Abstract
Biosynthesis of the anticancer drug Taxol involves 19 enzymatic steps from the universal diterpenoid progenitor geranylgeranyl diphosphate derived by the plastidial methylerythritol phosphate pathway for isoprenoid precursor supply. To gain further insight about Taxol biosynthesis relevant to the improved production of this drug and to draw inferences about the organization, regulation, and origins of this complex natural product pathway, random sequencing of a cDNA library derived from Taxus cuspidata cells (induced for taxoid biosynthesis with methyl jasmonate) was undertaken. This effort revealed surprisingly high abundances for transcripts of several of the 12 defined genes of Taxol biosynthesis, yielded cDNAs encoding two previously uncharacterized cytochrome P450 taxoid hydroxylases, and provided candidate genes for all but one of the remaining seven steps of this extended sequence of reactions.
Keywords: paclitaxel, Taxus cuspidata, expressed sequence tags, taxoid 10β-hydroxylase
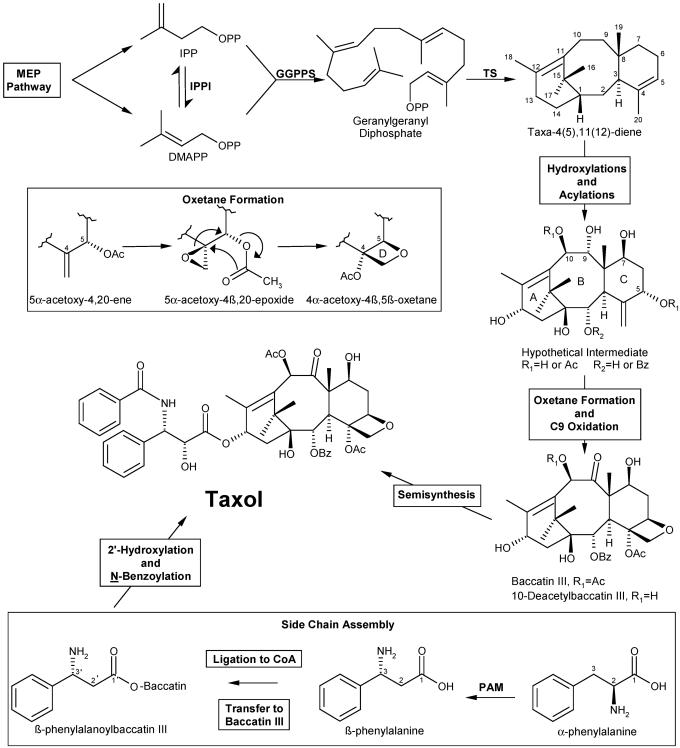
Taxol (Fig. 1) (Bristol-Myers Squibb) is a well established chemotherapeutic agent with increasing application against a range of cancers (1-3). This structurally complex taxane diterpenoid (taxoid) was first isolated from the bark of the Pacific yew (Taxus brevifolia) (4), but the principal source of the drug is now via semisynthesis from advanced taxoid intermediates, such as baccatin III and its precursor, 10-deacetylbaccatin III (Fig. 1), isolated from the leaves of various Taxus species (for recent reviews, see refs. 5 and 6). Because the supply of Taxol and its precursors for semisynthesis will continue to rely on biological sources for the foreseeable future (7), only knowledge of the biosynthetic pathway, the responsible enzymes, and the underlying genes can provide the foundation for improved production of the drug in yew trees or, more likely, in cell cultures derived therefrom (8). The latter offers an attractive production platform for the pharmaceutical industry, and cell suspension cultures have become the model system of choice for studies of Taxol biosynthesis, because of the ease of manipulation and because Taxol production by such cells is inducible with methyl jasmonate (9).
Fig. 1.
Outline of the Taxol biosynthetic pathway. IPPI, isopentenyl diphosphate isomerase; GGPPS, GGPP synthase; TS, taxadiene synthase.
All taxoids arise via the plastidial 2-C-methyl-D-erythritol phosphate (MEP) pathway for supply of the isoprenoid precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) (10). After their conversion to the universal C20 precursor of the diterpenes by plastidial geranylgeranyl diphosphate (GGPP) synthase (11), a series of 19 enzymatic steps yields Taxol (Fig. 1), the first of which is the committed step involving the cyclization of GGPP to taxadiene by plastidial taxadiene synthase (12, 13). This taxane core structure is then functionalized by eight cytochrome P450 oxygenases and decorated with two acetate groups and a benzoate group by acyl and aroyl CoA-dependent transferases (14, 15). The order of oxygenation beyond the initial C5α-hydroxylation (16) is uncertain; however, a survey of the relative abundances of the variously functionalized, naturally occurring taxoids (17) suggests the approximate progression of oxygenation from C5 to C10, C2, C9, C13, C7, and finally C1, to the level of an acylated heptaol of uncertain identity (Fig. 1). cDNA clones encoding the cytochrome P450 taxoid C2α-, C5α-, C7β-, C10β-, and C13α-hydroxylases have been isolated by a range of cloning strategies (18-22). The C1 and C9 oxygenases have not yet been identified, although a cytochrome P450 taxoid C14β-hydroxylase has been described that is responsible for diversion of the Taxol pathway to side-route metabolites (23). cDNA clones encoding the taxoid C5-O-and C10-O-acetyltransferases and the C2-O-benzoyltransferase also have been isolated (24-26). As with the hydroxylations, the precise timing of these acylation steps is not fully clear, although based on similar biochemical considerations the order is likely C5, C2 then C10, and finally C13 (side-chain) addition (14, 15).
