Abstract
The pink stem borer, Sesamia inferens (Walker), is a major pest of rice and is endemic in China and other parts of Asia. Small heat shock proteins (sHSPs) encompass a diverse, widespread class of stress proteins that have not been characterized in S. inferens. In the present study, we isolated and characterized three S. inferens genes that encode members of the α-crystallin/sHSP family, namely, Sihsp21.4, Sihsp20.6, and Sihsp19.6. The three cDNAs encoded proteins of 187, 183 and 174 amino acids with calculated molecular weights of 21.4, 20.6 and 19.6 kDa, respectively. The deduced amino acid sequences of the three genes showed strong similarity to sHSPs identified in other lepidopteran insects. Sihsp21.4 contained an intron, but Sihsp20.6 and Sihsp19.6 lacked introns. Real-time quantitative PCR analyses revealed that Sihsp21.4 was most strongly expressed in S. inferens heads; Whereas expression of Sihsp20.6 and Sihsp19.6 was highest in eggs. The three S. inferens sHSP genes were up-regulated during low temperature stress. In summary, our results show that S. inferens sHSP genes have distinct regulatory roles in the physiology of S. inferens.
Keywords: Sesamia inferens, small heat shock protein, cloning, expression pattern
1. Introduction
Small heat shock proteins (sHSPs) were first identified as a set of low molecular proteins (15–30 kDa) induced after heat shock in Drosophila melanogaster [1]. Compared to other HSPs, sHSPs exhibit a greater variation in sequence, structure, size, and function [2,3]. They are a superfamily of proteins that contain an α-crystallin domain and variable N- and C-terminal extensions [4]. sHSPs can exist as large oligomers comprised of ~50 subunits and can approach a mass of 1.2 MDa [5,6]. Structurally, sHSPs remain poorly understood, mainly because members of this protein family are extremely dynamic and heterogeneous [7,8]. Many function as molecular chaperones that block the aggregation of unfolded proteins and have cytoprotective functions under stressful conditions [9]. In addition to the stress response, sHSPs have been implicated in apoptosis and autophagy, actin and intermediate filament dynamics, organization of the cytoskeleton, and membrane fluidity [10,11,12,13]. They also function as therapeutic targets and biomarkers for many diseases [14]. In insects, they presumably perform important roles in heat/cold stress, metamorphosis, normal development, diapause, and immune responses [15,16,17,18,19,20,21]; However, these roles remain for many insect species.
The pink stem borer, Sesamia inferens (Walker) (Lepidoptera: Noctuidae), is a major pest of rice in China and other parts of Asia, and recently, damage incited by S. inferens has become more serious [22]. According to our previous surveys, this pest now occurs in the more northern regions of China. Many studies of S. inferens have focused on biological characteristics [23,24,25,26,27,28]. We previously demonstrated that S. inferens still survived during exposure to low temperatures [29].
The underlying mechanisms that explain sudden outbreaks and the widespread distribution of S. inferens remain obscure. Hence, expression analysis of relevant genes, such as those encoding sHSPs, may provide insight on the incidence of S. inferens. To investigate whether shsps expression regulates cold tolerance in S. inferens, we cloned three genes encoding sHSPs from this insect pest. The structure of these genes was examined, and we analyzed their expression in different tissues and stages of insect development. Our results indicate that expression of the three shsps is modulated in response to cold stress.
2. Results and Discussion
2.1. Results
2.1.1. Sequence Analysis of S. inferens Small Heat Shock Proteins (sHSPs) Genes
Three complete cDNA sequences were obtained and named Sihsp21.4, Sihsp20.6, and Sihsp19.6; These were deposited as GenBank accession nos. KM217075, KM217077, and KM217079, respectively. The sizes of full-length cDNAs were 1385 (Sihsp21.4), 835 (Sihsp20.6), and 798 bp (Sihsp19.6). Each gene contained three regions, a 5' untranslated region (5' UTR), the open reading frame (ORF), and a 3' UTR. Lengths of 5' UTRs were 118 (Sihsp21.4), 119 (Sihsp20.6), and 146 bp (Sihsp19.6), respectively. Lengths of 3' UTRs were 703 (Sihsp21.4), 164 (Sihsp20.6), and 127 bp (Sihsp19.6); Each 3' UTR contained a polyadenylation signal (AATAAA) located 16 (Sihsp21.4), 21 (Sihsp20.6), and 22 bp (Sihsp19.6) upstream with respect to the poly(A) tract.
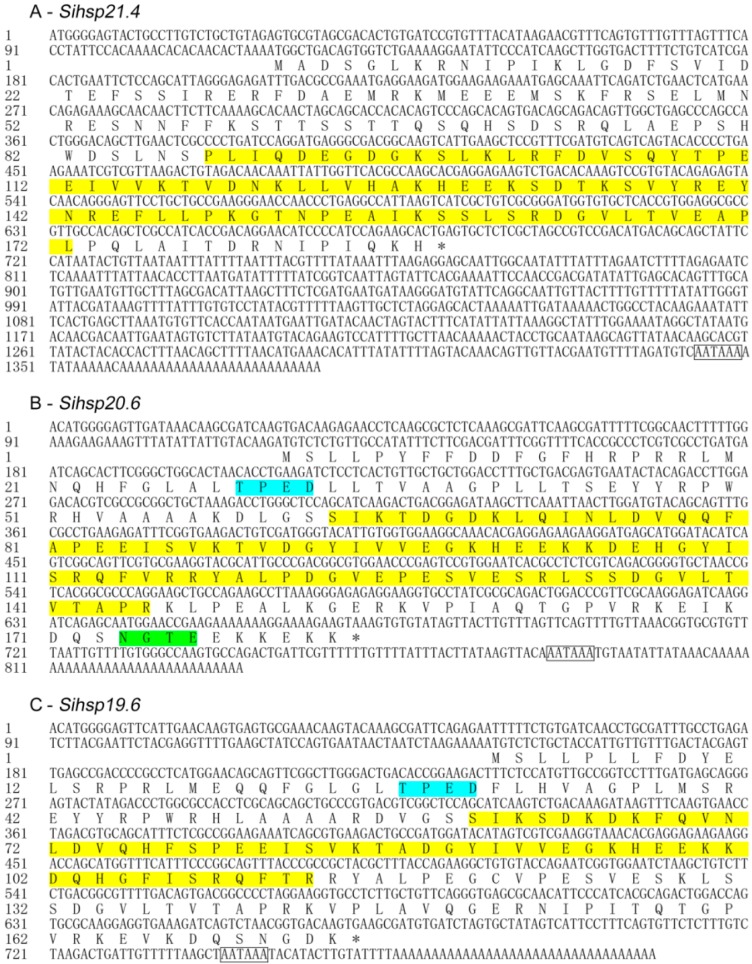
The three ORFs consisted of 564 (Sihsp21.4), 552 (Sihsp20.6), and 525 nucleotides (Sihsp19.6). The deduced proteins contained 187 (HSP21.4), 183 (HSP20.6), and 174 amino acids (HSP19.6), with predicted molecular weights of 21.4, 20.6, and 19.6 kDa, respectively. The theoretical isoelectric points (pI) of the predicted proteins were 5.79 (HSP21.4), 6.54 (HSP20.6), and 6.53 (HSP19.6). When compared with the NCBI GenBank and PROSITE databases, the three deduced proteins had high similarity to the small heat shock protein (HSP20) family. Each of the predicted proteins contained the typical α-crystallin domain at the following locations: HSP21.4, 88–172 aa (Figure 1A), HSP20.6, 63–145 aa (Figure 1B), and HSP19.6, 60–142 aa (Figure 1C). PROSITE analysis indicated that HSP20.6 has a casein kinase II phosphorylation site (TPED, 29–32 aa) and a N-glycosylation site (NGTE, 174–177 aa) (Figure 1B). Similarly, S. inferens HSP19.6 also contains a casein kinase II phosphorylation site (TPED, 27–30 aa) (Figure 1C).
