Abstract
Animal evolution is closely linked to the emergence of the nervous system. At present it is unknown how the basic mechanisms of neural induction and formation of central nervous systems evolved. We addressed this question in Nematostella vectensis, a member of cnidarians, the ancient sister group of bilaterians. We found that β-catenin signalling is crucial for the early induction of the embryonic nervous system. β-Catenin activity at the blastopore induces specific neurogenic genes required for development of the oral nervous system. β-Catenin signalling induces also Bmp signalling, which, at later larval stages, becomes indispensible for the maintenance and asymmetric patterning of the oral nervous system along the primary and secondary (directive) axes. We hypothesize that the consecutive and functionally linked involvement of β-catenin and Bmp signalling in the formation of the cnidarian oral nervous system reflects an ancestral mechanism that evolved before the cnidarian/bilaterian split.
 The bilaterian central nervous system is thought to have evolved from a cnidarian-like ancestor, but the mechanisms of neural induction in cnidarians are largely unknown. Here the authors study the cnidarian Nematostella vectensis and show that β-catenin signalling is crucial for the early induction of its embryonic nervous system, suggesting evolutionary roots for this pathway.
The bilaterian central nervous system is thought to have evolved from a cnidarian-like ancestor, but the mechanisms of neural induction in cnidarians are largely unknown. Here the authors study the cnidarian Nematostella vectensis and show that β-catenin signalling is crucial for the early induction of its embryonic nervous system, suggesting evolutionary roots for this pathway.
The emergence of nerve cells is one of the key novelties in animal evolution1. How cells of an embryo are committed to a neuronal fate is therefore a central question in biology. A related issue is the formation of a central nervous system (CNS), which is coordinating the action of sensory cells and neurons in higher metazoan animals.
The general view holds that the bilaterian CNS can be traced back through evolution to a nerve net in a cnidarian-like ancestor2. Cnidarians are simple diploblastic animals with a gastrula-like body plan, and they constitute the sister group of all bilaterians3,4,5,6,7,8,9. The cnidarian nervous system is organized as a nerve net with an increased density of neurons at the oral and aboral end of the body axis5,6,8. It also exhibits a remarkable sophistication of sensory cells and organs such as rhopalia8. Although the picture of the cnidarian nervous system is not complete, their neurons are known to be rich in neuropeptides, and comparative genomics in distant cnidarian species has revealed an almost complete set of homologous neurogenic transcription factors (TFs) and signalling molecules patterning the nervous system in bilaterians5,8. Cnidarian neurogenic TFs and neuropeptide-positive neurons exhibit a clear position dependency along the oral–aboral body axis5,6,8,9,10,11,12,13,14. However, the genetic mechanisms controlling the formation of the cnidarian nervous system are largely unknown.
In bilaterians, several secreted factors have been demonstrated to be crucial for the early segregation of the embryonic ectoderm into a neurogenic and a non-neurogenic tissue during neural induction. Bmp signalling is the critical factor for the early development of the CNS and for its anterior–posterior (AP) and dorsal–ventral (DV) patterning. Bmp signalling is suppressed by antagonists such as Chordin and Noggin on the anterior–dorsal side in chordates15 and on the ventral side in arthropods, flatworms and other gastroneuralians16,17,18. On the basis of experiments in Xenopus and Drosophila, which showed that an inhibition of Bmp signalling is sufficient to induce the neuroectoderm, it was proposed that the primary function of Bmp signalling in bilaterians is the inhibition of the neural fate, and accordingly, the default state of the ectoderm would be the neural fate15,16,19,20. This hypothesis was questioned for vertebrates because mutants lacking Bmp antagonists still develop neurons21. Also in other deuterostomes and in annelids, perturbation of Bmp signalling did not change the expression of neural makers22,23,24. Therefore, Bmp/Chordin signalling is not necessarily linked to neural induction in bilaterians21.
Wnt/β-catenin signalling has multiple and even opposite functions in neural development of deuterostomes, depending on the developmental stage21. Canonical Wnt signalling is required for neurogenesis at the dorsal side of early chordate embryos and for the AP and DV patterning of the CNS21. At later stages of chordate development, the pathway has a function in the posteriorization of the neural ectoderm26, which is similar to hemichordates and echinoderms27,28. The neurogenic function of Wnt signalling for the transition of proliferating neuronal precursor cells to differentiating neurons in deuterostomes25 has been recently described in lophotrochozoans29. Thus, a neurogenic function of Wnt/β-catenin signalling might be an ancestral bilaterian trait29. To understand which signalling pathway(s) controls neurogenesis in cnidarians, we focused our analysis on the signalling pathways and TFs that control the formation of the oral nervous system in N. vectensis. Many cnidarian polyps develop a conspicuous ring-like nerve plexus around their mouth8,10,30. This cnidarian nerve ring was repeatedly depicted as a beginning of the bilaterian CNS10,31,32.
The first sign for neurogenesis in N. vectensis is the salt-and-pepper-like expression of neural marker messenger RNAs (mRNAs) such as NvAchaete-scute homolog A (NvAshA), NvElav1 and Rfamide in blastula epithelium6,13,14. Recent studies provided evidence for their involvement in neural development of N. vectensis embryos. A whole-genome microarray analysis on NvAshA-overexpressing embryos has identified NvElav1, NvRfamide and genes involved neurotransmission as NvAshA targets13. It was demonstrated that NvElav1 is required at least ectodermal development of neuropeptides-expressing neurons16. The pervasive expression of NvAshA suggests that the early embryonic epithelium of N. vectensis has the potential to generate various neuronal cell types that form the larval nerve net13. In addition to this pervasive expression of neural genes, the presumptive blastopore region of the blastula expresses NvAshB, which is also involved in NvRfamide neuropeptide gene expression13. This suggests that the blastoporal side of the early embryos has distinct neurogenic feature(s). The blastopore of the gastrula in fact develops into a prominent neurogenic domain at the planula stage, which is characterized by some neural markers, NvAshC, NvSoxC, NvMusashi and NvRfamide that are expressed dominantly or exclusively around the blastopore6,12,13,14,33. This oral neurogenic domain of the embryo develops into an elaborate nerve plexus at the oral side of the planula larva and primary polyp that comprises a number of subsystems with separate physiological properties8.
Our study revealed that β-catenin signalling is essential for early neurogenesis during the development of the oral nervous system that starts at the blastula/gastrula transition. Wnt/β-catenin signalling is known to be active at the blastoporal side and defines the primary oral–aboral axis in N. vectensis blastulae34,35. By comparison, Bmp2/4 is also expressed at the blastoporal side, however, with a strong bias towards a secondary ‘directive’ body axis at later development36,37,38. We also show that in a subsequent developmental phase Bmp signalling has crucial influences on the regionalized development of the nervous system along both the primary and secondary (directive) body axes. Our data indicate that the sequential action of β-catenin and Bmp signalling in the cnidarian N. vectensis reflects the evolutionary emergence of these major signalling axes in the evolution of the nervous system.
Results
Early development of neuropeptide-positive neurons
The cnidarian nervous system is rich in neuropeptides8,11,39. Among these, the short amidated neuropeptides RFamide and GLWamide, belonging to R[F/Y]amide and [G/V/L]Wamide groups, respectively, are known to have deep evolutionary roots in the common Cnidaria/Bilateria ancestor40. These neuropeptides serve as specific markers for mature neurons in cnidarians8, and in bilaterians they are expressed in neuronal subpopulations of the CNS41,42.
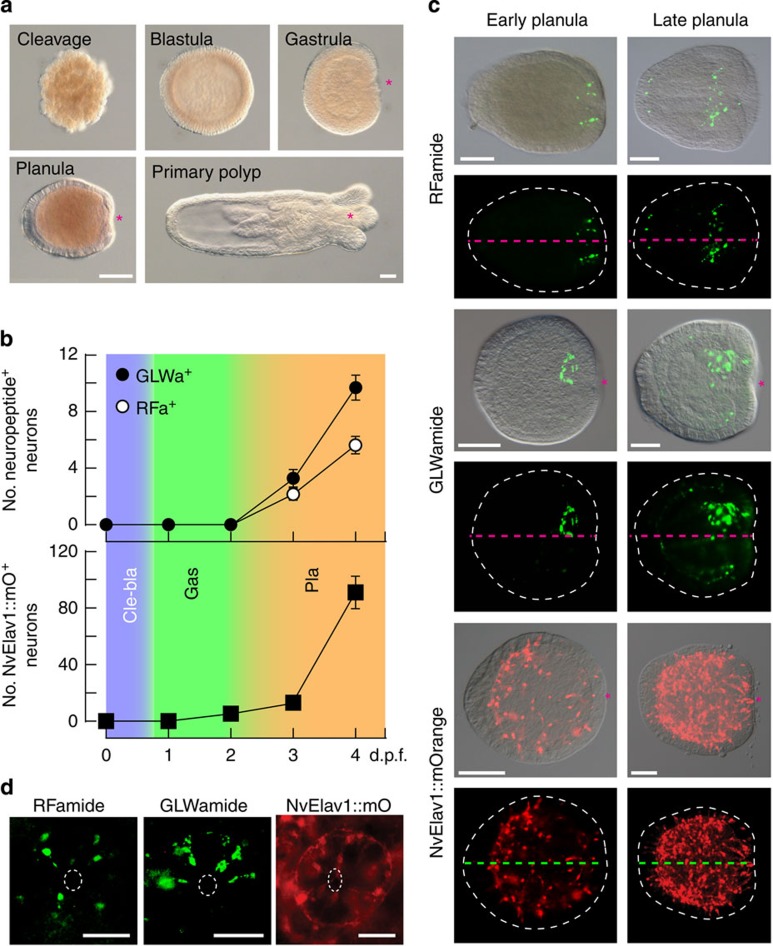
We studied various developmental stages of N. vectensis that belongs to anthozoans, the most basal class in cnidarians (Fig. 1a)7,43,44, and is known to show ‘bilateral organization of the endoderm45. Analyses using antibodies specific for the mature form of RFamide and GLWamide neuropeptides demonstrated that functional and peptidergic neurons are already present in early planulae (Fig. 1b,c)6,14. It has also recently been shown that NvElav1 is expressed in a substantial part of neurons during embryogenesis6,14. An NvElav1+ neuron-specific transgenic reporter line, where the mOrange fluorescent protein is expressed under the NvElav1 regulatory elements, demonstrated the development of the NvElav1::mOrange+ neurons at gastrula and planula stages14. A quantitative analysis revealed that the RFamidergic (RFa+) and GLWamidergic (GLWa+) neuronal subpopulations correspond to 10% of all neurons at the late planula stage (Fig. 1b). The RFa+ neurons develop in the entire ectoderm and form an elaborated nerve plexus at the oral side, but they did not form an aboral sensory cluster (Fig. 1c)6,14, which is often observed in planula larvae of hydrozoans46. In late planulae and after metamorphosis into a primary polyp, a domain rich in RFa+ perikaria was formed in the blastoporal and the hypostomal/tentacle region (Fig. 1c; Supplementary Fig. 1)6. GLWa+ neurons differentiate in the lateral ectoderm and in the oral endoderm of early planulae (Fig. 1d; Supplementary Fig. 2). The endodermal GLWa+ neurons formed a neuronal cluster in an asymmetric manner on one side at the oral region of the planula larvae (Fig. 1c,d), whereas the ectodermal neurons are distributed symmetrically and mainly in the midst body region (Supplementary Fig. 2). The asymmetry of the endodermal cluster of GLWa+ neurons completely vanished in primary polyps (Supplementary Fig. 1). The spatial arrangement of neurons at the oral region is not unique to the peptidergic neurons because Elav+ neurons form a ring-like sensory cell cluster around the blastopore at late planula stage (Fig. 1d). The formation of RFa+ and GLWa+ neurons at the oral side of early planula larvae suggests that those precursors were generated directly at the blastoporal region in early developmental stages, for example, gastrula. We therefore analysed the TFs expressed at the blastopore region and signalling pathways establishing the oral nervous system.
