Abstract
The human rhodopsin gene is the locus for numerous alleles linked to the neurodegenerative disease retinitis pigmentosa. To facilitate the study of retinal degeneration and to test reagents designed to alter the structure and function of this gene, we have developed strains of mice whose native rhodopsin gene has been replaced with the corresponding human DNA modified to encode an enhanced GFP fusion at the C terminus of rhodopsin. The human rhodopsin-GFP fusion faithfully mimics the expression and distribution of wild-type rhodopsin in heterozygotes and serves as a sensitive reporter of rod-cell structure and integrity. In homozygotes, however, the gene induces progressive retinal degeneration bearing many of the hallmarks of recessive retinitis pigmentosa. When the gene is flanked by recognition sites for Cre recombinase, protein expression is reduced ≈5-fold despite undiminished mRNA levels, suggesting translation inhibition. GFP-tagged human rhodopsin provides a sensitive method to monitor the development of normal and diseased retinas in dissected samples, and it offers a noninvasive means to observe the progress of retinal degeneration and the efficacy of gene-based therapies in whole animals.
The structure and function of rhodopsin and the gene encoding it have been the subjects of intense scrutiny for many years because rhodopsin serves as a useful model for understanding the largest receptor family in the human genome, the G protein-coupled receptors, and because defects in the rhodopsin gene are the most common cause of the most common inherited blinding disease, retinitis pigmentosa (1, 2). Retinitis pigmentosa is a progressive neurodegenerative disorder. It begins with death of rod photoreceptor cells, which are the only cells in the retina to express rhodopsin and which express it as their most abundant protein. Eventually, loss of rods leads to loss of cone photoreceptors, the mainstay of human vision. Animal models of retinal degeneration not only serve as models of human retinitis pigmentosa (2, 3), but they can also be instructive about more general features of progressive neurodegeneration (4).
An interesting and challenging feature of the link between the rhodopsin gene and retinal degeneration is that most alleles display dominant inheritance. Mice and human patients heterozygous for null alleles of the rhodopsin gene do not suffer from serious degeneration (5-7), even though mice with this genotype express only about half of the normal amount of rhodopsin (6). Thus, it is the presence of rhodopsin encoded by the dominant allele that leads to degeneration, but it remains an open question whether its deleterious consequences are due to toxic gain-of-function or interfering dominant-negative effects (3). In either case, repair, inactivation, or inhibition of the mutant allele would be desirable, and in some cases, gene-based therapy may be the only effective approach to slowing or preventing progression of the disease. Thus, the rhodopsin gene may also serve as a model for developing therapeutic approaches to dominant disorders based on targeting genes in terminally differentiated neurons.
We have shown (8) that sequence-specific reagents based on triple-helix formation can be used to inactivate the rhodopsin gene in cultured human cells. To facilitate the assessment of this approach and other gene-based strategies in living rod cells, we have generated lines of mice in which the endogenous mouse rhodopsin gene has been replaced with the corresponding portions of the human rhodopsin gene. The human gene includes all its introns and exons, as well as DNA encoding an enhanced GFP (EGFP) fusion at the C terminus of rhodopsin. By making knock-ins of the genomic human gene at the normal mouse locus, we hoped to ensure proper control of gene expression, in contrast to the variable expression often obtained by using transgenes with portions of the 5′ regulatory sequences (9, 10).
The human rhodopsin-GFP fusion used in ref. 8 allows easy visualization of expression in individual cells or through the lens of living mice. EGFP fusions with rhodopsin have been expressed (11, 12) in transgenic Xenopus laevis rods without apparent ill effect, but in these cases, expression was at tracer levels in the presence of normal amounts of endogenous rhodopsin (12). Here, we have tested the effect of expression of the rhodopsin-GFP fusion protein at 16% or 80% of the endogenous levels in the presence and absence of the normal mouse rhodopsin. We find that they produce a mouse model for autosomal recessive retinal degeneration. By creating two different knock-in lines, with different expression levels, we also have the opportunity to investigate the effects of protein levels on the severity and rapidity of disease progression and to address the mechanism of cell death.