Figure 1.
Nucleotide and deduced amino acid sequences of small heat shock proteins (sHSPs) genes from Sesamia inferens. sHSP genes include Sihsp21.4 (A); Sihsp20.6 (B); and Sihsp19.6 (C). HSP20 family profiles, typical α-crystalline domains, are shaded in yellow; Casein kinase II phosphorylation sites (TPED) are shaded in cyan; N-Glycosylation site (NGTE) is shaded in green; The putative polyadenylation signals are boxed.
2.1.2. Structural Prediction of S. inferens sHSPs
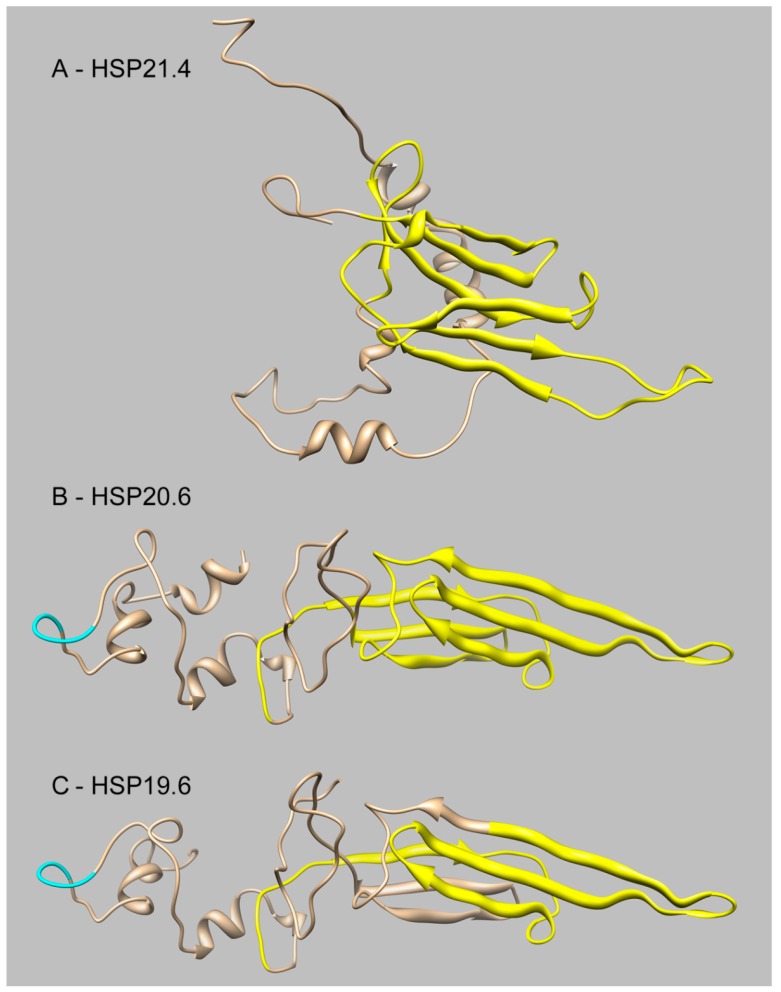
To investigate the structural characteristics of three sHSPs from S. inferens, we generated a homology model of S. inferens HSP21.4 with Phyre. In this analysis, we used a sHSP from Taenia saginata (PDB ID: 2BOL) as a template [30], for which the confidence and coverage are 100% and 88%, respectively (Figure 2A). Homology models of S. inferens HSP20.6 and HSP19.6 were deduced with Phyre using Homo sapiens αB-crystallin V * (PDB ID: 2YGD) as a template [31] (S. inferens HSP20.6: Confidence, 100%, coverage, 91%; HSP19.6: Confidence, 100%, coverage, 93%) (Figure 2B,C).
Figure 2.
The structural prediction of sHSPs from Sesamia inferens. sHSPs include HSP21.4 (A); HSP20.6 (B); and HSP19.6 (C). HSP20 family profiles; Typical α-crystalline domains are indicated in yellow, kinase II phosphorylation sites (TPED) are indicated in cyan, peptides are indicated in brown.
