Abstract
Mood regulation is known to be affected by the change of seasons. Recent research findings have suggested that mood regulation may be influenced by the function of circadian clocks. In addition, the activity of brown adipocytes has been hypothesized to contribute to mood regulation. Here, the overarching link to mood disorders might be the circadian clock protein nuclear receptor subfamily 1, group D, member 1.
Keywords: affective symptoms, brown fat, cryptochrome, depressive, diurnal, genetic variation, seasonal
There are seasonal variations in mood and behavior, including those in sleep duration, social activity, mood, appetite, weight, and energy levels that tend to reoccur year after year (1). These variations are common and present as a continuum: from individuals not having any marked variation, some having experienced the variations as a problem, and to those having the seasonal pattern of mood disorder known as seasonal affective disorder (2).
The current diagnostic criteria for seasonal affective disorder require that an episode of either recurrent major depressive disorder or bipolar disorder routinely occurs during a particular period of the year (3). Typically, in individuals with the winter type of seasonal affective disorder, or winter depression, the shortage of light during the autumn routinely induces depressive episodes, whereas the abundance of light during the spring routinely leads to remission. The treatment of choice for winter depression is bright light therapy, in which timed and repeated light exposures in the morning are administered via the eyes during the winter.
Concerning seasons and mood disorders, the clinical picture is more complex than these routine seasonal variations, as it has been noticed for centuries that a depressive episode tends to deepen and becomes life-threatening in the spring, but not to an equal extent in any other time of the year (4). There is no clear explanation to this phenomenon. A clue to understanding it may lie in reactions of the body to changes in daylight and ambient temperature during spring. Here, the circadian clocks that anticipate and try to adapt the body to the changes are in a key position. The function of brown fat being inducible in adult humans is another target to which attention is worth paying. In the following, I present my perspective on the circadian clocks and brown fat in mood disorders.
Circadian Clocks in Mood Disorders
Nearly all people suffering from mood disorders have disruptions in circadian rhythms (5). The circadian rhythms are generated in each cell, but maintained by the master circadian clock in the neurons that are located in the suprachiasmatic nucleus of the anterior hypothalamus in the brain (6). Because the sleep–wake rhythm is dictated by the circadian clock (7), these disruptions are often seen as sleeping problems. Documentation of circadian rhythm disruptions in patients with mood disorders relies on valid markers that are generated by the master circadian clock and display a reliable circadian rhythm, such as continuous recording of core body temperature and repeated assessments of melatonin concentration (8).
Dysfunction of the proteins encoded from the circadian clock genes is hypothesized to play a role in the etiology of mood disorders (9). Here, I consider those proteins that are repressors of transcription to be most important, since they are essential to the normal function of circadian clocks (10). Among them, nuclear receptor subfamily 1, group D, member 1 (NR1D1) has a key position as a connecting node in the transcriptional and translational loops that constitute the circadian clock in a cell (11–13). Further, CRY2 and CRY1 are the key repressors in the core of the circadian clock (14–20).
Thus far, genetic association studies have suggested that variants of some, not all, circadian clock genes associate with mood disorders. Of them, NR1D1 genetic variants have been demonstrated to associate with bipolar disorders (21–23) and depressive disorders (24, 25), CRY2 (cryptochrome 2) with depressive disorders (26, 27) and bipolar disorders (28), and CRY1 (cryptochrome 1) with depressive disorders (29). However, experimental studies elucidating the mechanisms of action by which the circadian clock proteins might contribute to mood disorders are missing.
Brown Fat in Mood Disorders
A hypothesis suggests that dysfunction of the brown adipose tissue contributes to mood regulation (30). This hypothesis was based on the original finding of brown adipose tissue being clearly over-activated in two suicide cases with depressive disorder (31). On the basis of only this data, it cannot be judged whether the finding was specific or whether it is reliable.
