involves the same RelB/p50 transcriptional dimer. This finding indicates mechanistic differences in the deregulation of transcriptional targets, probably involving components that affect retention of NF-κB in the nucleus and modification events such as histone acetylation and phosphorylation.
Recent studies have demonstrated that proliferative advantages, including p53 mutations, lead to progression into high-grade lymphomas and that MALT lymphoma patients with the t(11,18)(q21;q21) poorly respond to antibiotic therapy and are probably associated with more advanced stages of the disease (39). In accordance with this fact, our results strongly suggest that proliferative advantages due to API2/MALT1-mediated NF-κB activation, combined with their ability to inhibit apoptosis, may lead to clonal proliferation and tumor development. We further provide mechanistic insights into the involvement of NF-κB in the inhibition of p53-induced cell death, and the results of this study may well aid in the design of novel treatments that would combine appropriate chemotherapeutic agents with inhibitors of the NF-κB pathway for more efficient results.
Supplementary Material
Acknowledgments
We thank Dr. Alexander Tarakhovsky, Marc-Werner Dobenecker, Christine Walsh, Shenghan Jin, and Martin Teichmann for their thoughtful suggestions and valuable discussion throughout the course of this work. We also thank Ms. Caroline Jones for excellent technical assistance. The API2 construct was kindly provided by Dr. Eric LaCasse (Apoptogen), and the FUS2 construct was provided by Dr. V. Dixit (Genentech). cDNA microarray experiments were performed in the Memorial Sloan-Kettering Cancer Center Core Facility with Dr. Agnes Viale. This work was supported by the Lymphoma Research Foundation (A.S.) and the Robert and Harriet Heilbrunn Cancer Fund (A.J.L.).
Abbreviations: BIR, baculovirus IAP repeat; MALT, mucosa-associated lymphoid tissue; PARP, poly(ADP-ribose) polymerase; IκB-SR, IκB superrepressor.
References
- 1.Zucca, E., Roggero, E. & Pileri, S. (1998) Brit. J. Haematol. 100, 3-14. [DOI] [PubMed] [Google Scholar]
- 2.Ott, G., Katzenberger, D. D., Greiner, A., Kalla, J., Rosenwald, A., Heinrich, U., Ott, M. & Mueller-Hermelink, H. K. (1997) Cancer Res. 57, 3944-3998. [PubMed] [Google Scholar]
- 3.Willis, T. G., Jadayel, D. M., Du, M. Q., Peng, H., Perry, A. R., Abdul-Rauf, M., Price, H., Karran, L., Oluwatosin, M., Wlodarska, I., Pan, L., et al. (1999) Cell 96, 35-45. [DOI] [PubMed] [Google Scholar]
- 4.Zhang, Q., Siebert, R., Yan, M., Hinzmann, B., Cui, X., Xue, L., Rakestraw, K. M., Naeve, C. W., Beckmann, G., Weisenburger, D. D., et al. (1999) Nat. Genet. 22, 63-68. [DOI] [PubMed] [Google Scholar]
- 5.Ruland, J., Duncan, G. S., Elia, A, Barco Barrantes, I., Nguyen, L., Plyte, S., Millar, D. G., Bouchard, D., Wakeham, A., et al. (2001) Cell 104, 33-42. [DOI] [PubMed] [Google Scholar]
- 6.Stoffel, A. & Le Beau, M. M. (2001) Hum. Heredity 51, 1-7. [DOI] [PubMed] [Google Scholar]
- 7.Stoffel, A., Rao, P. H., Louie, D. C., Krauter, K., Liebowitz, D. N., Koeppen, H., Le Beau, M. M. & Chaganti, R. S. K. (1999) Genes Chrom. Cancer 24, 156-159. [PubMed] [Google Scholar]
- 8.Dierlamm, J., Baens, M., Wlodarska, I., Stefanova-Ouzounava, M., Hernandez, J., Hossfeld, D. K., DeWolf-Peeters, C., Hagemeijer, A., Van den Berghe, H. & Marynen, P. (1999) Blood 93, 3601-3609. [PubMed] [Google Scholar]
- 9.Akagi, T., Motegi, M., Tamura, A., Suzuki, R., Hosokawa, Y., Suzuki, H., Ota, H., Nakamura, S., Morishima, Y., Taniwaki, M. & Seto M. (1999) Oncogene 118, 5785-5794. [DOI] [PubMed] [Google Scholar]
- 10.Morgan, J. A., Yin, Y., Borowsky, A. D., Kuo, F., Nourman, N., Koontz, J. I., Reynolds, C., Soreng, L., Griffin, C. A., Graema-Cook, F., et al. (1999) Cancer Res. 59, 6205-6213. [PubMed] [Google Scholar]
- 11.Liston, P., Roy, N., Tamai, K., Lefebvre, C., Baird, S., Cherton, H. G., Farahani, R., McLean, M., Ikeda, J. E., MacKenzie, A. & Korneluk, R. G. (1996) Nature 379, 349-353. [DOI] [PubMed] [Google Scholar]
