Abstract
Loss-of-function DJ-1 mutations can cause early-onset Parkinson's disease. The function of DJ-1 is unknown, but an acidic isoform accumulates after oxidative stress, leading to the suggestion that DJ-1 is protective under these conditions. We addressed whether this represents a posttranslational modification at cysteine residues by systematically mutating cysteine residues in human DJ-1. WT or C53A DJ-1 was readily oxidized in cultured cells, generating a pI 5.8 isoform, but an artificial C106A mutant was not. We observed a cysteine-sulfinic acid at C106 in crystalline DJ-1 but no modification of C53 or C46. Oxidation of DJ-1 was promoted by the crystallization procedure. In addition, oxidation-induced mitochondrial relocalization of DJ-1 and protection against cell death were abrogated in C106A but not C53A or C46A. We suggest that DJ-1 protects against neuronal death, and that this is signaled by acidification of the key cysteine residue, C106.
Loss of vulnerable neuronal groups is a consistent feature of several familial forms of Parkinson's disease (PD). Mitochondrial dysfunction, oxidative stress, and proteasome failure are among the several hypotheses suggested to explain the molecular basis of neuronal damage (1). The discovery of different recessive genes for parkinsonism, where loss of protein function is associated with neuronal damage, may increase our understanding of the process, or processes, involved. For example, the recessive gene parkin, an E3 ubiquitin-protein ligase, protects neurons against a number of damaging insults, including α-synuclein mutations that also cause PD (2) and mitochondrial damage (3).
The finding of mutations in a second recessive gene associated with recessive PD, DJ-1 (4), provides an opportunity to understand cellular pathways that protect against neuronal damage. In support of the idea that recessive genes normally protect neurons, two recent reports have shown that DJ-1 limits cellular toxicity (5, 6). DJ-1 is part of a large superfamily, including diverse bacterial enzymes of unclear relevance to human metabolism (7). Therefore, the function(s) of DJ-1 that are relevant to protecting neurons have not been elucidated. Although initial reports suggested a preferential localization of mutant DJ-1 to mitochondria (4), we have found that WT or mutant DJ-1 localized to mitochondria in a proportion of transfected cells (8). These observations suggest that DJ-1 can be targeted to mitochondria in some circumstances.
DJ-1 is also known to be responsive to oxidative stress, with a shift of pI from 6.2 to 5.8 (9, 10). In the PD brain, there is an accumulation of acidic forms (11), but the nature of these modifications is unclear. Mitsumoto et al. (10) have suggested that DJ-1 may be directly oxidized by free radicals, because the pI shift after oxidation is consistent with formation of cysteine-sulfinic acid (10), an idea recently tested using mass spectrometry where cysteine-sulfonic acids were identified in oxidized DJ-1 (12). Formation of cysteine-sulfinic acid has been recognized as an important reversible posttranslational modification of proteins, such as the peroxiredoxins (13-15). Furthermore, DJ-1 protects against oxidative stress in vitro (5, 6).
If the pI shift of DJ-1 represents a formation of cysteine-sulfinic acid, then mutating these cysteine residues will block oxidation. C106 is a prime candidate for modification, because it is highly conserved and may be part of an active site in a deep crevice on the DJ-1 dimer (16-20). Although others have modified C106 (21, 22), no systematic analyses have been performed, and data on which mutations block oxidation are unclear (5). We show here that C106 is oxidized to a cysteine-sulfinic acid, consistent with other recent analyses (12). A C106A substitution, but not C46A or C53A, prevents formation of an oxidized isoform in intact cells. Furthermore, the C106A mutation also blocks oxidation-induced mitochondrial localization and protection against 1-methyl-4-phenylpyridinium (MPP+) toxicity in neuronal cells, suggesting that C106 also controls the previously reported (5, 6) neuroprotective function of DJ-1.
Materials and Methods
Cloning and Transfections. Full-length cDNA vectors for WT and L166P mutant human DJ-1 have been described previously (8). C46A, C53A, C106A, and C106D mutants were generated by using the QuikChange mutagenesis kit (Stratagene; primer sequences are available on request), and the inserts of all constructs were fully sequenced by using the BigDye terminator kit on an ABI 3100 sequencer (Applied Biosystems). Transient transfections of M17 human neuroblastoma cell lines were performed as described (8). Stable cell lines were established by selecting with 50 μg/ml zeocin.