Materials and Methods
Modification of the Rhodopsin Locus in Mouse Embryonic Stem (ES) Cells. The rhodopsin locus in the HPRT- ES cell line AB2.2 123, derived from mouse strain 129SvEv, was modified in two steps to contain a 7.4-kb SacI-HindIII fragment of the human rhodopsin gene fused to EGFP (8) in place of the mouse rhodopsin gene (Fig. 1A). Details are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. The human rhodopsin-GFP sequences in the three different constructs were flanked only by mouse sequences, loxP and lox511 sites, or loxH and lox511 sites. In the first targeting step, the mouse rhodopsin gene was replaced by an HPRT minigene (13), which was replaced in the second step by a human rhodopsin-GFP gene. Colonies that survived selection were screened by Southern blotting with 3′ and 5′ probes (Fig. 1B).
Fig. 1.
Gene targeting and analysis of ES cells and mice. (A) Strategy for knocking in a human rhodopsin-GFP fusion gene in place of the mouse rhodopsin gene. The mouse rhodopsin locus is shown in black, with exons and polyadenylation sites indicated by small rectangles and stars, respectively. The HPRT minigene (gray) is driven by the phosphoglycerate kinase (PGK) promoter. The human rhodopsin-GFP fusion gene is shown in white, with the GFP portion crosshatched. Small arrows show the points of fusion between the mouse sequences and the HPRT or human rhodopsin sequences. White inverted triangles indicate locations of lox sites. P/H refers to two different constructs: one construct carrying an upstream LoxP site and the other construct carrying an upstream LoxH site. HR, homologous recombination; SR, segmental replacement. (B) Southern-blot analysis of ES cells and germline mice. Restriction enzymes used to digest genomic DNA are given in parentheses, and fragment sizes are given in kilobases. Both 5′- and 3′-specific probes were used to verify the structure of the modified locus. (C) PCR analysis of tail DNA from mice. PCR product sizes are indicated in kilobases.
Preparation of Knock-In Mice. Modified ES cells were injected into blastocysts from albino C57BL/6-Tyrc-Brd mice (14) or C57BL/6J mice. Chimeric male mice were tested for germline transmission by crossing with C57BL/6-Tyrc-Brd or C57BL/6J mice and observing coat color. Knock-out and knock-in mice were bred to homozygosity by crossing F1 progeny. hrhoG/hrhoG mice were bred to rd/rd mice in an FVB background to generate hrhoG/mrho mice that were either heterozygous or homozygous for the rd allele (15). The original hrhoG/hrhoG mice were lost in tropical storm Allyson but were reconstituted by breeding mice that were heterozygous for hrhoG and rd and crossing out the rd allele. Mice were genotyped by Southern blotting or PCR analysis using tail DNA (Fig. 1 B and C), as described in Supporting Materials and Methods.
Histology and Fluorescence Microscopy. For frozen sections, eyes were collected from killed mice of different ages and genotypes, fixed, cryosectioned, and imaged unstained or after staining for cone sheaths with rhodamine peanut agglutinin by using an LSM 510 laser-scanning confocal microscope (Zeiss). For unstained retinal wholemounts, the retina was dissected from the eye cup, placed on a slide with the photoreceptors up, flattened by making radial incisions, and mounted with Gel/Mount (Biomeda, Foster City, CA). To visualize cones, the retinal wholemounts were stained with rhodamine peanut agglutinin. Details on slide preparation and imaging are provided in Supporting Materials and Methods.
For mice expressing rhodopsin-GFP, nuclei were counted by using the trace amounts of fluorescence in the nuclear layer. For other mice, nuclei were stained by using TO-PRO-3 iodide (Molecular Probes). Images were captured from different locations in the retina, excluding areas around the optic nerve and the periphery. We counted 30-100 columns of nuclei for each retina and averaged them for each time point.