Several more, as yet biochemically undefined, transformations are required to reach baccatin III (Fig. 1), including the oxidation to the ketone function at C9, potentially mediated by a dehydrogenase or by a cytochrome P450 oxygenase via the ketone hydrate, and the construction of the oxetane D-ring. This latter process (Fig. 1 Inset) most plausibly (27) involves epoxidation of the C4-C20 double bond (likely mediated by cytochrome P450) and ring expansion with intramolecular migration of the C5α-acetoxy group catalyzed by an isomerase (oxomutase) of unknown type. Oxetane formation likely precedes C9 oxidation and may precede the late pathway hydroxylation at C1 (17) such that the hypothetical intermediate of Fig. 1 would be a derivative of a hexaol not a heptaol.
The final steps of Taxol biosynthesis from baccatin III require the assembly of the C13 side chain. This process (Fig. 1 Inset) seemingly involves conversion of α-phenylalanine to β-phenylalanine by an aminomutase (28, 29), ligation to the corresponding CoA thioester and aroyl transfer to the baccatin III core, and hydroxylation at the C2′ side-chain position (to the phenylisoserinoyl moiety) and N-benzoylation to complete the sequence. cDNA clones encoding these two transferases are described in refs. 30 and 31.
By using induced taxoid production in Taxus cell cultures as a model system (9, 32), considerable progress has been made in understanding the pathway, enzymology, and molecular genetics of Taxol biosynthesis (14, 15). However, it is clear that gaps exist in defining several pathway steps and establishing the sequence of this extended series of reactions; additionally, seven genes encoding presumptive biosynthetic enzymes are still missing. To gain further insight about Taxol biosynthesis relevant to the improved production of this drug, we undertook the partial sequencing of anonymous cDNA clones from an induced Taxus cuspidata cell library (18). The resulting evaluation of these expressed sequence tags is herein described.
Materials and Methods
cDNA Library Construction and Analysis. Procedures for mRNA isolation from induced T. cuspidata cells (9) and for cDNA library construction are described in ref. 18 and 24. cDNA clones were mass excised in vivo as pBluescript SK(-) phagemids in Escherichia coli host strain SOLR (Stratagene), and purified plasmids from the resulting individual colonies were sequenced with the M13R vector-specific primer. Cytochrome P450 clones were fully sequenced by using T3, T7, and gene-specific primers. Of a total of 10,176 individual clones, 8,424 gave useable sequences. blastz (33) was used to compare each edited sequence with every other sequence to assemble consensus sequences that were annotated by using GenBank via blastz (33) and GenPept via fastx (34).
Phylograms based on protein sequences of cytochrome P450 enzymes and acyltransferases were generated by using phylip (J. Felsenstein, University of Washington, http://evolution.genetics.washington.edu/phylip.html). Alignments were made by using the GCG pileup function (gap creation penalty 3, gap extension penalty 1) (35). Aligned sequences were converted to phylip format by using genedoc (K. B. Nicholas, H. B. Nicolas, Jr., and D. W. Deerfield, II, www.psc.edu/biomed/genedoc) and entered into the seqboot (J. Felsenstein) algorithm to create 100 data sets by bootstrap resampling. These data were entered into the protml (protein maximum likelihood) (J. Adachi and M. Hasegawa) method where 100 multiple data sets were randomly entered three times based on the arbitrary number seven. The protml output file was then entered into the consense (J. Felsenstein) program to calculate an extended majority rule consensus tree with confidence intervals.
Functional Screening of Cytochrome P450 Clones. Protocols for the expression of C-terminally His-6-tagged cytochrome P450 oxygenases in Saccharomyces cerevisiae and for screening of activity by in vivo feeding of the transformed yeast with taxoid substrates are described in refs. 18, 20, and 21. Sequence-verified pYES2.1/V5-HIS-TOPO clones were transformed into S. cerevisiae strain WAT11 cells, which express an Arabidopsis thaliana NADPH-cytochrome P450 reductase (36). Control cells contained the same vector but with a β-galactosidase gene, and both control cells and cells harboring a Taxus cytochrome P450 cDNA were grown, induced, and harvested as described in refs. 18 and 23.