Western Blotting, Crosslinking, and Immunoprecipitations. Western blotting from whole-cell lysates was performed as described (8). Limited trypsinolysis of isolated mitochondrial fractions was performed as described by Darios et al. (3). Primary antibodies used were monoclonal anti-DJ1 (clone 3E8; 1:2,000; Stressgen Biotechnologies, San Diego), monoclonal anti-V5 (1:5,000, Invitrogen), monoclonal anti-Tim23 (clone 32; 1:2,500; BD Biosciences, San Jose, CA), monoclonal anti-β-actin (clone AC-15; 1:5,000; Sigma), and polyclonal anti-VDAC (1:2,500; Abcam, Cambridge, MA). Blots were developed with peroxidase-labeled secondary antibodies (1:5,000; Jackson Immunochemicals, West Grove, PA) and captured by using ECL-plus (Amersham Pharmacia Biosciences) with a Storm phosphorimager. Quantification was performed by using imagequant software (Amersham Pharmacia).
For crosslinking experiments, cells were rinsed with PBS and exposed to 100 μM disuccinimidyl suberate (DSS) in PBS for 30 min at room temperature. Excess DSS was quenched by rinsing twice in PBS containing 50 mM Tris·HCl (pH 7.5) before extraction and Western blotting as above. Immunoprecipitation experiments were performed as described (8). Immunoprecipitated complexes were analyzed by Western blotting with monoclonal anti-DJ-1.
2D Gel Electrophoresis. Soluble cytosolic fractions were prepared as in previously reported methods (23). Protein samples (20 μg) were loaded onto 4-7 linear pH gradient Immobiline DryStrip (Amersham Pharmacia Biosciences) and separated by isoelectric focusing in an Ettan IPGphor system (Amersham Pharmacia Biosciences) for 16,000 V·h. These first-dimension gels were subsequently separated on 12% Tris·HCl SDS/PAGE gels (Bio-Rad) and proteins detected by Western blotting (below). We used 2D SDS/PAGE standards (Bio-Rad) to calibrate pI and molecular weights. For experiments using recombinant DJ-1, we separated 1 μg of protein on 2D gels and stained with Coomassie brilliant blue (Pierce).
Crystal Growth, Data Collection, and Model Refinement. Recombinant human DJ-1 was expressed and purified as described (20). Crystals were grown by the hanging drop vapor diffusion method by mixing DJ-1 at 23 mg/ml with a reservoir solution consisting of 30% polyethylene glycol 400, 100 mM Tris·HCl (pH 7.5), and 200 mM sodium citrate. The crystals were cryo-cooled by direct immersion into liquid nitrogen.
A 1.2-Å resolution data set was collected from a single crystal maintained at 110 K at beamline 8.2.1 of the Advanced Light Source (Lawrence Berkeley National Laboratory, Berkeley, CA). Radiation damage, which was a problem in a previously reported 1.1-Å structure (20), was minimized by a combination of short incident wavelength (0.83 Å), diminished flux, and short exposure time (5 sec/1° oscillation). The data were indexed and scaled by using denzo and scalepack (24), respectively. See Table 1, which is published as supporting information on the PNAS web site, for data statistics.
Rigid body refinement of the starting model (PDB ID code 1P5F) (20) was followed by iterative cycles of restrained least squares refinement of both coordinates and anisotropic displacement parameters against all of the measured intensity data (excluding the reflections sequestered for the calculation of Rfree) in shelx-97 (25). Manual adjustments to the model were performed in O (26). See Table 1 for final model statistics.
Confocal Microscopy. Staining for V5-tagged DJ-1 and mitotracker was performed as described (8). Slides were examined by using a Zeiss LSM510 confocal microscope with independent excitation for both channels. Because the signal strength for the C46A variant was lower than that for WT, we increased the photomultiplier tube voltage on this channel. In this fashion, we maintained separate signals for both channels with minimal bleed through, assessed using controls as described.
Toxicity Assays. Cells were exposed to MPP+ (10-500 μM) for 48 h and viability assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay as described (2). For each experiment, eight wells were used per concentration, and each experiment was repeated three times with similar results. Statistical differences between cell lines were assessed by using two-way ANOVA.
