Abstract
In bacteria, yeast, and mammals, iron-sulfur (Fe-S) cluster-containing proteins are involved in numerous processes including electron transfer, metabolic reactions, sensing, signaling, and regulation of gene expression. In humans, iron-storage diseases such as X-linked sideroblastic anemia and ataxia are caused by defects in Fe-S cluster availability. The biogenesis of Fe-S clusters involves several pathways, and in bacteria, the SufABCDSE operon has been shown to play a vital role in Fe-S biogenesis and repair during oxidative stress. Although Fe-S proteins play vital roles in plants, Fe-S cluster biogenesis and maintenance and physiological consequences of dysfunctional Fe-S cluster assembly remains obscure. Here we report that Arabidopsis plants deficient for the SufC homolog AtNAP7 show lethality at the globular stage of embryogenesis. AtNAP7 is expressed in developing embryos and in apical, root, and floral meristems and encodes an ATP-binding cassette/ATPase that can partially rescue growth defects in an Escherichia coli SufC mutant during oxidative stress. AtNAP7 is plastid-localized, and mutant embryos contain abnormal developing plastids with disorganized thylakoid structures. We found that AtNAP7 can interact with AtNAP6, a plastidic Arabidopsis SufD homolog, and because Arabidopsis plastids also harbor SufA, SufB, SufS, and SufE homologs, plastids probably contain a complete SUF system. Our results imply that AtNAP7 represents a conserved SufC protein involved in the biogenesis and/or repair of oxidatively damaged Fe-S clusters and suggest an important role for plastidic Fe-S cluster maintenance and repair during Arabidopsis embryogenesis.
Iron-sulfur (Fe-S) clusters, derived from two of the most versatile and abundant elements on our planet, are important cofactors of Fe-S proteins involved in numerous important biological processes (1, 2). The process of Fe-S biosynthesis requires an intricate interplay of a large number of proteins (3, 4) that can be divided into three basic steps: formation of elemental sulfur, sulfur and iron cluster assembly, and cluster insertion into apoproteins.
Most research on Fe-S assembly has come from studies on bacteria, and three bacterial systems exist termed NIF (nitrogen fixation), ISC (iron-sulfur cluster), and SUF (mobilization of sulfur) (5, 6). The NIF system is most specific, involved in the assembly and maturation of Fe-S clusters in nitrogenase proteins (7) in nitrogen-fixing bacteria and ε-proteobacteria (8), whereas the ISC system is more general and is found in both bacteria and higher eukaryotes (9-13). Although classified as two distinct systems, similarities exist between the two in that the N-terminal domain of NifU corresponds to IscU (14-16), and NifS shows functional similarity to IscS (9). The suf operon (sufABCDSE) represents the third Fe-S system, and recent studies have shown that Suf proteins are involved in Fe-S cluster biosynthesis and repair. SufB, SufC, and SufD are conserved in nature, and in bacteria SufC interacts with SufB and SufD in the cytosol (17, 18). SufE interacts with SufS, involved in cysteine desulfuration (6), and SufA assembles Fe-S clusters transiently in vitro (19). SufC is probably the most essential Suf protein, because SufC deficiency in Escherichia coli results in phenotypes similar to mutants lacking the entire suf operon, related to oxidative stress and iron homeostasis (18, 20).
Bacterial SufC can hydrolyze ATP and probably acts as an energizer for the Suf system (6, 17, 18). SufC represents an ATP-binding cassette (ABC) protein with no membrane-spanning domains (18), implying that it is soluble, although conflicting evidence suggests that SufC may be membrane-localized in E. coli (17). Prokaryotic ABC/ATPases represent the nucleotide-hydrolyzing domains that interact with membrane proteins to generate a functional ABC protein (21, 22). In contrast, eukaryotic ABC proteins generally harbor an ABC domain and a membrane domain, although a subset of small eukaryotic ABC proteins, termed nonintrinsic ABC proteins (NAPs), seem to lack a membrane-spanning domain (21, 22).
In yeast and mammals, it has been suggested that all Fe-S clusters are generated in the mitochondria (3), and in plants, mitochondria harbor an Fe-S cluster biogenesis system involving the ABC protein Sta1 (23). However, the existence of the chloroplast-localized Nif proteins AtCpNIFS/AtNFS2, AtC-nfU-V and AtCnfU-IVb (24-26), and HCF101 (27) in Arabidopsis demonstrates that plant Fe-S cluster biogenesis also occurs in plastids. The identification of both mitochondrial and plastidic NFU Fe-S cluster biogenesis proteins in Arabidopsis (28) has further underlined this fact. In chloroplasts Fe-S clusters are required for a functional cytochrome b6/f complex, ferredoxin, and photosystem I, ensuring electron flow in the thylakoids (29, 30). Interestingly, the chloroplast import protein Tic55 contains an Fe-S cluster and may be involved in redox regulation during chloroplast protein import (31). Fe-S proteins are also involved in gene regulation (32), and the maize Fe-S protein LLS1 may act as a rheostat for cell death regulation (33).
Fe-S cluster biosynthesis and maintenance is clearly of great importance during plant development, and the requirement for Fe-S proteins in multiple chloroplast processes argues that Fe-S cluster assembly is an essential part of plastid functionality. However, to date, Fe-S cluster maintenance and in particular Fe-S cluster repair represents poorly understood processes in plastids. Here we show that AtNAP7 is a functional plastid-localized ABC/ATPase that can partially complement a SufC-deficient E. coli mutant during oxidative stress. Furthermore, AtNAP7 can interact with the plastid-localized SufD homolog AtNAP6. Loss of AtNAP7 in Arabidopsis leads to an embryolethal phenotype, and mutant embryonic plastids appear abnormal, with altered thylakoid structures. We have shown previously that Arabidopsis plastids contain the SufB homolog AtABC1/AtNAP1 (34), and together with the fact that Arabidopsis harbors plastid-localized SufA, SufD, SufS, and SufA homologs, our results suggest that Arabidopsis plastids contain a functional and complete Fe-S SUF system.