2.1.3. Phylogenetic Analysis of S. inferens sHSPs
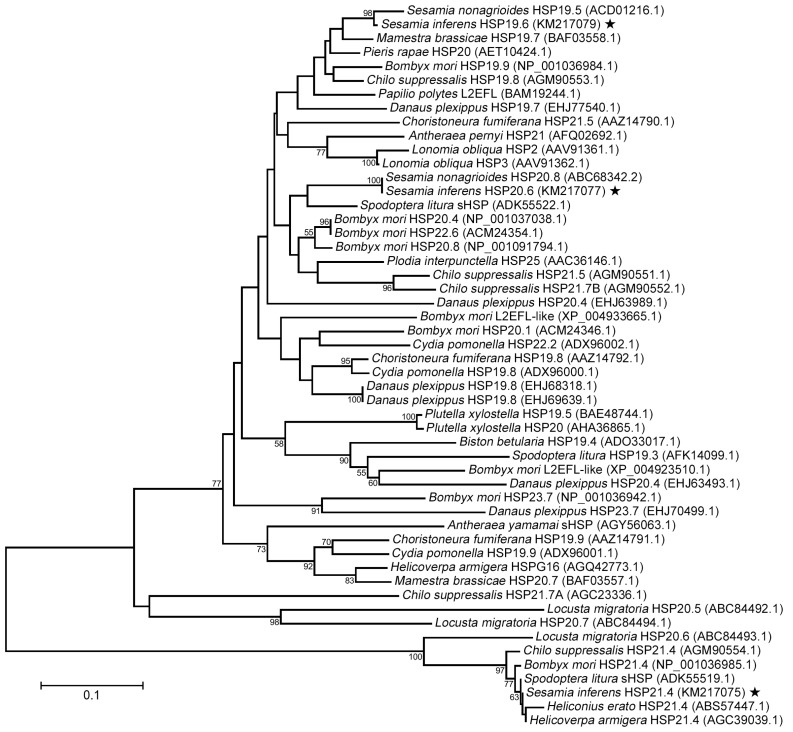
The deduced amino acid sequences of the three shsps displayed a high degree of relatedness with orthologous proteins reported in other insects. To compare S. inferens sHSPs with those from other insects, ClustalX and MEGA 6.06 were used to perform multiple phylogenetic analyses, including neighbor-joining, minimum evolution, maximum likelihood, and maximum parsiomony. The four resulting phylogenetic trees were similar; Thus, only the neighbor-joining tree is shown (Figure 3). The tree could be divided into two major clusters; S. inferens HSP19.6 and HSP20.6, which show high sequence similarity, were grouped together in a well-supported cluster (Figure 3). S. inferens HSP19.6 showed 97% amino acid identity with Sesamia nonagrioides HSP19.5, and S. inferens HSP20.6 exhibited 98% identity with S. nonagrioides HSP20.8 (Figure 3). S. inferens HSP21.4 showed 97%–99% identity with HSP21.4/sHSP from five other lepidopteran species (e.g., Chilo suppressalis HSP21.4, Bombyx mori HSP21.4, S. litura sHSP, Heliconius erato HSP21.4, and Helicoverpa armigera HSP21.4) (Figure 3).
Figure 3.
Phylogenetic tree of Sesamia inferens and other insect species sHSPs based on neighbor-joining method. The S. inferens shsps are labeled with five-pointed star. The Locusta migratoria HSP20.5, L. migratoria HSP20.6 and L. migratoria HSP20.7 were used as the out groups. Origin of sHSP proteins and their GenBank accession numbers are showed in the figure. Numbers on the branches are the bootstrap values obtained from 1000 replicates (only bootstrap values >50 are shown).
2.1.4. Genomic Structure of S. inferens sHSP Genes
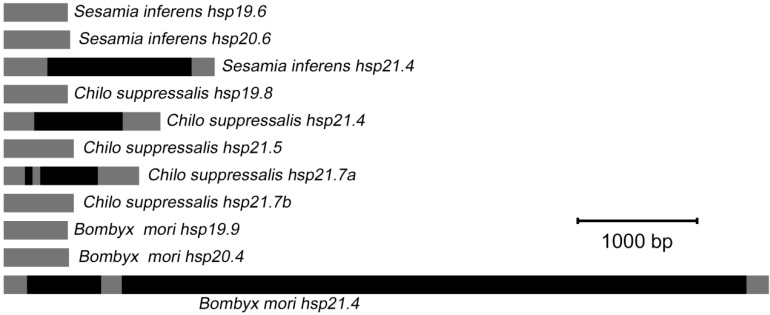
The genomic DNA sequences of the three S. inferens sHSP genes varied in length as follows: 2541 bp for Sihsp21.4 (GenBank accession no. KM217076), 559 bp for Sihsp20.6 (accession no. KM217078), and 692 bp for Sihsp19.6 (accession no. KM217080). The position and size of introns were noted by aligning cDNA with genomic DNA sequences. Sihsp21.4 had a single 1210 bp intron located within the coding region (472–1681 bp) (Figure 4), and the nucleotide sequences at the intron splice junctions were consistent with the canonical GT-AG rule. Although S. inferens HSP21.4, B. mori HSP21.4, and C. suppressalis HSP21.4 had high amino acid identity (Figure 3), the genomic DNAs differed in the size and/or number of introns (Figure 4). Unlike Sihsp21.4, which had a single, 1210 bp intron, Bmhsp21.4 had two introns (720 and 5531 bp). Cshsp21.4 had a single intron, but it was smaller than Sihsp21.4. The genomic DNAs of Sihsp20.6 and Sihsp19.6 lacked introns and were highly homologous to Cshsp19.8, Cshsp21.5, Cshsp21.7b, Bmhsp19.9, and Bmhsp20.4 (Figure 3 and Figure 4).
Figure 4.
Schematic representation of genomic structures of three lepidopteran insect shsps. Origin of shsps genomes and their GenBank accession numbers: Sesamia inferens hsp19.6 (KM217080), S. inferens hsp20.6 (KM217078), S. inferens hsp21.4 (KM217076), Chilo suppressalis hsp19.8 (KC210020), C. suppressalis hsp21.4 (KC210021), C. suppressalis hsp21.5 (KC210022), C. suppressalis hsp21.7A (KC210023), C. suppressalis hsp21.7B, (KC210024), Bombyx mori hsp19.9 (BGIBMGA004540-TA), B. mori hsp20.4 (BGIBMGA004541-TA), and B. mori hsp21.4 (BGIBMGA000944-TA). Gray and black rectangles are used to highlight the exons and introns, respectively.
2.1.5. Expression of Genes Encoding sHSPs in S. inferens Tissues
qRT-PCR was used to study the expression profiles of the three shsps in S. inferens. The presence of single, sharply defined peaks in melting curve analysis of the three shsps and six reference genes was confirmed. A standard curve was generated for each gene using eight ten-fold serial dilutions (1×, 10×, 102×, 103×, 104×, 105×, 106× and 107×) of the pooled cDNAs. The PCR efficiency (as calculated from the standard curve) and correlation coefficient (R2) for each standard curve are shown in Table 2, and the parameters satisfied the basic requirements for quantitative real-time PCR [32].
Table 2.