Figure 1. An asymmetry in the N. vectensis nervous system.
(a) Development of N. vectensis from cleavage to the primary polyp (differential interference contrast images). (b) Number of neurons expressing the mature neuropeptides RFamide and GLWamide (upper graph), and number of NvElav1::mOrange positive neurons (lower graph) at different days post fertilization (d.p.f.). Colours indicate cleavage/blastula (Cle-bla), gastrula (Gas) and planula (Pla) stage. The data represent the mean±s.e.m. of at least three experiments (n=486 RFamide, n=423 GLWamide and n=239 Elav). (c,d) Lateral view (c) and oral view (d) of RFa+, GLWa+ and NvElav1::mOrange+ neurons in planula larvae. The pink and green dotted lines in c denote the oral–aboral axis and the dotted white circles in d indicate the blastopore. Note that only GLWa+ neurons exhibit an additional bias along the orthogonal axis around the blastopore. Shown are representative images of at least three with similar results. The asterisks in a and c indicate the blastopore. Scale bars in a,c and d, 100 μm and 50 μm, respectively.
Identification and expression of oral neurogenic TFs
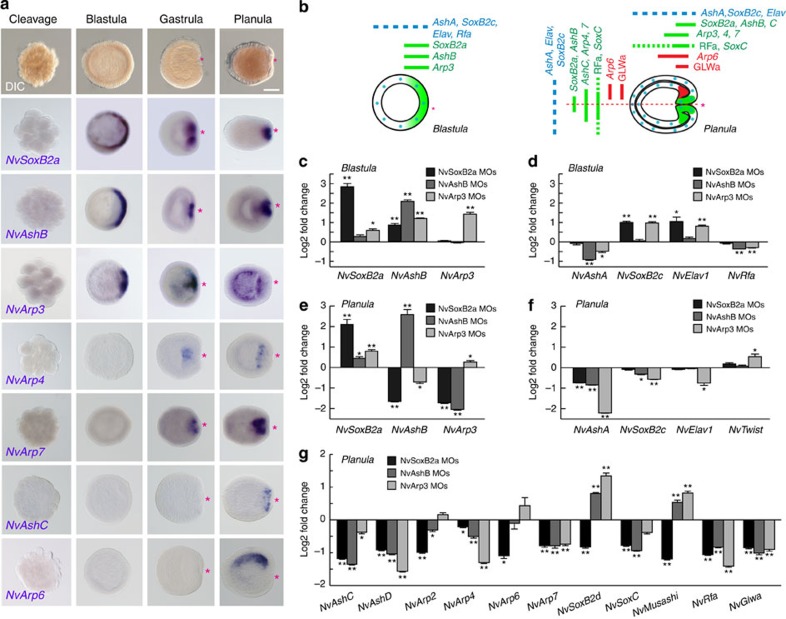
We screened the EST and genomic database of N. vectensis for TFs that have conserved neurogenic functions among bilaterians. We determined the temporal and spatial expression patterns of the TFs during N. vectensis embryogenesis and analysed their neurogenic activity. Our reverse transcriptase (RT)-PCR and whole-mount in situ hybridization (WISH) data demonstrate that several neurogenic genes were strongly upregulated during early embryonic development (Fig. 2a,b; Supplementary Fig. 3). We found that NvAtonal-related protein 3 (NvArp3) formed a synexpression group at the prospective and established blastopore site together with the previously described NvSoxB2a and NvAshB12,13 during the blastula and gastrula stage (Fig. 2a,b). NvSoxB2a was considered to be an unclassified NvSox1 (ref. 12), but has recently been identified as the orthologous gene to the bilaterian SoxB2 genes and was reclassified as NvSoxB2 (ref. 47). Since the C-terminal region of NvSoxB2a is similar to bilaterian SoxB2 (Supplementary Fig. 4), NvSoxB2a might have a conserved neurogenic function47. At the planula larva stage, NvSoxB2a and NvAshB were restricted to the oral side of the pharynx, while NvArp3 was expressed in cells distributed broadly at the endodermal layer. The concurrent expression of NvSoxB2a with bHLH genes in N. vectensis is in accord with results from bilaterians, where SoxB2 genes are expressed in neural precursors together with bHLH genes48. We identified additional bHLH TFs showing also distinct blastoporal expression patterns at the oral region, that is, NvArp4 in endoderm, NvArp7 in pharynx and NvAshC in ectoderm13. Furthermore, we were able to find NvArp6 as the first neurogenic TF being asymmetrically expressed along the secondary (directive) axis (Fig. 2a,b).
Figure 2. Action of early oral neurogenic genes.
(a) WISH assay of neural transcription factors. Red asterisks denote the blastopore and future oral pole. Shown are representative images of at least three with similar results. Note the late asymmetric expression of NvArp6 at the planula stage. Scale bar, 100 μm. (b) A schematic illustration of the expression patterns of oral and lateral neurogenic and marker genes at the blastula (left) and planula (right) stages. (c–g) qPCR analyses of the neural gene expression in the blastula (c,d) and planula (e,f) that were injected with MOs against NvSoxB2a, NvAshB or NvArp3. Bars represent the mean±s.e.m. of at least three experiments. The black asterisks denote the statistical significance using the Student’s t-test (*P<0.05, **P<0.01).
We next analysed the genetic relationships between these early oral neurogenic genes (that is, NvSoxB2a, NvAshB and NvArp3) and other early neurogenic and neural marker genes (NvAshA, NvSoxB2c (former SoxB2), NvElav1 and NvRfamide) as well as the late oral neurogenic genes (NvAshC, D, NvArp2, 4, 6, 7, NvSoxB2d, C, NvMusashi, NvRfamide and NvGlwamide) using morpholino (MO) antisense oligonucleotides against the early blastoporal neurogenic genes (Supplementary Fig. 5). Figure 2c,d show that in morphants of NvSoxB2a, NvAshB and NvArp3 an upregulation of all early genes at the blastula stage, except for NvAshA and NvRfamide, indicating that NvSoxB2a, NvAshB and NvArp3 are not required for the initial steps of most of these genes, but rather form a negative gene regulatory circuit. At the planula stage, however, a suppression of NvSoxB2a resulted in the downregulation of NvAshB and NvArp3 (Fig. 2e), indicating that NvSoxB2a maintains the expression of these early oral bHLH genes. At the planula stage, expression of the lateral genes (Fig. 2f) and the late oral genes (that is, neurogenic TFs and neuropeptides) (Fig. 2g) were affected in these morphants. This clearly demonstrated the crucial function of early oral neurogenic genes for the development of the oral nervous system. The MO experiments indicated also that these early oral neurogenic TFs are involved at least partially in the transcriptional control of lateral neural markers from the blastula to planula stage (Fig. 2d,f). The function of the early oral genes is specific to neurogenesis because expression of NvTwist, a marker for endoderm was not suppressed in these morphants (Fig. 2f).