Materials and Methods
Isolation of an AtNAP7 Transferred DNA Insertion Mutant and Complementation. Surface-sterilized Arabidopsis thaliana seeds (Columbia) were sown on Murashige and Skoog medium or soil and grown under 16-h light/8-h dark cycles at 22°C. The Arabidopsis atnap7 knockout mutant was isolated from the SALK Institute Genomic Analysis Laboratory transferred DNA (T-DNA) insertion lines (http://signal.salk.edu). The genotype of atnap7 was analyzed by PCR using primers (see Fig. 3A) specific for the T-DNA (primer 2: 5′-GCGTGGACCGCTTGCTGCACCT-3′) and specific for the AtNAP7 ORF (primer 1: 5′-GCGGTAATGGTTCTGCTTCCC-3′; primer 3: 5′-TGCAAACCAAAACTACGGGTCA-3′). For complementation analysis the AtNAP7 cDNA was PCR-amplified with primers NAP7/1 (5′-AATCTCGAGATGGCCGGCGTTAACCTAC-3′; XhoI is underlined) and NAP7/2 (5′-ATACTAGTTCAGAGCAAGCCTTTGACG-3′; SpeI is underlined), cloned into the cauliflower mosaic virus 35S promoter binary vector pBA002 (35), and transformed into heterozygous atnap7 plants by using the Agrobacterium-mediated floral-dip method (36).
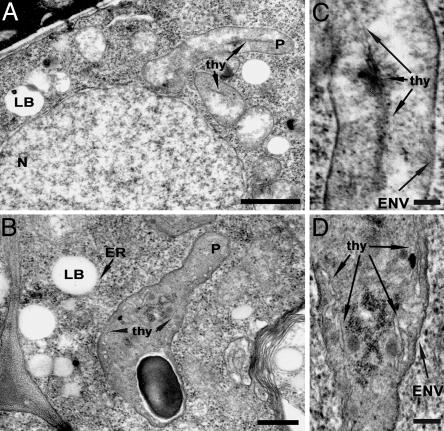
Fig. 3.
Electron micrographs of WT and atnap7 mutant embryos at the globular stage. (A and C) WT embryos showing the presence of plastids containing developing, organized thylakoid structures. (B and D) AtNAP7-deficient embryos containing electron-dense plastids with surface-rugged envelopes and single-thylakoid or bithylakoid structures. N, nucleus; LB, lipid body; P, plastid; thy, thylakoid; ENV, envelope. (Scale bars: A and B,1 μm; C and D, 100 nm.)
Heterologous Expression of AtNAP7 and ATPase Assay. A 1,017-bp full-length AtNAP7 cDNA was PCR-amplified by using primers NAP7/3 (5′-AATCTCGAGATGGCCGGCGTTAACCTAC-3′; XhoI is underlined) and NAP7/4 5′-AAGGTACCCTAACCCCATATCGCTTTGT-3′; KpnI is underlined) and ligated into pRSETA (Invitrogen) to generate pRSETA-AtNAP7. Protein expression and purification was performed following standard protocols. A 100-ml midlog-phase bacterial culture was induced with 1.5 mM isopropyl β-D-thiogalactoside for 3 h, and the insoluble fraction was purified by using TALON metal affinity resin (BD Biosciences) under denaturing conditions following the user manual. The purity of the protein was verified by SDS/PAGE and refolded by dialysis. All ATP hydrolysis assays were performed by using 1.5 μM protein and 0.1 μCi/μl [γ-32P]ATP (1 Ci = 37 GBq) (specific activity 10 mCi/mmol). For time-course assays, the reaction buffer contained 100 mM Tris·Cl (pH 7.4), 50 mM NaCl, 0.1 mM EDTA, 1.5 mM DTT, 10% glycerol, and 5 mM MgCl2, and for ion effects 5 mM MgCl2 was replaced with 5 mM CaCl2, FeSO4, MnSO4, or MnCl2. Samples were applied to TLC plates (Macherey and Nagel), developed in 0.5 M LiCl and 0.5 M formic acid buffer, and visualized by autoradiography.
AtNAP7 and AtNAP6 Localization Analysis. Full-length AtNAP7 and AtNAP6 (1,425 bp) cDNAs were PCR-amplified by using primer pairs NAP7/1 and NAP7/5 (5′-AAGGTACCACCGGATATCGCTTTGTAGCC-3′; KpnI is underlined) and NAP6/D1 (5′-AATCTCGAGATGGCGGCTGCCACAGTTCTC-3′; XhoI is underlined) and NAP6/D2 (5′ AAGGTACCGAGC AAGCCTTTGACGTGAT 3′; KpnI is underlined), respectively, and cloned into pWEN18 as N-terminal fusions to yellow fluorescence protein (YFP). pWEN18/AtNAP7 and pWEN18/AtNAP6 were transfected into tobacco leaves by particle bombardment (35), and YFP fluorescence analysis was performed on a Nikon TE-2000U inverted microscope, and image analysis was performed by using openlab software (Improvision, Coventry, U.K.).
Histochemical Staining and Embryo Analysis. For in planta AtNAP7 expression analysis, a 468-bp upstream AtNAP7 promoter region was PCR-amplified by using primers NAP7/6 (5′-GCTCTAGATATGTTTATATATAGATCACACTATATCAGGACAA: 3′; XbaI is underlined) and NAP7/7 (5′-GGAGACAGAAATTGCGGCGGAAG-3′) and cloned into pBI101 (CLONTECH). Histochemical β-glucuronidase (GUS) activity staining was performed by using 1 mg/ml 5 bromo-4-chloro-3-indolyl β-D-glucuronide in 100 mM sodium phosphate buffer (pH 7.0, 0.1% Triton X-100/5 mM potassium ferricyanide/5 mM potassium ferrocyanide) at 37°C for 1-12 h and cleared in 70% ethanol. To visualize embryos, seeds were cleared in lactophenol [H2O/glycerol/lactate/phenol, 1:1:1:2 (vol/vol)] overnight.