Microsomal proteins from recombinant yeast cells were subjected to SDS/PAGE and immunoblotting to monitor expression (23). Aliquots of these expression-verified cultures were then incubated overnight with 0.2 μCi (1 Ci = 37 GBq) of each taxoid test substrate. The preparations of the [20-3H]-labeled, racemic test substrates (all at 2 Ci/mol) taxa-4(5),11(12)-diene, taxa-4(20),11(12)-dien-5α-ol, taxa-4(20),11(12)-dien-5α-yl acetate, taxa-4(20),11(12)-dien-5α-acetoxy-10β-ol, and taxa-4(20),11(12)-dien-5α,13α-diol are described in refs. 23, 37, and 38. After incubation, the radiolabeled products were extracted and separated by reversed-phase radio-HPLC for subsequent GC-MS and 1H-NMR analysis by established procedures (21, 23).
Results and Discussion
Random Sequencing of a Taxus Cell cDNA Library. The mRNA used for λZAPII library construction (18, 24) was extracted from cultured T. cuspidata cells 16 h after treatment with methyl jasmonate, at the initiation of Taxol production (9) when mRNA levels were at maximum, and transcripts for all relevant biosynthetic enzymes should be present (11). This library has been used successfully in the isolation of several genes of taxoid metabolism, including six cytochrome P450 taxoid hydroxylases (18-22) and five acyl and aroyl CoA-dependent transferases (25-27, 30, 31). The attempted sequencing of 10,000 anonymous cDNA clones yielded 8,424 successful reactions that led to the identification of 3,563 unique transcripts.
Plastidial Precursor Supply and Early Steps of Taxol Formation. Data analysis revealed (Table 1) at least one EST encoding each of the seven enzymes of the plastidial MEP pathway for the supply of IPP and DMAPP from pyruvate and glyceraldehyde 3-phosphate (for recent reviews of this pathway, see refs. 39-42). Given that taxoid biosynthesis is substantially induced in the source Taxus cells (9), the up-regulation of this pathway for precursor supply might be anticipated. ESTs encoding 1-deoxy-D-xylulose-5-phosphate (DXP) reductoisomerase (GenBank accession no. AY575140) were substantially more abundant than acquisitions encoding other MEP pathway enzymes, although transcripts encoding all but MEP cytidyltransferase and hydroxymethylbutenyl diphosphate reductase were well represented. DXP reductoisomerase and DXP synthase have both been implicated as catalyzing slow steps in the biosynthesis of plastid-derived terpenoids (43, 44).
Table 1. T. cuspidata cell culture ESTs of the MEP and taxoid biosynthetic pathways.
| EST identification | No. of hits (of 8,424) | Abundance, % |
|---|---|---|
| DXP synthase | 4 | 0.5 |
| DXP reductoisomerase | 14 | 1.7 |
| MEP cytidyltransferase | 1 | 0.1 |
| CDP-ME kinase | 4 | 0.5 |
| MEcPP synthase | 8 | 0.9 |
| HMBPP synthase | 5 | 0.6 |
| HMBPP reductase | 1 | 0.1 |
| IPP isomerase | 1 | 0.1 |
| GGPP synthase | 14 | 1.7 |
| Taxadiene synthase | 41 | 4.9 |
| Taxoid 2α-hydroxylase | 6 | 0.7 |
| Taxoid 5α-hydroxylase | 7 | 0.8 |
| Taxoid 7β-hydroxylase | 10 | 1.2 |
| Taxoid 10β-hydroxylase* | 1 | 0.1 |
| Taxoid 13α-hydroxylase | 7 | 0.8 |
| Taxoid 14β-hydroxylase | 13 | 1.5 |
| Cytochrome P450 reductase | 5 | 0.6 |
| Taxoid 2-O-benzoyltransferase | 16 | 1.9 |
| Taxoid 5-O-acetyltransferase | 3 | 0.4 |
| Taxoid 10-O-acetyltransferase | 9 | 1.1 |
| Taxoid 13-O-side chain transferase | 21 | 2.4 |
| Taxoid 3′-N-benzoyltransferase | 10 | 1.2 |
| Phenylalanine aminomutase | 5 | 0.6 |
MEcPP, 2-C-methyl-d-erythritol 2,4-cyclopyrophosphate; HMBPP, hydroxymethylbutenyl diphosphate.
New taxoid 10β-hydroxylase described herein. For full description of the MEP pathway and enzymes see ref. 42.
ESTs encoding GGPP synthase were quite abundant, but only one hit for IPP isomerase was noted (Table 1). The construction of GGPP (C20) by GGPP synthase requires three IPP (C5) units for each DMAPP (C5) starter. Because the MEP pathway yields IPP and DMAPP in a 5:1 ratio (45), conversion of IPP to DMAPP by plastidial IPP isomerase could establish a more appropriate ratio for GGPP synthesis [i.e., at equilibrium IPP isomerase favors the allylic isomer (46)].
ESTs corresponding to taxadiene synthase (47), responsible for the committed step of Taxol biosynthesis (12), were very abundant with 41 acquisitions. These sequences represent two previously identified, catalytically comparable variants (20) of 36 ESTs and 5 ESTs. Despite the relatively high level of expression of this gene, in vivo studies indicate that this cyclization is a slow step of the pathway (12, 48), perhaps reflecting the low turnover rate of this enzyme (0.01 s-1) (49).
