Abstract
NATs (natural antisense transcripts) are important regulators of eukaryotic gene expression. Interference between the expression of protein-coding sense transcripts and the corresponding NAT is well documented. In the present review, we focus on an additional, higher-order role of NATs that is currently emerging. The recent discovery of endogenous siRNAs (short interfering RNAs), as well as NAT-induced transcriptional gene silencing, are key to the proposed novel function of NATs.
Keywords: endogenous short interfering RNA, monoallelic gene expression, natural antisense transcript, non-protein-coding RNA, random imprinting, transcriptional silencing
Introduction
The sequencing of the human genome revealed that the number of protein-coding genes does not scale with organismal complexity [1]. Interestingly, most of the coding genes are well-conserved in animal evolution and orthologous proteins fulfil similar biological roles in different species. The difference in biological complexity in animals is therefore unlikely to be entwined in protein-coding genes. Emerging evidence from several areas of research strongly supports the view that non-protein-coding RNA is essential in establishing organismal complexity [2]. Driven by rapid technical advances in transcriptome analysis and high-throughput sequencing, the overwhelming complexity of the non-protein-coding transcriptome is currently emerging. One of the formidable challenges will be to separate non-protein-coding transcripts with a regulatory role in gene expression from transcriptional noise. Considering the shear amount of information that has recently been published in the field of non-protein-coding RNAs, this review attempts to not miss the forest for the trees, but also avoid cherry-picking.
Genome-wide transcription
Tiling arrays and high-throughput sequencing strategies have revealed a plethora of non-protein-coding transcripts from both genic and intergenic regions, indeed most of the genome seems to be sporadically transcribed [3]. Based on a 200 nucleotide cut-off according to RNA purification protocols, the transcripts are classified as short RNAs and long non-protein-coding RNAs [4]. Structural and functional features can be used to classify transcripts into broad groups with common biological roles or raisons d’être. A brief summary of selected RNA groups that are often discussed in the context of NATs (natural antisense transcripts) will be given. These include various classes of short RNAs, promoter-associated transcripts and long non-protein-coding RNAs (lincRNAs). The short profiles of the selected RNA groups are based on findings from animal model systems. This limitation is to avoid the dilemma of studying drivers of organismal complexity (non-protein-coding RNAs) in systems that score comparably low in that very discipline.
Short RNAs
Short RNAs derive from double-stranded RNA precursors by the processing of type III RNA endonucleases. The resulting oligonucleotides of 20–24 nucleotides promote post-transcriptional gene silencing. The two main classes of short RNAs are miRNAs (microRNAs) and siRNAs (short interfering RNAs) [5]. A third class of short RNAs, piRNAs (piwi-interacting RNAs) are slightly longer (26–32 nucleotides) and derive from single-stranded precursors [6].
miRNAs are important regulators of gene expression at the level of translation, with an essential input in developmental processes. Inhibitory complexes are guided to target mRNAs that are largely, but not fully, complementary to the miRNA. Inhibition is not necessarily linked to mRNA degradation [5].
siRNAs are perfectly complementary to the target sequence and result in endonucleic cleavage of the mRNA. In plants, siRNAs represent a powerful defence strategy against viruses. Accordingly, plant cells produce virus-derived siRNAs upon infection, whereas animal cells do not [7]. Defence against viruses is largely covered by the immune system and the biological role of siRNAs in animals is speculative [8]. siRNAs that derive from endogenous sources (endo-siRNAs) will be discussed in conjunction with NATs below.
piRNAs are germ-line-specific short RNAs, and male mice that lack key enzymes in piRNA processing become sterile [9]. In contrast with the other short RNAs mentioned so far, the biogenesis of piRNAs does not depend on RNase type III enzymes [10]. They derive from distinct non-coding regions of the genome and suppress transposon activity by transcriptional silencing [11].