For embryo analysis, seeds were removed from siliques and cleared for 24 h in a chloral hydrate solution [chloral hydrate/H2O/glycerol, 8:2:1 (wt/vol/vol)].
Viability tests were performed by soaking seeds in a 1% 2,3,5-triphenyl tetrazolium chloride solution (Sigma) for 2 days in the dark at 30°C.
All samples were examined by using differential interference contrast microscopy on a Nikon TE-2000U inverted microscope.
Electron microscopy of embryos was performed by using standard protocols, and ultrathin sections were examined by using a JEOL 1220 transmission electron microscope.
Generation of an E. coli SufC Mutant and Complementation by atNAP7. The SufC gene in E. coli strain MG1655 was disrupted by the insertion of a chloramphenicol resistance cassette (see ref. 37 and Supporting Materials and Methods, which is published as supporting information on the PNAS web site, for further details). The resulting SufC mutant strain (MG1655ΔsufC) was transformed with the pRSETA-AtNAP7 vector to generate the strain MG1655ΔsufC/AtNAP7.WT E. coli MG1655, MG1655ΔsufC, and MG1655ΔsufC/AtNAP7 strains were grown overnight in LB medium at 37°C. Cultures were used to inoculate minimal A medium containing 0.2% gluconate and 1 μg/ml thiamine. During the exponential growth phase, phenazine methosulfate (PMS) was added at a final concentration of 15 μM, and growth was followed by measuring OD600 values of the cultures.
Yeast Two-Hybrid Analysis. Full-length AtNAP7 and AtNAP6 cDNAs were PCR-amplified by using primer pairs NAP7/C1 (5′-GGAATTCATGGCCGGCGTTAACCTAC-3′; EcoRI is underlined) and NAP7/C2 (5′-AATCCCGGGCTAACCGGATATCGCTTTGT-3′; SmaI is underlined) and NAP6/D3 (5′-ATGAATTCATGGCGGCTGCCACAGTTCTC-3′; EcoRI is underlined) and NAP6/D4 (5′-ATGGATCCGAGCAAGCCTTTGACGTGAT-3′; BamHI is underlined), respectively, and cloned into pGADT7 (GAL4 activation domain) and pGBKT7 (GAL4 DNA-binding domain). The resulting plasmids, together with empty vector controls (AD and BD), were transformed into HF7c yeast cells in different combinations (see Fig. 7B, which is published as supporting information on the PNAS web site) and tested for restoration of His auxotrophy (+++) as recommended by the manufacturer (Matchmaker two-hybrid system, version 3, Clontech).
Results
The NAP AtNAP7 Shows Similarity to Fe-S ABC Proteins. We cloned a full-length (1,017 nt) cDNA encoding the NAP AtNAP7 (21, 22) from Arabidopsis. AtNAP7 is a single-copy nuclear gene (At3g10670) located on chromosome 3 encoding a 338-aa protein with a Walker A and Walker B domain, an ABC signature motif, and a conserved His residue (Fig. 1A) found in many ABC proteins (38, 39). AtNAP7 contains no predicted transmembrane domain, suggesting that AtNAP7 represents a peripheral ATP-binding subunit found in prokaryotic ABC transporter complexes (22).
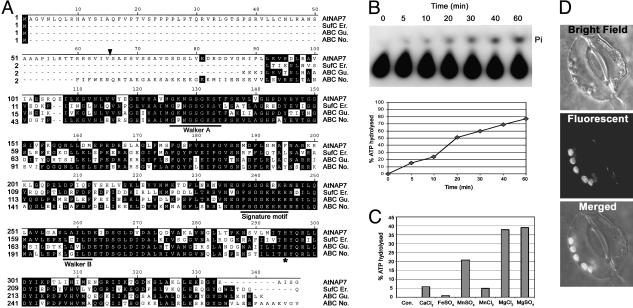
Fig. 1.
AtNAP7 contains ABC signature motifs and is an ABC/ATPase. (A) Comparison of the AtNAP7 amino acid sequence with E. chrysanthemi SufC (SufC Er.), a G. theta sulfate ABC protein (ABC Gu.), and an N. punctiforme ABC protein (ABC No.). The Walker A and B domains and the ABC signature motif are underlined. The conserved His residue is indicated by an asterisk. (B) ATP hydrolysis by purified AtNAP7 protein. Autoradiography and quantification of a time-course experiment showing an increase in released radioactively labeled phosphate (Pi) with time. (C) Ion effect on AtNAP7 ATPase activity showing a marked stimulation by MgCl2 and MgSO4. All experiments were performed in triplicate. (D) Subcellular localization of an AtNAP7/YFP fusion protein in tobacco stomata.
AtNAP7 shows 53% similarity to SufC from Erwinia chrysanthemi (20) and in bacteria SufC is an ABC/ATPase, part of the sufABCDSE operon, acting together with SufB and SufD, playing a vital role in Fe-S cluster biogenesis and repair (17, 18). AtNAP7 also shows 55% similarity to the ycf16-encoded sulfate ABC protein found on the plastid genome of Guillardia theta (40) and an ABC protein from the cyanobacterium Nostoc punctiforme (Fig. 1 A). AtNAP7 contains an N-terminal extension predicted to be a chloroplast-targeting transit peptide, and phylogenetic analysis shows that AtNAP7 has the closest relationship to the ABC protein from N. punctiforme (data not shown), suggesting that AtNAP7 is an evolutionary descendant of a cyanobacterial ABC protein.