Cytochrome P450 Oxygenases. Cytochrome P450-mediated oxygenations constitute nearly half of the reactions of the Taxol biosynthetic pathway (Fig. 1). Previously identified cytochrome P450 taxoid oxygenase clones were acquired by differential display methods or more traditional homology-based screening of this same induced T. cuspidata cell cDNA library, with functional assessment of activity by expression in yeast or insect cells. This group of cDNA clones, including the taxoid 2α-, 5α-, 7β-, 10β-, and 13α-hydroxylases (18-22), represents a family of cytochrome P450 sequences that display high similarity (>70%) within the group but appear to be only distantly related to other cytochrome P450s of plant origin (<35% similarity). The selection of new candidate clones as potential taxoid oxygenases could thus be made by sequence relatedness.
Database searches identified 285 ESTs of the 8,424 T. cuspidata ESTs as sequences showing good homology to cytochrome P450 monooxygenases, and these corresponded to 98 unique cytochrome P450 transcripts. The most abundant cytochrome P450 (28 ESTs) represents a gene of unknown function that shows significant similarity (52%) to (S)-N-methylcoclaurine 3′-hydroxylase involved in the biosynthesis of benzylisoquinoline alkaloids (50). Several other cytochrome P450s were readily assigned to hydroxylases of known function, including cinnamate 4-hydroxylases (11 ESTs) and flavanone 3-hydroxylase (7 ESTs).
In addition to the ESTs that were identical to previously identified taxoid hydroxylases, and that were generally well represented (Table 1), 70 ESTs, corresponding to 19 different genes, were identified that showed high similarity (>75%) to these defined taxoid hydroxylases. Nine of these 19 ESTs had been encountered in previous cloning attempts (18, 20). Thus, the EST project has added 10 new candidate taxoid hydroxylases to the group. At present, six of these 19 cytochrome P450 clones are established as encoding a relevant enzyme of taxoid metabolism. It is presumed that the remaining regioselective oxygenases of Taxol biosynthesis (i.e., the C1β- and C9α-taxane core hydroxylases, the C4-C20 double bond β-epoxidase, the β-phenylalanyl side-chain 2′-hydroxylase, and, possibly, a hydroxylase responsible for oxidizing the C9-hydroxyl function to a carbonyl) reside within this family of clones. Although not strictly a component of the Taxol pathway, the NADPH:cytochrome P450 reductase (required for electron transfer to the various cytochrome P450 oxygenases) was represented in the library by five ESTs.
Most of the previously identified regiospecific hydroxylases of taxoid metabolism were well represented in the EST library (Table 1), including the taxoid 14β-hydroxylase (13 ESTs) involved in the production of a family of side-route metabolites (23) that are abundant in Taxus cell cultures (9, 32). Curiously, not a single EST was identified that corresponded to the taxoid 10β-hydroxylase clone previously isolated (18, 20). This enzyme converts taxadien-5α-yl-acetate to the corresponding 10β-hydroxy derivative (18, 37).
To search for an alternative taxoid 10β-hydroxylase, the previously uncharacterized candidate cytochrome P450 oxygenases were subcloned into the yeast expression vector pYES2.1 TOPO for expression in the S. cerevisiae strain WAT11 host (36) as described in refs. 18 and 23. Transgene expression was confirmed by immunoblot analysis (23) by taking advantage of the appended C-terminal epitope tag, and test of function was conducted by established protocols for in vivo feeding of the transformed yeast by using the available taxoid substrates. These same functional screening procedures have been used in the isolation of five other cytochrome P450 taxoid hydroxylases (18, 20-23).
Of the previously uncharacterized cytochrome P450 clones evaluated by this method, only one (designated clone 1054) was capable of transforming [20-3H]taxadien-5α-yl acetate to a more polar product as demonstrated by radio-HPLC analysis. Control experiments in which yeast cells were transformed with the same vector harboring a β-galactosidase gene, instead of the cytochrome P450 ORF, showed that these cells were inactive with taxadien-5α-yl acetate (or with any of the other taxoid test substrates). The more polar metabolite was produced in relatively high yield from taxadienyl acetate (>50% conversion), and it possessed the same retention time (by HPLC and GC-MS analysis) and the identical mass spectrum as that of authentic taxa-4(20),11(12)-dien-5α-acetoxy-10β-ol (18). Preparative scale conversion of taxadien-5α-yl acetate by the intact yeast harboring cytochrome P450 clone 1054 allowed the isolation of ≈300 μg of the biosynthetic product by HPLC (>99% pure as judged by GC-MS), and the 1H-NMR spectrum of this material was also shown to be identical to that of authentic taxa-4(20),11(12)-dien-5α-acetoxy-10β-ol (18). Thus, a second taxoid 10β-hydroxylase gene was confirmed. Although the regioselectivity and substrate specificity of the recombinant enzyme have not been fully assessed (however, the other test substrates described above were inactive), these results do indicate the presence in Taxus of at least two distinct 10β-hydroxylases that are capable of mediating this presumptive early step of the Taxol biosynthetic pathway.