However, it provides a basis for a view that the activation of brown adipose tissue improves cold tolerance at the cost of heat tolerance, triggering anxiety, and psychomotor agitation, and affects mood in a negative way during the spring. It provides some evidence to stimulate not only replication studies but also experimental studies to demonstrate the mechanisms of action by which the brown adipose tissue might affect mood and contribute to mood disorders.
Role of Orphan Nuclear Receptors
Nuclear receptor subfamily 1, group D, member 1 is one of the so-called orphan nuclear receptors, while it seems to be a molecular link between the circadian clocks and mood regulation (32). Studies with Nr1d1-knockout mice agree with and support this finding, as there is up-regulation of tyrosine hydroxylase in the hippocampus (33) and increased proliferation of hippocampal neurons (34) in these mice. In these experiments, their mood-related behaviors were manifested as less anxious and less depressive.
Among the circadian clock genes, NR1D1 is the only one that maintains its oscillation on time at the light–dark transitions as well as under constant darkness in organs throughout the body (35). Therefore, NR1D1 seems to be the principal metronome of the body. NR1D1 regulates the transcription of “the long-day gene” TSHB (thyroid stimulating hormone, beta), and through this action NR1D1 is also a link between the effects of light and the seasonal variation in behavior (36). Transcription of TSHB is induced to a greater extent about 14 h after dawn of the first long day in the spring by the increasing exposure to light (37).
Intriguingly, the circadian clock protein NR1D1 has recently been demonstrated to link the body’s circadian and thermogenic networks through the regulation of the function of brown adipose tissue (38). The physiological induction of uncoupling protein in the mitochondria by cold temperature is preceded by rapid down-regulation of NR1D1 gene in brown adipose tissue, or in other words, the high levels of NR1D1 protein must fall before cold ambient temperature can induce uncoupling protein 1 to start producing heat and warm up the body. This switching off of the NR1D1-dependent repression is a key to the acute thermogenic response to cold and to subsequent cold tolerance.
Switching the NR1D1-dependent repression on again after it has once been switched off, however, is challenged in the spring, when the days are already long but may still be cold. Combination of long light exposure together with cold ambient temperature gives a conflicting signal of seasonal mismatch to the body (30). Having such conflict, the body is likely to continue producing heat and building up improvement in cold tolerance. If the activity of brown adipose tissue were not to be shut down as normal in the spring, it would easily become over-activated (39) and would produce excessive heat load that would give abnormal feedback from brown adipose tissue to the brain (40, 41).
Role of Cryptochromes
NR1D1 responds to a switch to longer days but does not immediately reset to the long-day state (42). During the resetting, the readouts of the circadian clock genes shift further away from the signal of NR1D1, and the magnitude of this escape is greater in Cry2-deficient than Cry1-deficient mice (43). Of the two cryptochromes, CRY2 opposes the actions of CRY1, thereby denying CRY1 from accessing to DNA targets too early (44), and in addition CRY2 opposes the actions of PER1 (45). It is the timing of peaks of PER1 and CRY2 expression, in particular, that varies directly with the length of the photoperiod (46, 47), and it is therefore the PER1–CRY2 and period 2 (PER2)–CRY2 protein complexes (48, 49) that control for their downstream targets during the resetting.
In addition to actions in the nucleus of a cell, the two cryptochromes act as inhibitors of adenylyl cyclase and thereby limit cyclic adenosine monophosphate production (50, 51). Interferon regulatory factor 4 is induced by cold as well as by cyclic adenosine monophosphate in adipocytes, driving up the activity of uncoupling protein 1 for heat production (52). Cryptochromes also inhibit the G protein coupled receptors activity, receptive to ligands such as vasoactive intestinal peptide and glucagon, through a direct interaction with the stimulatory G(s)alpha subunit (50). By these mechanisms, the cryptochromes might protect the individual from a depression-like state seen in conditions where dysfunction in control of the mesolimbic dopaminergic tracts leads to increased cyclic adenosine monophosphate production and increased depression-like behavior (53).