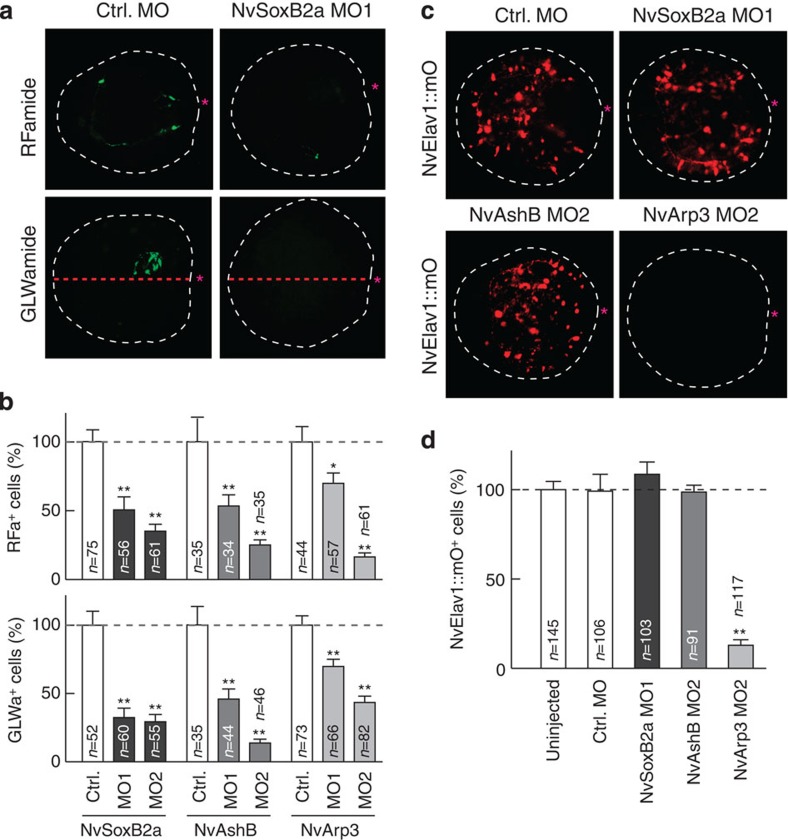
We further analysed the neurogenic activity of the early oral TFs by monitoring the mature RFa+ and GLWa+ neurons. For all morphants, we confirmed that these genes are required for ectodermal and endodermal development of RFa+ and GLWa+ neurons (Fig. 3a,b; Supplementary Figs 6 and 7). Interestingly, development of NvElav1::mOrnage+ neurons were inhibited only in NvArp3 MO-injected planula larvae (Fig. 3c,d), which is consistent with the specific involvement of NvArp3 on NvElav1 expression at the planula stage (Fig. 2f). These results clearly indicate that NvSoxB2a, NvAshB and NvArp3 have neurogenic function and that the presumptive blastopore region of the blastula represents an early neurogenic field, which gives rise to neuronal populations of the oral (RFa+/GLWa+) and endodermal (NvElav1+) nervous systems. As NvSoxB2a and NvAshB are required for the expression of many early and late oral neuronal genes, they might mainly be involved in the commitment of embryonic cells that gives rise to neural precursor cells, whereas NvArp3 might have a selective function to specify certain neuronal cell types such as Elav1+ neurons.
Figure 3. Oral neurogenic genes are required for RFa+, GLWa+ and Elav1+neurons.
Development of RFa+ and GLWa+ neurons (a,b) and NvElav1::mOrange+ neurons (c,d) in the planula injected with MO against NvSoxB2a, NvAshB and NvArp3. (a) RFa+ and GLWa+ neurons in control MO or NvSoxB2a MO-injected planulae (3 d.p.f.). The red dotted lines denote the oral–aboral axis. (c) NvElav1::mOrange+ neurons in MO-injected planulae (3 d.p.f.). The red asterisks indicate the blastopore. (b,d) Number of RFa+ and GLWa+ neurons (4 d.p.f.) (b) and NvElav1::mOrange+ neurons (3 d.p.f.) (d). The data in b and d represent the relative number of neurons. Bars represent the mean±s.e.m. The black asterisk denotes the statistical significance using the Student’s t-test (*P<0.05, **P<0.01).
β-Catenin signalling in early neurogenesis
To identify signals required for expression of the early oral neurogenic genes, we searched for factors turned on in the blastula at the site of presumptive blastopore region. RT-PCR and WISH analyses indicated that NvBrachyury (NvBra), a target of canonical Wnt/β-catenin signalling in cnidarians49, is strongly upregulated at the blastula stage (Supplementary Fig. 8). Although β-catenin protein seems to be expressed in all blastomeres at the early-cleavage stages, it has been described to become stabilized and translocated to the nuclei at the embryonic region where gastrulation will occur34 and NvTcf expression is maintained (Supplementary Fig. 8)50. Wnt/β-catenin activity is maintained in later developmental stages (planula and polyp) at the blastopore/mouth35,50 and was postulated to pattern gene expression (including NvRfamide) along the oral–aboral (primary) body axis51,52. The β-catenin function on the early phase of neurogenesis, however, has not yet been clarified. The concurrent temporal and spatial expression pattern of early neurogenic genes with downstream target genes of the β-catenin pathway at the blastula stage prompted us to investigate the involvement of β-catenin signalling in the early development of the oral nervous system.
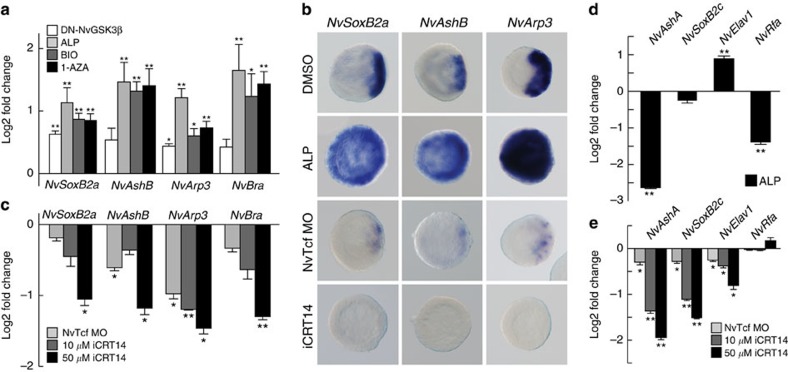
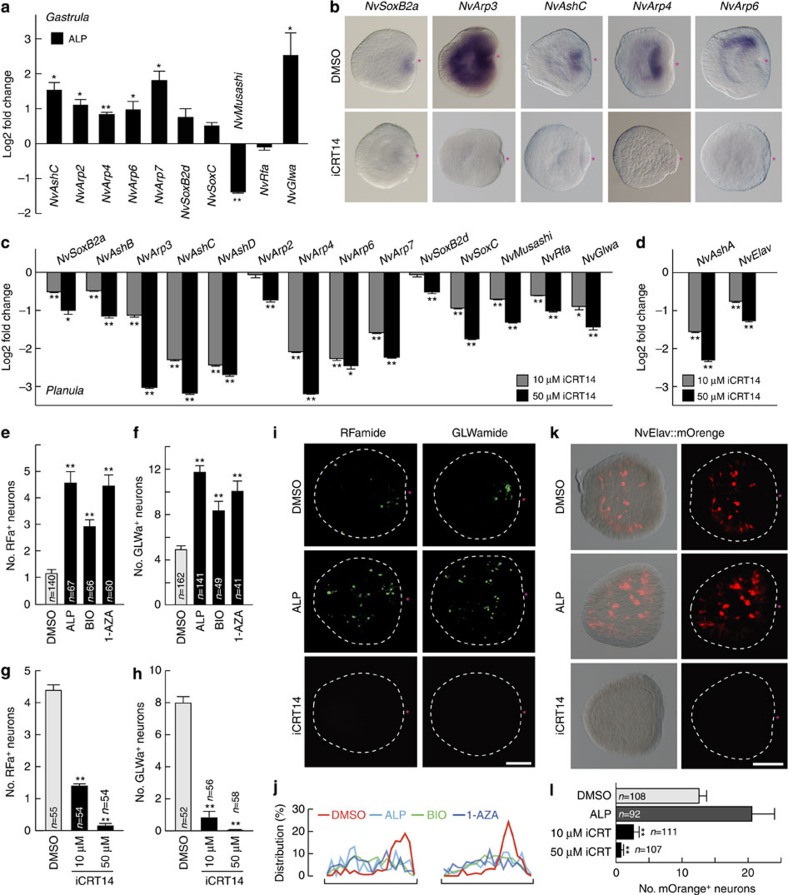
To examine the function of the β-catenin pathway in the neural induction, we stimulated β-catenin signalling by injecting mRNA for a dominant-negative GSK3-β (DN-GSK3β) into fertilized eggs. The augmentation of β-catenin signalling induced increased expression of the neurogenic genes NvSoxB2a, NvAshB and NvArp3, as well as a β-catenin target gene NvBra at the blastula stage (Fig. 4a). A more prominent and ectopic upregulation of neurogenic gene expression could be observed when early embryos were treated with the GSK3-β inhibitors Alsterpaullone (ALP), BIO or 1-Azakenpaullone (1-AZA) (Fig. 4a,b; Supplementary Figs 8b and 9). To analyse β-catenin regulation of the neurogenic gene expression at early embryonic stages, we treated embryos only for a short time (that is, from the egg to the blastula stage). The temporal activation of β-catenin signalling with GSK3β inhibitors was also needed to circumvent potential toxic effects caused by a long-term treatment with these inhibitors51, so that the mortality rate of planulae remained unchanged (Supplementary Fig. 10). Under these conditions, we observed an ‘oralized’ phenotype with an ectopic expression of early neurogenic TFs (Fig. 4; Supplementary Fig. 8b). In blastula embryos, the injection of NvTcf MO or treatment of Tcf/β-catenin inhibitor iCRT14 (ref. 53) suppressed the expression of NvSoxB2a, NvAshB and NvArp3 (Fig. 4b,c). These data therefore indicate that the β-catenin pathway positively regulates expression of the oral neurogenic TFs at the blastula stage.
Figure 4. β-Catenin signalling regulates early neurogenesis.
(a-c) Expression analyses of early neurogenic genes NvSoxB2a, NvAshB and NvArp3 at the blastula stage after DN-GSK3β mRNA injection or treatment with several GSK3β inhibitors (2.5 μM ALP, 0.5 μM BIO and 2.5 μM 1-AZA) (a,b) and in blastulae injected with NvTcf MO or treated with an increasing concentrations of iCRT14 (b,c). NvBra was used as positive control in the qPCR analyses (a,c). In the WISH analysis (b), ALP and iCRT14 were used at 2.5 μM and 50 μM concentrations, respectively. (d,e) qPCR analyses of early lateral neural genes NvAshA, NvSoxB2c, NvElav1 and NvRfamide in 2.5 μM ALP-treated blastulae (d), in Tcf morphants and in 50 μM iCRT14-treated blastulae (e). Shown in b are representative images of at least three with similar results. Bars represent the mean±s.e.m. of at least three experiments. Asterisks denote statistical significance using the Student’s t-test (*P<0.05; **P<0.01).
