Abstract
NATs (natural antisense transcripts) are widespread in eukaryotic genomes. Experimental evidence indicates that sense and antisense transcripts interact, suggesting a role for NATs in the regulation of gene expression. On the other hand, the transcription of a gene locus in both orientations and RNA hybrid formation can also lead to transcriptional interference, trigger an immune response or induce gene silencing. Tissue-specific expression of NATs and the compartmentalization of cells ensure that the regulatory impact of NATs prevails. Consequently, NATs are now acknowledged as important modulators of gene expression. New mechanisms of action and important biological roles of NATs keep emerging, making regulatory RNAs an exciting and quickly moving area of research.
Keywords: natural antisense transcript, RNA masking, transcription interference
Introduction
NATs were first described in bacteria as early as in 1981 and found to control plasmid numbers [1]. Since then, bacterial regulatory RNAs have been extensively studied. In general, they interfere with translation initiation and both inhibitory and stimulatory effects can be observed. Inhibitory NATs block or melt hairpin structures in mRNAs that are required for ribosome binding. Activation is observed when NATs counteract inhibitory structures and enable efficient translation initiation or elongation. The significance of RNA-mediated control mechanisms in maintaining homoeostasis in bacteria is unclear. An attractive hypothesis suggests that NAT-mediated regulation is not essential in single cells, but enables decisions that control collective behaviour such as biofilm formation, motility or virulence [2]. Biological roles of NATs and the related enzymatic mechanisms differ fundamentally between bacteria and higher eukaryotic systems. The present chapter focuses on the latter, although excellent reviews cover bacterial regulatory RNAs in detail [3].
Between 1986 and 2002, sporadic NATs were also discovered in eukaryotes, including mice and humans. The significance of the serendipitous findings was unclear and NATs were clearly thought to be an exception rather than the rule [4]. This image changed drastically with the start of the genomic age at the beginning of this century. Large-scale sequencing approaches, tiling arrays and data mining projects all identified staggering numbers of NATs, particularly in the transcriptomes of higher eukaryotes. The most comprehensive study was performed in mice by the FANTOM Consortium and reported that up to 72% of transcriptional units are transcribed in both orientations [5]. The figures reported for humans are considerably lower (40%) depending on different cell types [6,7]. In this context it is important to note that somewhat arbitrary parameters set to distinguish real NATs from experimental noise can skew the perceived scale of NATs. However, there is a consensus that natural antisense transcription is a pervasive and highly relevant phenomenon.
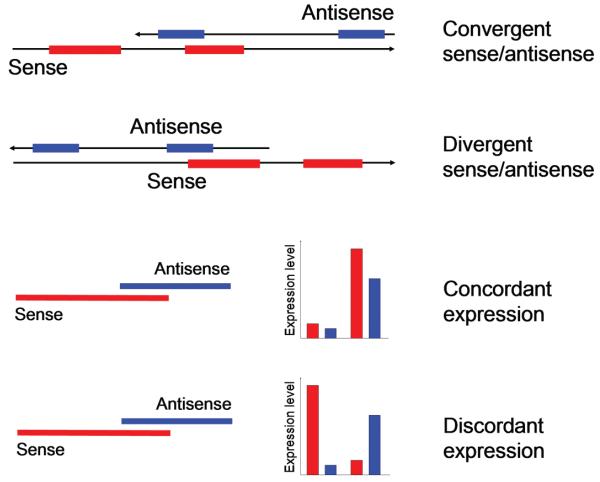
NATs can be categorized according to their mode of action or structure. Quite generally, NATs can act in cis and trans. Cis describes a regulatory impact that affects predominantly the corresponding sense transcript. Alternatively, NATs can regulate transcripts from other genomic loci, thus they act in trans. Trans-regulation is largely observed with short RNAs such as miRNAs (microRNAs) and endogenous siRNAs (small interfering RNAs) as well as with NATs transcribed from pseudogenes, which may interfere with their highly similar parent gene transcript [8]. The present chapter focuses on cis-acting NATs as the other groups of ncRNAs (non-coding RNAs) are discussed in Chapters 2, 3 and 8. Cis-NATs are transcribed by RNA Pol II (RNA polymerase II) and show hallmarks of mRNA processing such as splicing, a poly-A tail and a cap structure. The direction of transcription can either be convergent or divergent, resulting in sense/antisense transcript pairs that overlap either at the 3′- or the 5′-end respectively (Figure 1). In rare cases, NATs can be fully embedded within the corresponding sense gene. These transcripts may or may not share complementarity with the fully processed sense transcript [9,10].