Results
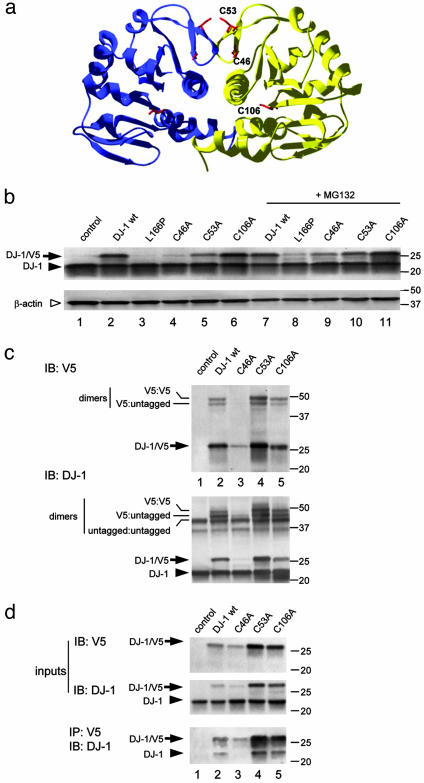
Oxidation of DJ-1 Cysteine Mutants. To establish whether cysteine oxidation can occur in DJ-1, we systematically mutated all cysteine residues. In the 1.1-Å crystal structure, C46 and C53 are on two consecutive β strands that form part of the dimer interface (Fig. 1a), with symmetry-related C53 within 4 Å of each other at the dimer interface. C106 is at the bottom of a narrow cleft but still solvent accessible. We made C46A, C53A, and C106A constructs with a C-terminal V5 tag to discriminate transfected DJ-1 from endogenous protein. Given that C46 and C53 are near the dimer interface, we considered that either might affect protein stability and/or dimer formation. C46A consistently produced lower steady-state protein levels than WT protein when transfected into cells, although the effect was less dramatic than L166P (Fig. 1b). This appeared to be due to increased protein turnover, because mRNA levels were equivalent (data not shown) and the proteasome inhibitor MG132 partially rescued the steady-state protein levels (Fig. 1b). C53A and C106A were as stable as the WT protein. These stable variants retained their capacity to form dimers, because we were able to both crosslink endogenous DJ-1 with V5-tagged WT, C53A, and C106A variants (Fig. 1c) and coimmunoprecipitate endogenous (WT) DJ-1 with the same V5-tagged variants (Fig. 1d).
Fig. 1.
Generation of stable dimeric cysteine mutants of human DJ-1. (a) The crystal structure of human DJ-1 with cysteine residues highlighted. Two cysteine residues, C46 and C53, whose sidechains are shown in red, are present in a β-turn close to the dimer interface. A third cysteine, C106, is within the center of the dimer. (b) Transient transfections of V5-tagged cysteine mutants into M17 cells. Because some variants, notably C46A, were unstable, samples in lanes 7-11 were treated with 5 μM MG132 for 24 h. Western blots were probed with anti-DJ-1 (Upper) to visualize both transfected DJ-1 (arrows) and endogenous DJ-1 (arrowhead). The blot was reprobed with β-actin as a loading control (Lower, open arrowhead). Markers on the right of the blot are in kilodaltons. (c) To assess whether these variants form dimers, transfected cells were treated with the crosslinker disuccinimidyl suberate before extraction and Western blotting for V5 (Upper) or DJ-1 (Lower). Monomeric V5-tagged protein is indicated with an arrow, and endogenous DJ-1 in the cells indicated by an arrowhead. The V5-tagged proteins formed dimers with either endogenous or transfected DJ-1, indicated by lines on the left of the blot. Blots are representative of at least duplicate experiments. To confirm these results, we coimmunoprecipitated V5-tagged DJ-1 with endogenous protein (d). (Upper) Lysates of transfected cells probed with V5 or DJ-1 to demonstrate input levels of proteins. (Lower) Lysates immunoprecipitated withV5 antibody and blotted with DJ-1 antibody to show interaction of endogenous DJ-1 with the tagged proteins.
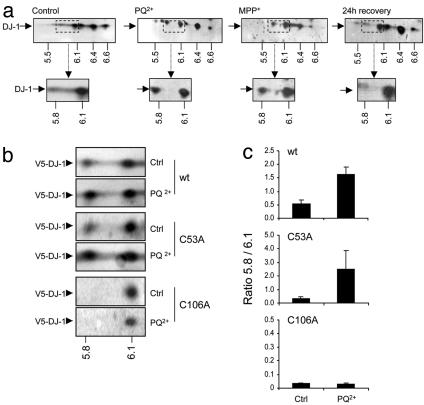
We next examined how endogenous and transfected DJ-1 mutants responded to oxidative stress. Previous reports (9, 10) have described two main isoforms at pI = 5.8 and 6.2. However, endogenous DJ-1 in M17 neuroblastoma cells is present as several isoforms with isoelectric points between 6 and 7 (Fig. 2a), in agreement with data from human brain samples (11). Subsequently, cells were exposed to paraquat (PQ2+) or the parkinsonian toxin, MPP+, which induces free radicals as a consequence of its inhibition of mitochondrial complex I activity. The most consistent change in DJ-1 was the appearance of a pI 5.8 species, similar to that reported in other studies as being oxidation responsive (9, 10). A low level of this isoform is present without exposure to oxidative stress. Oxidation is reversible, because the pattern of isoforms returned to control levels when cells were placed in fresh media for 24 h after PQ2+ treatment (Fig. 2 a).
Fig. 2.