- 12.Deveraux, Q. L. & Reed, J. (1999) Genes Dev. 13, 239-252. [DOI] [PubMed] [Google Scholar]
- 13.Rothe, M., Pan, M. G., Henze, W. J., Ayres, T. M. & Goedel, D. V. (1995) Cell 83, 1243-1252. [DOI] [PubMed] [Google Scholar]
- 14.Uren, A. G., O'Rourke, K., Aravind, L., Pisabarro, M. T., Seshagiri, S., Koonin, E. V. & Dixit, V. M. (2000) Mol. Cell 6, 961-967. [DOI] [PubMed] [Google Scholar]
- 15.Lucas, P. C., Yonezumi, M., Inohara, N., McAllister-Lucas, L. M., Abazeed, M. E., Chen, F. F., Yamaoka, S., Seto, M. & Nunez, G. (2001) J. Biol. Chem. 276, 19012-19019. [DOI] [PubMed] [Google Scholar]
- 16.Ruefli-Brasse, A. A., French, D. M. & Dixit, V. M. (2003) Science 302, 1581-1584. [DOI] [PubMed] [Google Scholar]
- 17.Ruland, J., Duncan, G. S., Wakeham, A. & Mak, T. W. (2003) Immunity 19, 749-758. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez-Beato, M., Sanchez-Aquilera, A. & Piris, M. A. (2003) Blood 15, 1220-1235. [DOI] [PubMed] [Google Scholar]
- 19.Singh, B., Reddy, P. G., Goberdhan, A., Walsh, C., Dao, S., Ngai, I., Chou, T. C., Charoenrat, P. O., Levine, A. J., Rao, P. H. & Stoffel, A. (2002) Genes Dev. 16, 984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shih, D. Q., Bussen, M., Sehayek, E., Ananthanarayanan, M., Shneider, B. L., Suchy, F. J., Shefer, S., Bollileni, J. S., Gonzalez, F. J., Breslow, J. L. & Stoffel, M. (2001) Nat. Genet. 27, 375-382. [DOI] [PubMed] [Google Scholar]
- 21.Lockhart, D. J., Dong, H., Byrne, M. C., Follettie, M. T., Gallo, M. V., Chee, M. S., Mittmann, M., Wang, C., Kobayashi, M., Horton, H. & Brown, E. L. (1996) Nat. Biotechnol. 14, 1675-1680. [DOI] [PubMed] [Google Scholar]
- 22.Schmittgen, T. D. & Zakrajsek, B. A. (2000) J. Biochem. Biophys. Methods 46, 69-81. [DOI] [PubMed] [Google Scholar]
- 23.Chu, Z. L., McKinsey, T. A., Liu, L., Gentry, J. J., Malim, M. H. & Ballard, D. W. (1997) Proc. Natl. Acad. Sci. USA 94, 10057-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauerle, P. A. & Baltimore, D. (1996) Cell 87, 13-20. [DOI] [PubMed] [Google Scholar]
- 25.Karin, M., Cao, Y., Greten, F. R. & Li, Z. W. (2002) Nat. Rev. Cancer 2, 301-310. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh, S. & Karin, M. (2002) Cell 109, S81-S96. [DOI] [PubMed] [Google Scholar]
- 27.Cogswell, P. C., Guttridge, D. C., Funkhauser, W. K. & Baldwin, A. S. (2000) Oncogene 19, 1123-1131. [DOI] [PubMed] [Google Scholar]
- 28.Pahl, H. L. (1999) Oncogene 18, 6852-6866. [Google Scholar]
- 29.Taniura, H., Matsumoto, K. & Yoshikawa, K. (1999) J. Biol. Chem. 274, 1242-16248. [DOI] [PubMed] [Google Scholar]
- 30.Levine, A. J. (1997) Cell 88, 323-331. [DOI] [PubMed] [Google Scholar]
- 31.Prives, C. & Hall, P. A. (1999) J. Pathol. 187, 112-126. [DOI] [PubMed] [Google Scholar]
- 32.Weih, D. S., Yilmaz, Z. B. & Weih, F. (2001) J. Immunol. 167, 1909-1919. [DOI] [PubMed] [Google Scholar]
- 33.Bahler, D. W., Miklos, J. A. & Swerdlow, S. H. (1997) Blood 16, 3822-3831. [PubMed] [Google Scholar]
- 34.Maruyama, K., Usami, M., Aizawa, T. & Yoshikawa, K. (1991) Biochem. Biophys. Res. Commun. 178, 291-296. [DOI] [PubMed] [Google Scholar]
- 35.Giaccia, A. J. & Kastan, M. B. (1998) Genes Dev. 12, 2973-2983. [DOI] [PubMed] [Google Scholar]
- 36.Lin, Y., Ryan, J., Lewis, J., Wani, M. A., Lingrel, J. B. & Liu, Z. G. (2003) Mol. Cell. Biol. 23, 5849-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lyden, D., Young, A. Z., Zagzag, D., Yan, W., Gerald, W., O'Reilly, R., Bader, B. L., Hynes, R. O., Zhuang, Y., Manova, K. & Benezra, R. (1999) Nature 401, 670-677. [DOI] [PubMed] [Google Scholar]
- 38.Wei, Y., Renard, C. A., Labalette, C., Wu, Y., Levy, L., Neuveut, C., Prieur, X., Flajolet, M., Prigent, S. & Buendia, M. A. (2003) J. Biol. Chem. 278, 5188-5194. [DOI] [PubMed] [Google Scholar]
- 39.Liu, H., Ruskon-Fourmestraux, A., Lavergne-Slove, A., Ye, H., Molina, T., Bouhnik, Y., Hamoudi, R. A., Diss, T. C., Dogan, A., Megraud, F., et al. (2001) Lancet 357, 39-40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.