Figure 1. General genomic configuration of bi-directionally transcribed genes.
Top two panels: sense is indicated in red, antisense in blue. Bottom panels: NATs can affect the expression of the corresponding sense transcript in a concordant (NAT ↑, sense transcript ↑) or discordant (NAT ↑, sense transcript ↓) manner.
From a functional point of view, NATs can stimulate or reduce the expression of the sense transcript. The terms concordant and discordant regulation are used to describe stimulatory or inhibitory effects of NATs respectively (Figure 1) [11]. Although the mechanisms involved in both concordant and discordant regulation are under intense investigation, key questions remain. For example, the finding that NATs have very low expression levels begs the question: why are sense transcripts expressed in orders of magnitude greater than their regulatory antisense counterparts? The next section of the present chapter discusses the possibilities of how antisense transcripts regulate gene expression and also point to the limitations of our current understanding.
Two biological phenomena in humans and mice that have been established to depend on expression of specific NATs are parental imprinting and compensatory X chromosome inactivation in females. Interestingly in these cases, antisense transcription from one allele is essential to silence the corresponding sense transcript in cis-NAT while the sense transcript on the other allele stays active. These are arguably the best studied examples of epigenetic gene silencing that involve a NAT. There are additional aspects to imprinting and X chromosome inactivation such as allele choice or the spreading of the silencing mark to neighbouring genes that do not necessarily apply to the majority of NATs. For this reason it is unclear to what extent paradigms from imprinting and X chromosome inactivation translate to the general field of NATs. The present chapter focuses on this majority of NATs and we would like to refer the reader to excellent review articles for insights into imprinting and X chromosome dosage compensation [12].
How do NATs regulate gene expression?
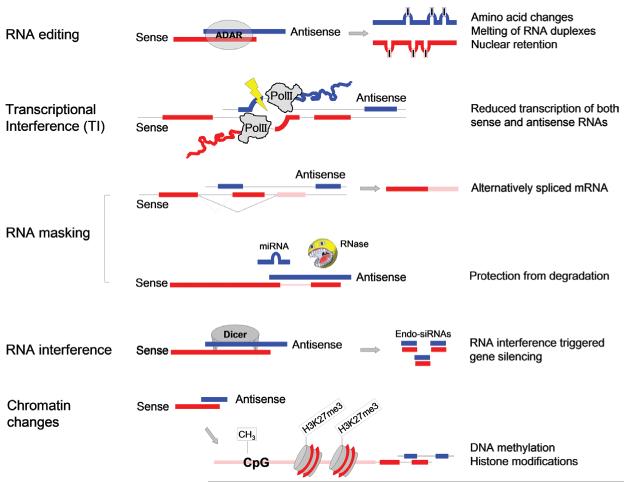
Although bi-directionally transcribed genes are often manipulated experimentally to investigate sense-antisense interactions, the underlying mechanisms are still poorly understood. Research has implicated NATs in a range of mechanisms with various complexities, from the simple creation of a physical barrier against modifying factors to the direction of an intricate web of dynamic chromatin remodelling. Some of these processes are better established than others with underlying theories on the basis of a more sturdy foundation of evidence. It is important, however, to discuss them all to gauge the extent of this convoluted regulatory picture (Figure 2).
Figure 2. Schematic representation of cellular mechanisms induced by bi-directional transcription or NATs.