Oxidation of DJ-1 is prevented in the C106A mutant. (a) Endogenous DJ-1 in M17 cells responds to oxidative stress. (Upper) From left to right, blots of 2D gels from extracts from untreated cells, cells exposed to PQ2+ or MPP+; on the right, from cells exposed to PQ2+ for 24 h then placed in fresh media for 24 h. Blots were probed with monoclonal anti-DJ1 (arrow), which shows multiple isoforms: numbers below the blots show approximate pI values for each of the major isoforms. (Inset) Area around the pI 5.8 isoform, which accumulates after oxidative stress. (b) Response to oxidative stress in cysteine mutants. Samples were extracted from cells transfected with V5-tagged WT, C53A, or C106A DJ-1, from either untreated cells or cells exposed to PQ2+. Blots were probed with monoclonal anti-V5 and the pI 5.8 isoform identified (indicated below). The pI 5.8 isoform accumulated with both WT and C53A variants but not with the C106A variant (Bottom). Quantification of this and three similar experiments in c shows the ratio of pI 5.8:pI 6.1 isoform (n = 4; bars represent SEM). Note that the y axis scales are different for C106A; we included the faint immunoreactivity in the pI 5.8 region, but the ratio was still substantially lower than for the other variants.
To identify which of the different Cys residues are responsible for this pI shift, M17 cells stably expressing V5-tagged versions of WT, C53A, or C106A DJ-1 were treated with PQ2+ (Fig. 2b). Because C46A DJ-1 was unstable (see Fig. 1c), we were unable to distinguish the multiple pI isoforms, so we did not analyze this mutant further. Basal amounts of the pI 5.8 isoform were present in both WT and C53A DJ-1, and both responded to oxidation with an increase in the pI 5.8 isoform. C106A, however, failed to respond in this way and did not support the formation of the pI 5.8 isoform. Quantification of the amount of the pI 5.8 isoform relative to the pI 6.1 isoform (the other major isoform of the V5 transfected protein) in several independent experiments is shown in Fig. 2c. Exposure of cells to hydrogen peroxide also induced the formation of the pI 5.8 isoform for WT or C53A variants but not for C106A (data not shown). Total DJ-1 protein was quantified on 1D blots, and exposure to PQ2+ had no significant effect on the total amount of DJ-1 expression (data not shown).
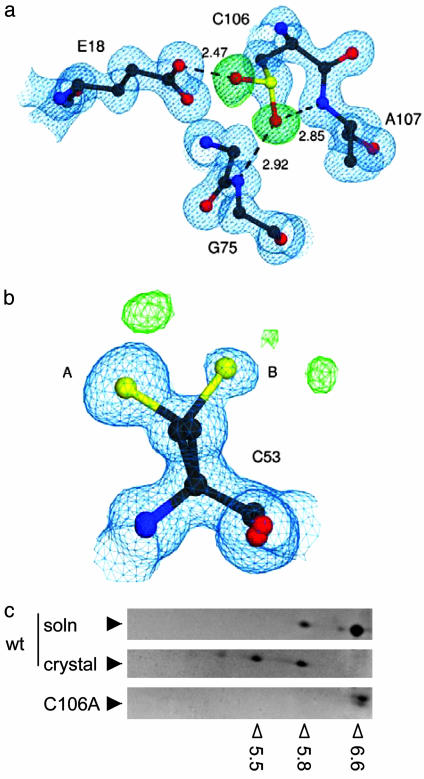
Direct Observation of Cysteine-Sulfinic Acid in the Crystal. The 1.2-Å resolution crystal structure of DJ-1 shown in Fig. 3 indicates that C106 is oxidized to cysteine sulfinic acid (C106-SO2H) in the crystal. The electron density around C106 unambiguously indicates that the oxidized species is sulfinic acid, even before this moiety is introduced into the model (Fig. 3a). Outside of this modification, no significant structural changes occur in response to oxidation at C106. The two oxygens of C106-SO2H make three direct hydrogen bonds with the protein; a 2.47-Å hydrogen bond with Glu-18 and two hydrogen bonds with the backbone amide hydrogens of G75 and A107 (Fig. 3a), possibly stabilizing the C106-SO2H modification.
Fig. 3.
Identification of C106-SO2H in the crystal. (a) Immediate environment of C106-SO2H, with 2mFO-DFC electron density contoured at 1.0 σ (blue) and mFO-DFC electron density contoured at 4.0 σ (green) m and D are weights for the Fourier coefficients. Both electron density maps were calculated before the introduction of the Oδ1 and Oδ2 oxygen atoms of C106-SO2H. Dotted lines indicate hydrogen bonds between C106-SO2H and the surrounding residues E18, G75, and A107, with lengths in angstroms. (b) C53 is not modified to the same extent as C106, although this residue can adopt two conformations (A and B). Difference electron densities (green) represent probable minor modifications. This figure was made with povscript+ (28). (c) Accumulation of acidic pI isoforms after crystallization. Recombinant WT DJ-1 before (Top) or after crystallization (Middle) were separated on 2D gels and different isoforms identified by Coomassie staining. Note that the crystallization process induces a pI shift in the recombinant protein, although a basal level of oxidized protein is seen in solution. In contrast, C106A is present only as the most basic isoform (pI 6.6-6.8). Open arrowheads below the gels show experimentally determined pI values, which were calibrated with 2D gel standards; filled arrowheads indicate the position of DJ-1.