Promoter associated transcripts
Pioneering studies by Kapranov and others [12,13] identified short RNAs and unstable transcripts of up to 600 nucleotides which mapped to gene boundaries. Several distinct RNA species were observed that originated either from transcription start sites [PASRs (promoter-associated short RNAs)], terminator sites [TASRs (termini-associated short RNAs)] or promoter regions [PROMPTs (promoter upstream transcripts)]. PASRs and TASRs are between 20 and 90 nucleotides long, whereas PROMPTs are usually longer than 200 nucleotides [13]. Interestingly, all of the different RNAs occur in sense and antisense orientations and are generally short lived [14]. The transcripts are assumed to reflect inherent properties of RNA polymerase II, such as promiscuous promoter binding and transcription initiation, as well as discontinuous elongation [15,16]. The fact that both gene boundaries display these spurious RNAs refers to a putative loop structure of transcribed genes, but also raises the intriguing perspective of synthesizing stable natural antisense transcripts [17].
lincRNAs
lincRNAs play a well-described role in X chromosome inactivation and parental imprinting. The transcripts Air, Kcnq1ot1 or Xist, for example, recruit repressive regulatory complexes and result in chromatin silencing of the corresponding locus or X allele respectively [18]. Another example of RNA-directed repressive chromatin modifications has recently been reported for the developmental regulation of Hox gene expression [19]. Interestingly, the non-protein-coding RNA essential to this process, called HOTAIR, is transcribed from an intergenic region and acts in trans. The idea of regulatory RNAs from intergenic regions was further developed: based on chromatin marks, over a thousand lincRNAs were identified and experimentally confirmed [20]. Many of these transcripts recruit chromatin-modifying complexes; however, other mechanisms of action also apply [18,21].
NATs
A particularly intriguing group of non-protein-coding RNAs are NATs. These are generally non-protein-coding, but fully processed, mRNAs that are transcribed from the opposite strand of protein-coding sense transcripts [22]. After splicing, the sense and the corresponding antisense transcripts share complementary exons and potentially form RNA–RNA hybrids. To highlight the potential regulatory impact on the cognate sense transcript, NATs are also referred to as cis-NATs. This definition of NATs originates from the pre-genomic area when the vast majority of non-protein-coding RNAs included the well-characterized transfer, ribosomal and spliceosomal RNAs, plus a few antisense transcripts from genic loci. Experiments involving NATs usually used the criterion of mRNA processing to distinguish between breakdown products of sense transcripts and ‘real’ NATs. These constraints applied to both large-scale cDNA sequencing projects pioneered by the FANTOM consortium as well as early bioinformatic approaches [23-25].
The focus on genic transcripts gives an incomplete account of cellular non-protein-coding and complementary RNAs; nevertheless, these early studies revealed a number of important findings. Despite the rather stringent limitations to qualify as a NAT (evidence of mRNA-like processing), up to 72% of all mouse genes show evidence of bi-directional transcription [26]. Comparable numbers are found in human and other animals. Caenorhabditis elegans represents an exception with a markedly decreased number of NATs [27]. Phylogenetic comparison of NATs in human, mouse and puffer fish indicated conservation of antisense transcription [28]. Neither coding potential nor splice structure, however, are under stringent selective pressure. The most striking feature of NATs represents their significant under-representation on the human and murine X chromosome [23,29]. Interestingly, NATs that are expressed from mammalian X chromosomes tend to be clustered in areas that escape inactivation [29]. In addition, flies and worms that adjust the output from the X chromosomes, but keep both alleles active, show equal distribution of NATs on the X chromosomes and autosomes [27]. These findings indicate that the expression of NATs is favourable on a bi-allelic background, but counter-indicated if monoallelically expressed. Interestingly, the bias only applies to NATs with exon complementarity to the sense transcript, but not antisense transcription as such. This observation indicates that an RNA–RNA intermediate is essential in the biological role of NATs. Taken together, these findings point towards a higher-order biological function of NATs that is not linked to the ontology of bi-directionally transcribed genes [30]. An emerging hypothesis describing this higher-order role will be presented below.
On the other hand, there is a wealth of well-documented examples of NATs that regulate the expression of their corresponding protein-coding sense transcripts [31]. Generally, increased levels of a specific NAT have an inhibitory influence on the sense transcript. However, NATs that protect the sense transcript against nuclease degradation have also been reported [32]. These findings are of clinical relevance because genes that are linked to cancer and other diseases are regulated by NATs. Excellent recent reviews discuss the gene-specific effects of NATs in detail, therefore this aspect will not feature prominently in the present review [33].