Next, we analysed the β-catenin regulation of early genes, NvAshA, NvSoxB2c, NvElav1 and NvRfamide that show lateral expression patterns at blastula. Most of these genes except for NvElav1 were not upregulated under augmented β-catenin signalling (Fig. 4d), whereas NvAshA, NvSoxB2c and NvElav1 were strongly suppressed by the β-catenin inhibitor (Fig. 4e). These data indicate that β-catenin signalling is globally required at an early developmental stage for the expression of oral and lateral neural genes and neurogenic TFs.
The wide spectrum of β-catenin dependency of neurogenic TFs suggests that rather for specifying certain neuronal cell types, β-catenin signalling is required for the early fate decision of orally and laterally localized neural progenitors giving rise to multiple neuronal cell types in the planula nervous system. Consistent with this idea, the activation of β-catenin signalling at early embryogenesis (egg-blastula) also induced strong upregulation of other neural genes that show a later oral expression at the gastrula stage (Fig. 5a). All neurogenic and neural marker genes analysed were suppressed at planula stage after the iCRT14 inhibitor treatment (Fig. 5b–d). The early stage-specific activation of the β-catenin pathway resulted in an ectopic development and increase in the number of both, RFa+ and GLWa+ neurons at the planula stage (Fig. 5e,f,i,j; Supplementary Fig. 11). The iCRT14 treatment confirmed the β-catenin requirement for the development of RFa+, GLWa+ and Elav::mOrange+ neurons (Fig. 5g–i,k,l). Thus, global and localized actions of β-catenin signalling are essential for the neural commitment of early embryonic cells and for the formation of a region with a high neurogenic potential that then develops into the oral nervous system.
Figure 5. β-Catenin signalling positions neuronal genes to the oral site.
(a) qPCR analysis of late oral neural genes in 2.5 μM ALP-treated gastrula (16 h.p.f.). (b) WISH analysis of oral neurogenic transcription factors in the planula treated without (dimethyl sulfoxide (DMSO)) or with 50 μM iCRT14. (c,d) qPCR analyses of the oral and lateral neural gene expression in the iCRT14-treated planulae. (e–h) Quantification of number of RFa+ (e,g) and GLWa+ (f,h) neurons in the planula larvae treated with GSK3β inhibitors (2.5 μM ALP, 0.5 μM BIO or 2.5 μM 1-AZA) up to the blastula stage (e,f) or treated with increasing concentrations of iCRT14 up to the planula stage (g,h). Neurons were counted at 3 d.p.f. (e,f) or 4 d.p.f. (g,h). (i–l) Development of the RFa+ and GLWa+ neurons (I,j) and NvElav1::mOrange+ neurons (k,l) in the planula treated with 2.5 μM ALP (up to blastula) or 50 μM iCRT14. (j) Distributions of RFa+ (left) and GLWa+ (right) neurons along the OA axis of planulae treated with GSK3β inhibitors (2.5 μM ALP, 0.5 μM BIO or 2.5 μM 1-AZA) up to the blastula stage were plotted (n=9 (ctrl.), n=9 (ALP), n=6 (BIO) and n=6 (1-AZA) for RFa; n=21 (ctrl.), n=10 (ALP), n=9 (BIO) and n=7 (1-AZA) for GLWa). (l) Quantification of number of NvElav1::mOrange+ neurons in the planula larvae treated with 2.5 μM ALP increasing concentrations of iCRT14. Shown in i and k are representative images of at least three with similar results. Scale bars, 100 μm. The red asterisks in b,i and k denote the blastopore. Bars represent the mean±s.e.m. of three experiments. Black asterisks denote statistical significance using the Student’s t-test (*P<0.05; **P<0.01).
Bmp signalling in late neural patterning at the oral side
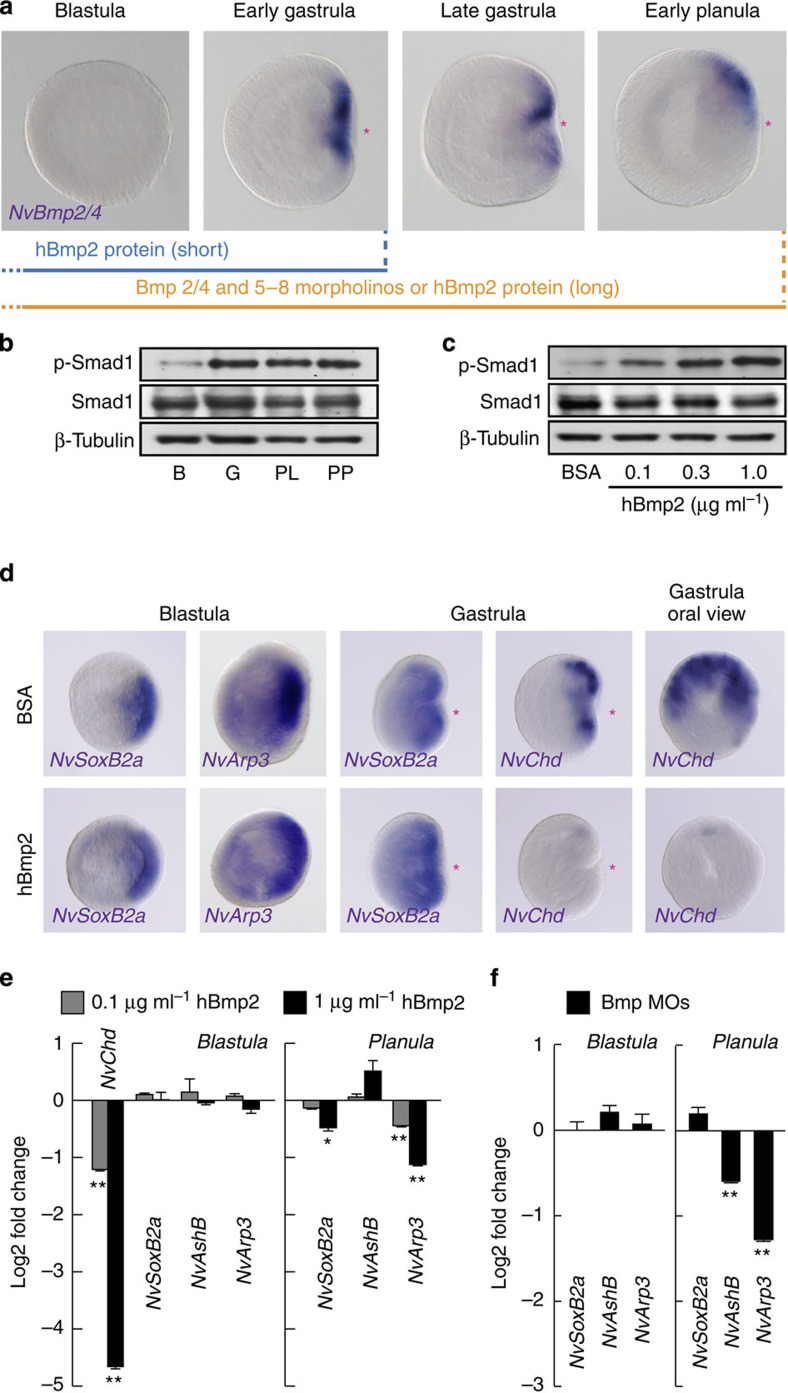
We next analysed the effect of Bmp signalling on the development of the oral nervous system in N. vectensis. The asymmetric expression of NvBmp2/4, NvBmp5–8 and their antagonist NvChordin became visible around the blastoporal lip only after the gastrula stage (Fig. 6a)37,38,54. Although an early expression of NvBmp2/4 and NvChordin was reported at the blastula stage by RT-PCR and microarray analyses37,55, no regionalized expression was detectable at this stage. The oral expression of NvBmp2/4, NvBmp5–8 and NvChordin was depending on the β-catenin signalling, as the expression was suppressed by iCRT14 (Supplementary Fig. 12). An augmentation of β-catenin signalling by 1-AZA induced upregulation of NvBmp2/4 and NvBmp5–8 expression, whereas NvChordin expression was inhibited, suggesting that the dual role of β-catenin signalling on the NvChordin expression. When we analysed the level of activated Bmp signalling by determining phosphorylation levels of the NvSmad1 C terminus, we found a strong signal at the gastrula stage (Fig. 6b), suggesting that the local expression of NvBmp2/4 and NvBmp5–8 in the gastrula is required for an activation of Bmp signalling. To test whether Bmp signalling functions in the delimited expression of early neurogenic TFs at the prospective blastoporal side of the blastula, we exposed early embryos to increasing concentrations of exogenous human Bmp2 protein (hBmp2) to globally elevate the Bmp pathway. hBmp2 treatment induced the phosphorylation of NvSmad1 in a dose-dependent manner (Fig. 6c). The expression of NvChordin was strongly suppressed by the augmented Bmp signalling (Fig. 6d)—as has been demonstrated in bilaterians and N. vectensis36—whereas the polarized expression of early oral neurogenic TFs at the future oral side of the blastula and gastrula was not altered (Fig. 6d). Consistently, development of the RFa+ and GLWa+ neurons was not affected by the early hBmp2 treatment (Supplementary Fig. 13). The hBmp2 suppression of NvChordin in the early embryos was confirmed by quantitative PCR (qPCR), whereas no significant effect was observed on the expression level of the early oral TFs (Fig. 6e). Elimination of Bmp signalling by co-injection of MOs for NvBmp2/4 and NvBmp5–8 (ref. 36) confirmed that the expression of the early neurogenic TFs is independent of Bmp signalling at the blastula stage (Fig. 6f).
Figure 6. Bmp-independent early neurogenesis.
(a) WISH analysis of orally expressed NvBmp2/4 and scheme of hBmp2 treatment and of knockdown using morpholinos against Bmp2/4 and Bmp5–8. (b,c) NvSmad1 phosphorylation at the blastula (B), gastrula (G), planula (P) and primary polyp (PP) stages (b) and in blastula treated with increasing concentrations of exogenous hBmp2 protein (c). Smad1 and β-tubulin proteins were used as internal control. (d) WISH analysis of the effect of hBmp2 treatment (1 μg ml−1) on the expression of NvSoxB2a, NvArp3 and NvChordin (NvChd). (e) qPCR analysis of the early neurogenic genes at the blastula (left) and planula (right) treated with increasing concentrations of hBmp2. (f) qPCR analysis of early neurogenic genes at the blastulae and planula larvae injected with MOs against NvBmp2/4 and NvBmp5–8. Red asterisks denote the blastopore and future oral pole. Bars represent the mean±s.e.m. of three experiments. Black asterisks denote statistical significance using the Student’s t-test (*P<0.05; **P<0.01).