In contrast to the robust oxidation of C106, neither C46 nor C53 is comparably modified, although there is some prominent positive difference electron density around both of the alternate side chain positions of C53 (Fig. 3b). These difference peaks are ≈1.9 Å away from the Sγ atom of C53 and are therefore consistent with covalent modification. However, these difference electron density peaks are much weaker than those at C106 and represent minor modifications.
To determine whether oxidation at C106 occurs prior or subsequent to crystallization, we ran recombinant DJ-1 before and after crystallization on 2D gels (Fig. 3c). WT recombinant protein was present as a pI 6.6 isoform before crystallization, with minor amounts of a more oxidized pI 5.8 isoform. After crystallization, several more acidic isoforms accumulated as the pI 6.6 isoform was lost. Recombinant C106A protein was present only as the most basic isoform (Fig. 3c), but C53A was able to oxidize (data not shown). These data indicate the recombinant protein behaves as the transfected protein in mammalian cells, and that crystallization facilitated oxidation in vitro (see Discussion).
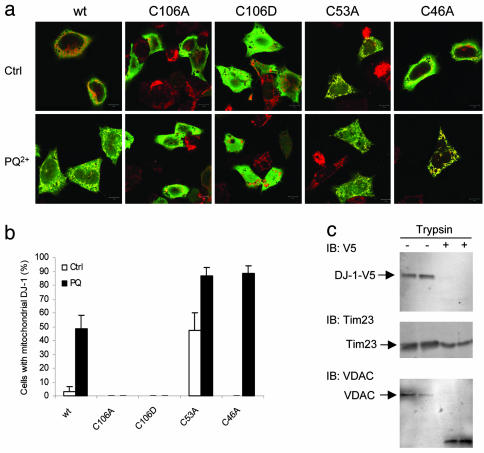
C106A Blocks Mitochondrial Localization and Protection Against Mitochondrial Damage. We explored the functional consequences of oxidation of DJ-1 in intact cells using C106A to dissect those phenomena that depend on cysteine-sulfinic acid formation. We stained stably transfected M17 cells for transfected DJ-1 and counterstained with either nuclear (data not shown) or mitochondrial (Fig. 4a) markers. Similar to our previous results (8), WT DJ-1 showed little specific localization to mitochondria. However, treatment with PQ2+ induced a localization of DJ-1 to mitochondria (Fig. 4a). In separate experiments, we saw that endogenous, untagged DJ-1 also redistributed to mitochondria under oxidative conditions (data not shown).
Fig. 4.
DJ-1 localizes to the outer mitochondrial membrane after oxidative stress. (a) M17 cells were transfected with V5-tagged DJ-1 variants as indicated and stained for DJ-1 (V5, green) and mitotracker (red). After exposure to PQ2+ (Lower), DJ-1 colocalized with mitochondria much more extensively than under control conditions (Upper), as evidenced by the yellow color where staining overlaps. The C106A and C106D variants did not relocalize to mitochondria, whereas C53A and C46A did. This finding is quantified in b by counting the proportion of V5-positive cells that showed strong colocalization with mitotracker. Representative data are shown from at least three experiments for each construct, performed in duplicate. (c) DJ-1 is not imported into mitochondria. Cells transfected with WT DJ-1 were exposed to PQ2+ and mitochondrial fractions prepared. Duplicate samples were either untreated (-, lanes 1 and 2) or subjected to limited proteolysis with trypsin (+, lanes 3 and 4) and blotted for V5 (Top), the outer mitochondrial membrane protein VDAC1 (Middle), or the inner mitochondrial membrane protein Tim23 (Bottom).
We next looked at the response of C106A-DJ1 to oxidation, and included an additional mutant where C106 was substituted with a negatively charged aspartate. In stark contrast to WT protein, neither C106A nor C106D localized to mitochondria basally or after PQ2+ treatment (Fig. 4 a and b). The staining patterns for both C106 variants were similar to WT DJ-1 under unstressed conditions, although an increase in nuclear staining was seen. To show specificity for C106 variants, we also examined C46A and C53A. C46A immunostaining was relatively low, but we were able to detect sufficient signal in transfected cells by increasing sensitivity on this channel and found that this variant acted as WT, moving toward mitochondria after PQ2+ treatment. C53A showed mitochondrial localization under control conditions, although PQ2+ still increased the proportion of cells where mitochondrial localization occurred. Using subcellular fractionation (27), we confirmed that, although C53A moved from cytosol to mitochondria after PQ2+ treatment, C106A did not (data not shown).