A higher-order function of NATs
Expression of NATs in specific tissues provides clues about their physiological role. Unfortunately, a comprehensive atlas of NATs is yet to be established. Genome-wide expression of NATs has been reported; however, the data were generated using human cell lines and is therefore of limited use to inquire tissue-specific expression patterns [34]. We used Affymetrix gene expression arrays to assess the level of NATs in selected tissues [35]. The commercial arrays contain probe sets that hybridize to the opposite strand of annotated exons and will therefore bind to antisense transcripts complementary to this region [35-37]. We identified the probe sets and used them to mine datasets from different mouse tissues. NATs, as represented by the unbiased selection of probes, were most prominently expressed in testis, more specifically in haploid spermatids [38]. This finding was confirmed using other approaches [39]. Low signals were found in all tissues [25,35] and, interestingly, NATs were strongly correlated with the expression of the corresponding sense transcript. These studies do not specify whether sense and antisense transcripts are expressed in the same cell, but co-expression has been confirmed for specific bi-directionally transcribed genes [40]. Consequently, the hypothesized higher-order function for NATs is likely to be important during spermogenesis, but may also be relevant in other organs or specific cell populations. In addition, the expression of NATs seems to be linked to the expression of the sense transcripts and involve the formation of RNA hybrids.
NATs as a source of endo-siRNAs
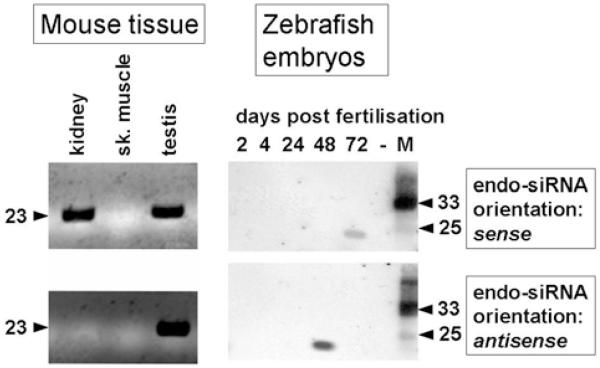
Long perfect RNA-hybrids in the cytoplasm of a cell are a sign of viral infection and trigger a stress response through protein kinase R. In the nucleus, however, RNA duplexes may have other functions. The sense/antisense hybrids could become edited by the enzyme Adar (adenosine deaminase acting on RNA) or alternatively feed into an RNAi (RNA interference)-related pathway [41]. A bioinformatic survey revealed that RNA editing was prevalent in introns of primary transcripts, but absent from sense/antisense complementary exons [42]. The connection between complementary transcripts and RNAi was initially challenged due to the lack of NAT-related clones in short RNA-sequencing projects. More recent RNA-Seq results and our own work, however, corroborate a link between NATs and RNAi [38,43-45]. The term ‘endo-siRNAs’ was coined to highlight the endogenous origin of these short RNA species. Most interestingly, only one strand of the original duplex was usually detected, indicating non-random processing of the double-stranded RNA precursor. We observed a switch in orientation of gene-specific endo-siRNAs that was developmentally induced during zebrafish embryogenesis (Figure 1). The orientation correlated with the expression level of the corresponding sense or antisense transcript [43]. Detection of both strands of siRNAs and miRNAs confirms that strand selection of short RNAs may not be merely driven by the physico-chemical properties of the RNA hybrid, but also by the amount of sense or the antisense transcript [46-49].
Figure 1. Endo-siRNAs from sense/antisense transcripts in mouse and zebrafish.
The sense transcript encodes a sodium/phosphate transporter, the antisense transcript is most likely non-protein-coding. Northern blot analysis of RNA extracted from different mouse tissues (left-hand panel) and zebrafish embryos (right-hand panel) shows the expression of endo-siRNAs. The orientation of the oligonucleotides is tissue- or development-specific. The molecular mass in kDa is indicated. M, molecular mass marker lane.