The mechanisms include RNA editing, transcriptional interference, RNA masking, RNAi and chromatin modifications. Sense is indicated in red, antisense in blue. More details are given in the text.
RNA masking
The first mechanism to be discussed centres on the formation of duplexes with sense and antisense pairs providing a physical barrier against post-transcriptional interactions. This ‘masking’ of a transcript blocks out factors that would otherwise induce splicing, influence stability or direct the RNA to specific cellular compartments (Figure 2) [9]. An increasing number of physiologically important sense-antisense pairs have recently been shown to act via RNA masking.
Sense-antisense pairs tend to cross either one or more exon-intron border, the NAT therefore carries the potential to alter splicing of the sense transcript. This begs the question of whether there is an indirect relationship between alternative splicing and antisense regulation. Interesting as this theory may be, there are only two reported studies that demonstrate this link. The first example includes a NAT complementary to Zeb2 mRNA and its expression promotes the retention of a large intron at the 5′-end of the sense transcript. The intron contains an internal ribosome-binding site that enhances the expression of Zeb2. Being a transcriptional repressor, Zeb2 down-regulates E-cadherin (epithelial cadherin) expression, thereby inducing epithelial-mesenchymal transition [13].
The second example, TRα2 (thyroid hormone receptor α2), expresses two isoforms, TRα2a and TRα2b. The spliceform TRα2a is fully active, whereas the TRα2b only binds to the hormone without triggering a downstream response. The splicing of the primary transcript is regulated by a NAT RevErbAα whose expression correlates with the synthesis of the inactive receptor isoform. Interestingly, alternative splicing can also be induced by an oligonucleotide complementary to the crucial splice site [14].
These two examples seem to represent relatively isolated findings and little evidence so far suggests that NATs are heavily involved in the regulation of alternative splicing. Splice sites are not the only regulatory sites that can be affected by NATs; indeed, masking of regulatory sequences as well as interference with miRNA-binding sites by NATs has been reported in previous studies [15,16].
A case of RNA masking thought to have pathological significance is β-secretase mRNA. β-Secretase 1 is a key enzyme in the formation of amyloid protein fragments (Aβ1–40 and Aβ1–42) and therefore closely linked to the pathophysiology of Alzheimer’s disease. The expression of β-secretase is concordantly regulated by a processed and spliced antisense transcript. Neuroblastoma-derived cells (SH-SY5Y) show increased levels of both the antisense transcript and β-secretase in response to stressors known to promote Alzheimer’s disease. Importantly, the antisense transcript was also found to be elevated in the brains of Alzheimer’s patients [17]. A mechanism for how the NAT contributes to stabilization of the sense transcript was proposed by Faghihi et al. [16]. It is thought that the NAT competes with a repressive miRNA (miR-485-5p) for the same binding site, therefore stabilizing β-secretase mRNA [16]. miRNA–NAT competition could well be of general importance since a large proportion of NATs are complementary to the 3′-end of the corresponding sense transcript. On the other hand, the 3′-end of protein-coding transcripts also harbours most of the miRNA-binding sites.
Finally, a case of exposure rather than masking can be seen with aHIF (antisense hypoxiainducible factor), a transcript complementary to HIF-1α mRNA. HIF-1α is a transcription factor induced by hypoxia and its expression is linked to tumour growth and progression. aHIF regulates HIF-1α in a discordant manner by exposing AU-rich sequences that destabilizes the protein-coding sense transcript [15]. A general problem with RNA masking that is yet to be mentioned is in the formation of extended RNA duplexes. If found in the cytoplasm, these structures may be interpreted by the cell as signs of a viral infection and through PKR (double-stranded-RNA-dependent protein kinase) an immune response is triggered [18]. As a consequence, the cell containing the RNA hybrids will be destroyed, a result that cannot be the intention of a NAT-related ‘regulatory’ mechanism.