Primers used for qRT-PCR.
| Gene | Primer Sequences (5'–3') | Amplicon Size (bp) | PCR Efficiency | Tm (°C) | R2 |
|---|---|---|---|---|---|
| hsp21.4qRT-F | TGGCTGACAGTGGTCTGAAAA | 196 | 91.6% | 60.1 | 0.996 |
| hsp21.4qRT-R | GTGGTGCTGCTAGTTGTGCTT | ||||
| hsp20.6qRT-F | GCATCAAGACTGACGGAGATAAG | 111 | 99.5% | 60.1 | 0.994 |
| hsp20.6qRT-R | GTTTGCCTTCCACCACAATG | ||||
| hsp19.6qRT-F | CGAAGGTAAACACGAGGAGAAG | 132 | 102.7% | 58.2 | 0.970 |
| hsp19.6qRT-R | GTCAAAACGCCGTCAGAAGA | ||||
| RPS13qRT-F | TGGTAAGGGTATCTCCCAATCA | 75 | 93.5% | 60.1 | 0.994 |
| RPS13qRT-R | TCGTCAGCAGTCAGTTTCAGC | ||||
| RPS20qRT-F | CTCATCAATGGAGCCAAGAAAC | 162 | 102.0% | 60.1 | 0.986 |
| RPS20qRT-R | GTGCAGGTCAATGACACGCT | ||||
| EF1qRT-F | GTCGCTTTCGTACCCATTTCT | 86 | 97.4% | 56.6 | 0.994 |
| EF1qRT-R | ACAGTCCATCCCTTGAACCAG | ||||
| 18SqRT-F | CAACACGGGAAATCTCACCA | 115 | 107.3% | 55.6 | 0.996 |
| 18SqRT-R | GACAAATCGCTCCACCAACTAA | ||||
| GAPDHqRT-F | GGTCATCTCCAACGCTTCCT | 166 | 95.0% | 56.6 | 0.993 |
| GAPDHqRT-R | ACGTCCATCACGCCACAAT | ||||
| TUBqRT-F | TTGCTACAGAACCCTCAAAGTGC | 159 | 104.4% | 59.2 | 0.985 |
| TUBqRT-R | AGACGTGGGAACGGAACCAT |
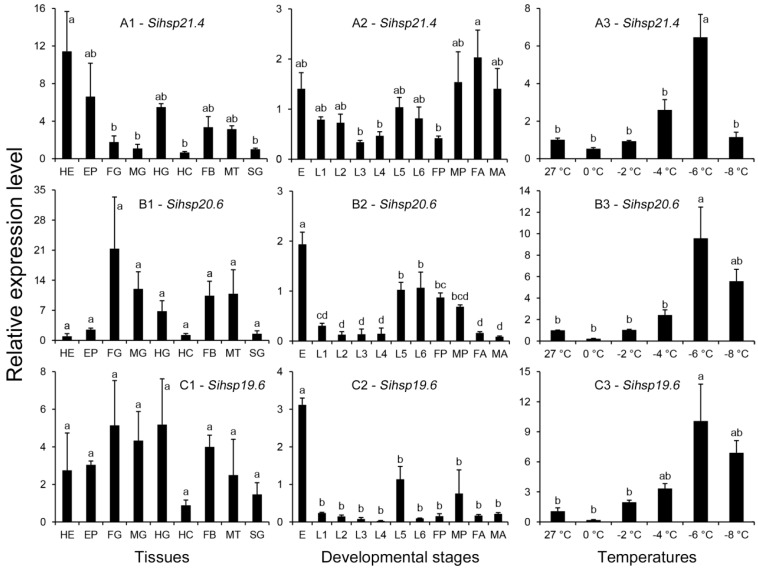
The three S. inferens sHSP genes were expressed in all insect tissues examined, although expression patterns differed amongst tissues. For example, Sihsp21.4 was expressed more highly in heads than other tissues (F8,18 = 3.387, p = 0.015) (Figure 5A1). However, the expression of Sihsp20.6 (F8,18 = 2.006, p = 0.105) and Sihsp19.6 (F8,18 = 0.935, p = 0.512) was not significantly different in the various insect tissues examined (Figure 5B1,C1).
Figure 5.
Relative mRNA expression levels of Sihsp21.4 (A); Sihsp20.6 (B); and Sihsp19.6 (C) in different tissues of the 5th instar larvae (1), different developmental stages (2), and under low-temperature stress (3). HE: Head; EP: Epidermis; FG: Foregut; MG: Midgut; HG: Hindgut; HC: Haemocytes; FB: Fat body; MT: Malpighian tubules; SG: Salivary glands; E: Egg; L1: First instar larva; L2: Second instar larva; L3: Third instar larva; L4: Forth instar larva; L5: Fifth instar larva; L6: Six instar larva; FP: Female pupa; MP: Male pupa; FA: Female adult; and MA: Male adult. Each treatment includes three replicates. The data are denoted as mean ± SE. Different letters above bars represent significant differences in the relative expression levels at the 0.05 level (Tukey’s multiple range test).
2.1.6. Expression of Genes Encoding sHSPs in Different Developmental Stages
The three S. inferens sHSP genes were expressed in all stages of S. inferens development, although the expression levels varied widely. Expression of Sihsp21.4 was highest in female adults (Figure 5B2) and lowest in third instar larvae (F10,22 = 3.027, p = 0.015). Expression of Sihsp20.6 and Sihsp19.6 was highest in eggs, and this difference was significant when compared to expression in other developmental stages (Sihsp20.6, F10,22 = 17.301, p < 0.001; Sihsp19.6, F10,22 = 16.518, p < 0.001) (Figure 5B2,C2).
2.1.7. Expression of Genes Encoding sHSPs in Response to Cold Temperatures
The three S. inferens sHSP genes showed similar expression patterns in response to low temperatures. Sihsp21.4, Sihsp20.6 and Sihsp19.6 were induced in response to cold temperatures. For example, expression levels were significantly higher (6.42–9.56-fold higher) in S. inferens exposed to −6 °C for 2 h as compared to insects maintained at 27 °C (Sihsp21.4, F5,11 = 14.706, p < 0.001; Sihsp20.6, F5,11 = 7.024, p = 0.004; Sihsp19.6, F5,11 = 5.035, p = 0.012) (Figure 5A3,B3,C3).