Northern and Western Blotting. Total RNA from retinas was analyzed by Northern blotting, as described in Supporting Materials and Methods, by using a bovine rhodopsin cDNA probe that was labeled by random priming. For normalization, blots were stripped and rehybridized with labeled probe from β-actin cDNA or GAPDH cDNA. Samples were quantified by PhosphorImager analysis with imagequant 5.2 software (Molecular Dynamics). Immunoblotting of retinal homogenates separated by SDS/PAGE was carried out as described in Supporting Materials and Methods by using the B6-30N mAb (gift from P. A. Hargrave, University of Florida, Gainesville), which binds the N termini of mouse and human rhodopsins. Bands were detected with enhanced chemiluminescence Western blotting detection reagents (Amersham Biosciences) and x-ray film. Bands were quantified by densitometry using a Personal Densitometer SI and imagequant 5.2 software.
Results and Discussion
Knock-Ins of Human Rhodopsin-GFP at the Mouse Rhodopsin Locus. The human rhodopsin-GFP fusion gene encodes the complete rhodopsin sequence linked through the peptide APVAT at its C terminus to the complete sequence for the EGFP gene (8). Two strategies were used to replace the mouse rhodopsin gene with the human rhodopsin-GFP gene (Fig. 1 A). For both strategies, the entire coding region of the mouse rhodopsin gene, from the middle of the upstream untranslated region to immediately before the first polyadenylation signal, was first replaced by the HPRT minigene with or without the flanking site-specific recombination signals loxP and lox511 or loxH and lox511 (16, 17). In a second step, the HPRT minigene was replaced by the human rhodopsin-GFP gene either by homologous recombination or by site-specific recombination (segmental replacement) mediated by Cre recombinase (Fig. 1 A).
The anticipated advantages of segmental replacement were ease of plasmid construction and higher targeting efficiencies (16). We did not know, however, whether loxP and loxH, which differ by 3 bp in one 13-bp inverted repeat (17), would affect targeting efficiency in ES cells or rhodopsin expression levels in mice. In yeast, loxP is 4-fold more efficient than loxH for site-specific recombination, but it reduces protein expression 4-fold more than loxH when present in an upstream untranslated region of an mRNA (17).
ES cells were generated for all three first-step mouse rhodopsin knock-outs and for all three second-step human rhodopsin-GFP knock-ins (Fig. 1 A). The targeting efficiency (targeted clones/HPRT- clones) for segmental replacement by loxH-lox511 was ≈3-fold higher than for homologous recombination; for loxP-lox511, it was ≈20-fold higher. These results confirm the anticipated higher targeting efficiency of segmental replacement versus homologous recombination and suggest that Cre-mediated recombination of loxP sites is more efficient than that of loxH sites, as it is in yeast (17).
Mice were generated from the modified ES cells for the loxP-HPRT-lox511 knock-out [HPRT(P)], for the human rhodopsin-GFP homologous knock-in (hrhoG), and for the human rhodopsin-GFP loxH-lox511 segmental knock-in [hrhoG(H)]. We were unable to generate mice from ES cells modified to carry human rhodopsin-GFP by loxP-lox511 segmental replacement [hrhoG(P)], even though the loxP-HPRT-lox511 ES cells were competent for germline transmission. Analysis of the effects of loxP on expression of human rhodopsin-GFP in mice was carried out in chimeric mice, as described below.