A-to-I editing
ADARs (adenosine deaminases that act on RNA) target dsRNAs (double-stranded RNAs). Any RNA that is double-stranded could be a potential substrate, revealing a mechanism for possible modifications of sense–antisense RNA duplexes. If such RNA pairs were to undergo A-to-I editing, it might follow that an antisense strand could manipulate the primary sequence of the sense partner as well as its localization or stability (Figure 2) [19,20].
Nevertheless, several studies have found that almost all A-to-I editing sites lie within inverted elements, mostly Alu repeats, in folded hairpin-like structures. Moreover, antisense transcripts showed few signs of editing sites other than in inverted repeats [21]. Despite these findings, it is important to acknowledge the chance of evidence being experimentally overlooked; edited sequences may swiftly degrade or remain in the nucleus and not be seen in expressed sequence datasets [22]. Nevertheless, it seems improbable that RNA editing plays a leading role in the regulatory functioning of NATs.
RNAi (RNA interference)
If sense and antisense RNAs are co-expressed in the same cell RNAi may be triggered. RNAi describes both an intrinsic method of post-transcriptional gene regulation and a process that is exploited experimentally to knock out genes of interest [23]. Central to this process are the type III ribonucleases Drosha and/or Dicer that cleave the dsRNA precursors (Figure 2). Whereas miRNA production involves a stem-loop RNA precursor and the action of Drosha and Dicer, the processing of sense–antisense RNA hybrids is likely to be performed by Dicer alone resulting in endo-siRNAs. The products are short oligonucleotides 21–23 nt in length. The short RNAs pair with a member of the Argonaute protein family to form the core of the RISC (RNA-induced silencing complex) [24].
In terms of the pairing with Argonautes, only one strand of the double-stranded short RNA precursor is selected as guide RNA, whereas the other so-called passenger strand unwinds and becomes degraded. The question of which strand becomes the guide and which takes the passenger role is particularly important in the context of endo-siRNAs. Strand selection dictates whether the sense or the antisense transcripts become the prime target for the newly formed RISC.
The involvement RNAi in the biology of NATs is still contentious owing to the elusive nature of naturally produced endo-siRNAs. Only the latest transcriptome sequencing efforts are starting to find abundant endo-siRNAs in selected tissues, most prominently in testis [25,26]. Moreover, the simple expression of a naturally occurring sense-antisense pair from plasmids in a cell line does not necessarily result in the formation of siRNAs [27,28]. Therefore it could well be that NATs only trigger RNAi in selected cell types or at specific stages of development and differentiation [28].
TI (transcriptional interference)
Antisense transcripts do not necessarily require formation of a dsRNA duplex with their sense partner to exert a regulatory effect; the process of their transcription alone may be sufficient. There are a few mechanisms proposed for how transcription of one strand could suppress transcription of another in cis. These theories are collectively named TI [29]. TI in the initiation phase is proposed to involve the two promoters competing for use of regulatory elements and RNA Pol II. During the elongation phase, interference occurs in the form of a physical blockage: an oncoming RNA Pol complex from one strand halts progress of an RNA Pol II complex on the other stand (Figure 2). Alternatively, it may clear the promoter on the opposite strand [9]. Much of the complex web of regulatory mechanisms is still unclear. Since theories of TI are on the basis of limited experimental evidence, it is likely they only apply to a small number of NATs.