AtNAP7 Is a Plastid-Localized ABC/ATPase. To test whether AtNAP7 is a bona fide ABC protein with ATPase activity, we expressed a (His)6-AtNAP7 fusion protein in E. coli. AtNAP7 was insoluble, and the protein was purified under denaturing conditions by using Ni2+ affinity chromatography followed by refolding by dialysis. The purity of the refolded AtNAP7 protein was verified by SDS/PAGE (Fig. 7A). The AtNAP7 protein then was incubated with radiolabeled [γ-32P]ATP at pH 7.2 in the presence of 5 mM MgCl2 and analyzed by TLC for the release of inorganic phosphate (Pi). Analysis showed a clear linear increase in AtNAP7-induced ATP hydrolysis with time (Fig. 1B). To ensure that the measured ATPase activity was not caused by a contaminating E. coli ATPase, we also expressed and purified a nonfunctional (His)6-ATPase from E. coli (Fig. 7A) alongside AtNAP7 and found that this purified protein had no detectable ATPase activity (Fig. 7B). We also tested the effect of different ions on AtNAP7 ATPase activity. In the absence of ions we could not detect any measurable ATP hydrolysis, whereas magnesium ions (MgCl2 and MgSO4) had a dramatic effect on activity (Fig. 1C). Although not as pronounced, MnSO4 also had an effect on AtNAP7 activity (Fig. 1C).
The presence of an AtNAP7 N-terminal extension suggested that AtNAP7 may localize to plastids. To test this we generated a construct containing the AtNAP7 cDNA fused to YFP and transiently expressed this construct in tobacco leaves. From these experiments it was evident that AtNAP7 is localized to chloroplasts (Fig. 1D). Combined, these data demonstrate that AtNAP7 represents a plastid-localized ABC/ATPase.
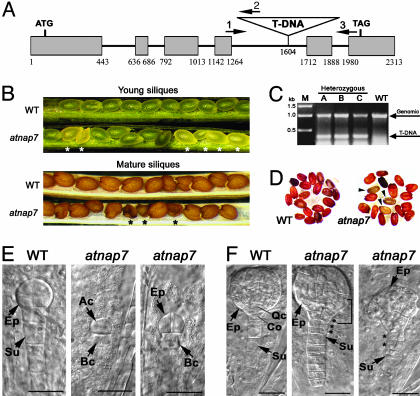
AtNAP7 Is Essential for Seed Viability and Normal Embryo Development. To gain insight into the function of AtNAP7, we analyzed a SALK T-DNA insertion line N576086 (41) carrying a T-DNA in intron four of AtNAP7 (Fig. 2A). To confirm the presence of the T-DNA in atnap7 plants we used AtNAP7-specific and T-DNA-specific primers (Fig. 2A) to PCR-amplify the flanking region (Fig. 2C). Segregation analysis on nonselective media of five T3 progeny derived from a heterozygous atnap7 plant revealed a germination ratio of 3:1, indicating lethality of the homozygous insertion mutant (Table 1, which is published as supporting information on the PNAS web site). To verify this we dissected individual siliques from heterozygous atnap7 mutants and WT plants at different developmental stages (Fig. 2B). In young WT siliques, maturing seeds were of uniform size and shape containing developing green embryos, whereas in segregating atnap7 plants a number of immature and pale seeds were observed (Fig. 2B). The immature and pale homozygous atnap7 seeds failed to develop in mature siliques, resulting in a dark and wrinkled appearance (Fig. 2B). To test for seed viability, we further stained seeds from individual siliques with tetrazolium, which demonstrated that the aborted seeds were not viable (Fig. 2D). These results were consistent with a recessive lethal segregation of embryos homozygous for the AtNAP7 T-DNA insertion.
Fig. 2.
AtNAP7 deficiency in Arabidopsis results in a seed abortion. (A) Exon-intron structure of AtNAP7 showing the site of T-DNA insertion. Primers used for genotyping in C are indicated. (B) WT young siliques show uniformly maturing seeds with green embryos, whereas siliques from a T2 segregating heterozygous atnap7 mutant plant show the presence of white lethal seeds (asterisk). WT mature siliques contain uniform brown seeds, whereas atnap7 mature siliques show the presence of aborted dark brown seeds (asterisk). (C) PCR amplification using primers 1-3 of heterozygous atnap7 and WT plants demonstrating the presence of the T-DNA insertion in atnap7 plants, its absence in WT plants, and the presence of the WT AtNAP7 copy (genomic) in both backgrounds. (D) Tetrazolium staining of viable seeds. (E) WT embryo at the four-cell stage and retarded embryos homozygous for the AtNAP7 insertion (atnap7). (F) WT embryo at the globular stage and abnormal embryos homozygous for the AtNAP7 insertion (atnap7). The asterisks indicate abnormal cell divisions in the suspensor, and the bracket indicates abnormal cell division around the quiescent center. Ep, embryo proper; Su, suspensor; Ac, apical cell; Bc, basal cell; Qc, prospective quiescent center; Co, prospective columella. (Scale bars, 20 μm.)
Although analysis of a second AtNAP7 insertion mutant showed a homozygous embryo-lethal phenotype (data not shown), we tested whether an AtNAP7 cDNA could rescue the atnap7 mutant phenotype. We transformed heterozygous atnap7 plants with a cauliflower mosaic virus 35S promoter-driven WT AtNAP7 cDNA, and four of five independent transgenic lines tested showed a germination rate ratio of 15:1 (germinating seeds to aborted seeds), demonstrating that the homozygous embryo-lethal phenotype is caused by AtNAP7 disruption.