Expression of Human Rhodopsin-GFP in Mice. Mice with a gene for human rhodopsin-GFP had eyes that fluoresced bright green when illuminated with UV light. Expression of human rhodopsin-GFP was quantified by Western blot analysis using a primary antibody that recognizes an epitope common to mouse and human rhodopsin (Fig. 2). Heterozygous mice were used so that expression of hrhoG and hrhoG(H) could be measured relative to endogenous mouse rhodopsin. Expression of hrhoG(P) was measured in the retinas from a mouse that was ≈75% chimeric, as judged by area of green fluorescence. Compared with expression of the single mouse gene (100%), hrhoG expressed at 80%, hrhoG(H) expressed at 16%, and hrho(P) expressed at 14%. Comparable expression by hrhoG(H) and hrhoG(P) was confirmed by analysis of the intensity of green fluorescence in rod outer segments of hrhoG(H) and hrhoG(P) chimeric mice (data not shown).
Fig. 2.
Western blot analysis of mice expressing human rhodopsin-GFP. Positions of mouse rhodopsin (filled triangles) and human rhodopsin-GFP (open triangles) are indicated. The lower two triangles show the position of rhodopsin monomers; the upper two triangles show the position of rhodopsin dimers. Numbers below each lane indicate microliters of retinal protein added to the gel. The +/hrhoG(P) lanes were generated by using the pair of retinas from a mouse that was 75% chimeric, as judged by area of retinal green fluorescence; all other preparations were from pairs of retinas as well.
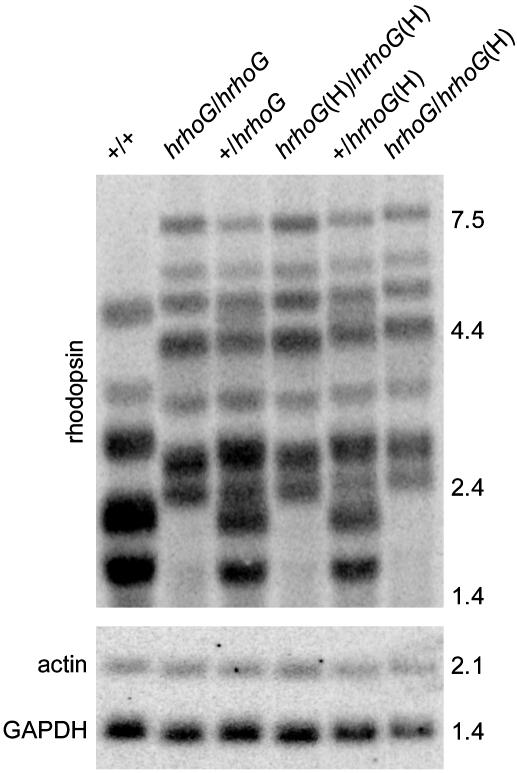
To determine whether the reduced expression of hrhoG(H) resulted from decreased mRNA levels or from decreased mRNA translation, we performed Northern blot analysis of heterozygous and homozygous hrhoG and hrhoG(H) mice (Fig. 3). Wild-type mice gave the multiband pattern that is typical for rhodopsin (18), which was shifted to longer lengths in the knock-in mice, as predicted from their structure (Fig. 1 A). Relative to two internal mRNA controls, expression of rhodopsin mRNA was approximately equal in all genotypes (Fig. 3). Thus, it is likely that the lower expression of human rhodopsin-GFP protein in hrhoG(H) and hrhoG(P) mice is due to effects of the lox sites on efficiency of mRNA translation. The equal effects of loxP and loxH on rhodopsin expression differ from the results in yeast, in which loxP inhibits translation 4-fold more effectively than loxH (17).
Fig. 3.
Northern blot analysis of rhodopsin mRNA in mice. Membranes were probed for rhodopsin mRNA, stripped, and reprobed for GAPDH and β-actin mRNA. Each mRNA sample was prepared from six retinas. Images obtained by using a PhosphorImager are shown. Marker sizes are given in kilobases.