NAT-induced chromatin changes
Epigenetics describe the phenomenon of altered gene expression with inheritable consequences to the phenotype that do not involve changes to the DNA sequence. The best studied epigenetic marks are DNA methylation at cytosine (usually in a CG context) and histone modifications at various sites, notably methylation or acteylation of lysine residues. The modification of histones affect chromatin packaging and, depending on whether structures are tightened or loosened, repression or activation of gene expression are observed. Methylated DNA, on the other hand, interferes with the transcriptional machinery and also binds to repressor complexes containing enzymes that enable chromatin remodelling. Sequence specificity to these modifications is thought to be brought about by ncRNA. Accordingly, many of the mechanisms discussed above have been linked to epigenetic modifications at bi-directionally transcribed genomic loci. For example, hyper-edited RNA binds to a protein called vigilin, which in turn orchestrates a set of proteins that participate in chromatin silencing [30]. The link between antisense transcription and chromatin changes has been particularly well studied in parentally imprinted genes. Such genes are only expressed from one allele depending on the parental origin and characteristically express an antisense transcript. The importance of the antisense RNA was convincingly shown for the imprinted locus Igf2r (insulin-like growth factor 2 receptor)/Airn. A mouse was generated that expressed only a truncated short piece of the NAT by introducing an artificial polyadenylation site [31]. The transgenic animals show bialleic expression of the locus and reduced birth weight. The exact mechanism of how the antisense transcript Airn induces allelic silencing is still elusive. In this respect, another imprinted locus, Kcnq1 and its antisense transcript Kcnq1ot, are better characterized: a model predicts that the antisense transcripts recruit a silencing complex that locally modifies chromatin. Interestingly, the process only works efficiently in the perinuclear space [32]. Epigenetic changes induced by NATs are not restricted to imprinted loci: for example in human leukaemic cells, the tumour suppressor gene p15 (CDKN2B; cyclin-dependent kinase 2 inhibitor B) has been found to be silenced by p15–NAT. A study using various reporter constructs demonstrated that p15–NAT expression led to heterochromatin formation and transcriptional silencing of the sense transcript [33]. Indeed, there is a rapidly growing list of specific NATs that repress the expression of the corresponding sense RNA at the transcriptional level. Silencing is brought on by repressive chromatin marks such as di- or tri-methylated histones (H3K9 and H3K27) or methylated DNA at CG residues (CpG methylation) in promoter regions (Figure 2) [34]. How exactly RNA recruits the different modifying complexes is not well understood, but it is an area of intense research.
NATs as drug targets
The link between NATs and epigenetic repression of protein-coding sense transcripts has prompted novel strategies to up-regulate clinically relevant genes. The idea is to knock down NATs to increase sense transcript levels and stimulate subsequent protein expression. The proof-of-concept for this approach has recently been delivered by Modarresi et al. [35] in a study focusing on BDNF (brain-derived neurotrophic factor). Knock down of the BDNF–NAT using siRNAs targeting areas outside the sense–antisense overlap lead to a transient stimulation of both BDNF mRNA and protein. A similar effect was observed with RNA oligonucleotides containing locked nucleic acid modifications, so-called antagoNATs. They were demonstrated to be effective in both cell culture models and in vivo. AntagoNATs were administered directly into the brain of mice by a small peristaltic pump, resulting in increased BDNF expression and neuronal outgrowth [35]. A comparable approach was also used to stimulate the expression of β-secretase [17]. Aside from the considerable difficulties surrounding the delivery of active agents to the correct cells, antagoNATs may prove a novel and versatile weapon against a broad range of diseases.
Is there a bigger picture?
The number of regulatory NATs known to have physiological or pathophysiological significance is ever increasing. There is also substantial evidence to indicate a role of NATs in the evolution of highly complex organisms. The existence of a bigger picture is underpinned by the observation that NATs are selectively under-represented on mammalian X chromosomes [10,36]. The X chromosome is only present in one active copy in mammalian cells: in females, the other X copy is epigenetically silenced and males carry a Y chromosome. The under representation of NATs on the X chromosome may therefore reflect a strategy to avoid antisense-induced gene silencing. Autosomal genes, in contrast, are expressed from both alleles and aberrant expression of a NAT would only cause one allele to be silenced. The accuracy of this scenario is corroborated by the observations that first, such examples of monoallelic gene expression are quite frequent [37] and secondly, that these genes tend to express NATs [38].