To examine the effect of AtNAP7 deficiency on Arabidopsis embryo development, we compared WT and homozygous atnap7 mutant embryos at the same developmental stages. In contrast to normal WT embryo development (Fig. 8 A-E, which is published as supporting information on the PNAS web site) (42), atnap7 embryos showed retarded and abnormal development and did not progress beyond the late globular stage (Fig. 8 F-J). When WT embryos had reached the four-cell stage, atnap7 embryos showed either signs of initial zygote division or had only reached the two-cell stage (Fig. 2E), demonstrating a role for AtNAP7 during early embryogenesis. At later stages of development when WT embryos had reached the globular stage, atnap7 embryos were either developmentally retarded or showed abnormal phenotypes (Fig. 2F). Some atnap7 mutant embryos had altered cell-division characteristics, showing vertical divisions in the upper part of the suspensor and in the embryo proper, whereas others were severely distorted in shape (Fig. 2F).
To further examine the ultrastructural changes that take place in mutant embryos at the stage of developmental arrest, we examined WT and mutant embryos at the globular stage by using electron microscopy. Although the overall cellular structures appeared similar, there were several differences between the WT and mutant. In mutant embryos (Fig. 3B and Fig. 9B, which is published as supporting information on the PNAS web site), cells contain more lipid bodies compared with WT cells (Fig. 3A and 9A). Mutant cells were also more vacuolated, and the vacuoles contained electron-dense material that seemed to form precipitates (Fig. 9B). The main difference, however, between WT and mutant cells relates to the plastid structure. In WT cells, the plastids are generally ellipsoidal in shape with a smooth envelope surface (Fig. 3 A and C). In contrast, AtNAP7-deficient plastids show slight shape abnormalities and, more strikingly, an uneven envelope surface (Fig. 3 B and D). AtNAP7-deficient plastids were also more electron-dense than WT. Although plastids at this stage of embryogenesis remain relatively undifferentiated, plastids in WT embryos do contain developing thylakoid structures (Fig. 3 A and C). Analysis of large numbers of mutant embryos failed to detect such ordered structures, although disorganized single-thylakoid or bithylakoid structures could be observed (Fig. 3 B and D).
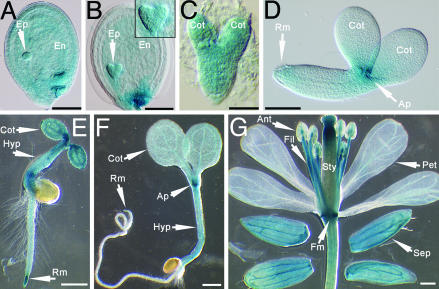
AtNAP7 Is Expressed in Embryos, Meristems, and Flowers. To determine spatial and temporal AtNAP7 expression patterns, we analyzed three independent transgenic Arabidopsis lines expressing an AtNAP7 promoter-GUS fusion. During the early globular stage, GUS staining was observed in the developing endosperm, the embryo proper, and the suspensor (Fig. 4A). At the heart stage, staining became more restricted to the embryo proper (Fig. 4B), and at the torpedo stage, GUS activity was evident in the entire developing embryo (Fig. 4C). Interestingly, at the bent-cotyledon stage, GUS staining was limited to the apical meristem, and faint staining could be observed in the root meristem also (Fig. 4D). In developing seedlings, the highest GUS activity was observed in the root meristem and in the apical meristem, although staining was also detected in vascular tissue in young seedlings (Fig. 4 E and F). Adult plants also showed AtNAP7 expression in the vascular tissue of rosette leaves (data not shown) and in flowers (Fig. 4G). In flowers, the most intense staining was observed in the floral meristem, in the filaments of the stamen, and in the style (Fig. 4G). The ubiquitous expression of AtNAP7 is in agreement with Northern blot data showing AtNAP7 expression in all aerial plant regions but with lower expression in roots (data not shown).
Fig. 4.
Histochemical analysis of plants expressing an AtNAP7 promoter-GUS fusion. (A) Globular-stage embryo. (B) Heart-stage embryo. (C) Torpedo-stage embryo. (D) Bent-cotyledon-stage embryo. (E and F) GUS staining in young developing seedlings. (G) GUS staining in adult flowers. (B Inset) Magnification of the embryo. Ep, embryo proper; En, endosperm; Cot, cotyledons; Ap, apical meristem; Rm, root meristem; Hyp, hypocotyl; Fm, floral meristem; Sep, sepal; Pet, petal; Sty, style; Fil, filament; Ant, anthers. (Scale bars: A and B, 100 μm; C, 30 μm; D, 60 μm; E-G, 0.5 mm.)
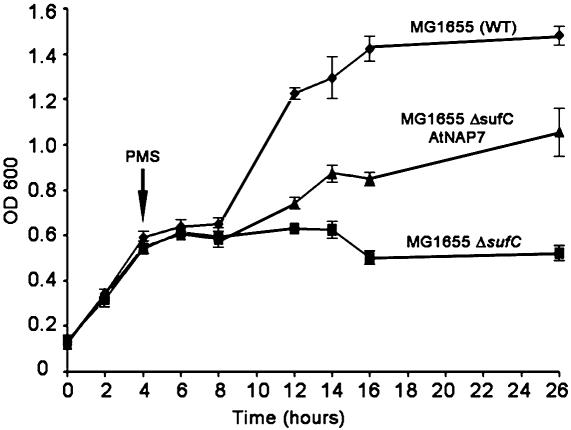
AtNAP7 Is an Evolutionarily Conserved SufC Protein Able to Partially Complement a SufC-Deficient E. coli Mutant During Oxidative Stress. In bacteria, SufC is vital for the protection of enzymes with oxygen-labile Fe-S clusters, and E. coli lacking SufC is unable to grow in minimal A medium under oxidative stress conditions (18). With respect to this fact, we tested whether AtNAP7 could functionally complement a SufC-deficient E. coli strain. We generated an E. coli knockout mutant lacking SufC (MG1655ΔSufC) and compared its growth characteristics with WT (MG1655) under oxidative stress conditions, with gluconate as a sole carbon source. In the absence of oxidative stress, both strains grew equally well (Fig. 5). After exposure to the oxidative agent PMS, WT cells were able to recover and resume exponential growth after a 4-h lag period, whereas mutant MG1655ΔSufC cells were unable to resume growth (Fig. 5) (18). We then analyzed the effect of AtNAP7 expression in MG1655ΔSufC cells after PMS addition. MG1655ΔSufC transformants expressing AtNAP7 (MG1655ΔSufC/AtNAP7) showed exponential growth before PMS addition and the characteristic initial 4-h lag phase after PMS addition (Fig. 5). However, after 4 h, MG1655ΔSufC/AtNAP7 cells resumed growth (Fig. 5), demonstrating that AtNAP7 can partially complement SufC deficiency in E. coli. These results demonstrate that AtNAP7 encodes an evolutionarily conserved plastidic SufC protein in Arabidopsis.