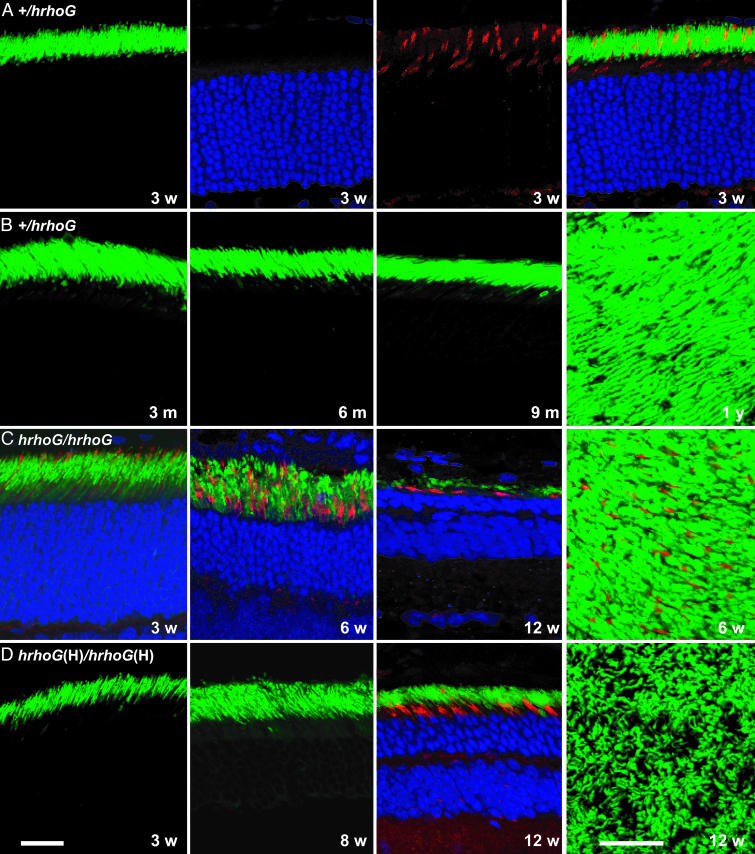
Localization of Human Rhodopsin-GFP in the Mouse Retina. By analyzing eye sections for green fluorescence, we demonstrated that human rhodopsin-GFP localizes properly to the rod outer segments in heterozygous and homozygous mice (Fig. 4). Moreover, as shown for 21-day-old +/hrhoG mice in Fig. 4A, the number of nuclei in the outer nuclear layer and the distribution of cones are normal, indicating that the overall morphology of the retina is preserved. The correct localization of human rhodopsin-GFP in hrhoG and hrhoG(H) homozygous mice (Fig. 4 C and D) indicates that the signals responsible for membrane targeting must be accessible and functional in the fusion protein. Localization to the rod outer segments cannot, for example, result from cotransport with normal rhodopsin, as has been suggested for the P23H mutant form of rhodopsin (19). Correct localization in the homozygote also implies that human rhodopsin-GFP folds properly so that the targeting signals can be recognized.
Fig. 4.
Images of retinal sections from mice expressing human rhodopsin-GFP. (A) Sections of a 3-week-old +/hrhoG mouse retina. Left to right, GFP fluorescence of rod outer segments, nuclei in the outer nuclear layer, rhodamine peanut agglutinin staining of cone sheaths, and a composite image are shown. (B) Retinas from +/hrhoG mice. Eye sections at 3, 6, and 9 months of age are shown in the first three images. The last image is a confocal image of a retinal wholemount from a 1-year-old mouse. (C) Retinal degeneration in hrhoG/hrhoG mice. Eye sections at 3, 6, and 12 weeks of age, showing rods, cones, and the outer nuclear layer, are shown in the first three images. The last image is a confocal image of a retinal wholemount, showing rods and cones. (D) Retinal degeneration in hrhoG(H)/hrhoG(H) mice. Eye sections at 3, 8, and 12 weeks of age are shown in the first three images, and the last image is a confocal image of a retinal wholemount. (Bars = 20 μm.)