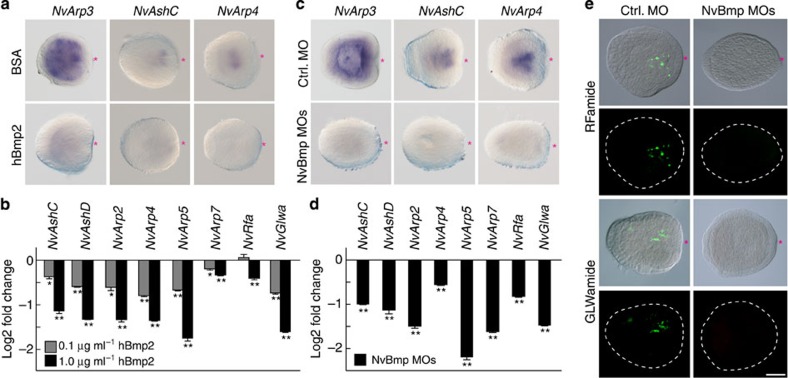
Finally, we extended the hBmp2 treatment up to the planula stage to test the effect of Bmp signalling on TF genes that are also expressed in late neurogenesis. Surprisingly, under these conditions, the expression of early oral NvArp3 and most of the late oral neurogenic TFs as well as NvGlwamide were largely reduced by exogenous hBmp2 in a concentration-dependent manner (Figs 6e and 7a,b). When we further analysed the expression of the same set of genes in Bmp morphants, we also observed a significant suppression of these early and late oral neurogenic genes and of NvRfamide and NvGlwamide (Figs 6f and 7c,d), which resulted in a decreased number of RFa+ and GLWa+ neurons at planula stage (Fig. 7e; Supplementary Fig. 14). These data indicate that Bmp signalling is positively and negatively involved in the development of the oral nervous system. This dual function of Bmp signalling is reminiscent to the Bmp-dependent transcriptional regulation of other oral genes36.
Figure 7. Bmp-dependent late neurogenesis.
(a–d) Expression analyses of oral neurogenic genes in the planulae treated with hBmp2 protein (a,b) or injected with MOs against NvBmp2/4 and NvBmp5–8 (c,d). WISH analysis shows the expression NvArp3, NvAshC and NvArp4 after hBmp2 (1.0 μg ml−1) treatment (a) or after Bmp MOs injection (c) at the planula stage. qPCR analyses show the expression of the oral neural genes at the planula larvae treated with increasing concentrations of hBmp2 (b) or injected with Bmp MOs (d). (e) Development of the RFa+ and GLWa+ neurons in NvBmp morphants. Shown in a,c and e are representative images of three experiments with similar results. Red asterisks indicate the blastopore. Scale bars, 100 μm. Bars represent the mean±s.e.m. of at least three experiments. Black asterisks denote statistical significance using the Student’s t-test (*P<0.05; **P<0.01).
NvArp6 induces the asymmetric pattern of NvGLWa+ neurons
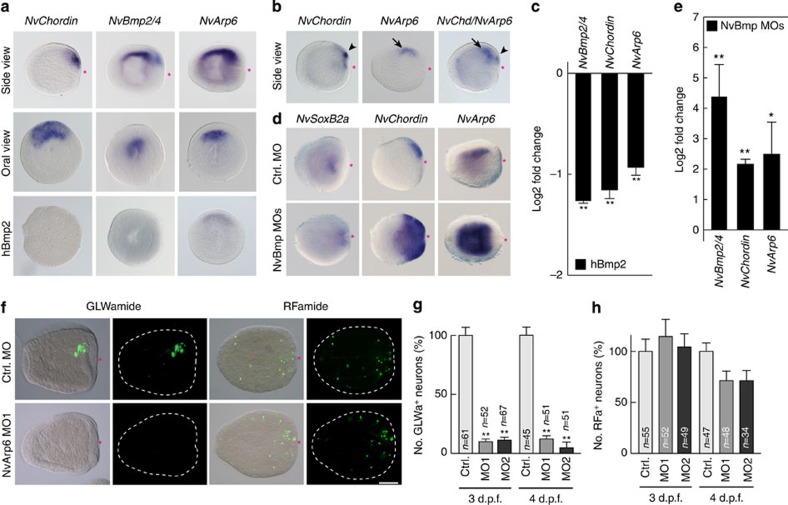
Among the TFs with a blastoporal expression pattern, NvArp6 was exceptional in that it exhibits a distinct asymmetric pattern along both body axes, that is, the oral–aboral and the directive axis (Fig. 8a). Double WISH analyses indicated that NvArp6 is expressed at the same side of the directive axis as NvChordin (Fig. 8b), and NvBmp2/4 and NvBmp5–8 (ref. 37). Different from NvChordin, which was restricted to the oral ectoderm, the gradient of NvBmp2/4, NvBmp5–8 and NvArp6 expression along the primary axis was reaching the aboral side of endoderm. It was proposed that the overlapping expression of the NvBmp ligands and its antagonist NvChordin on the same side of the embryo creates a complex gradient of Bmp activity along the primary and directive axes36. Accordingly, the overlapping expression domains of NvBmp2/4 and NvArp6 indicate a Bmp-mediated suppression of NvArp6 transcription. To test this hypothesis, we either activated or suppressed Bmp signalling and determined NvArp6 expression levels at the planula stage. NvArp6 expression was significantly suppressed by hBmp2 treatment and activated in NvBmp2/4/5–8 double morphants (Fig. 8a,c–e). Note that the NvSoxB2a was left unchanged in the Bmp-depleted planula larvae (Fig. 8d). These data clearly indicate that Bmp signalling has a suppressive function in inducing NvArp6 expression along the primary and directive axes.
Figure 8. Asymmetric differentiation of GLWa+ neurons by Bmp and NvArp6.
(a–e) WISH (a,b) shows the expression of NvChordin, NvBmp2/4 and NvArp6 on the same site along the directive axis at the planula stage. Arrowheads and arrows in b indicate expression of NvChordin and NvArp6, respectively. Influence of 1 μg ml−1 hBmp2 treatment on the expression of NvArp6, NvBmp2/4 and NvChordin is shown in the lower panel of a. (c) qPCR analysis of NvBmp2/4, NvChordin and NvArp6 in planulae treated with 1 μg ml−1 hBmp2. (d) WISH analysis of NvSoxB2a, NvChordin and NvArp6 expression in Bmp morphants. (e) qPCR analysis of NvBmp2/4, NvChordin and NvArp6 in Bmp morphants. Bars in c and e represent the mean±s.e.m. of three experiments. (f–h) NvArp6-dependent asymmetric formation of GLWa+ neurons. Immunocytochemical analysis of GLWa+ and RFa+ neurons (f) and quantification of GLWa+ (g) and RFa+ (h) neuron formation in NvArp6 morphants. The data in g and h represent the relative number of neurons. The bars represent the mean±s.e.m. Black asterisks denote statistical significance (*P<0.05; **P<0.01). Shown in a,b,d and f are representative images of three experiments with similar results. Red asterisks indicate blastopore. Scale bars, 100 μm.
We next analysed whether the asymmetric development of the GLWa+ neuronal subset along the directive axis was NvArp6 dependent. In NvArp6 morphants, we found an almost complete inhibition of GLWa+ neurons (Fig. 8f), whereas RFa+ neurons were not affected. A quantitative analysis using NvArp6 morphants confirmed this principal finding (Fig. 8g,h). These data clearly show that NvArp6 activity is specifically required for differentiation of GLWa+ neurons. Our data also indicate that the localized induction of NvArp6 by Bmp suppression demarcates a specific neurogenic region in the oral nervous system along directive axis to give rise to the GLWa+ neuronal subdomain.
Discussion
Here, we analysed neural development in embryos of the cnidarian sea anemone N. vectensis. We focused our study on the genetic mechanisms neuralizing embryonic cells and regulating the development of the oral nervous system, which forms a conspicuous nerve plexus around the mouth of many polyps8,10,30 and was repeatedly depicted as the precursor of the bilaterian CNS10,31,32. We used neuronal markers that are abundantly or exclusively expressed during the early development of the oral nervous system at the side of the blastopore of the embryo and planula larva8,12,13,14,33. By deciphering the consecutive actions of β-catenin and Bmp signalling in the development of the oral nervous system in N. vectensis embryos, our findings replicate the evolutionary emergence of these major signalling axes in animal evolution.
β-Catenin signalling is essential for gastrulation and oral development in N. vectensis34,35. In this study, we demonstrate that β-catenin is the early inducer of the oral nervous system of N. vectensis. β-Catenin signalling is highly activated at the future oral side at the blastula stage and even before (Supplementary Fig. 8)34, which causes an upregulation of early and late neurogenic and neural marker genes at the oral side, while inhibition of β-catenin showed opposite effects. Thus, β-catenin signalling is essential for early cell fate decisions of progenitor cells giving rise to multiple neural cell types. Since the target genes of β-catenin signalling, for example, NvBra and NvAshB, start being upregulated before canonical Wnt ligands NvWnt3 and NvWnt4 become detectable55, the higher activity of β-catenin signalling at the future oral side of early embryos might include NvDishevelled-mediated stabilization of β-catenin protein at the oral region56.
Another intriguing finding revealed in this study is the β-catenin dependency of expression of the early lateral neural genes NvAshA, NvSoxB2c and NvElav1 at the blastula stage. Although it is yet unknown how the locally activated β-catenin signalling contributes to the early expression of these genes at the blastula lateral epithelium (for example, we cannot rule out that the effects of β-catenin in neuronal cell formation are indirect), our findings may suggest that β-catenin protein that is expressed uniformly in early blastomeres has a role to predispose the blastula epithelium to become neurogenic, as it has been suggested by previous studies on vertebrates57. Our data support observations made in distinct bilaterians showing a function of β-catenin signalling in early neurodevelopment29,57,58,59,60.