Fig. 5.
AtNAP7 can partially complement SufC deficiency in E. coli under oxidative stress. WT E. coli (MG1655), an E. coli SufC mutant (MG1655ΔSufC), and E. coli MG1655ΔSufC expressing AtNAP7 (MG1655 ΔSufC AtNAP7) were grown in minimal A medium with 0.2% gluconate and 1 μg/ml thiamine. During exponential growth, PMS was added to the cultures. Experiments were performed in triplicate, and standard deviations are shown.
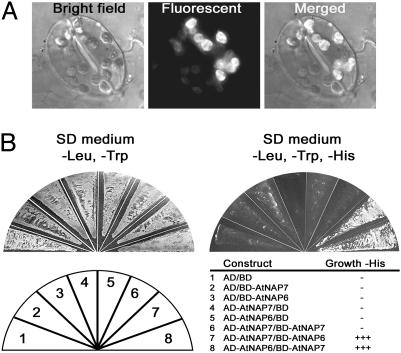
AtNAP7 Interacts with the Plastid-Localized SufD Homolog AtNAP6. Bacterial SufC interacts with SufD in the cytosol (17, 18), and through blast searches we identified a SufD homolog in Arabidopsis named AtNAP6 (22) (Table 2, which is published as supporting information on the PNAS web site). AtNAP6 contains an N-terminal extension predicted to encode a chloroplast-targeting transit peptide, and we analyzed the subcellular localization of an AtNAP6/YFP fusion protein in tobacco leaf cells. Epifluorescence analysis revealed that, as observed for AtNAP7, AtNAP6 is localized exclusively to chloroplasts (Fig. 6 A-C).
Fig. 6.
AtNAP6 is plastid-localized and interacts with AtNAP7. (A) Subcellular localization of an AtNAP6/YFP fusion protein in tobacco stomata. (B) Yeast two-hybrid analysis of AtNAP7-AtNAP6 interactions. HF7c yeast cells cotransformed with different vector combinations were plated on synthetic dropout medium lacking Leu and Trp, and positive interactions were scored on synthetic dropout medium plates lacking Leu, Trp, and His. Only yeast cells containing AtNAP7 and AtNAP6 were able to restore His auxotrophy. The different vector combinations are shown, and the experiments were performed in triplicate.
To further examine whether AtNAP7 could interact with AtNAP6, we made use of the yeast two-hybrid system. Full-length AtNAP7 and AtNAP6 proteins were fused to the GAL4 activation domain (AD-AtNAP7 and AD-AtNAP6) and to the GAL4 DNA-binding domain (BD-AtNAP7 and BD-AtNAP6) and expressed in yeast HF7c cells. As a protein-protein interaction marker, we made use of the ability of HF7c to only grow in the absence of His after positive protein-protein interactions. We found that His auxotrophy was restored in yeast cells coexpressing AD-AtNAP7/BD-AtNAP6 and BD-AtNAP7/AD-AtNAP6, demonstrating that AtNAP7 and AtNAP6 can interact (Fig. 6D). In contrast, His auxotrophy was not restored in yeast cells coexpressing the empty AD vector together with BD-AtNAP7 or BD-AtNAP6 or the empty BD vector together with AD-AtNAP7 or AD-AtNAP6. We also tested whether AtNAP7 could form homodimers but found that yeast cells coexpressing AD-AtNAP7/BD-AtNAP7 did not restore His auxotrophy (Fig. 6D).
This result further implied that Arabidopsis plastids may harbor a complete SUF system. Although Arabidopsis contains the plastid-localized SufB homolog AtABC1/AtNAP1 (34) and the SufS homolog AtCpNifS/AtNSF2 (24), we wanted to examine whether Arabidopsis contains other Suf homologs. To this end we identified a SufA and SufE homolog. We cloned full-length cDNAs encoding SufA and SufE proteins, and sequence-alignment and -prediction analyses revealed that both SufA and SufS contain N-terminal extensions predicted to encode plastid-targeting transit peptides (Table 2). Based on the data we believe that Arabidopsis plastids harbor a complete SUF system.
Discussion
For Fe-S proteins to function, they require both Fe-S cluster biogenesis and Fe-S cluster repair. Fe-S cluster formation and repair are therefore important areas of study, because Fe-S proteins are involved in vital processes in organisms ranging from bacteria to mammals (1, 2, 43). Similarly, in plants, Fe-S proteins have been identified as playing important roles (29-31, 33). Despite this, little is known about Fe-S cluster biogenesis in plants and in particular the repair of damaged clusters. We have demonstrated that AtNAP7 encodes a plastidic nonintrinsic ABC/ATPase that plays an essential role during Arabidopsis embryogenesis, ensuring developmental progression beyond the globular stage. AtNAP7 shows high similarity to bacterial SufC, and the present study demonstrates that AtNAP7 represents an evolutionarily conserved plastidic SufC protein probably involved in the maintenance and repair of oxidatively damaged Fe-S clusters in plastids.