To determine whether DJ-1 was imported into the mitochondrial space or remained on the cytoplasmic side of the organelle, mitochondrial preparations from WT transfected, PQ2+ treated cells were subjected to limited trypsin digestion (Fig. 4c). DJ-1 was digested after a 10-min treatment with trypsin, as was the mitochondrial outer membrane voltage-dependent anion channel (Fig. 4c). In contrast, the mitochondrial inner membrane protein Tim23, remained intact (Fig. 4c).
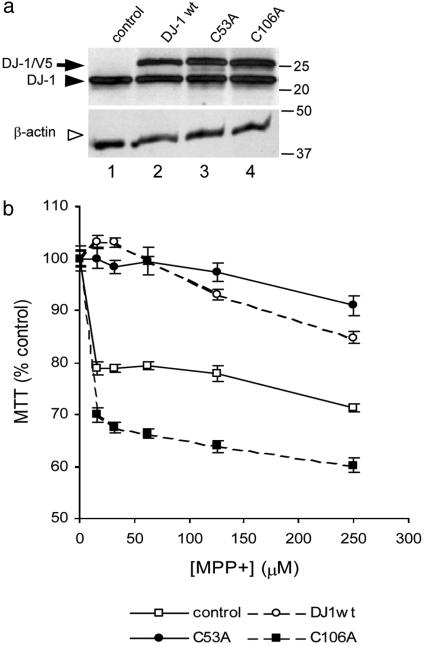
Given that the C106A variant was resistant to oxidation and mitochondrial localization under oxidative conditions, we considered that cysteine mutations might affect the ability of DJ-1 to protect against cell death (5, 6). Using stable cell lines (Fig. 5a), we screened several toxins, including PQ2+ (100-2,000 μM), H2O2 (100-1,000 μM), and MPP+ (20-250 μM), and found that overexpressing WT DJ-1 had the largest protective effects against MPP+ (Fig. 5b and data not shown). Cells expressing WT DJ-1 showed an ≈20% increase in cell viability against all tested concentrations of MPP+. We confirmed this in multiple clonal lines and found that expression levels correlated with degree of protection (data not shown). Cells expressing C53A showed a marginally greater resistance to MPP+. However, expression of C106A increased sensitivity to MPP+ compared to control cells by ≈15% at all doses tested (see Fig. 5b). Using two-way ANOVA, differences between concentrations of MPP+ were significant (P < 0.05), as were differences between cell lines (P < 0.01).
Fig. 5.
Cysteine mutants affect the ability of DJ-1 to protect against mitochondrial damage. For toxicity assays, we generated stable cell lines that expressed either WT, C53A, or C106A DJ-1 (a). Western blots show expression of the V5-tagged constructs (arrow) and endogenous DJ-1 (filled arrowhead) and were reprobed with β-actin (Lower, open arrowhead). (b) Exposure to MPP+ for 48 h resulted in cell death, assessed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assays. Cells expressing WT or C53A DJ-1 were resistant to MPP+ toxicity, whereas cells expressing C106A DJ-1 showed increased sensitivity to this toxin. Data are shown from one set of experiments with all four cell lines performed on the same day, expressed as a percentage of untreated cells (n = 8 wells per dose, error bars indicate SEM) and are representative of at least triplicate experiments per cell line.
Discussion
The identification of PD-associated mutations in DJ-1, a protein responsive to oxidative stress (10), is consistent with the hypothesis that reactive oxygen and nitrogen species contribute to PD (29). In the present study, we have shown that oxidative conditions induce a modification at C106 of DJ-1, because the pI shift of DJ-1 is blocked in the C106A variant, and C106 is oxidized to cysteine-sulfinic acid in vitro. Recently, a cysteine-sulfonic acid has been identified at C106 by mass spectrometry of DJ-1 isolated from hydrogen-peroxide-treated human umbilical vein endothelial cells (12). In the present study, C106 readily oxidized, whereas C46 and C53 remained in their reduced state under the same conditions, consistent with other recent data (12).
Using 1.2-Å resolution x-ray diffraction data, we have identified a sulfinic acid at C106 of crystalline DJ-1. Lee et al. (18) have also noted a modification at C106 in hydrogen peroxide treated crystals at 3.0 Å. This residue was a favored site of radiation damage in the 1.1-Å structure of DJ-1, and a similar modification of the equivalent Cys was observed in YDR533Cp, a protein from Saccharomyces cerevisiae related to DJ-1 (30). However, other structures of (nonoxidized) DJ-1 (17, 19) did not show oxidation at C106. Furthermore, we did not intentionally oxidize DJ-1 in the crystal. A possible explanation is the presence of peroxide decomposition products in the polyethylene glycol (PEG) 400 used as a precipitant for crystal growth. Other crystallization conditions for DJ-1 have used either higher molecular-weight PEGs (16) or sodium citrate (31), both of which are less prone to degradation. Supporting this hypothesis, crystallized DJ-1 was more acidic than DJ-1 before crystallization. The formation of C106-SO2H appears to be facile and likely contributes to the appearance of the acidic isoform of DJ-1 upon oxidative stress.