When we tested the function of Bmp signalling on the early induction of oral neuronal genes, we could not detect any inhibitory Bmp effect, which was unexpected since Bmp/Chordin signalling is frequently considered as the primary and general CNS inducer15,20. Our findings are, however, in accord with data on early neural development in several slow-evolving bilaterians. In annelids22 and in hemichordates24, Bmp treatments did not inhibit early neurogenic gene expression. We therefore conclude that the ancestral mode of neural induction is probably not the Bmp-mediated event, which was described for Xenopus and Drosophila21. Instead, the β-catenin signalling dependency of neural development at the blastoporal site appears to represent the evolutionary conserved and ancestral mode of neural development with a deep root in the common ancestor of cnidarians and bilaterians.
Treatment of gastrulae and planula larvae with inhibitors of GSK3 (for example, ALP or 1-AZA) triggered β-catenin activation and the expansion of oral markers including RFa+ neurons6, or even the formation of multiple polyp heads52. This clearly suggests an additional function of Wnt/β-catenin signalling for maintaining and/or patterning the cnidarian nervous system. Since the oral end of the cnidarian body plan is considered to represent the posterior end of the bilaterian AP body axis61,62,63, Wnt/β-catenin signalling might also be involved in the oral–aboral patterning of the pervasive cnidarian nerve net. Bmp signalling was also postulated, however, to have a role in maintaining RFa+ neurons at the planula stage36. How can these observations fit with our principal finding that Bmp signalling is indispensable for development of the oral nervous system in N. vectensis? When we analysed Bmp signalling at the planula stage, we discovered that RFa+ and GLWa+ neurons as well as early and late oral neurogenic TFs (NvArp2, -3, -4, -5, -7, NvAshB, -C, -D) exhibited indeed a distinct Bmp sensitivity. Only for NvAshB this Bmp dependency was weak, and it was absent for NvSoxB2a. Therefore, Bmp signalling is an important factor in the development of the oral nervous system. However, this Bmp regulation is complex. When Bmp signalling was either suppressed or over-activated at gastrula and planula stages, neurogenic TFs and GLWamide neuropeptide became downregulated under both conditions. This is reminiscent to the blastoporal regulation of NvSoxB1 and NvNk2.1 that are likewise downregulated in NvChordin and NvBmp2/4 morphants36. Thus, a balanced level of Bmp signalling seems to be crucial for the development of the oral nervous system. Because the expression of NvBmp2/4, NvBmp5–8 and NvChordin genes is dependent on β-catenin signalling, the Wnt/β-catenin regulation of neural patterning might be mediated, at least in part, by the Bmp activity. This point is of interest for future studies.
A striking function of Bmp signalling at the planula stage is the asymmetric induction of neurons along the (secondary) directive axis. NvArp6-expressing cells and GLWa+ neurons form a distinct asymmetric pattern along the directive axis, which is Bmp dependent. This asymmetric suppression of Bmp is reminiscent to an evolutionarily conserved Bmp function in the mediolateral patterning of the trunk nervous system in bilaterians22. It should be pointed out that Bmp inhibition (which activates NvArp6 but suppresses other oral neuronal TFs) did not result in more and symmetric development of GLWa+ neurons. Therefore, GLWa+ neurons depend on additional neurogenic TFs, which are similarly restricted to the oral site.
Our data provide a new view on the evolution of the nervous system, and we propose that the cnidarian oral nervous system exhibits some important features that are essential for the evolution of the bilaterian CNS (Fig. 9). (i) The oral nervous system is demarcated from other parts of the diffused nervous system and exhibits a distinct condensation of neuronal perikaria with neuronal processes extending to the periphery (tentacles and aboral side). The condensations of neuronal perikaria in the oral nervous system is an anatomical feature related to the formation of a CNS in bilaterians with distinct and functionally specialized neuronal domains (ganglia)64. (ii) Development of the cnidarian oral nervous system is dependent on early β-catenin- and late Bmp signalling activities. The function of these signalling pathways in the oral nervous system development is highly reminiscent to their function in the specification of neural cell types in the CNS along bilaterian body axes. (iii) There is an evidence for a common set of orthologous neuronal genes expressed in the cnidarian oral nervous system and the bilaterian CNS. The neuropeptides belonging to R[F/Y]amide and [G/V/L]Wamide groups have been shown to form clusters in bilaterian CNS41,42. In addition to neurogenic TFs used in this study, some of marker genes for bilaterians CNS (that is, SoxB1 and Nk2.1) are exclusively expressed in the oral nervous system in N. vectensis12,36. These findings suggest that the genetic signature of the cnidarian oral nervous system is shared, at least in part, by that of the bilaterian CNS.
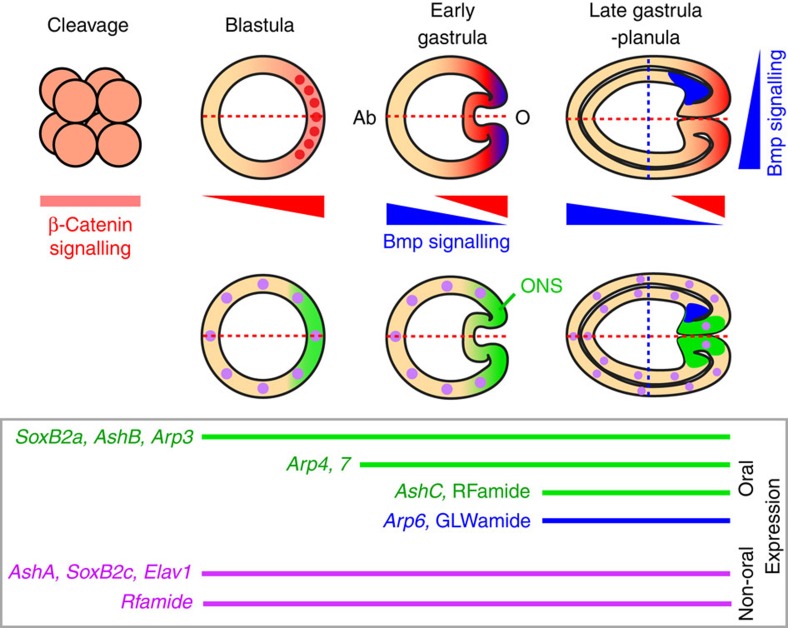
Figure 9. β-Catenin and Bmp regulate early Nematostella neurogenesis.
The embryonic regions of the gradient of β-catenin activation (red), Bmp activation (blue) and oral nervous system (ONS) (green) in different embryonic stages of N. vectensis are shown. β-Catenin protein is observed in all blastomeres at the early-cleavage stage, and stabilized more at the prospective oral side (O) than aboral side (Ab) along the primary axis (red dashed line) of the blastula, which induces the expression of the early neurogenic genes (NvSoxB2a, NvAshB and NvArp3) as well as NvBmp2/4, NvBmp5–8 and NvChordin at the oral region. β-Catenin-translocated nuclei at the presumptive blastopore region are shown in red dots. During gastrulation, Bmp signalling starts to be involved in the development of the oral nervous system with respect to the primary and secondary (blue dashed line) body axes. At the planula stage, asymmetric suppression of Bmp activity along the secondary axis becomes indispensable for the expression of NvArp6 and development of GLWa+ neurons at the one side of the oral endoderm.
Most recently the genome of comb jellies (ctenophores), another pre-bilaterian clade with a primitive nervous system, was published65,66. Many neuronal genes are missing in ctenophores, for example, those for the synthesis and transmission of the classical neurotransmitters GABA and acetylcholine. However, several of the ‘neurogenic’ TFs (for example, SoxB and bHLH) and RNA-binding proteins (Elav and Musashi) are present in ctenophore genomes65,66. In the ctenophore, SoxB and RFamide are strongly expressed at neuron-enriched body regions, that is, the aboral sensory organ and the polar fields67,68. In addition, members of Bmp and β-catenin signalling are also highly expressed at this region during development69,70. The fact that these markers are expressed during the neural development in ctenophores is questioning, whether the nervous system of ctenophores had really a different evolutionary origin from the cnidarian-bilaterian clade as recently proposed66.
To understand the emergence of the bilaterian CNS from simple neural clusters, deeper insights into the anatomical and molecular features of the cnidarian nervous systems are very helpful. Although this knowledge is still far from being complete, our data on the development of the oral nervous system in N. vectensis give an insight how a first step in the evolution of nervous system centralization may have been accomplished. The cnidarian nervous system is also an intriguing example, how basic positional information, which is organized in bilaterians in a Cartesian coordinate system26, can be translated into the development of the nervous system.
Methods
Culture of N. vectensis
N. vectensis were cultured at a salinity one-third of normal seawater (that is, brackish water, 108.32 g l−1 Tropic Marine sea salt, pH 7.6). Adult males and females were separately cultured at 18 °C in the dark and fed two to three times per week. On 2 days after the feeding, the culture boxes were washed. For spawning the release of eggs and sperms, the culture boxes were incubated at 26 °C under the light for at least 14 h. After fertilization, embryos were cultured at 20 °C.
Gene isolation from N. vectensis and plasmid construction
Gene-specific primers were then designed to recover both 3′ and 5′ rapid amplification of complementary DNA (cDNA) ends (RACE) fragments using the GeneRacer kit (Invitrogen) with annealing temperatures between 55 and 65 °C for 1st and nested PCR. Gene-specific primer sequences are listed in Supplementary Table 1. RACE products were cloned using pGEM-T (Promega) and sequenced. Overlapping 5′and 3′ RACE fragments were aligned to obtain mRNA sequences. The DN form of GSK3β (K83R) was made by two-step PCR-based mutagenesis. The PCR product was inserted into the pCS2 plasmid and sequenced. For the TnT assays, NvSoxB2a, NvAshB, NvArp3 and NvArp6 genes were subcloned with partial 5′ untranslated region (UTR) and the complete open-reading frame into pGEM-T vectors and sequenced. The insert was cut using EcoRI and inserted into the pCS2 plasmid.