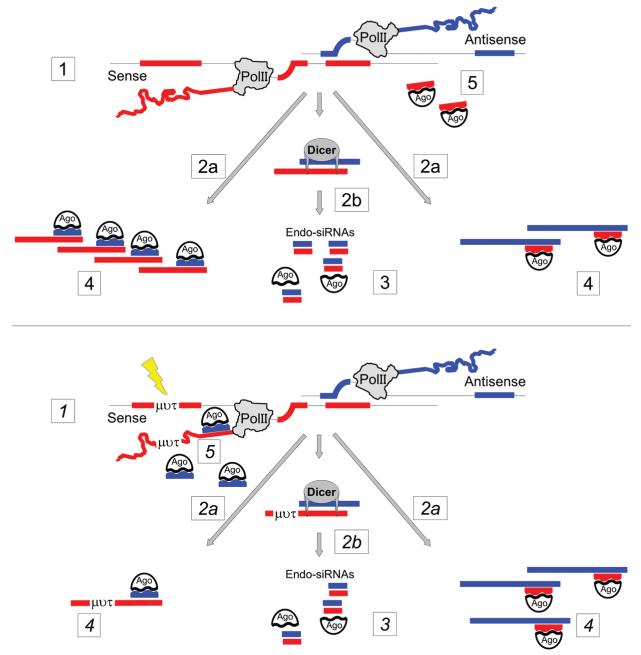
In order to understand the bigger picture and to acknowledge the benefit of NAT-induced gene silencing, one has to focus on haploid developing sperm cells where NATs are prominently expressed [38]. In a previously published theory [39], it was suggested that NATs play a role in the genomic quality control of developing sperm cells (Figure 3). According to this hypothesis, sense and antisense transcripts are co-expressed in haploid sperm cells and a proportion of the full-length transcripts is processed into endo-siRNAs. The ratio of proteincoding sense and non-coding antisense transcripts present is thought to determine strand selection of endo-siRNAs in the RISC. Accordingly, the RISC then targets the less abundant of the two transcripts which, in most cases, is the NAT. Consequently, an epigenetic signature is established that favours the expression of the protein-coding sense transcript and represses NAT. Any mutation that reduces the level of sense transcript might switch the balance in endo-siRNA strand selection and lead to the silencing of the protein-coding sense gene [39]. This will impinge on the development of the sperm cell and eventually positively select for cells without genomic damage.
Figure 3. NAT-mediated genomic quality control.
Top panel: the bi-directionally transcribed locus is intact. To run the control cycle, both sense and antisense transcripts are produced (1). Since the transcripts become fully processed, they can both persist as mRNAs (2a) or form RNA hybrids and feed into the RNAi pathway (2b). The resulting endo-siRNAs combine with Argonautes (most probably a non-slicing isoform such as Ago4) (3). According to the availability of target sequence, here the full-length sense and antisense transcripts, RISCs are formed (4). The spare Argonaute–endo-siRNA complexes (pre-RISC) will find the target sequence at the transcribed locus and induce transcriptional silencing (5). Bottom panel: the control cycle with a mutated sense transcript (μντ). The initial steps are identical with the top panel (1, 2 and 3); however, the mutation causes reduced levels of the full-length sense transcript (4). This could be the result of reduced transcription or transcript instability for example. As a consequence, pre-RISCs against the mutated transcript will reach the site of transcription and induce the silencing of the sense RNA (5) [39].
Conclusions
NATs are arguably rising stars in the as yet rather dark universe of ncRNAs. This is an astonishing promotion, considering that only 8 years ago NATs were widely perceived as mere transcriptional noise. This dismissal was fuelled by the elusive nature of NATs. They are notoriously difficult to handle not only owing to their usually low expression level, but also because sense–antisense RNA pairs are tightly balanced and any experimental intervention distorts the system. Establishing physiological relevance of the observed effects often proves the greatest challenge. Nevertheless, a wealth of elegant studies have recently underpinned the biological importance of NATs and opened exciting and novel avenues for further research.
Summary.
Natural antisense transcripts constitute a distinct group within the ever increasing family of non-coding RNAs.
Natural antisense transcripts influence the expression of the corresponding sense transcript.