cDNA clone 1054 (GenBank accession no. AY563635) of 1,788 bp bears an ORF of 1,458 bp encoding a 485-residue protein of calculated molecular weight of 55,329, in agreement with the size of the heterologously expressed enzyme observed by SDS/PAGE. The deduced amino acid sequence revealed all of the characteristics typical of cytochrome P450 enzymes (51), and it displayed 68% identity and 82% similarity compared with the previously isolated taxoid 10β-hydroxylase (18); scores relative to the other defined taxoid hydroxylases (19-23) were somewhat lower (56-58% identity and 74-77% similarity).
It seemed reasonable to assume that this family of taxoid hydroxylases derived from a common progenitor by gene duplication and differentiation (52) to evolve alternative substrate selectivities and new regio- and stereochemistries of the hydroxylation reaction on the core taxadiene structure. It also seemed possible that the pattern of descent from the parental gene (as determined by sequence relatedness) might reflect the order of hydroxylations in the Taxol biosynthetic pathway starting from the presumptive initial hydroxylation at C5α of the committed taxadiene precursor (see Fig. 1).
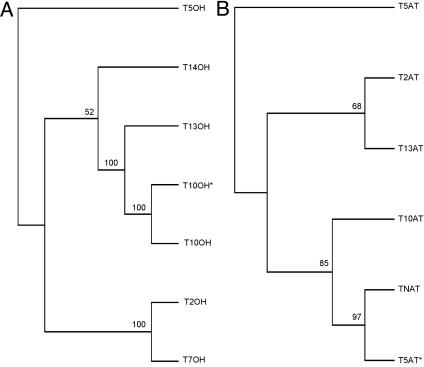
To evaluate this possibility, a cladogram of the taxoid oxygenases was constructed with rooting at the C5α-hydroxylase as the first oxygenation of the pathway (Fig. 2A). The pattern is consistent with the preliminary hydroxylation steps leading from C5 hydroxylation to C10 or C13 hydroxylation (37) and with the early emergence of C14 hydroxylation after C5 hydroxylation as a major side route (23). However, phylogenic considerations place the taxoid C7 hydroxylase closer to the C5 hydroxylase than might have been anticipated based on the proposed sequence of hydroxylations deduced from the relative abundances of taxoid metabolites functionalized at the various positions (17).
Fig. 2.
Cladograms for cytochrome P450 taxoid hydroxylases (TOH, with the position of oxygenation indicated) and taxoid acyl/aroyl transferases (TAT, with the position of addition indicated) based on amino acid alignments. Asterisks denote new sequences of recently defined function. Bootstrap values are indicated on the branches.
Acyl and Aroyl Transferases. The CoA-dependent acyl and aroyl transferases are also prominent members of the Taxol biosynthetic enzymes, constituting 5 of the 19 pathway steps. These defined transferases are remarkably abundant in the library (Table 1). In addition, 10 other distinct transferases were observed (60 total ESTs), bringing the total of this enzyme type to 15 (119 total ESTs). A very large number of taxoid side-chain variants are known that differ in position on the hydroxylated taxane core and in the type of acyl/aroyl substitution, including tiglate, hexanoate, cinnamate, and other aromatic and aminoacyl esters (53). The remaining transferase genes are probably responsible for these regiochemical and side-chain structural variants.
As with the cytochrome P450 taxoid oxygenases, it seemed reasonable to assume that this family of taxoid acyl and aroyl transferases (showing >65% similarity within the 15-member group) also derived from a common ancestral gene by duplication and differentiation (52) to evolve alternative acyl/aroyl CoA substrate selectivities and new regiochemistries for ester synthesis at the various hydroxylated positions on the taxane core and amidation of the C13-phenylisoserinoyl side chain. Here also, it might be expected that the pattern of descent as gauged by sequence relatedness could reflect the order of acylation in the Taxol biosynthetic pathway. Assessment of the acylation patterns of extant taxoid metabolites, feeding studies, and evaluation of the apparent selectivity of the recombinant acyl/aroyl transferases (14, 30, 31, 37, 53) suggest that initial acetylation at C5 of the taxane core is followed by benzoylation at C2, acetylation at C10 (ultimately leading to baccatin III), addition of the side chain at C13, and N-benzoylation of the C13 side chain.
To evaluate the possibility of such a relationship, a cladogram of the six acyltransferases was constructed with rooting at the 5α-O-acetyl transferase as the first probable acylation (Fig. 2B). The relative phylogenic placement of these transferases is generally consistent with the predicted order of acylation in Taxol biosynthesis, but the placement of a recently acquired 5-O-acetyltransferase is apparently anomalous. The specificity of this newly acquired transferase for taxoid and acyl CoA cosubstrates and its regioselectivity have not yet been fully explored, however, and its preliminary designation as a 5-O-acetyltransferase may be premature.