Quantitative and semi-quantitative RT-PCR
For semi-quantitative RT-PCR, the level of amplified product was compared in the exponential range of amplification. qRT-PCR was performed on the Bio-Rad DNAEngine equipped with Chromo4 real-time PCR detector (Bio-Rad) using Absolute QPCR SYBR Green mix (Thermo Scientific). The primer sequences are listed in Supplementary Table 1.
Embryo treatment with Bmp protein or GSK3β inhibitors
Embryos were treated with recombinant hBmp2 (a gift from Dr Wölfl) at concentrations from 0.1 to 1 μg ml−1. As a control, 1 μg ml−1 bovine serum albumin (BSA) was applied. hBmp2 protein was added at one-two-cell-stage embryos and cDNAs were prepared at the blastula (12–14 h.p.f.), gastrula (30 h.p.f.) or planula (74 h.p.f.) stages. For WISH experiments, hBmp-treated blastulae and gastrulae (12 h.p.f. (blastula) and 24 h.p.f. (gastrula) were fixed. For experiments using the GSK3β inhibitors to enhance β-catenin signalling, embryos were treated with ALP, BIO or 1-AZA (Sigma) at the concentrations indicated. cDNAs were prepared at the blastula and early gastrula stages (12–18 h.p.f.) for the qPCR analyses. For immunostaining of neuropeptides and mOrange protein, embryos were treated with GSK3β inhibitors from one- and two-cell stage to blastula stage. After being washed, the embryos were cultured in Nematostella medium until early (72 h.p.f.) or late (98 h.p.f.) planula stages. To pharmacologically inhibit β-catenin signalling, embryos were treated with 10 μM or 50 μM iCRT14 (Sigma), a Tcf/β-catenin inhibitor, for 14 h (blastula), 3 days (early planula) or 4 days (late planula).
Microinjection of mRNA and MO antisense oligonucleotides
pCS2 constructs, pCS2-MT-β-Eng (DN-β-catenin) (a gift from Dr McCrea), pCS2-NvBmp2/4 and DN-GSK3β, were linearized using appropriate restriction enzymes. Capped mRNAs were synthesized using the SP6 mMessage mMachine mRNA synthesis kit (Ambion). Capped mRNAs or MO antisense oligonucleotide for control (5′- CCTCTTACCTCAGTTACAATTTATA ), neurogenic TFs NvSoxB2a (MO1: 5′- TCTAAATCCCGTAGAAGTTCTAGGT -3′, MO2: 5′- CGTCAAGTTTACTCGTTCCGGCACT -3′), NvAshB (MO1: 5′- AGGAAGCCTCCATCGGAATCTCCAT -3′, MO2: 5′- TTGTAAGCGAGAAGCACTCACATGC -3′), NvArp3 (MO1: 5′- TTTCTGTACCTTCGCCTTTTGTCAT -3′, MO2: 5′- CTGAGAGCCGGTGAGTTTTCTTGAT -3′), NvArp6 (MO1: 5′- TCATTGAGTGAGTGCATCCGGCTTC -3′, MO2: 5′- GCGCACTCTCTGTTTGATCGTAAGT -3′), NvBmp2/4 (5′- GTAAGAAACAGCGTAAGAGAAGCAT -3′)36, NvBmp5–8 (5′- GTAACAGGTCTCGTATTCTCCGCAT -3′)36 and NvTcf (5′- CTGAGGCATACTACCGTCTCATGTG -3′)55 were injected at 250 μM concentration into fertilized eggs with 0.2 μg μl−1 rhodamine- or Alxa488-dextran (Invitrogen). Embryos were then rinsed with N. vectensis medium and incubated until desired stages.
TnT assay
The translation-blocking MOs were tested using the TnT SP6 quick coupled transcription/translation system (Promega). pCS2 plasmids (250–500 ng) containing MO-target sites (5′ UTR and first ATG) were incubated with control or gene-specific MOs (50 μM) at 30 °C for 90 min. The biotinylated proteins synthesized were detected by western blotting using horseradish peroxidase-conjugated streptavidin (1:10,000 dilution).
Whole-mount in situ hybridization
In situ hybridization was performed as described previously37 with following modifications: specimens were fixed with 4% paraformaldehyde/PBST (PBS, 0.1% Tween 20) for 1 h, then washed with methanol for 3 times and stored at −20 °C. Hybridization of 0.6- to 1.4-kb digoxygenin-labelled antisense RNA probes were carried out using a hybridization solution containing 1% SDS at 50–65 °C for at least 22 h. For post-hybridization washes, specimens were washed by serial dilutions (75, 50 and 25%) of hybridization solution with 2 × SSC at 55 °C. After processing of the digoxygenin-labelled probe with BM purple (Roche), specimens were washed with PBST.
Western blotting
N. vectensis embryos were solubilized with 2 × SDS–polyacrylamide gel electrophoresis sample buffer and immediately boiled at 90 °C for 5 min. After being sonicated, proteins separated by SDS–polyacrylamide gel electrophoresis were transferred to a PVDF (polyvinylidene difluoride) membrane (Millipore). The membranes were blocked with 5% BSA in buffer consisting of 50 mM Tris-HCl, pH 8.0, 2 mM CaCl2, 80 mM NaCl and 0.2% Nonidet P-40, incubated with the blocking solution containing anti-phospho-Smad1/5/8 antibody (Cell Signaling Technology) (1:200 dilution), anti-Smad1/5/8 (N-18) antibody (Santa Cruz Biotechnology) (1:300 dilution) or anti-β-tubulin (2-28-33) antibody (Sigma) (1:1,000 dilution) followed by incubation with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch) (1:5,000 dilution) in blocking solution. Immunoreactive proteins were detected with an ECL immunoblotting detection reagent (Amersham Pharmacia Biotech). Full images of western blots are shown in Supplementary Fig. 15.
Immunostaining and fluorescent microscopy
The NvElav1::mOrange transgenic embryos were fixed with 4% paraformaldehyde/PBT for 1 h at room temperature and stained using anti-DsRed antibody (1:300 dilution)(Clontech), as was previously reported14. For neuropeptide staining, embryos and primary polyps were fixed by Zamboni’s fixative o/n at 4 °C. After permeabilization in PBS+0.1% Triton X-100, specimens were blocked with blocking solution (10 mg ml−1 BSA+5% normal goat serum in PBS), incubated with anti-FMRFamide antibody (1:600 dilution) (Sigma) or anti-GLWamide antibody (1:400 dilution) (a gift from Dr Koizumi) in the blocking solution containing 0.1% Triton X-100, and then with Alexa488- or Alexa568-conjugated anti-Rabbit antibody (1:800 dilution) (Invitrogen). Specimens were imaged using a Nikon ECLIPSE 80i fluorescent microscope equipped with DIGITAL SIGH DSU1 (Nikon). For confocal microscopy, a fluorescent microscope ECLIPSE Ti (Nikon) equipped with an A1R MP confocal scanner (Nikon) was used.
Author contributions
H.W. performed most of the experiments. A.K. and M.F. performed some WISH analyses. H.W., K.A., S.Ö., T.F. and T.W.H. designed the project. H.W. and T.W.H. wrote the manuscript.
Additional information
How to cite this article: Watanabe, H. et al. Sequential actions of β-catenin and Bmp pattern the oral nerve net in Nematostella vectensis. Nat. Commun. 5:5536 doi: 10.1038/ncomms6536 (2014).
Supplementary Material
Supplementary Figures 1-15 and Supplementary Table 1
Acknowledgments
We thank Fabian Rentzsch and Ulrich Technau for the NvElav1::mOrange transgenic animals, Pierre McCrea for the pCS2-MT-β-Eng plasmid, Arne Kusserow for the pCS2-NvBmp2/4 plasmid, Stefan Wölfl for hBmp2 protein, Osamu Koizumi for the anti-GLWamide antibody; Bianca Bertulat, Barbara Kostron, Chiemi Nishimiya-Fujisawa, Mihaela Zigman, Martin Ritthaler and Ildiko Somorjai for discussions and comments on the manuscript; Hiroe Yuzawa-Watanabe for technical assistance. H.W. was supported by an initial fellowship from the TOYOBO Biotechnology Foundation and the Alexander von Humboldt Foundation. This study was also supported by the Heidelberg Excellence Cluster CellNetworks and funded by grants from the German Science Foundation (DFG) to S.Ö. (FOR1036/TP2) and to T.W.H. (SFB 488/A12; HO-1088/6; and FOR 1036/TP1).