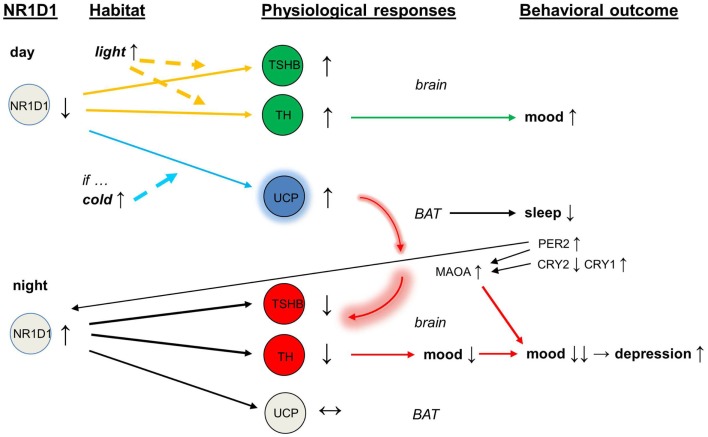
With abnormal expression of CRY2, the circadian protein PER2 and the enzyme monoamine oxidase A (MAOA) would become overactive (54). The over-expression of NR1D1 inhibits the activity of tyrosine hydroxylase (32), and the over-activity of MAOA depletes dopamine release and impairs further the dopaminergic transmission. In addition, the direct interaction of PER2 with NR1D1 (55) may feedback to this vicious circle that was initiated by the overactive brown adipose tissue. In the end, mood is lowered and there is a deepening of depressive episode (see Figure 1).
Figure 1.
Schematic of mood regulation affected by the circadian clock proteins (NR1D1, PER2, CRY2, CRY1) and by the activity of brown adipose tissue. Abbreviations: NR1D1, nuclear receptor subfamily 1, group D, member 1; PER2, period 2; CRY2, cryptochrome 2; CRY1, cryptochrome 1.
Conclusion
The loss of cryptochromes does change physiology, and dysfunction of cryptochromes may change mood. On the basis of the data presented above, CRY2 appears to be “a mood gene.” Success in the resetting has been hypothesized to improve lowered mood in the depressed (56), whereas failure in the resetting may deepen a depressive episode any time of the year, especially in the spring. Here, the overarching link might be the circadian clock protein NR1D1.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Rosenthal NE, Sack DA, Gillin JC, Lewy AJ, Goodwin FK, Davenport Y, et al. Seasonal affective disorder: a description of the syndrome and preliminary findings with light therapy. Arch Gen Psychiatry (1984) 41:72–80. 10.1001/archpsyc.1984.01790120076010 [DOI] [PubMed] [Google Scholar]
- 2.Kasper S, Wehr TA, Bartko JJ, Gaist PA, Rosenthal NE. Epidemiological findings of seasonal changes in mood and behavior: a telephone survey of Montgomery County, Maryland. Arch Gen Psychiatry (1989) 46:823–33. 10.1001/archpsyc.1989.01810090065010 [DOI] [PubMed] [Google Scholar]
- 3.Partonen T, Lönnqvist J. Seasonal affective disorder. Lancet (1998) 352:1369–74. 10.1016/S0140-6736(98)01015-0 [DOI] [PubMed] [Google Scholar]
- 4.Wehr TA, Rosenthal NE. Seasonality and affective illness. Am J Psychiatry (1989) 146:829–39 10.1176/ajp.146.7.829 [DOI] [PubMed] [Google Scholar]
- 5.McClung CA. How might circadian rhythms control mood? Let me count the ways. Biol Psychiatry (2013) 74:242–9. 10.1016/j.biopsych.2013.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ukai H, Kobayashi TJ, Nagano M, Masumoto KH, Sujino M, Kondo T, et al. Melanopsin-dependent photo-perturbation reveals desynchronization underlying the singularity of mammalian circadian clocks. Nat Cell Biol (2007) 9:1327–34. 10.1038/ncb1653 [DOI] [PubMed] [Google Scholar]
- 7.Kawato M, Fujita K, Suzuki R, Winfree AT. A three-oscillator model of the human circadian system controlling the core temperature rhythm and the sleep-wake cycle. J Theor Biol (1982) 98:369–92 10.1016/0022-5193(82)90125-4 [DOI] [PubMed] [Google Scholar]
- 8.Coogan AN, Thome J. Chronotherapeutics and psychiatry: setting the clock to relieve the symptoms. World J Biol Psychiatry (2011) 12(Suppl 1):40–3. 10.3109/15622975.2011.598389 [DOI] [PubMed] [Google Scholar]
- 9.Bunney JN, Potkin SG. Circadian abnormalities, molecular clock genes and chronobiological treatments in depression. Br Med Bull (2008) 86:23–32. 10.1093/bmb/ldn019 [DOI] [PubMed] [Google Scholar]