2.2. Discussion
Overwintering insects are exposed to temperature stress in nature and must adopt specialized adaptive mechanisms to survive low temperatures. The induction of small heat shock proteins is a potential survival mechanism during temperature stress [19]; However, to our knowledge, the study of sHSPs in S. inferens has not been previously undertaken. In the present study, we cloned three members of the sHSP family from S. inferens. Analysis of the cDNA sequences and deduced ORFs indicated that Sihsp21.4, Sihsp20.6, and Sihsp19.6 encoded proteins containing 187, 183, and 174 amino acids, respectively. The predicted amino acid sequences shared considerable sequence similarity with sHSP from other insects and α-crystallin proteins from vertebrate eye lenses. Phylogenetic analysis indicated that S. inferens HSP19.6 and S. nonagrioides HSP19.5 clustered with the same group; Whereas S. inferens HSP20.6 and S. nonagrioides HSP20.8 clustered together in another group (Figure 3). S. inferens HSP21.4 and five other lepidopteran HSP21.4 orthologs grouped together in a well-supported cluster, which supports the accuracy of the sequence analysis conducted in the present study. S. inferens HSP19.6 and HSP20.6, which show high sequence similarity, grouped together in a well-supported cluster, whereas S. inferens HSP21.4 sorted to a different cluster. Thus, Sihsp21.4 may have evolved differently from Sihsp20.6 and Sihsp19.6. It is interesting to note that sHSP orthologs from S. litura and C. suppressalis show phylogenetic similarities to the S. inferens proteins identified in the present study [19,33].
A negative correlation between intron size and the level of gene expression has been suggested previously; In other words, genes containing smaller introns or lacking introns were more highly expressed than genes containing large or multiple introns [34]. It is also possible that genes either lacking or containing shorter introns may be more sensitive to environmental stresses. Based on chromosomal location and intron number, sHSP genes could be subdivided into two types: Orthologous or species-specific [35]. Thus, the three shsps from S. inferens could be classified into two groups: Orthologous that contained introns (Sihsp21.4) and species-specific forms lacking introns (Sihsp20.6 and Sihsp19.6).
The various functions of small heat shock proteins in insect tissues are not well-understood. One possibility is that sHSPs play important, specialized roles in maintaining normal functioning in different tissues [15]. In this study, Sihsp21.4 was highly expressed in S. inferens heads, which is similar to the high expression of hsp19.1 and hsp22.6 reported in B. mori heads [35]. The primary nerve center in insects, e.g., the supraoesophageal ganglion and suboesophageal ganglion, is located within the head; Thus, it is possible that sHSPs could protect the nerve centers from external injury. However, shsps of S. litura, Apis cerana cerana, and C. suppressalis were expressed at very high levels in malpighian tubules and hindgut tissues [19,33,36]. Malpighian tubules and hindguts function by reabsorbing water, salts, and other substances prior to excretion by the insect; Thus, it remains unclear why shsps were highly expressed in these tissues. One hypothesis is that sHSPs protect these tissues from potentially toxic substances [19]. Taken together, our data suggest that different shsps play distinct roles in the physiology of S. inferens.
sHSPs play important roles in development, including the regulation of insect development [33,37,38]. For instance, l2efl, a type of shsp, reached a maximum level of expression in the third instar larvae of D. melanogaster [39]. However, in Lucilia cuprina, expression of hsp24 was lowest in third instar larvae [37]. Lu et al. reported a high level of Cshsp21.7a expression in first instar larvae of C. suppressalis, whereas the highest expression of Cshsp19.8, Cshsp21.4, Cshsp21.5, and Cshsp21.7b was observed in C. suppressalis adults; In this study, expression of Sihsp20.6 and Sihsp19.6 was highest in insect eggs, whereas Sihsp21.4 expression was highest in female adults [19]. Thus our data support the hypothesis that sHSPs have evolved specific roles in different stages of insect development.
Sihsp21.4, Sihsp20.6 and Sihsp19.6 were dramatically up-regulated during low temperature stress, which is similar to hsp20.4 and hsp20.8 in S. litura and hsp19.8, hsp21.5, hsp21.7b in C. suppressalis [19,33]. Previous studies have emphasized the role of sHSPs in modulating thermo-tolerance of insects. However, not all shsps are up-regulated during cold stress. For example, the expression of hsp21.4 and hsp21.7b (C. suppressalis), hsp20 and hsp21.4 (S. litura), and hsp21.4 (B. mori) was insensitive to cold stress [19,33,35]. Because the overproduction of HSPs may cause deleterious effects, regulatory controls are critical with respect to maintaining the cost/benefit ratio in organisms expressing these genes [40,41].
More in-depth studies are needed to clarify the role of sHSPs in insect behavior and development. Future investigations will help reveal the underlying physiological mechanisms of shsps in S. inferens, thus enhancing our ability to implement more effective control measures for this significant pest.
3. Experimental Section
3.1. Insects
Populations of S. inferens were collected from a suburb of Yangzhou (32°39'N, 119°42'E), located in the Jiangsu province. The pink stem borers were reared for more than 3 generations in environmental chambers maintained at 27 ± 1 °C with a 16:8 (light/dark) photoperiod and 60%–70% relative humidity as described previously [42].
3.2. Reverse Transcription Polymerase Chain Reaction (PCR) and Rapid-Amplification of cDNA Ends (RACE)
Total RNA was extracted from S. inferens using the SV Total RNA isolation system (Promega, Madison, WI, USA) and then treated with DNase I. The integrity of RNA was verified by comparing RNA bands in gels stained with ethidium bromide. RNA purity was analyzed at 260 and 280 nm using a spectrophotometer (Eppendorf BioPhotometer plus, Eppendorf, Germany). cDNA copies of genes encoding sHSPs were synthesized using oligo(dT)18 primers (Fermentas, Helsingborg, Sweden). Degenerate primers for PCR were designed using consensus sequences of shsps obtained from several lepidopteran insects; These sequences were previously deposited in GenBank (Table 1). Degenerate primers of Sihsp21.4 were designed using consensus sequences of H. armigera hsp21.4, Spodoptera litura hsp21.4 and C. suppressalis hsp21.4; Degenerate primers of Sihsp20.6 were designed using consensus sequences of S. nonagrioides hsp20.8, S. litura hsp20.4 and B. mori hsp20.4; Degenerate primers of Sihsp19.6 were designed using consensus sequences of S. nonagrioides hsp19.5, B. mori hsp19.5 and Plutella xylostella hsp19.5. And the amino acid regions used to design degenerate primers are conserved regions of each gene. Full-length cDNAs were obtained using 5' and 3' RACE (SMARTer™ RACE, Clontech, Palo Alto, CA, USA). Primers for RACE were designed based on partial sequence information derived from shsps cDNA fragments (Table 1). Complete sequences of intact ORFs were confirmed by 5' RACE cDNA. Products were purified using the AxyPrep™ DNA Gel Extraction Kit (Axygen, Union City, CA, USA), cloned into pGEM-T Easy Vector (Promega), and then transformed into Escherichia coli DH5α cells for subsequent sequence analysis.