Long-Term Status of Retinas Expressing Human Rhodopsin-GFP. Mice heterozygous for hrhoG or hrhoG(H) maintain a healthy retinal morphology for up to 1 year, as shown for +/hrhoG mice in Fig. 4B. By contrast, the retinas in homozygous hrhoG and hrhoG(H) mice degenerate within a few months (Fig. 4 C and D). Rates of retinal degeneration were measured by counting the number of nuclei in the outer nuclear layer over time (Fig. 5). Heterozygous +/hrhoG and +/hrhoG(H) mice showed a slight degeneration over the course of 1 year, with the number of nuclei in the +/hrhoG mice reduced to about half of their normal number in that time. Retinas in +/hrhoG(H) mice retained ≈80% of their outer nuclear layer at 1 year, which is about the same as the loss reported for +/null heterozygotes (19). Retinas in homozygous hrhoG and hrhoG(H) mice and in hrhoG/hrhoG(H) mice degenerated within 3-5 months (Fig. 5).
Fig. 5.
Loss of cells in outer nuclear layer. Nuclei in individual rows in the outer nuclear layers were counted in retinal sections at the indicated ages for mice of different genotypes. ♦, +/+; ▴, +/hrhoG(H); ⋄, +/hrhoG; •, +/hrhoG (modified Fvb background); ▵, hrhoG(H)/hrhoG(H); ▿, hrhoG/hrhoG(H); ○, hrhoG/hrhoG.
The rates of degeneration correlated with expression levels of human rhodopsin-GFP, with hrhoG/hrhoG (80%) > hrhoG/hrhoG(H) (48%) > hrhoG(H)/hrhoG(H) (16%). In all cases, the time course of degeneration could be fit as a simple exponential-decay curve, consistent with a uniform probability of cell death over time and with the level of expression of human rhodopsin-GFP in some way setting the probability constant (4). These results indicate that hrhoG and hrhoG(H) behave as recessive alleles for retinal degeneration, and that the severity of the degeneration depends on the expression levels of the gene products. Because expression of human rhodopsin at similar levels does not induce retinal degeneration in mice (10), we presume that some aspect of the structure of the rhodopsin-GFP fusion protein is responsible for the degeneration seen in our studies.
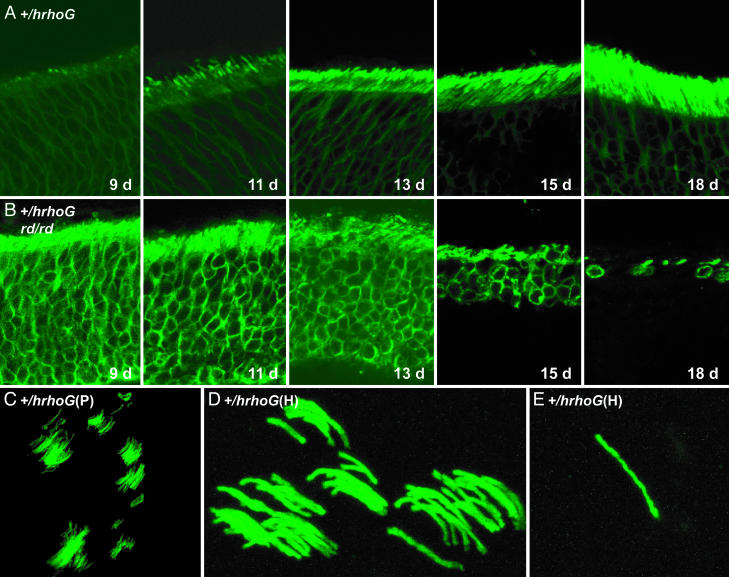
Human Rhodopsin-GFP as a Visible Marker of Rhodopsin Expression. The principal goal of knocking in human rhodopsin-GFP was to use it as a visible marker of rhodopsin expression. As an example, we show the normal formation of rod outer segments in the first few days after birth as visualized by the human rhodopsin-GFP marker (Fig. 6A). Rhodopsin gene expression is easily detectable at early stages when there is almost no development of outer segments, and the emergence of nascent outer segments can be clearly visualized by their bright green fluorescence.