- 10.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell (2011) 144:268–81. 10.1016/j.cell.2010.12.019 [DOI] [PubMed] [Google Scholar]
- 11.Lazar MA, Jones KE, Chin WW. Isolation of a cDNA encoding human Rev-ErbA alpha: transcription from the noncoding DNA strand of a thyroid hormone receptor gene results in a related protein that does not bind thyroid hormone. DNA Cell Biol (1990) 9:77–83. 10.1089/dna.1990.9.77 [DOI] [PubMed] [Google Scholar]
- 12.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell (2002) 110:251–60. 10.1016/S0092-8674(02)00825-5 [DOI] [PubMed] [Google Scholar]
- 13.Robinson I, Reddy AB. Molecular mechanisms of the circadian clockwork in mammals. FEBS Lett (2014) 588:2477–83. 10.1016/j.febslet.2014.06.005 [DOI] [PubMed] [Google Scholar]
- 14.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, Todo T, et al. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry (1996) 35:13871–7. 10.1021/bi962209o [DOI] [PubMed] [Google Scholar]
- 15.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature (1999) 398:627–30. 10.1038/19323 [DOI] [PubMed] [Google Scholar]
- 16.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A (1999) 96:12114–9. 10.1073/pnas.96.21.12114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science (1999) 286:768–71. 10.1126/science.286.5440.768 [DOI] [PubMed] [Google Scholar]
- 18.Dardente H, Fortier EE, Martineau V, Cermakian N. Cryptochromes impair phosphorylation of transcriptional activators in the clock: a general mechanism for circadian repression. Biochem J (2007) 402:525–36. 10.1042/BJ20060827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, et al. A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell (2009) 139:199–210. 10.1016/j.cell.2009.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem (2011) 286:25891–902. 10.1074/jbc.M111.254680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etain B, Milhiet V, Bellivier F, Leboyer M. Genetics of circadian rhythms and mood spectrum disorders. Eur Neuropsychopharmacol (2011) 21(Suppl 4):S676–82. 10.1016/j.euroneuro.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 22.Partonen T. Clock gene variants in mood and anxiety disorders. J Neural Transm (2012) 119:1133–45. 10.1007/s00702-012-0810-2 [DOI] [PubMed] [Google Scholar]
- 23.Nováková M, Praško J, Látalová K, Sládek M, Sumová A. The circadian system of patients with bipolar disorder differs in episodes of mania and depression. Bipolar Disord (2014). 10.1111/bdi.12270 [DOI] [PubMed] [Google Scholar]
- 24.Utge SJ, Soronen P, Loukola A, Kronholm E, Ollila HM, Pirkola S, et al. Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS One (2010) 5:e9259. 10.1371/journal.pone.0009259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, et al. Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proc Natl Acad Sci U S A (2013) 110:9950–5. 10.1073/pnas.1305814110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavebratt C, Sjöholm LK, Soronen P, Paunio T, Vawter MP, Bunney WE, et al. CRY2 is associated with depression. PLoS One (2010) 5:e9407. 10.1371/journal.pone.0009407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovanen L, Kaunisto M, Donner K, Saarikoski ST, Partonen T. CRY2 genetic variants associate with dysthymia. PLoS One (2013) 8:e71450. 10.1371/journal.pone.0071450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sjöholm LK, Backlund L, Cheteh EH, Ek IR, Frisén L, Schalling M, et al. CRY2 is associated with rapid cycling in bipolar disorder patients. PLoS One (2010) 5:e12632. 