Table 1.
Primers used for gene clone and verification.
| Primer Name | Primer Sequences (5'–3') | Amplicon Size (bp) | Purpose |
|---|---|---|---|
| hsp21.4DP-F | ATGGARGAAGAAATGASAARTT | 241 | Intermediate fragment amplification |
| hsp21.4DP-R | TCGACHGTCTTVACRACGAT | ||
| hsp20.6DP-F | CCTMGCCGYCTGDTGGAYCARC | 467 | |
| hsp20.6DP-R | TCCTTGATCTCCTTGCGVACGG | ||
| hsp19.6DP-F | AAGTBAACCTDGACGTGCAGCATT | 296 | |
| hsp19.6DP-R | TTTCACCTCCTTGCGCACTGGT | ||
| hsp21.4RACE-5' | CTTCAATGACTTGCCGTCGCCCTC | 418 | Rapid-amplification of cDNA ends (RACE) |
| hsp21.4RACE-3' | AGCACAGTGACAGCAGACAGTTGGC | 1062 | |
| hsp20.6RACE-5' | ATTCCACGGACTCGGGTTCCACGC | 510 | |
| hsp20.6RACE-3' | TTGCTGCTGGACCTTTGCTGACGA | 610 | |
| hsp19.6RACE-5' | TCTGGTAAAGCGTAGCGGCGGGT | 502 | |
| hsp19.6RACE-3' | GGCAGTTTACCCGCCGCTACGCT | 327 | |
| hsp21.4cDNA-F | ATGGGGAGTACTGCCTTG | 963 | Verification of open reading frame (ORF) |
| hsp21.4cDNA-R | TGCCTGAATACATCCCTTA | ||
| hsp20.6cDNA-F | AAGATGTCTCTGTTGCCA | 559 | |
| hsp20.6cDNA-R | CACTTTACTTCTTTTCCTTT | ||
| hsp19.6cDNA-F | GTGCGAAACAAGTACAAAGC | 692 | |
| hsp19.6cDNA-R | CAAAGAGAACACTGAAAGGAAT | ||
| hsp21.4DNA1-F | TGTCTGCTGTAGAGTGCGTAG | 372 | Verification of genome |
| hsp21.4DNA1-R | TTGAACTCGCCCCTGATC | ||
| hsp21.4DNA2-F | TGGGACAGCTTGAACTCG | 1529 | |
| hsp21.4DNA2-R | AGTGCTTCTGGATGGGGA | ||
| hsp21.4DNA3-F | CGACAGGAACATCCCCATCCAGAA | 697 | |
| hsp21.4DNA3-R | TATTGACATCTAAAACATTCGTAAC | ||
| hsp20.6DNA-F | AAGATGTCTCTGTTGCCA | 559 | |
| hsp20.6DNA-R | CACTTTACTTCTTTTCCTTT | ||
| hsp19.6DNA-F | GTGCGAAACAAGTACAAAGC | 692 | |
| hsp19.6DNA-R | CAAAGAGAACACTGAAAGGAAT |
3.3. Characterization of Genomic DNA
The genomic DNA of S. inferens was extracted using the AxyPrep™ Multisource Genomic DNA Kit (Axygen Biosciences, Union City, CA, USA). Specific primer pairs (Table 1) were designed to amplify genomic fragments based on analysis of full-length cDNAs. The products were purified using the AxyPrep™ DNA Gel Extraction Kit (Axygen), cloned into pGEM-T EasyVector (Promega), and transformed into E. coli DH5α for sequence analysis.
3.4. Tissues Samples
The larvae selected for analysis were similar in size and randomly assigned to experimental groups. Each group contained ten larvae, and each experiment was repeated three times. Larvae were anesthetized on ice prior to dissection. The head, epidermis, fat body, foregut, midgut, hindgut, malpighian tubules, haemocytes, and salivary glands were collected from larvae and rinsed with a 0.9% sodium chloride solution. The samples were frozen immediately in liquid nitrogen and stored at −70 °C prior to real-time PCR analyses.
3.5. Samples Representing Developmental Stages and Sex
Samples included egg masses, the first, second, third, fourth, fifth and sixth instar larvae, male and female pupae, and one-day-old male and female adults; Samples were randomly selected for the experiment. The samples were frozen immediately in liquid nitrogen and stored at −70 °C until needed for analyses.
3.6. Cold Tolerance Samples
In this experiment, larvae representing the fifth instar were placed individually in glass tubes, and groups of ten were then exposed to various temperatures (−8, −6, −4, −2 and 0 °C) for 2 h in a constant-temperature incubator (DC-3010, Jiangnan Equipment, Changzhou, China). The larvae were recovered at 27 ± 1 °C for 2 h, after which surviving larvae were frozen in liquid nitrogen and stored at −70 °C. A set of larvae maintained at 27 ± 1 °C was regarded as a control group. Each treatment included at least three surviving larvae.
3.7. Quantitative Real-Time PCR
Total RNA was extracted using the methods described above for reverse transcription PCR and RACE. RNA (0.5 μg) was reverse-transcribed into first-strand cDNA using the Bio-Rad iScript™ cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). Real-time PCR reactions were performed in a 20 μL reaction volume comprised of 10 μL Bio-Rad iTaq™ Universal SYBR® Green supermix (Bio-Rad, 2×), 1 μL of each gene-specific primer (10 μM) (Table 2), 2 μL of cDNA template, and 6 μL of ddH2O. Reactions were carried out using a CFX-96 real-time PCR system (Bio-Rad) under the following conditions: 3 min at 95 °C, 40 cycles of denaturation at 95 °C for 30 s, and annealing at the Tm for each gene (30 s; Table 2). Each treatment included three replicates, and each reaction was run in triplicate.
3.8. Data Analysis
ORFs were identified using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The deduced amino acid sequences were aligned using CLUSTAL X1.83 [43]. Sequence analysis tools of the ExPASy Molecular Biology Server (Swiss Institute of Bioinformatics, Basel, Switzerland) were used to analyze the deduced sHSP sequences, including Translate, Compute pI/MW, and Blast. Amino acid sequences were used to estimate phylogeny using neighbor-joining, minimum evolution, maximum likelihood, and maximum parsimony methods. Phylogenetic trees were constructed with 1000 bootstrap replicates using MEGA version 6.06 (Tempe, AZ, USA) [44].