Remaining Pathway Steps. cDNA clones encoding several presumptive cytochrome P450 oxygenases of the Taxol biosynthetic pathway remain to be acquired. These clones include the C1β-and C9α-hydroxylases and the putative C4,C20-epoxidase, which modify the taxane core en route to baccatin III. Uncertainties about the timing of these reactions within the pathway provide little guidance as to the true substrates of these reactions. Such uncertainties have limited synthetic efforts directed to the preparation of potential “surrogate” substrates for test of function of these genes by expression in yeast and in vivo feeding studies; this general approach has been successful recently in the acquisition of both the taxoid 2α-hydroxylase (22) and the taxoid 7β-hydroxylase (21).
The oxidation of the taxane C9-hydroxyl function to the corresponding ketone could also be mediated by a cytochrome P450 hydroxylase (via the ketone hydrate). This step occurs relatively late in the pathway, where uncertainties about the preceding steps (e.g., the timing of oxetane ring formation) again constrain substrate design for test of function of the remaining candidate clones. This oxidation step may also be catalyzed by a more typical pyridine nucleotide-dependent dehydrogenase. The EST acquisitions revealed many undefined candidate dehydrogenases for this step, and it is clear that biochemical studies to define the type of enzyme involved in C9-oxidation must precede a cloning effort.
The last remaining step in the formation of baccatin III is the presumed ring expansion of the oxirane to the oxetane function with migration of the 5α-acetoxy group (see Fig. 1). Nothing is known about this reaction or its precise timing (or substrate) late in the pathway sequence, and the EST project provided no insight. Biochemical studies to define the substrate(s) and product(s) of this unusual reaction must precede the effort to acquire the responsible gene.
As regards side-chain assembly, two genes are still missing, that encoding the β-phenylalanoyl CoA ligase required as a prelude to side-chain transfer to baccatin III, and the presumed cytochrome P450 C2′ side-chain hydroxylase for the conversion of β-phenylalanoyl-baccatin III to phenylisoserinoyl-baccatin III as the penultimate step of Taxol biosynthesis (Fig. 1). Database searching revealed 22 total ESTs encoding seven distinct CoA ligases of unknown function, in addition to the well known coumaroyl CoA ligase (14 ESTs). Given the variety of acyl and aroyl substitutions found in the naturally occurring taxoids (53), it is not surprising that a number of CoA ligases are expressed in this system for the purpose of activating the various acyl and aroyl groups before transfer to the taxane core or C13 side chain.
Conclusions
The utilization of induced T. cuspidata cell transcripts to generate a cDNA library has provided a productive source of ESTs related to taxoid metabolism and terpenoid precursor supply. EST evaluation revealed 10 previously uncharacterized, putative cytochrome P450 taxoid hydroxylases, bringing this family of related (>60% identity) clones to 29. Functional testing of two of these provided a taxoid 2α-hydroxylase (22) and the second taxoid 10β-hydroxylase reported here. This group of cytochrome P450 clones almost certainly contains the remaining four oxygenases of Taxol biosynthesis. Six new acyl/aroyl transferases were observed in the induced cell library, bringing the total number to 15, of which the 5 involved in Taxol biosynthesis have been defined. The remaining transferases are likely responsible for the formation of the many taxoid side-chain structural variants, such as taxol C and cephalomannine, that differ from Taxol only in the C13 side-chain N-substitution (i.e., hexanoyl and tigloyl, respectively, rather than N-benzoyl; see Fig. 1). Also abundant in the library was the phenylalanine aminomutase (GenBank accession no. AY582743) involved in C13 side chain construction that resembles phenylalanine ammonia lyase (51% identity) but differs in mechanism in involving intramolecular migration, rather than loss, of the amino group. Seven CoA ester synthetases (ligases) of various abundances were also revealed as candidates for C13 side chain (β-phenylalanine) activation and for the synthesis of other relevant CoA esters for side-chain additions in taxoid metabolism. Although the phylogenetic tree for the acyltransferases was largely consistent with prediction based on pathway sequence, that for the cytochrome P450 oxygenases (Fig. 2) was less consistent with prediction than anticipated (albeit in the absence of confirmatory biochemical evidence), and reconstruction of the tree based on predicted substrate recognition sites (54) did not significantly alter the phylogeny. Continued exploitation of the functional genomics approach in parallel with biochemical studies should provide the necessary information to more rigorously test the hypothesis that phylogeny reflects the pathway sequence of oxygenation and acylation reactions leading to Taxol. The functional genomics approach has clearly accelerated gene discovery and characterization and has provided many promising targets for genetically engineering the production of Taxol and its precursors.