10.1371/journal.pone.0012632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soria V, Martínez-Amorós E, Escaramís G, Valero J, Pérez-Egea R, García C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology (2010) 35:1279–89. 10.1038/npp.2009.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Partonen T. Hypothesis: cryptochromes and brown fat are essential for adaptation and affect mood and mood-related behaviors. Front Neurol (2012) 3:157. 10.3389/fneur.2012.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huttunen P, Kortelainen ML. Long-term alcohol consumption and brown adipose tissue in man. Eur J Appl Physiol Occup Physiol (1990) 60:418–24. 10.1007/BF00705030 [DOI] [PubMed] [Google Scholar]
- 32.Chung S, Lee EJ, Yun S, Choe HK, Park SB, Son HJ, et al. Impact of circadian nuclear receptor REV-ERBα on midbrain dopamine production and mood regulation. Cell (2014) 157:858–68. 10.1016/j.cell.2014.03.039 [DOI] [PubMed] [Google Scholar]
- 33.Jager J, O’Brien WT, Manlove J, Krizman EN, Fang B, Gerhart-Hines Z, et al. Behavioral changes and dopaminergic dysregulation in mice lacking the nuclear receptor Rev-erbα. Mol Endocrinol (2014) 28:490–8. 10.1210/me.2013-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnell A, Chappuis S, Schmutz I, Brai E, Ripperger JA, Schaad O, et al. The nuclear receptor REV-ERBα regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS One (2014) 9:e99883. 10.1371/journal.pone.0099883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazzoccoli G, Cai Y, Liu S, Francavilla M, Giuliani F, Piepoli A, et al. REV-ERBα and the clock gene machinery in mouse peripheral tissues: a possible role as a synchronizing hinge. J Biol Regul Homeost Agents (2012) 26:265–76. [PubMed] [Google Scholar]
- 36.Aninye IO, Matsumoto S, Sidhaye AR, Wondisford FE. Circadian regulation of Tshb gene expression by Rev-Erbα (NR1D1) and nuclear corepressor 1 (NCOR1). J Biol Chem (2014) 289:17070–7. 10.1074/jbc.M114.569723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dardente H, Wyse CA, Birnie MJ, Dupré SM, Loudon AS, Lincoln GA, et al. A molecular switch for photoperiod responsiveness in mammals. Curr Biol (2010) 20:2193–8. 10.1016/j.cub.2010.10.048 [DOI] [PubMed] [Google Scholar]
- 38.Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, et al. The nuclear receptor Rev-erbα controls circadian thermogenic plasticity. Nature (2013) 503:410–3. 10.1038/nature12642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zukotynski KA, Fahey FH, Laffin S, Davis R, Treves ST, Grant FD, et al. Seasonal variation in the effect of constant ambient temperature of 24°C in reducing FDG uptake by brown adipose tissue in children. Eur J Nucl Med Mol Imaging (2010) 37:1854–60. 10.1007/s00259-010-1485-2 [DOI] [PubMed] [Google Scholar]
- 40.Contreras C, Gonzalez F, Fernø J, Diéguez C, Rahmouni K, Nogueiras R, et al. The brain and brown fat. Ann Med (2014). 10.3109/07853890.2014.919727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaughan CH, Bartness TJ. Anterograde transneuronal viral tract tracing reveals central sensory circuits from brown fat and sensory denervation alters its thermogenic responses. Am J Physiol Regul Integr Comp Physiol (2012) 302:R1049–58. 10.1152/ajpregu.00640.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazlerigg DG, Andersson H, Johnston JD, Lincoln G. Molecular characterization of the long-day response in the Soay sheep, a seasonal mammal. Curr Biol (2004) 14:334–9. 10.1016/S0960-9822(04)00081-8 [DOI] [PubMed] [Google Scholar]