Homology models were generated using Protein Homology/analogy Recognition Engine software version 2.0 (http://www.sbg.bio.ic.ac.uk/~phyre2/html) [45]. The Chimera tool was used to visualize the 3D coordinates for the atoms in the predicted protein models [46].
qRT-PCR data were analyzed using the Bio-Rad CFX Manager™ 3.1 software (Bio-Rad). The threshold cycle (Ct value) denotes the cycle at which the fluorescent signal first shows significant difference with respect to the background. All biological replicates were used to calculate the average Ct values. Relative expressions were calculated using the 2−ΔΔCt method [47]. Three genes (RPS13, RPS20 and EF1) were used for normalizing gene expression in different tissues. 18S rRNA, EF1 and GAPDH were used as reference genes in different developmental stages and sexes. 18S rRNA, RPS20 and TUB were used for normalizing gene expression at different temperatures. The means of the reference genes were used as normalization under different experimental conditions [48]. The above-mentioned reference genes were previously validated in a research study that has been submitted elsewhere. Tukey’s test was conducted for statistical analysis using PASW Statistics 18.0 (SPSS Inc., Chicago, IL, USA).
4. Conclusions
In conclusion, we cloned three genes encoding sHSPs from S. inferens. The structure of these genes was examined, and we analyzed their expression in different tissues and stages of insect development. Our results also indicate that expression of the three shsps is modulated in response to cold stress.
Acknowledgments
This research was funded by the National Basic Research Program of China (973 Program) (2012CB114100), the National Natural Science Foundation of China (31371937) and Youth Science Foundation of Yangzhou. The authors also appreciate the support and help of the Testing Center of Yangzhou University on experimental apparatus.
Author Contributions
Ming-Xing Lu and Yu-Zhou Du conceived and designed the experiments; Meng Sun and Xiao-Tian Tang carried out the experiments and analysed the data; Meng Sun wrote the main manuscript text. All authors reviewed the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tissières A., Mitchell H.K., Tracy U.M. Protein synthesis in salivary glands of Drosophila melanogaster: Relation to chromosome puffs. J. Mol. Biol. 1974;84:389–398. doi: 10.1016/0022-2836(74)90447-1. [DOI] [PubMed] [Google Scholar]
- 2.De Jong W.W., Caspers G.J., Leunissen J.A. Genealogy of the α-crystallin-small heat-shock protein superfamily. Int. J. Biol. Macromol. 1998;22:151–162. doi: 10.1016/s0141-8130(98)00013-0. [DOI] [PubMed] [Google Scholar]
- 3.Franck E., Madsen O., van Rheede T., Ricard G., Huynen M.A., de Jong W.W. Evolutionary diversity of vertebrate small heat shock proteins. J. Mol. Evol. 2004;59:792–805. doi: 10.1007/s00239-004-0013-z. [DOI] [PubMed] [Google Scholar]
- 4.Stromer T., Fischer E., Richter K., Haslbeck M., Buchner J. Analysis of the regulation of the molecular chaperone Hsp26 by temperature-induced dissociation: The N-terminal domail is important for oligomer assembly and the binding of unfolding proteins. J. Biol. Chem. 2004;279:11222–11228. doi: 10.1074/jbc.M310149200. [DOI] [PubMed] [Google Scholar]
- 5.Gusev N.B., Bogatcheva N.V., Marston S.B. Structure and properties of the small heat shock proteins (sHsp) and their interaction with cytoskeleton proteins. Biochemistry. 2002;67:511–519. doi: 10.1023/a:1015549725819. [DOI] [PubMed] [Google Scholar]
- 6.Horwitz J. α-Crystallin can function as a molecular chaperone. Proc. Natl. Acad. Sci. USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haslbeck M., Franzmann T., Weinfurtner D., Buchner J. Some like it hot: The structure and function of small heat-shock proteins. Nat. Struct. Mol. Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 8.Horwitz J. α-Crystallin. Exp. Eye Res. 2003;76:145–153. doi: 10.1016/S0014-4835(02)00278-6. [DOI] [PubMed] [Google Scholar]
- 9.Garrido C., Paul C., Seigneuric R., Kampinga H.H. The small heat shock proteins family: The long forgotten chaperones. Int. J. Biochem. Cell B. 2012;44:1588–1592. doi: 10.1016/j.biocel.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Haslbeck M. sHsps and their role in the chaperone network. Cell Mol. Life Sci. 2002;59:51–60. doi: 10.1007/PL00012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinlan R. Cytoskeletal competence requires protein chaperones. Prog. Mol. Subcell. Biol. 2002;28:219–234. doi: 10.1007/978-3-642-56348-5_12. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y., MacRae T.H. Small heat shock proteins: Molecular structure and chaperone function. Cell Mol. Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsvetkova N.M., Horvath I., Torok Z., Wolkers W.F., Balogi Z., Shigapova N., Crowe L.M., Tablin F., Vierling E., Crowe J.H., et al. Small heat shock proteins regulate membrane lipid polymorphism. Proc. Natl. Acad. Sci. USA. 2002;99:13504–13509. doi: 10.1073/pnas.192468399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arrigo A.P., Simon S., Gibert B., Kretz-Remy C., Nivon M., Czekalla A., Guillet D., Moulin M., Diaz-Latoud C., Vicart P. Hsp27 (HspB1) and α B-crystallin (HspB5) as therapeutic targets. FEBS Lett. 2007;581:3665–3674. doi: 10.1016/j.febslet.2007.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Gu J., Huang L.X., Shen Y., Huang L.H., Feng Q.L. Hsp70 and small Hsps are the major heat shock protein members involved in midgut metamorphosis in the common cutworm, Spodoptera litura. Insect Mol. Biol. 2012;5:535–543. doi: 10.1111/j.1365-2583.2012.01158.x. [DOI] [PubMed] [Google Scholar]
- 16.Hayward S.A.L., Pavlidesb S.C., Tammariellob S.P., Rineharta J.P., Denlinger D.L. Temporal expression patterns of diapause associated genes in flesh fly pupae from the onset of diapause through post-diapause quiescence. J. Insect Physiol. 2005;51:631–640. doi: 10.1016/j.jinsphys.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Huang L.H., Kang L. Cloning and inter-specific altered expression of heat shock protein genes in two leaf miner species in response to thermal stress. Insect Mol. Biol. 2007;16:491–500. doi: 10.1111/j.1365-2583.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- 18.Jakob U., Buchner J. Assisting spontaneity: The role of Hsp90 and small Hsps as molecular chaperones. Trends Biochem. Sci. 1994;19:205–211. doi: 10.1016/0968-0004(94)90023-X. [DOI] [PubMed] [Google Scholar]