Fig. 6.
Expression of human rhodopsin-GFP in various mice. (A) Eye sections of +/hrhoG mice at 9, 11, 13, 15, and 18 days after birth, showing expression of human rhodopsin-GFP during early development of rod cells. (B) Eye sections of +/hrhoG rd/rd mice at 9, 11, 13, 15, and 18 days, showing rapid retinal degeneration. (C) Confocal image of retinal wholemount from a mouse chimeric for cells of the genotype +/hrhoG(P). (D) Confocal image of retinal wholemount from a mouse chimeric for cells of the genotype +/hrhoG(H). (E) Individual rod cell expressing human rhodopsin-GFP in a retinal wholemount from a mouse chimeric for cells of the genotype +/hrhoG(H).
This marker also allows one to follow the progress of development and degeneration in diseased retinas. To demonstrate this capability, we bred mice to be heterozygous for hrhoG and homozygous for rd, which is deficient in the β subunit of rod cGMP phosphodiesterase (15, 20). The early expression of rhodopsin is clearly visible, as is the beginning development of outer segments (Fig. 6B). As the rods begin to die, the shortening of outer segments and loss of cells are revealed strikingly by fluorescence in unstained samples. Even at advanced stages of degeneration, the high sensitivity of rhodopsin-GFP allows the recognition of remaining rods and rod-cell debris. This bright fluorescence should allow noninvasive monitoring in live mice of the progression of inherited retinal degenerative disorders and the efficacy of experimental treatments.
Rhodopsin-GFP also provides insight into cell lineage during retinal development. By analyzing the retinas of chimeric mice generated by injecting modified ES cells into blastocysts, we were able to observe clusters of rod photoreceptors (Fig. 6 C and D), which are derived from retinal progenitor cells during mosaic retinal development (21). Much more rarely, we observe isolated rod cells (Fig. 6E), which may result from tangential dispersion, which is much less common for rods than it is for other retinal neurons (21). It is remarkable that an individual rod cell with a functional hrhoG(H) allele can be detected unambiguously in a retinal wholemount. In experiments designed to correct a nonfunctional allele by gene repair, it should be possible to detect initial success even at the level of one rod cell among the few million rod cells occupying the back of the retina.
Conclusions
The results presented here demonstrate that knock-ins of human rhodopsin-GFP fusion genes produce expression patterns that are consistently identical to the expression patterns of normal mouse rhodopsin. Moreover, expression can be tuned by manipulation of the 5′ untranslated region of the gene to produce amounts ranging from 16% to 80% of endogenous levels. In the homozygous state, these alleles make models available for studying recessive retinal degeneration and the role of protein levels in disease progression. In the heterozygous state, at which they do not perturb rod-cell structure, these alleles provide an exquisitely sensitive and specific signal for rod-cell status and for the structure and function of the gene itself. Thus, they provide a sensitive way to monitor the progress of retinal degeneration and the efficacy of gene-based therapies in dissected retinas and through noninvasive techniques in the retinas of living mice.
Supplementary Material
Acknowledgments
We thank Paul A. Hargrave for providing the rhodopsin mAb B6-30N; Art Beaudet (Baylor College of Medicine) for mouse 129SvEv genomic libraries; and Wolfgang Baehr (University of Utah, Salt Lake City) for SE6 and MOPS1 plasmid DNAs. This work was supported by National Institutes of Health Grant EY11731 (to J.H.W.) and Vision Research Core Grant EY002520. T.G.W. was supported by the Robert A. Welch Foundation.
Abbreviations: EGFP, enhanced GFP; ES, embryonic stem.