Acknowledgments
We thank R. Ketchum for helpful input, J. Tamura for typing the manuscript, D. Pompon (Gif-sur-Yvette, France) for providing WATII cells, and R. M. Williams and C. D. Rithner (Colorado State University, Fort Collins) for preparing taxoid substrates and obtaining the NMR spectrum. This work was supported by National Institutes of Health Grant CA-55254 and McIntire-Stennis Project 0967 from the Washington State University Agricultural Research Center.
Abbreviations: DMAPP, dimethylallyl diphosphate; DXP, 1-deoxy-D-xylulose-5-phosphate; GGPP, geranylgeranyl diphosphate; IPP, isopentenyl diphosphate; MEP, 2-C-methyl-D-erythritol phosphate.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY563635, AY575140, and AY582743).
References
- 1.Goldspiel, B. R. (1997) Pharmacotherapy 17, 1105-1255. [PubMed] [Google Scholar]
- 2.Skeel, R. T. (1999) in Handbook of Chemotherapy, 5th Ed., ed. Skeel, R. T. (Lippincott Williams & Wilkins, Baltimore), pp. 63-144.
- 3.Michaud, L. B., Valero, V. & Hortobagyi, G. (2000) Drug Saf. 23, 401-428. [DOI] [PubMed] [Google Scholar]
- 4.Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P. & McPhail, A. T. (1971) J. Am. Chem. Soc. 93, 2325-2327. [DOI] [PubMed] [Google Scholar]
- 5.Wuts, P. G. M. (1998) Curr. Opin. Drug Discov. Dev. 1, 329-337. [PubMed] [Google Scholar]
- 6.Kingston, D. G. I., Jagtap, P. G., Yuan, H. & Samala, L. (2002) Prog. Chem. Org. Nat. Prod. 84, 53-225. [DOI] [PubMed] [Google Scholar]
- 7.Suffness, M. (1995) Am. Chem. Soc. Symp. Ser. 583, 1-17. [Google Scholar]
- 8.Ketchum, R. E. B. & Gibson, D. M. (1996) Plant Cell Tissue Organ Cult. 46, 9-16. [Google Scholar]
- 9.Ketchum, R. E. B., Gibson, D. M., Croteau, R. B. & Shuler, M. L. (1999) Biotechnol. Bioeng. 62, 97-105. [DOI] [PubMed] [Google Scholar]
- 10.Eisenreich, W., Menhard, B., Hylands, P. J., Zenk, M. H. & Bacher, A. (1996) Proc. Natl. Acad. Sci. USA 93, 6431-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hefner, J., Ketchum, R. E. B. & Croteau, R. (1998) Arch. Biochem. Biophys. 360, 62-74. [DOI] [PubMed] [Google Scholar]
- 12.Koepp, A. E., Hezari, M., Zajicek, J., Stofer Vogel, B., LaFever, R. E., Lewis, N. G. & Croteau, R. (1995) J. Biol. Chem. 270, 8686-8690. [DOI] [PubMed] [Google Scholar]
- 13.Hezari, M., Lewis, N. G. & Croteau, R. (1995) Arch. Biochem. Biophys. 322, 437-444. [DOI] [PubMed] [Google Scholar]
- 14.Walker, K. D. & Croteau, R. (2001) Phytochemistry 58, 1-7. [DOI] [PubMed] [Google Scholar]
- 15.Jennewein, S. & Croteau, R. (2001) Appl. Microbiol. Biotechnol. 57, 13-19. [DOI] [PubMed] [Google Scholar]
- 16.Hefner, J., Rubenstein, S. M., Ketchum, R. E. B., Gibson, D. M., Williams, R. M. & Croteau, R. (1996) Chem. Biol. 3, 479-489. [DOI] [PubMed] [Google Scholar]
- 17.Floss, H. G. & Mocek, U. (1995) in Taxol: Science and Applications, ed. Suffness, M. (CRC, Boca Raton, FL), pp. 191-208.
- 18.Schoendorf, A., Rithner, C. D., Williams, R. M. & Croteau, R. (2001) Proc. Natl. Acad. Sci. USA 98, 1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennewein, S., Rithner, C. D., Williams, R. M. & Croteau, R. (2001) Proc. Natl. Acad. Sci. USA 98, 13595-13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jennewein, S., Long, R. M., Williams, R. M. & Croteau, R. (2004) Chem. Biol. 11, 379-387. [DOI] [PubMed] [Google Scholar]
- 21.Chau, M., Jennewein, S., Walker, K. & Croteau, R. (2004) Chem. Biol. 11, 663-672. [DOI] [PubMed] [Google Scholar]
- 22.Chau, M. & Croteau, R. (2004) Arch. Biochem. Biophys., in press. [DOI] [PubMed]
- 23.Jennewein, S., Rithner, C. D., Williams, R. M. & Croteau, R. (2003) Arch. Biochem. Biophys. 413, 262-270. [DOI] [PubMed] [Google Scholar]
- 24.Walker, K., Schoendorf, A. & Croteau, R. (2000) Arch. Biochem. Biophys. 374, 371-380. [DOI] [PubMed] [Google Scholar]
- 25.Walker, K. & Croteau, R. (2000) Proc. Natl. Acad. Sci. USA 97, 583-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker, K. & Croteau, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guéritte-Voegelein, F., Guénard, D. & Potier, P. (1987) J. Nat. Prod. 50, 9-18. [DOI] [PubMed] [Google Scholar]
- 28.Walker, K. D. & Floss, H. G. (1998) J. Am. Chem. Soc. 120, 5333-5334. [Google Scholar]
- 29.Steele, C. L., Chen, Y., Dougherty, B., Hofstead, S., Lam, K. S., Li, W. & Xing, Z. (2000) International Patent WO 03/066871.