NATs and transcriptional gene silencing
RNA-directed transcriptional gene silencing is well-established in yeast and plants. To what extent these findings were relevant to mice and humans was unclear because mammals lack one of the key enzymes of the process which enables amplification of the RNA signal [50]. Few endogenous antisense RNAs were found to silence the corresponding sense transcripts, indicating that a comparable mechanism may also apply in mammals. For example, a rare form of α-thalassaemia was shown to be caused by a rearranged, constitutively expressed LUC7 gene. The LUC7 mRNA is antisense to HBA2 and resulted in methylation and concomitant inactivation of the HBA2 promoter [51]. In addition, the tumour suppressor gene p15 was found to be repressed by a NAT in leukaemia [52]. The specific role of the (antisense) RNAs in these processes, however, was unclear. Pioneering work from K. Morris’s laboratory and others has now deciphered the process in some detail [53,54]. Using exogenous siRNAs they showed that the gene encoding the elongation factor 1α (EF1A) could be transcriptionally silenced. Transcriptional repression was associated with methylation of the EF1A promoter and required targeting of the siRNAs to the nucleus [54]. In addition, AGO1 (Argonaute 1), TRBP2 (TAR RNA-binding protein-2) and Polycomb protein EZH2 were shown to mediate transcriptional silencing of CCR5 (human immunodeficiency virus-1 co receptor) and tumour suppressor RASSF1A [55]. Interestingly, comparable studies revealed the unexpected finding that short RNAs could not only silence genes, but also activate their expression [56,57]. Activation was associated with increased di- and tri-methylation of histone H3K4 and reduced acetylation at histones H3K9 and H3K14. When the targets for several activating siRNAs were examined (progesterone receptor gene, E-cadherin, p21) it emerged that all genes were bi-directionally transcribed and the activating oligonucleotides targeted in fact the respective antisense transcripts [49,58]. Indeed, suppression of the p21 antisense transcript resulted in a loss of suppressive histone modifications in the sense promoter [49]. These findings were recently shown to apply to other mammals, such as monkeys and rodents [59]. A strong link between NATs and transcriptional gene silencing is suggested by the correlation of NATs and genes that show random monoallelic expression [38].
This, to a certain extent, closes a circle and explains the initial observation that NATs are under-represented on the X chromosome. Because only one copy of the X chromosome is usually active, NAT-induced silencing would trigger complete gene knockdown with potentially fatal consequences for the affected cell. If a comparable incident happened on autosomes, the second allele would compensate for the loss of function. This argument is supported by the fact that NATs expressed from mammalian X chromosomes tend to be clustered in areas that escape inactivation [29].
What good are NATs for? A hypothesis
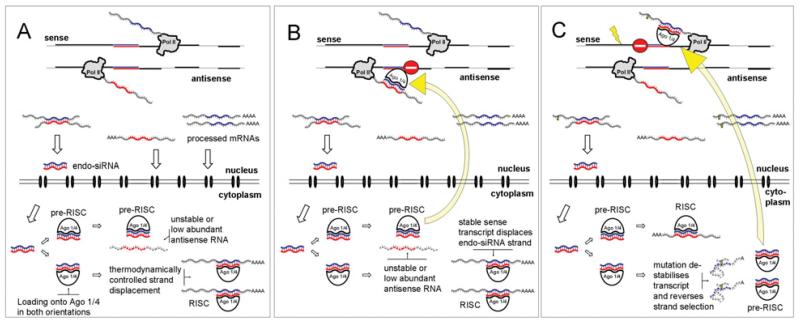
To conclude this review, we attempt to answer the initial question and suggest a hypothesis of a higher-order biological role for NATs (Figure 2). The model predicts that sense and antisense transcripts are transiently co-expressed. The transcripts either persist as mRNAs or, alternatively, hybridize and initiate the processing into endo-siRNAs. Based on indirect evidence we hypothesize that the selection of a specific endo-siRNA strand is related to the formation of the RISC (RNA-induced silencing complex) with a non-slicing Argonaute protein [60]. Interestingly, the expression of the non-slicing Ago4 protein in testis mirrors the pattern established for NATs [61]. Recent investigations into RISC assembly revealed that the unwinding of the siRNA duplex after Ago1 binding is thermodynamically controlled [60]. We therefore hypothesize that double-stranded endo-siRNAs bind in random orientation to Ago4 to form a ‘pre-RISC’. If the fully processed mRNA is accessible in the cytoplasm, a RISC will be formed and contained in cytoplasmic compartments. If the target is unavailable in the cytoplasm, the pre-RISC may diffuse into the nucleus, target the nascent complementary transcript and induce a silencing response. It is conceivable that the RISC complexes in cytoplasm and nucleus have different stabilities and fates resulting in the detection of skewed single-stranded endo-siRNA ratios. To summarize, strand selection will be determined by the availability of stable mRNAs complementary to the endo-siRNAs in the cytoplasm. Any mutation that reduces cytoplasmic mRNA levels by, for example, affecting stability or nuclear export may induce selection of the opposite endo-siRNA strand and re-direction of transcriptional silencing.