References
- 1.Rivolta, C., Sharon, D., DeAngelis, M. M. & Dryja, T. P. (2002) Hum. Mol. Genet. 11, 1219-1227. [DOI] [PubMed] [Google Scholar]
- 2.Menon, S. T., Han, M. & Sakmar, T. P. (2001) Physiol. Rev. 81, 1659-1688. [DOI] [PubMed] [Google Scholar]
- 3.Wilson, J. H. & Wensel, T. G. (2003) Mol. Neurobiol. 28, 149-158. [DOI] [PubMed] [Google Scholar]
- 4.Clarke, G., Collins, R. A., Leavitt, B. R., Andrews, D. F., Hayden, M. R., Lumsden, C. J. & McInnes, R. R. (2000) Nature 406, 195-199. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld, P. J., Cowley, G. S., McGee, T. L., Sandberg, M. A., Berson, E. L. & Dryja, T. P. (1992) Nat. Genet. 1, 209-213. [DOI] [PubMed] [Google Scholar]
- 6.Lem, J., Krasnoperova, N. V., Calvert, P. D., Kosaras, B., Cameron, D. A., Nicolo, M., Makino, C. L. & Sidman, R. L. (1999) Proc. Natl. Acad. Sci. USA 96, 736-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphries, M. M., Rancourt, D., Farrar, G. J., Kenna, P., Hazel, M., Bush, R. A., Sieving, P. A., Sheils, D. M., McNally, N., Creighton, P., et al. (1997) Nat. Genet. 15, 216-219. [DOI] [PubMed] [Google Scholar]
- 8.Intody, Z., Perkins, B. D., Wilson, J. H. & Wensel, T. G. (2000) Nucleic Acids Res. 28, 4283-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung, C.-H., Makino, C., Baylor, D. & Nathans, J. (1994) J. Neurosci. 14, 5818-5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsson, J. E., Gordon, J. W., Pawlyk, B. S., Roof, D., Hayes, A., Molday, R. S., Mukai, S., Cowley, G. S., Berson, E. L. & Dryja, T. P. (1992) Neuron 9, 815-830. [DOI] [PubMed] [Google Scholar]
- 11.Tam, B. M., Moritz, O. L., Hurd, L. B. & Papermaster, D. S. (2000) J. Cell Biol. 151, 1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moritz, O. L., Tam, B. M., Papermaster, D. S. & Nakayama, T. (2001) J. Biol. Chem. 276, 28242-28251. [DOI] [PubMed] [Google Scholar]
- 13.Harriman, G. R., Bradley, A., Das, S., Rogers-Fani, P. & Davis, A. C. (1996) J. Clin. Invest. 97, 477-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng, B., Sage, M., Sheppeard, E. A., Jurecic, V. & Bradley, A. (2000) Mol. Cell. Biol. 20, 648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pittler, S. J. & Baehr, W. (1991) Proc. Natl. Acad. Sci. USA 88, 8322-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soukharev, S., Miller, J. L. & Sauer, B. (1999) Nucleic Acids Res. 27, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, Q., Li, M. Z., Leibham, D., Cortez, D. & Elledge, S. J. (1998) Curr. Biol. 8, 1300-1309. [DOI] [PubMed] [Google Scholar]
- 18.al-Ubaidi, M. R., Pittler, S. J., Champagne, M. S., Triantafyllos, J. T., McGinnis, J. F. & Baehr, W. (1990) J. Biol. Chem. 265, 20563-20569. [PubMed] [Google Scholar]
- 19.Frederick, J. M., Krasnoperova, N. V., Hoffmann, K., Church-Kopish, J., Ruther, K., Howes, K., Lem, J. & Baehr, W. (2001) Invest. Ophthalmol. Visual Sci. 42, 826-833. [PubMed] [Google Scholar]
- 20.Bowes, C., Li, T., Danciger, M., Baxter, L. C., Applebury, M. L. & Farber, D. B. (1990) Nature 347, 677-680. [DOI] [PubMed] [Google Scholar]
- 21.Reese, B. E. & Galli-Resta, L. (2002) Prog. Retin. Eye Res. 21, 153-168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.