We demonstrate that AtNAP7 can hydrolyze ATP, and in bacteria the Suf BCD complex acts as an ATP-dependent energizer via its SufC ATPase activity directly involved in the repair of oxidatively damaged Fe-S clusters (18). Our observation that AtNAP7 can restore growth of a SufC-deficient E. coli mutant during oxidative stress (Fig. 5), a condition known to damage Fe-S clusters, implies that AtNAP7 has retained its ability to repair oxygen-labile Fe-S clusters and that AtNAP7 may be involved in Fe-S repair in Arabidopsis plastids. During photosynthesis the generation of high-energy radicals can result in photooxidative damage, and it is therefore possible that the observed embryo arrest in the AtNAP7-deficient plant is caused by a loss of this repair ability during the development of photosynthetic competence. Developing plastids in AtNAP7-deficient embryos contain disorganized thylakoids, forming single-thylakoid or bithylakoid structures (Fig. 3D), suggesting that the maintenance and repair of Fe-S clusters mediated by AtNAP7 are important for correct thylakoid development during embryo maturation.
A recent report has shown that the plastidic NifU-like protein AtCnfU-V is required for the biogenesis of ferredoxin and photo-system I (26). Moreover, homozygous mutants lacking AtCnfU-V exhibit a dwarfed phenotype (26), which is in contrast to the embryo-lethal phenotype of atNAP7-deficient mutants, supporting the notion that AtNAP7 is most probably involved in Fe-S cluster maintenance and repair rather than Fe-S cluster biogenesis.
In prokaryotes, SufC interacts with SufD (6) and AtNAP7 interacts with the plastid-localized SufD homolog AtNAP6 (Fig. 6D), which implies that Arabidopsis plastids contain a functional SufC/SufD complex and further underlines the evolutionary conservation of the SufC mode of action. Arabidopsis plastids also harbor plastid-localized SufB (34) and SufS (24) homologs, and we have provided evidence that SufA and SufE proteins are also present in Arabidopsis plastids. Our findings demonstrate, therefore, that Arabidopsis plastids not only contain a functional SufC protein but most probably a complete SUF system.
The requirement for AtNAP7 during early embryogenesis is intriguing. AtNAP7-deficient embryo development is severely retarded during early cell divisions; however, development is not arrested until the late globular stage. Although this implies that Fe-S maintenance and repair are required, AtNAP7 is not absolutely essential during early embryonic stages, possibly because of less Fe-S cluster damage. This notion is supported by the relatively weak AtNAP7 expression in young embryos (Fig. 4A).
We can only speculate on the precise mechanics of AtNAP7-mediated Fe-S cluster maintenance and repair in developing Arabidopsis plastids during embryogenesis. Because AtNAP7-deficient Arabidopsis do not progress beyond the globular stage of embryogenesis, the analysis of Fe-S cluster-containing proteins in arrested embryos represents a difficult task. Although little is known regarding Fe-S cluster repair in plants, this study reveals that a functional plastid-specific SUF system exists in Arabidopsis. The demonstration that AtNAP7 is an evolutionarily conserved SufC protein interacting with the plastid-localized SufD homolog AtNAP6 and that AtNAP7 plays an essential role during embryogenesis has shed light on the importance of plastidic Fe-S proteins during early stages of plant development. Given the complexity of the SUF system in other organisms, the characterization of AtNAP7 is an important step toward understanding the intricate nature of Fe-S cluster maintenance and repair in plants.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants and Nottingham Arabidopsis Stock Centre for providing atnap7 seeds. We also thank the E. coli Genetics Stock Center at Yale University for providing MG1655/pKD46, BW25141/pKD3, and BT340/pCP20; Donna Sharples and David Capenerhurst for technical assistance; and Natalie Allcock and Stefan Hyman for electron microscopy. This work was supported by a grant from the Biotechnology and Biological Sciences Research Council (91/P16510) and The Royal Society (574006.G503/23280/SM) to S.G.M.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NIF, nitrogen fixation; ISC, iron-sulfur cluster; SUF, mobilization of sulfur; ABC, ATP-binding cassette; NAP, nonintrinsic ABC protein; T-DNA, transferred DNA; YFP, yellow fluorescence protein; GUS, β-glucuronidase; PMS, phenazine methosulfate.