- 43.Stratmann M, Stadler F, Tamanini F, van der Horst GT, Ripperger JA. Flexible phase adjustment of circadian albumin D site-binding protein (DBP) gene expression by cryptochrome1. Genes Dev (2010) 24:1317–28. 10.1101/gad.578810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anand SN, Maywood ES, Chesham JE, Joynson G, Banks GT, Hastings MH, et al. Distinct and separable roles for endogenous CRY1 and CRY2 within the circadian molecular clockwork of the suprachiasmatic nucleus, as revealed by the Fbxl3(Afh) mutation. J Neurosci (2013) 33:7145–53. 10.1523/JNEUROSCI.4950-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richards J, All S, Skopis G, Cheng KY, Compton B, Srialluri N, et al. Opposing actions of Per1 and Cry2 in the regulation of Per1 target gene expression in the liver and kidney. Am J Physiol Regul Integr Comp Physiol (2013) 305:R735–47. 10.1152/ajpregu.00195.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner GC, Johnston JD, Clarke IJ, Lincoln GA, Hazlerigg DG. Redefining the limits of day length responsiveness in a seasonal mammal. Endocrinology (2008) 149:32–9. 10.1210/en.2007-0658 [DOI] [PubMed] [Google Scholar]
- 47.Lincoln G, Messager S, Andersson H, Hazlerigg D. Temporal expression of seven clock genes in the suprachiasmatic nucleus and the pars tuberalis of the sheep: evidence for an internal coincidence timer. Proc Natl Acad Sci U S A (2002) 99:13890–5. 10.1073/pnas.212517599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lincoln GA, Andersson H, Hazlerigg D. Clock genes and the long-term regulation of prolactin secretion: evidence for a photoperiod/circannual timer in the pars tuberalis. J Neuroendocrinol (2003) 15:390–7. 10.1046/j.1365-2826.2003.00990.x [DOI] [PubMed] [Google Scholar]
- 49.Padmanabhan K, Robles MS, Westerling T, Weitz CJ. Feedback regulation of transcriptional termination by the mammalian circadian clock period complex. Science (2012) 337:599–602. 10.1126/science.1221592 [DOI] [PubMed] [Google Scholar]
- 50.Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med (2010) 16:1152–6. 10.1038/nm.2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narasimamurthy R, Hatori M, Nayak SK, Liu F, Panda S, Verma IM. Circadian clock protein cryptochrome regulates the expression of proinflammatory cytokines. Proc Natl Acad Sci U S A (2012) 109:12662–7. 10.1073/pnas.1209965109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell (2014) 158:69–83. 10.1016/j.cell.2014.04.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park SK, Nguyen MD, Fischer A, Luke MP, Affarel B, Dieffenbach PB, et al. Par-4 links dopamine signaling and depression. Cell (2005) 122:275–87. 10.1016/j.cell.2005.05.031 [DOI] [PubMed] [Google Scholar]
- 54.Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, Perreau-Lenz S, et al. Regulation of monoamine oxidase A by circadian-clock components implies clock influence on mood. Curr Biol (2008) 18:678–83. 10.1016/j.cub.2008.04.012 [DOI] [PubMed] [Google Scholar]
- 55.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev (2010) 24:345–57. 10.1101/gad.564110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bunney BG, Bunney WE. Mechanisms of rapid antidepressant effects of sleep deprivation therapy: clock genes and circadian rhythms. Biol Psychiatry (2013) 73:1164–71. 10.1016/j.biopsych.2012.07.020 [DOI] [PubMed] [Google Scholar]