Figure 2. Working model of gene regulation by NATs.
(A and B) Representation of the physiological situation where a stable, correctly folded, protein-coding sense transcript is produced. Upon transcription and processing of both sense and antisense transcripts, the RNAs are either processed into endo-siRNAs or transported into the cytoplasm. In the cytoplasm, the endo-siRNAs are loaded on to a non-slicing Argonaute protein (most probably Ago4 or Ago1) in both orientations (pre-RISC). The availability of a target sequence will drive RISC formation [60]. The resulting complex will stay in the cytoplasm and the mRNA may eventually become translated. If no target RNA molecules are available, the pre-RISC complex is transported back into the nucleus and induces silencing of the complementary transcript. (C) Situation where a mutation changes the stability or the folding of the protein-coding sense transcript. As a consequence, RISC is not formed with the sense but with the now more abundant antisense RNA, and the mutated sense transcript will become silenced. We can only speculate on the allele-specificity of the process; however, it is likely that the timing of the entire process, transcription→processing→feedback, is essential.
The impact of the suggested model becomes tangible when the hypothesis outlined in the previous paragraph is considered in conjunction with the expression pattern of NATs. NATs are most prominently expressed in testis and, at low levels, in other tissues. In haploid cells in testis (spermatids), mutations that trigger NAT-induced gene silencing will cause a complete gene knockdown. As a consequence, mutated (‘unfit’) cells will be preferentially eliminated and a positively selected sperm population will emerge. In diploid cells, on the other hand, the mutagenic environment will induce progressive monoallelic gene expression. The random but cumulative nature of these changes in slowly regenerating somatic cells may contribute to senescence.
Essential questions remain to be answered; for example, how allele specificity of the entire process is achieved. In addition, elements of the model that are based on indirect evidence require robust experimental scrutiny. Nevertheless, endo-siRNAs and transcriptional gene silencing seem to make sense in the context of antisense.
Acknowledgments
Funding: This work is supported by the Dunhill Medical Trust.
Abbreviations used
- Ago
Argonaute
- lincRNA
long non-protein-coding RNA
- miRNA
microRNA
- NAT
natural antisense transcript
- PASR
promoter-associated short RNA
- piRNA
piwi-interacting RNA
- PROMPT
promoter upstream transcript
- RISC
RNA-induced silencing complex
- RNAi
RNA interference
- siRNA
short interfering RNA
- endo-siRNA
endogenous siRNA
- TASR
termini-associated short RNA
References
- 1.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 2008;20:274–280. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol. Cell. 2007;26:603–609. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [DOI] [PubMed] [Google Scholar]
- 10.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 11.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 13.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 14.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 16.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 17.Saiz L, Vilar JM. DNA looping: the consequences and its control. Curr. Opin. Struct. Biol. 2006;16:344–350. doi: 10.1016/j.sbi.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: a critical look in both directions. Cell. Mol. Life Sci. 2009;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yelin R, Dahary D, Sorek R, Levanon EY, Goldstein O, Shoshan A, Diber A, Biton S, Tamir Y, Khosravi R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 25.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 26.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Liu XS, Liu QR, Wei L. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 2006;34:3465–3475. doi: 10.1093/nar/gkl473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahary D, Elroy-Stein O, Sorek R. Naturally occurring antisense: transcriptional leakage or real overlap? Genome Res. 2005;15:364–368. doi: 10.1101/gr.3308405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner A, Berdal A. Natural antisense transcripts: sound or silence? Physiol. Genomics. 2005;23:125–131. doi: 10.1152/physiolgenomics.00124.2005. [DOI] [PubMed] [Google Scholar]