These observations support the hypothesis that DJ-1 is an oxidative stress sensor within cells, and that a chemically stable C106-SO2H modification controls function. Furthermore, this modification can be reversed in whole cells, although the mechanism(s) involved in generating and controlling cysteine modification in DJ-1 have yet to be established. For other mammalian proteins that can be modified to produce cysteine-sulfinic acids, such as the peroxiredoxins, multiple mechanisms are proposed for the regeneration of the nonoxidized isoforms. These include ATP-dependent reduction (14, 15, 32) or selective degradation of the oxidized isoform. It will be important to determine whether there are similar mechanisms that control DJ-1 oxidation.
Our data indicate a correlation between ability of DJ-1 mutants to oxidize, translocate to mitochondria in response to oxidation and protect against toxicity. In particular, toxicity induced by the mitochondrial complex I inhibitor MPP+ was abrogated by overexpression of DJ-1 WT and C53A variant but not by C106A. Oxidative conditions promoted the localization of WT, C53A, and C46A DJ-1 toward the mitochondria, whereas C106A and C106D variants were prevented from translocating in this way. C106A, which dimerizes effectively with endogenous DJ-1, in fact provided a partial dominant negative effect on cell survival, suggesting that it may sequester the WT protein away from its site of action. In addition, DJ-1 is not imported into the inner mitochondrial space but remains on the cytoplasmic side of the organelle. The outer mitochondrial membrane (OMM) is the site of action of several cell death factors that control opening of the permeability transition pore PTP (33). The binding partner(s) of DJ-1 at the OMM are still under investigation in our lab. Further work is also needed to establish the function of DJ-1 after translocation to the OMM. However, our observations strongly suggest the hypothesis that DJ-1 protects against mitochondrial damage, signaled by the oxidation of C106. Therefore, it is reasonable to suggest that DJ-1 may protect neurons from various stressful stimuli like parkin, another recessive gene that causes PD (2).
Supplementary Material
Acknowledgments
G.A.P., D.R., and M.A.W. thank the Ellison Medical Foundation and the American Parkinson Disease Association for support.
Abbreviations: PD, Parkinson's disease; MPP+, 1-methyl-4-phenylpyridinium; PQ2+, paraquat.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1SOA).
References
- 1.Dawson, T. M. & Dawson, V. L. (2003) Science 302, 819-822. [DOI] [PubMed] [Google Scholar]
- 2.Petrucelli, L., O'Farrell, C., Lockhart, P. J., Baptista, M., Kehoe, K., Vink, L., Choi, P., Wolozin, B., Farrer, M., Hardy, J., et al. (2002) Neuron 36, 1007-1019. [DOI] [PubMed] [Google Scholar]
- 3.Darios, F., Corti, O., Lucking, C. B., Hampe, C., Muriel, M. P., Abbas, N., Gu, W. J., Hirsch, E. C., Rooney, T., Ruberg, M., et al. (2003) Hum. Mol. Genet. 12, 517-526. [DOI] [PubMed] [Google Scholar]
- 4.Bonifati, V., Rizzu, P., van Baren, M. J., Schaap, O., Breedveld, G. J., Krieger, E., Dekker, M. C., Squitieri, F., Ibanez, P., Joosse, M., et al. (2003) Science 299, 256-259. [DOI] [PubMed] [Google Scholar]
- 5.Taira, T., Saito, Y., Niki, T., Iguchi-Ariga, S. M., Takahashi, K. & Ariga, H. (2004) EMBO Rep. 5, 213-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokota, T., Sugawara, K., Ito, K., Takahashi, R., Ariga, H. & Mizusawa, H. (2003) Biochem. Biophys. Res. Commun. 312, 1342-1348. [DOI] [PubMed] [Google Scholar]