References
- Kirschner M. & Gerhart J. Evolvability. Proc. Natl Acad. Sci. USA 95, 8420–8427 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland L. Z. et al. Evolution of bilaterian central nervous systems: a single origin? Evodevo 4, 27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. J. et al. Estimating metazoan divergence times with a molecular clock. Proc. Natl Acad. Sci. USA 101, 6536–6541 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam N. H. et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (2007). [DOI] [PubMed] [Google Scholar]
- Galliot B. et al. Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2–24 (2009). [DOI] [PubMed] [Google Scholar]
- Marlow H. Q., Srivastava M., Matus D. Q., Rokhsar D. & Martindale M. Q. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev. Neurobiol. 69, 235–254 (2009). [DOI] [PubMed] [Google Scholar]
- Technau U. & Steele R. E. Evolutionary crossroads in developmental biology: Cnidaria. Development 138, 1447–1458 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Fujisawa T. & Holstein T. W. Cnidarians and the evolutionary origin of the nervous system. Dev. Growth Differ. 51, 167–183 (2009). [DOI] [PubMed] [Google Scholar]
- Chapman J. A. et al. The dynamic genome of Hydra. Nature 464, 592–596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi O. Nerve ring of the hypostome in hydra: is it an origin of the central nervous system of bilaterian animals? Brain Behav. Evol. 69, 151–159 (2007). [DOI] [PubMed] [Google Scholar]
- Fujisawa T. Hydra peptide project 1993-2007. Dev. Growth Differ. 50, (Suppl 1)): S257–S268 (2008). [DOI] [PubMed] [Google Scholar]
- Magie C. R., Pang K. & Martindale M. Q. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev. Genes Evol. 215, 618–630 (2005). [DOI] [PubMed] [Google Scholar]
- Layden M. J., Boekhout M. & Martindale M. Q. Nematostella vectensis achaete-scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development 139, 1013–1022 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N., Renfer E., Technau U. & Rentzsch F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 139, 347–357 (2012). [DOI] [PubMed] [Google Scholar]
- De Robertis E. M. & Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu. Rev. Cell Dev. Biol. 20, 285–308 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E. Anti-neural-inhibition: a conserved mechanism for neural induction. Cell 89, 681–684 (1997). [DOI] [PubMed] [Google Scholar]
- Mizutani C. M., Meyer N., Roelink H. & Bier E. Threshold-dependent BMP-mediated repression: a model for a conserved mechanism that patterns the neuroectoderm. PLoS Biol. 4, e313 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina M. D., Salo E. & Cebria F. The BMP pathway is essential for re-specification and maintenance of the dorsoventral axis in regenerating and intact planarians. Dev. Biol. 311, 79–94 (2007). [DOI] [PubMed] [Google Scholar]
- Saudemont A. et al. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 6, e1001259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R. Neural induction. Curr. Opin. Genet. Dev. 10, 357–362 (2000). [DOI] [PubMed] [Google Scholar]
- Stern C. D. Neural induction: old problem, new findings, yet more questions. Development 132, 2007–2021 (2005). [DOI] [PubMed] [Google Scholar]
- Denes A. S. et al. Molecular architecture of annelid nerve cord supports common origin of nervous system centralization in bilateria. Cell 129, 277–288 (2007). [DOI] [PubMed] [Google Scholar]
- Darras S. & Nishida H. The BMP signaling pathway is required together with the FGF pathway for notochord induction in the ascidian embryo. Development 128, 2629–2638 (2001). [DOI] [PubMed] [Google Scholar]
- Lowe C. J. et al. Dorsoventral patterning in hemichordates: insights into early chordate evolution. PLoS Biol. 4, e291 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa F. & Marti E. Wnt won the war: antagonistic role of Wnt over Shh controls dorso-ventral patterning of the vertebrate neural tube. Dev. Dyn. 239, 69–76 (2010). [DOI] [PubMed] [Google Scholar]
- Niehrs C. On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137, 845–857 (2010). [DOI] [PubMed] [Google Scholar]
- Wei Z., Yaguchi J., Yaguchi S., Angerer R. C. & Angerer L. M. The sea urchin animal pole domain is a Six3-dependent neurogenic patterning center. Development 136, 1179–1189 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darras S., Gerhart J., Terasaki M., Kirschner M. & Lowe C. J. Beta-catenin specifies the endomesoderm and defines the posterior organizer of the hemichordate Saccoglossus kowalevskii. Development 138, 959–970 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demilly A., Steinmetz P., Gazave E., Marchand L. & Vervoort M. Involvement of the Wnt/beta-catenin pathway in neurectoderm architecture in Platynereis dumerilii. Nat. Commun. 4, 1915 (2013). [DOI] [PubMed] [Google Scholar]
- Davis L. E., Burnett A. L. & Haynes J. F. Histological and ultrastructural study of the muscular and nervous system in Hydra. 2. Nervous system. J. Exp. Zool. 167, 295–331 (1968). [DOI] [PubMed] [Google Scholar]
- Hyman H. L. The Invertebrates: Protozoa Through Ctenophora Vol. 1 McGraw-Hill (1940). [Google Scholar]
- Arendt D., Denes A. S., Jékely G. & Tessmar-Raible K. The evolution of nervous system centralization. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1523–1528 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C. et al. Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol. Biol. 8, 311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake A. H. et al. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature 426, 446–450 (2003). [DOI] [PubMed] [Google Scholar]
- Kusserow A. et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 433, 156–160 (2005). [DOI] [PubMed] [Google Scholar]
- Saina M., Genikhovich G., Renfer E. & Technau U. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc. Natl Acad. Sci. USA 106, 18592–18597 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentzsch F. et al. Asymmetric expression of the BMP antagonists chordin and gremlin in the sea anemone Nematostella vectensis: Implications for the evolution of axial patterning. Dev. Biol. 296, 375–387 (2006). [DOI] [PubMed] [Google Scholar]
- Matus D. Q., Thomsen G. H. & Martindale M. Q. Dorso/ventral genes are asymmetrically expressed and involved in germ-layer demarcation during cnidarian gastrulation. Curr. Biol. 16, 499–505 (2006). [DOI] [PubMed] [Google Scholar]
- Grimmelikhuijzen C. J. & Hauser F. Mini-review: the evolution of neuropeptide signaling. Regulatory peptides 177, S6–S9 (2012). [DOI] [PubMed] [Google Scholar]
- Jekely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc. Natl Acad. Sci. USA 110, 8702–8707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessmar-Raible K. et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: insights into hypothalamus evolution. Cell 129, 1389–1400 (2007). [DOI] [PubMed] [Google Scholar]
- Conzelmann M. & Jekely G. Antibodies against conserved amidated neuropeptide epitopes enrich the comparative neurobiology toolbox. Evodevo 3, 23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. G. Recent insights into cnidarian phylogeny. Smithson. Contrib. Mar. Sci. 38, 139–149 (2009). [Google Scholar]
- Schierwater B. et al. Concatenated analysis sheds light on early metazoan evolution and fuels a modern "urmetazoon" hypothesis. PLoS Biol. 7, e20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berking S. Generation of bilateral symmetry in Anthozoa: a model. J. Theor. Biol. 246, 477–490 (2007). [DOI] [PubMed] [Google Scholar]
- Seipp S. et al. Neuronal cell death during metamorphosis of Hydractina echinata (Cnidaria, Hydrozoa). Invert. Neurosci. 10, 77–91 (2010). [DOI] [PubMed] [Google Scholar]
- Royo J. L. et al. Transphyletic conservation of developmental regulatory state in animal evolution. Proc. Natl Acad. Sci. USA 108, 14186–14191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg M., Kallstrom M. & Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 8, 995–1001 (2005). [DOI] [PubMed] [Google Scholar]
- Technau U. Brachyury, the blastopore and the evolution of the mesoderm. Bioessays 23, 788–794 (2001). [DOI] [PubMed] [Google Scholar]
- Lee P. N., Pang K., Matus D. Q. & Martindale M. Q. A. WNT of things to come: Evolution of Wnt signaling and polarity in cnidarians. Semin. Cell Dev. Biol. 17, 157–167 (2006). [DOI] [PubMed] [Google Scholar]
- Marlow H., Matus D. Q. & Martindale M. Q. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis. Dev. Biol. 380, 324–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guder C. et al. The Wnt code: cnidarians signal the way. Oncogene 25, 7450–7460 (2006). [DOI] [PubMed] [Google Scholar]
- Gonsalves F. C. et al. An RNAi-based chemical genetic screen identifies three small-molecule inhibitors of the Wnt/wingless signaling pathway. Proc. Natl Acad. Sci. USA 108, 5954–5963 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnerty J. R., Pang K., Burton P., Paulson D. & Martindale M. Q. Origins of bilateral symmetry: Hox and dpp expression in a sea anemone. Science 304, 1335–1337 (2004). [DOI] [PubMed] [Google Scholar]
- Rottinger E., Dahlin P. & Martindale M. Q. A framework for the establishment of a cnidarian gene regulatory network for "endomesoderm" specification: the inputs of ss-catenin/TCF signaling. PLoS Genet. 8, e1003164 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P. N., Kumburegama S., Marlow H. Q., Martindale M. Q. & Wikramanayake A. H. Asymmetric developmental potential along the animal-vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Dishevelled. Dev. Biol. 310, 169–186 (2007). [DOI] [PubMed] [Google Scholar]
- Stern C. D. Neural induction: 10 years on since the ‘default model’. Curr. Opin. Cell Biol. 18, 692–697 (2006). [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y. et al. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131, 2791–2801 (2004). [DOI] [PubMed] [Google Scholar]
- Valenta T. et al. Probing transcription-specific outputs of beta-catenin in vivo. Genes Dev. 25, 2631–2643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H., Wessely O. & De Robertis E. M. Neural induction in Xenopus: requirement for ectodermal and endomesodermal signals via Chordin, Noggin, beta-Catenin, and Cerberus. PLoS Biol. 2, E92 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein T. W., Watanabe H. & Ozbek S. Signaling pathways and axis formation in the lower metazoa. Curr. Top. Dev. Biol. 97, 137–177 (2011). [DOI] [PubMed] [Google Scholar]
- Holstein T. W. The Evolution of the Wnt pathway. Cold Spring Harb. Perspect. Biol. 4, a007922 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce J. C. & Holstein T. W. inWnt Signaling in Development and Disease: Molecular Mechanisms and Biological Functions eds Hoppler S., Moon C. S. 161–176John Wiley & Sons (2014). [Google Scholar]
- Northcutt R. G. Evolution of centralized nervous systems: two schools of evolutionary thought. Proc. Natl Acad. Sci. USA 109, (Suppl 1)): 10626–10633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan J. F. et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science 342, 1242592 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz L. L. et al. The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager M., Queinnec E., Chiori R., Le Guyader H. & Manuel M. Insights into the early evolution of SOX genes from expression analyses in a Ctenophore. J. Exp. Zool. B Mol. Dev. Evol. 310, 650–667 (2008). [DOI] [PubMed] [Google Scholar]
- Jager M. et al. New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Muller, 1776). J. Exp. Zool. B Mol. Dev. Evol. 316B, 171–187 (2011). [DOI] [PubMed] [Google Scholar]
- Pang K., Ryan J. F., Mullikin J. C., Baxevanis A. D. & Martindale M. Q. Genomic insights into Wnt signaling in an early diverging metazoan, the ctenophore Mnemiopsis leidyi. Evodevo 1, 10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang K., Ryan J. F., Baxevanis A. D. & Martindale M. Q. Evolution of the TGF-beta signaling pathway and its potential role in the ctenophore, Mnemiopsis leidyi. PloS ONE 6, e24152 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-15 and Supplementary Table 1