- 31.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, III, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1α and HIF-2α expression in lung epithelial cells: implication of natural antisense HIF-1α. J. Biol. Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 33.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werner A, Schmutzler G, Carlile M, Miles CG, Peters H. Expression profiling of antisense transcripts on DNA arrays. Physiol. Genomics. 2007;28:294–300. doi: 10.1152/physiolgenomics.00127.2006. [DOI] [PubMed] [Google Scholar]
- 36.Ge X, Rubinstein WS, Jung YC, Wu Q. Genome-wide analysis of antisense transcription with Affymetrix exon array. BMC Genomics. 2008;9:27. doi: 10.1186/1471-2164-9-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oeder S, Mages J, Flicek P, Lang R. Uncovering information on expression of natural antisense transcripts in Affymetrix MOE430 datasets. BMC Genomics. 2007;8:200. doi: 10.1186/1471-2164-8-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlile M, Swan D, Jackson K, Preston-Fayers K, Ballester B, Flicek P, Werner A. Strand selective generation of endo-siRNAs from the Na/phosphate transporter gene Slc34a1 in murine tissues. Nucleic Acids Res. 2009;37:2274–2282. doi: 10.1093/nar/gkp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada Y, Tashiro C, Numata K, Watanabe K, Nakaoka H, Yamamoto N, Okubo K, Ikeda R, Saito R, Kanai A, et al. Comparative expression analysis uncovers novel features of endogenous antisense transcription. Hum. Mol. Genet. 2008;17:1631–1640. doi: 10.1093/hmg/ddn051. [DOI] [PubMed] [Google Scholar]
- 40.Petit S, Meary F, Pibouin L, Jeanny JC, Fernandes I, Poliard A, Hotton D, Berdal A, Babajko S. Autoregulatory loop of Msx1 expression involving its antisense transcripts. J. Cell Physiol. 2009;220:303–310. doi: 10.1002/jcp.21762. [DOI] [PubMed] [Google Scholar]
- 41.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neeman Y, Dahary D, Levanon EY, Sorek R, Eisenberg E. Is there any sense in antisense editing? Trends Genet. 2005;21:544–547. doi: 10.1016/j.tig.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Carlile M, Nalbant P, Preston-Fayers K, McHaffie GS, Werner A. Processing of naturally occurring sense/antisense transcripts of the vertebrate Slc34a gene into short RNAs. Physiol. Genomics. 2008;34:95–100. doi: 10.1152/physiolgenomics.00004.2008. [DOI] [PubMed] [Google Scholar]
- 44.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 46.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35:5944–5953. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wei JX, Yang J, Sun JF, Jia LT, Zhang Y, Zhang HZ, Li X, Meng YL, Yao LB, Yang AG. Both strands of siRNA have potential to guide posttranscriptional gene silencing in mammalian cells. PLoS One. 2009;4:e5382. doi: 10.1371/journal.pone.0005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Werner A, Carlile M, Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 49.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 51.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 52.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Janowski BA, Huffman KE, Schwartz JC, Ram R, Hardy D, Shames DS, Minna JD, Corey DR. Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat. Chem. Biol. 2005;1:216–222. doi: 10.1038/nchembio725. [DOI] [PubMed] [Google Scholar]
- 54.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 55.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 56.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 57.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, Janowski BA. Antisense transcripts are targets for activating small RNAs. Nat. Struct. Mol. Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS One. 5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez-Gonzalez E, Lopez-Casas PP, del Mazo J. The expression patterns of genes involved in the RNAi pathways are tissue-dependent and differ in the germ and somatic cells of mouse testis. Biochim. Biophys. Acta. 2008;1779:306–311. doi: 10.1016/j.bbagrm.2008.01.007. [DOI] [PubMed] [Google Scholar]