- 7.Bandyopadhyay, S. & Cookson, M. R. (2004) BMC Evol. Biol. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller, D. W., Ahmad, R., Hague, S., Baptista, M. J., Canet-Aviles, R., McLendon, C., Carter, D. M., Zhu, P.-P., Stadler, J., Chandran, J., et al. (2003) J. Biol. Chem. 278, 36588-36595. [DOI] [PubMed] [Google Scholar]
- 9.Mitsumoto, A., Nakagawa, Y., Takeuchi, A., Okawa, K., Iwamatsu, A. & Takanezawa, Y. (2001) Free Radical Res. 35, 301-310. [DOI] [PubMed] [Google Scholar]
- 10.Mitsumoto, A. & Nakagawa, Y. (2001) Free Radical Res. 35, 885-893. [DOI] [PubMed] [Google Scholar]
- 11.Bandopadhyay, R., Kingsbury, A. E., Cookson, M. R., Reid, A. R., Evans, I. M., Hope, A. D., Pittman, A. M., Lashley, T., Canet-Aviles, R., Miller, D. W., et al. (2004) Brain 127, 420-430. [DOI] [PubMed] [Google Scholar]
- 12.Kinumi, T., Kimata, J., Taira, T., Ariga, H. & Niki, E. (2004) Biochem. Biophys. Res. Commun. 317, 722-728. [DOI] [PubMed] [Google Scholar]
- 13.Woo, H. A., Chae, H. Z., Hwang, S. C., Yang, K. S., Kang, S. W., Kim, K. & Rhee, S. G. (2003) Science 300, 653-656. [DOI] [PubMed] [Google Scholar]
- 14.Chevallet, M., Wagner, E., Luche, S., van Dorsselaer, A., Leize-Wagner, E. & Rabilloud, T. (2003) J. Biol. Chem. 278, 37146-37153. [DOI] [PubMed] [Google Scholar]
- 15.Biteau, B., Labarre, J. & Toledano, M. B. (2003) Nature 425, 980-984. [DOI] [PubMed] [Google Scholar]
- 16.Huai, Q., Sun, Y., Wang, H., Chin, L. S., Li, L., Robinson, H. & Ke, H. (2003) FEBS Lett. 549, 171-175. [DOI] [PubMed] [Google Scholar]
- 17.Honbou, K., Suzuki, N. N., Horiuchi, M., Niki, T., Taira, T., Ariga, H. & Inagaki, F. (2003) J. Biol. Chem. 278, 31380-31384. [DOI] [PubMed] [Google Scholar]
- 18.Lee, S. J., Kim, S. J., Kim, I. K., Ko, J., Jeong, C. S., Kim, G. H., Park, C., Kang, S. O., Suh, P. G., Lee, H. S., et al. (2003) J. Biol. Chem. 278, 44552-44559. [DOI] [PubMed] [Google Scholar]
- 19.Tao, X. & Tong, L. (2003) J. Biol. Chem. 278, 31372-31379. [DOI] [PubMed] [Google Scholar]
- 20.Wilson, M. A., Collins, J. L., Hod, Y., Ringe, D. & Petsko, G. A. (2003) Proc. Natl. Acad. Sci. USA 100, 9256-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olzmann, J. A., Brown, K., Wilkinson, K. D., Rees, H. D., Huai, Q., Ke, H., Levey, A. I., Li, L. & Chin, L. S. (2004) J. Biol. Chem. 279, 8506-8515. [DOI] [PubMed] [Google Scholar]
- 22.Gorner, K., Holtorf, E., Odoy, S., Nuscher, B., Yamamoto, A., Regula, J. T., Beyer, K., Haass, C. & Kahle, P. J. (2004) J. Biol. Chem. 279, 6943-6951. [DOI] [PubMed] [Google Scholar]
- 23.Allen, S., Heath, P. R., Kirby, J., Wharton, S. B., Cookson, M. R., Menzies, F. M., Banks, R. E. & Shaw, P. J. (2003) J. Biol. Chem. 278, 6371-6383. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 25.Sheldrick, G. M. & Schneider, T. R. (1997) Methods Enzymol. 277, 319-343. [PubMed] [Google Scholar]
- 26.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 27.Chua, B. T., Volbracht, C., Tan, K. O., Li, R., Yu, V. C. & Li, P. (2003) Nat. Cell Biol. 5, 1083-1089. [DOI] [PubMed] [Google Scholar]
- 28.Fenn, T. D., Ringe D., Petsko, G. A. (2003) J. Appl. Crystallogr. 36, 944-947. [Google Scholar]
- 29.Zhang, Y., Dawson, V. L. & Dawson, T. M. (2000) Neurobiol. Dis. 7, 240-250. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, M. A., Amour, C. V. S., Collins, J. L., Ringe, D. & Petsko, G. A. (2004) Proc. Natl. Acad. Sci. USA 101, 1531-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honbou, K., Suzuki, N. N., Horiuchi, M., Taira, T., Niki, T., Ariga, H. & Inagaki, F. (2003) Acta Crystallogr. D 59, 1502-1503. [DOI] [PubMed] [Google Scholar]
- 32.Wood, Z. A., Poole, L. B. & Karplus, P. A. (2003) Science 300, 650-653. [DOI] [PubMed] [Google Scholar]
- 33.Kim, J. S., He, L. & Lemasters, J. J. (2003) Biochem. Biophys. Res. Commun. 304, 463-470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.