- 19.Lu M.X., Hua J., Cui Y.D., Du Y.Z. Five small heat shock protein genes from Chilo suppressalis: Characteristics of gene, genomic organization, structural analysis, and transcription profiles. Cell Stress Chaperones. 2014;19:91–104. doi: 10.1007/s12192-013-0437-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinehart J.P., Li A.Q., Yocum G.D., Robich R.M., Hayward S.A.L., Denlinger D.L. Up-regulation of heat shock proteins is essential for cold survival during insect diapause. Proc. Natl. Acad. Sci. USA. 2007;104:11130–11137. doi: 10.1073/pnas.0703538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song K.H., Jung S.J., Seo Y.R., Kang S.W., Han S.S. Identification of up-regulated proteins in the hemolymph of immunized Bombyx mori larvae. Comp. Biochem. Phys. D. 2006;1:260–266. doi: 10.1016/j.cbd.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Xu L.N., Li C.C., Hu B.J., Zhou Z.Y., Li X.X. Review of history, present situation and prospect of pink stem borer in China. Chin. Agric. Sci. Bull. 2011;27:244–248. [Google Scholar]
- 23.Areekul S., Chamchanya T. Effects of humidity, temperature, and light on the growth and development of Sesamia inferens (Walker) Kasetsart J. 2004;7:65–75. [Google Scholar]
- 24.Chouraddi M., Mallapur C.P., Goud K.B., Patil R.H. Status of stem borers in major maize growing areas of Karnataka, India. J. Exp. Zool. 2012;15:505–511. [Google Scholar]
- 25.Joshi G., Ram L., Singh R. Biology of pink borer, Sesamia inferens (Walker) on Taraori Basmati rice. Ann. Biol. 2009;25:41–45. [Google Scholar]
- 26.Mahesh P., Chandran K., Srikanth J., Nisha M., Manjunatha T. Natural incidence of Sesamia inferens Walker, in sugarcane germplasm. Sugar Technol. 2013;15:384–389. doi: 10.1007/s12355-013-0212-2. [DOI] [Google Scholar]
- 27.Rahman M., Khalequzzaman M. Temperature requirements for the development and survival of rice stemborers in laboratory conditions. Insect Sci. 2004;11:47–60. doi: 10.1111/j.1744-7917.2004.tb00179.x. [DOI] [Google Scholar]
- 28.Singh B. Incidence of the pink noctuid stem borer, Sesamia inferens (Walker), on wheat under two tillage conditions and three sowing dates in north-western plains of India. J. Entomol. 2012;9:368–374. doi: 10.3923/je.2012.389.395. [DOI] [Google Scholar]
- 29.Sun M., Tang X.T., Lu M.X., Yan W.F., Du Y.Z. Cold tolerance characteristics and overwintering strategy of Sesamia inferens (Lepidoptera: Noctuidae) Fla. Entomol. 2014;97:1544–1553. [Google Scholar]
- 30.Stamler R., Kappé G., Boelens W., Slingsby C. Wrapping the α-crystallin domain fold in a chaperone assembly. J. Mol. Biol. 2005;353:68–79. doi: 10.1016/j.jmb.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 31.Braun N., Zacharias M., Peschek J., Kastenmüller A., Zou J., Hanzlik M., Haslbeck M., Rappsilber J., Buchner J., Weinkauf S. Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proc. Natl. Acad. Sci. USA. 2011;108:20491–20496. doi: 10.1073/pnas.1111014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huggett J., Dheda K., Bustin S., Zumla A. Real-time RT-PCR normalisation; Strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y., Gu J., Huang L.H., Zheng S.C., Liu L., Xu W.H., Feng Q.L., Kang L. Cloning and expression analysis of six small heat shock protein genes in the common cutworm, Spodoptera litura. J. Insect Physiol. 2011;57:908–914. doi: 10.1016/j.jinsphys.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 34.Comeron J.M. Selective and mutational patterns associated with gene expression in humans: Influences on synonymous composition and intron presence. Genetics. 2004;167:1293–1304. doi: 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z.W., Li X., Yu Q.Y., Xiang Z.H., Kishino H., Zhang Z. The small heat shock protein (sHSP) genes in the silk worm, Bombyx mori, and comparative analysis with other insect sHSP genes. BMC Evol. Biol. 2009;9:215. doi: 10.1186/1471-2148-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Z.H., Xi D.M., Kang M.J., Guo X.Q., Xu B.H. Molecular cloning and characterization of Hsp27.6: The first reported small heat shock protein from Apis cerana cerana. Cell Stress Chaperones. 2012;17:539–551. doi: 10.1007/s12192-012-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Concha C., Edman R.M., Belikoff E.J., Schiemann A.H., Carey B., Scott M.J. Organization and expression of the Australian sheep blowfly (Lucilia cuprina) hsp23, hsp24, hsp70 and hsp83 genes. Insect Mol. Biol. 2012;21:169–180. doi: 10.1111/j.1365-2583.2011.01123.x. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K.H., Rako L., Takano-Shimizu T., Hoffmann A.A., Lee S.F. Effects of small Hsp genes on developmental stability and micro-environmental canalization. BMC Evol. Biol. 2010;10:284. doi: 10.1186/1471-2148-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurzik-Dumke U., Lohmann E. Sequence of the new Drosophila melanogaster small heat-shock-related gene, lethal(2) essential for life [l(2)efl], at locus 59F4, 5. Gene. 1995;154:171–175. doi: 10.1016/0378-1119(94)00827-F. [DOI] [PubMed] [Google Scholar]
- 40.Krebs R.A., Feder M.E. Hsp70 and larval thermotolerance in Drosophila melanogaster: How much is enough and when is more too much? J. Insect Physiol. 1998;44:1091–1101. doi: 10.1016/S0022-1910(98)00059-6. [DOI] [PubMed] [Google Scholar]
- 41.Sørensen J.G., Kristensen T.N., Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003;6:1025–1037. doi: 10.1046/j.1461-0248.2003.00528.x. [DOI] [Google Scholar]
- 42.Han C., Peng Y.F., Hou M.L., Chen F.J., Zhai B.P., Han L.Z. A preliminary study on artificial rearing of the pink stem borer, Sesamia inferen. Chin. J. Appl. Entomol. 2012;49:281–285. [Google Scholar]
- 43.Chenna R., Sugawara H., Koike T., Lopez R., Gibson T.J., Higgins D.G., Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley L.A., Sternberg M.J.E. Protein structure prediction on the Web: A case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 46.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Pfaffl M. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandesompele J., de Preter K., Pattyn F., Poppe B., van Roy N., de Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]