Several cellular mechanisms are triggered by natural antisense transcripts, most of them involving co-expression of sense–antisense transcripts and RNA hybrid formation.
RNA–RNA and/or RNA–DNA hybrids may lead to RNA editing, RNA masking, RNA interference and eventually chromatin modifications.
Strategies to knock down the natural antisense transript with concomitant stimulation of protein encoding sense transcripts show great potential for medical applications.
The abundance of natural antisense transcripts in the genomes of complex organisms may be related to a control mechanism that assesses the integrity of the coding transcriptome during sperm development.
References
- 1.Lacatena RM, Cesareni G. Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature. 1981;294:623–626. doi: 10.1038/294623a0. [DOI] [PubMed] [Google Scholar]
- 2.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EG. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–2626. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanhee-Brossollet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene. 1998;211:1–9. doi: 10.1016/s0378-1119(98)00093-6. [DOI] [PubMed] [Google Scholar]
- 5.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 6.Engstrom PG, Suzuki H, Ninomiya N, Akalin A, Sessa L, Lavorgna G, Brozzi A, Luzi L, Tan SL, Yang L, et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beiter T, Reich E, Williams RW, Simon P. Antisense transcription: a critical look in both directions. Cell. Mol. Life Sci. 2008;66:94–112. doi: 10.1007/s00018-008-8381-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 13.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hastings ML, Ingle HA, Lazar MA, Munroe SH. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. J. Biol. Chem. 2000;275:11507–11513. doi: 10.1074/jbc.275.15.11507. [DOI] [PubMed] [Google Scholar]
- 15.Uchida T, Rossignol F, Matthay MA, Mounier R, Couette S, Clottes E, Clerici C. Prolonged hypoxia differentially regulates hypoxia-inducible factor (HIF)-1α and HIF-2α expression in lung epithelial cells: implication of natural antisense HIF-1α. J. Biol. Chem. 2004;279:14871–14878. doi: 10.1074/jbc.M400461200. [DOI] [PubMed] [Google Scholar]
- 16.Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St Laurent G, 3rd, Wahlestedt C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010;11:R56. doi: 10.1186/gb-2010-11-5-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004;68:432–452. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 21.Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 22.Neeman Y, Dahary D, Levanon EY, Sorek R, Eisenberg E. Is there any sense in antisense editing? Trends Genet. 2005;21:544–547. doi: 10.1016/j.tig.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 24.Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat. Rev. Genet. 2011;12:19–31. doi: 10.1038/nrg2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 27.Faghihi MA, Wahlestedt C. RNA interference is not involved in natural antisense mediated regulation of gene expression in mammals. Genome Biol. 2006;7:R38. doi: 10.1186/gb-2006-7-5-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gullerova M, Proudfoot NJ. Convergent transcription induces transcriptional gene silencing in fission yeast and mammalian cells. Nat. Struct. Mol. Biol. 2012;19:1193–1201. doi: 10.1038/nsmb.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shearwin KE, Callen BP, Egan JB. Transcriptional interference: a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr. Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 31.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 32.Kanduri C. Functional insights into long antisense noncoding RNA Kcnq1ot1 mediated bidirectional silencing. RNA Biol. 2008;5:208–211. doi: 10.4161/rna.7113. [DOI] [PubMed] [Google Scholar]
- 33.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr. Opin. Mol. Ther. 2010;12:214–222. [PMC free article] [PubMed] [Google Scholar]
- 35.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP, Wahlestedt C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012;30:453–459. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- 38.Carlile M, Swan D, Jackson K, Preston-Fayers K, Ballester B, Flicek P, Werner A. Strand selective generation of endo-siRNAs from the Na/phosphate transporter gene Slc34a1 in murine tissues. Nucleic Acids Res. 2009;37:2274–2282. doi: 10.1093/nar/gkp088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner A, Swan D. What are natural antisense transcripts good for? Biochem. Soc. Trans. 2010;38:1144–1149. doi: 10.1042/BST0381144. [DOI] [PMC free article] [PubMed] [Google Scholar]