- 30.Walker, K., Long, R. & Croteau, R. (2002) Proc. Natl. Acad. Sci. USA 99, 9166-9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker, K., Fujisaki, S., Long, R. & Croteau, R. (2002) Proc. Natl. Acad. Sci. USA 99, 12715-12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ketchum, R. E. B., Rithner, C. D., Qiu, D., Kim, Y. S., Williams, R. M. & Croteau, R. B. (2003) Phytochemistry 62, 901-909. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz, S., Kent, W. J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R. C., Haussler, D. & Miller, W. (2003) Genome Res. 13, 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson, W. & Lipman, D. J. (1988) Proc. Natl. Acad. Sci. USA 85, 2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genetics Computer Group (2000) Program Manual for the Wisconsin Package (Genetics Computer Group, Madison, WI), Version 10.0.
- 36.Pompon, D., Louerat, B., Bronine, A. & Urban, P. (1996) Methods Enzymol. 272, 51-64. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler, A.L., Long, R. M., Ketchum, R. E. B., Rithner, C. D., Williams, R. M. & Croteau, R. (2001) Arch. Biochem. Biophys. 390, 265-278. [DOI] [PubMed] [Google Scholar]
- 38.Rubenstein, S. M., Vazquez, A., Sanz-Cervera, J. F. & Williams, R. M. (2000) J. Labelled Comp. Radiopharm. 43, 481-491. [Google Scholar]
- 39.Lichtenthaler, H. K., Rohmer, M. & Schwender, J. (1997) Physiol. Plant. 101, 643-652. [Google Scholar]
- 40.Rohmer, M. (1999) Nat. Prod. Rep. 16, 565-574. [DOI] [PubMed] [Google Scholar]
- 41.Eisenreich, W., Rohdich, F. & Bacher, A. (2001) Trends Plant Sci. 6, 78-84. [DOI] [PubMed] [Google Scholar]
- 42.Kuzuyama, T. & Seto, H. (2003) Nat. Prod. Rep. 20, 171-183. [DOI] [PubMed] [Google Scholar]
- 43.Mahmoud, S. S. & Croteau, R. (2001) Proc. Natl. Acad. Sci. USA 98, 8915-8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Estevez, J. M., Cantero, A., Reindl, A., Reichler, S. & Leon, P. (2001) J. Biol. Chem. 276, 22901-22909. [DOI] [PubMed] [Google Scholar]
- 45.Rohdich, F., Hecht, S., Gärtner, K., Adam, P., Krieger, C., Amslinger, S., Arigoni, D., Bacher, A. & Eisenreich, W. (2002) Proc. Natl. Acad. Sci. USA 99, 1158-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramos-Valdivia, A. C., van der Heijden, R. & Verpoorte, R. (1997) Nat. Prod. Rep. 14, 591-603. [DOI] [PubMed] [Google Scholar]
- 47.Wildung, M. R. & Croteau, R. (1996) J. Biol. Chem. 271, 9201-9204. [DOI] [PubMed] [Google Scholar]
- 48.Hezari, M., Ketchum, R. E. B., Gibson, D. M. & Croteau, R. (1997) Arch. Biochem. Biophys. 337, 185-190. [DOI] [PubMed] [Google Scholar]
- 49.Williams, D. C., Wildung, M. R., Jin, A. Q., Dalal, D., Oliver, J. S., Coates, R. M. & Croteau, R. (2000) Arch. Biochem. Biophys. 379, 137-146. [DOI] [PubMed] [Google Scholar]
- 50.Pauli, H. H. & Kutchan, T. M. (1998) Plant J. 13, 793-801. [DOI] [PubMed] [Google Scholar]
- 51.von Wachenfeldt, C. & Johnson, E. F. (1995) in Cytochrome P450: Structure, Mechanism, and Biochemistry, ed. Ortiz de Montellano, P. R. (Plenum, New York), pp. 183-223.
- 52.Pichersky, E. & Gang, D. R. (2000) Trends Plant Sci. 5, 439-445. [DOI] [PubMed] [Google Scholar]
- 53.Baloglu, E. & Kingston, D. G. I. (1999) J. Nat. Prod. 62, 1448-1472. [DOI] [PubMed] [Google Scholar]
- 54.Gotoh, O. (1992) J. Biol. Chem. 267, 83-90. [PubMed] [Google Scholar]