References
- 1.Beinert, H., Holm, R. H. & Munck, E. (1997) Science 277, 653-659. [DOI] [PubMed] [Google Scholar]
- 2.Beinert, H. & Kiley, P. J. (1999) Curr. Opin. Chem. Biol. 3, 152-157. [DOI] [PubMed] [Google Scholar]
- 3.Lill, R. & Kispal, G. (2000) Trends Biochem. Sci. 25, 352-356. [DOI] [PubMed] [Google Scholar]
- 4.Frazzon, J. & Dean, D. R. (2003) Curr. Opin. Chem. Biol. 7, 166-173. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi, Y. & Tokumoto, U. (2002) J. Biol. Chem. 277, 28380-28383. [DOI] [PubMed] [Google Scholar]
- 6.Loiseau, L., Ollagnier-de-Choudens, S., Nachin, L., Fontecave, M. & Barras, F. (2003) J. Biol. Chem. 278, 38352-38359. [DOI] [PubMed] [Google Scholar]
- 7.Jacobson, M. R., Brigle, K. E., Bennett, L. T., Setterquist, R. A., Wilson, M. S., Cash, V. L., Beynon, J., Newton, W. E. & Dean, D. R. (1989) J. Bacteriol. 171, 1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olson, J. W., Agar, J. N., Johnson, M. K. & Maier, R. J. (2000) Biochemistry 39, 16213-16219. [DOI] [PubMed] [Google Scholar]
- 9.Zheng, L., Cash, V. L., Flint, D. H. & Dean, D. R. (1998) J. Biol. Chem. 273, 13264-13272. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi, Y. & Nakamura, M. (1999) J. Biochem. (Tokyo) 126, 917-926. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz, C. J., Djaman, O., Imlay, J. A. & Kiley, P. J. (2000) Proc. Natl. Acad. Sci. USA 97, 9009-9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokumoto, U. & Takahashi, Y. (2001) J. Biochem. (Tokyo) 130, 63-71. [DOI] [PubMed] [Google Scholar]
- 13.Muhlenhoff, F. & Lill, R. (2000) Biochim. Biophys. Acta 1459, 370-382. [DOI] [PubMed] [Google Scholar]
- 14.Garland, S. A., Hoff, K., Vickery, L. E. & Culotta, V. C. (1999) J. Mol. Biol. 294, 897-907. [DOI] [PubMed] [Google Scholar]
- 15.Agar, J. N., Krebs, C., Frazzon, J., Huynh, B. H., Dean, D. R. & Johnson, M. K. (2000) Biochemistry 39, 7856-7862. [DOI] [PubMed] [Google Scholar]
- 16.Kato, S., Mihara, H., Kurihara, T., Takahashi, Y., Tokumoto, U., Yoshimura, T. & Esaki, N. (2002) Proc. Natl. Acad. Sci. USA 99, 5948-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rangachari, K., Davis, C. T., Eccleston, J. F., Hirst, E. M., Saldanha, J. W., Strath, M. & Wilson, R. J. (2002) FEBS Lett. 514, 225-228. [DOI] [PubMed] [Google Scholar]
- 18.Nachin, L., Loiseau, L., Expert, D. & Barras, F. (2003) EMBO J. 22, 427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ollagnier-de-Choudens, S., Lascoux, D., Loiseau, L., Barras, F., Forest, E. & Fontecave, M. (2003) FEBS Lett. 555, 263-267. [DOI] [PubMed] [Google Scholar]
- 20.Nachin, L., El Hassouni, M., Loiseau, L., Expert, D. & Barras, F. (2001) Mol. Microbiol. 39, 960-972. [DOI] [PubMed] [Google Scholar]
- 21.Holland, I. B. & Blight, M. A. (1999) J. Mol. Biol. 293, 381-399. [DOI] [PubMed] [Google Scholar]
- 22.Sanchez-Fernandez, R., Davies, T. G., Coleman, J. O. & Rea, P. A. (2001) J. Biol. Chem. 276, 30231-30244. [DOI] [PubMed] [Google Scholar]
- 23.Kushnir, S., Babiychuk, E., Storozhenko, S., Davey, M. W., Papenbrock, J., De Rycke, R., Engler, G., Stephan, U. W., Lange, H., Kispal, G., et al. (2001) Plant Cell 13, 89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilon-Smits, E. A., Garifullina, G. F., Abdel-Ghany, S., Kato, S., Mihara, H., Hale, K. L., Burkhead, J. L., Esaki, N., Kurihara, T. & Pilon, M. (2002) Plant Physiol. 130, 1309-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leon, S., Touraine, B., Briat, J. F. & Lobreaux, S. (2002) Biochem. J. 366, 557-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yabe, T., Morimoto, K., Kikuchi, S., Nishio, K., Terashima, I. & Nakai, M. (2004) Plant Cell 16, 993-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lezhneva, L., Amann, K. & Meurer, J. (2004) Plant J. 37, 174-185. [DOI] [PubMed] [Google Scholar]
- 28.Leon, S., Touraine, B., Ribot, C., Briat, J. F. & Lobreaux, S. (2003) Biochem. J. 371, 823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raven, J. A., Evans, M. C. & Korb, R. E. (1999) Photosynth. Res. 60, 111-149. [Google Scholar]
- 30.Kapazoglou, A., Mould, R. M. & Gray, J. C. (2000) Eur. J. Biochem. 267, 352-360. [DOI] [PubMed] [Google Scholar]
- 31.Caliebe, A., Grimm, R., Kaiser, G., Lubeck, J., Soll, J. & Heins, L. (1997) EMBO J. 16, 7342-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hidalgo, E., Ding, H. & Demple, B. (1997) Trends Biochem. Sci. 22, 207-210. [DOI] [PubMed] [Google Scholar]
- 33.Gray, J., Close, P. S., Briggs, S. P. & Johal, G. S. (1997) Cell 89, 25-31. [DOI] [PubMed] [Google Scholar]
- 34.Møller, S. G., Kunkel, T. & Chua, N. H. (2001) Genes Dev. 15, 90-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kost, B., Spielhofer, P. & Chua, N. H. (1998) Plant J. 16, 393-401. [DOI] [PubMed] [Google Scholar]
- 36.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 37.Datsenko, K. A. & Wanner, B. L. (2000) Proc. Natl. Acad. Sci. USA 97, 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linton, K. J. & Higgins, C. F. (1998) Mol. Microbiol. 28, 5-13. [DOI] [PubMed] [Google Scholar]
- 39.Higgins, C. F. (1992) Annu. Rev. Cell. Biol. 8, 67-113. [DOI] [PubMed] [Google Scholar]
- 40.Douglas, S. E. & Penny, S. L. (1999) J. Mol. Evol. 48, 236-244. [DOI] [PubMed] [Google Scholar]
- 41.Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., Stevenson, D. K., Zimmerman, J., Barajas, P., Cheuk, R., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 42.Jurgens, G., Mayer, U., Busch, M., Lukowitz, W. & Laux, T. (1995) Philos. Trans. R. Soc. London B 350, 19-25. [DOI] [PubMed] [Google Scholar]
- 43.Johnson, M. K. (1998) Curr. Opin. Chem. Biol. 275, 15955-15961. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.