Abstract
Messenger RNA (mRNA) degradation is an important element of gene expression that can be modulated by alterations in translation, such as reductions in initiation or elongation rates. Reducing translation initiation strongly affects mRNA degradation by driving mRNA toward the assembly of a decapping complex, leading to decapping. While mRNA stability decreases as a consequence of translational inhibition, in apparent contradiction several external stresses both inhibit translation initiation and stabilize mRNA. A key difference in these processes is that stresses induce multiple responses, one of which stabilizes mRNAs at the initial and rate-limiting step of general mRNA decay. Because this increase in mRNA stability is directly induced by stress, it is independent of the translational effects of stress, which provide the cell with an opportunity to assess its response to changing environmental conditions. After assessment, the cell can store mRNAs, reinitiate their translation or, alternatively, embark on a program of enhanced mRNA decay en masse. Finally, recent results suggest that mRNA decay is not limited to non-translating messages and can occur when ribosomes are not initiating but are still elongating on mRNA. This review will discuss the models for the mechanisms of these processes and recent developments in understanding the relationship between translation and general mRNA degradation, with a focus on yeast as a model system.
How to cite this article: WIREs RNA 2014, 5:747–763. doi: 10.1002/wrna.1244
INTRODUCTION
Messenger RNA (mRNA) degradation, which is intimately coupled to translation, is important in the control of gene expression. Studies examining the interrelatedness of translation and decay suggest that translational repression is a major entryway into, if not a prerequisite for, the mRNA decay process. For example, mutations in proteins that bind and protect the mRNA cap serve to promote mRNA degradation. Supporting the hypothesis of the initiation of mRNA decay upon translational repression, trans-acting factors in mRNA degradation often include in their repertoire the ability to repress translation. Furthermore, even factors that do not affect translation interact with and recruit factors that do affect translation, as they are embedded within a dense web of interactions.1
However, recent studies suggest that the inverse relationship between translation and mRNA degradation may be more nuanced. One example is that in response to translational inhibition from stresses, yeast and other eukaryotes inhibit the mRNA decay process, which could allow cells to assess new environmental conditions. Another set of studies has suggested that mRNA degradation occurs on translating mRNA and that mRNA decay factors can inhibit translational elongation as well as affect mRNA degradation.
Much of our understanding of mRNA decay and the relationship between translation and decay has come from studies in the yeast, Saccharomyces cerevisiae. To limit the scope of this review, we will concentrate primarily on work in yeast and how it relates to the central understanding of mRNA translation and degradation.
mRNA translation not only affects general mRNA decay but is also linked to destruction of mRNA with defects in translation. These defects feed into the quality control of translational processes, such as Nonsense-Mediated (NMD), Non-Stop, and No-Go decay.2–6 We will concentrate on mRNAs undergoing decay in the general mRNA degradation pathway and their relationship to their translation status. Due to the extensive literature on the processes that affect translation and decay, we will focus on two major areas: the relationship between translational initiation and mRNA stability as well as mRNA decay and translation elongation. Due to the limited scope, we will refer to more detailed discussions when appropriate.
The Mechanism of mRNA Degradation
The linkage between translation and mRNA degradation begins with the mRNA itself. At the 3′ end of an mRNA is the poly-adenosine or poly(A) tail. Similarly, linked to the 5′ end is a methyl-7-guanosine cap (Figure 1). Integrating these ends is the translation initiation factor eIF4F, which is the cap-binding complex (see below). These structures are crucial for the mRNA translation and degradation processes.7,8 They serve as assembly points for initiation factors which when combined promote translation in a synergistic manner.7,8 The major steps of mRNA degradation are presented below, and for a recent comprehensive review, see Ref 9.
Fig 1.

Mechanism of messenger RNA (mRNA) degradation. The degradation of mRNA begins on the newly cytoplasmic mRNA. The decay process is regulated by elements on each end of the mRNA. The mRNA has a 7-methyl-guanosine cap at the 5′ end and a poly(A) tail at the 3′ end. The poly(A) length is approximately 60 nucleotides upon export. In the cytoplasm, the mRNA is initially deadenylated by Pan2/3 and then primarily by the Ccr4/Pop2/Not deadenylase complex, resulting in a median poly(A) length of approximately 30 nucleotides. Once the poly(A) tail is shortened to a length that is no longer able to bind Pab1p, the mRNA can be degraded from the 3′→5′ direction by the exosome. The 5′→3′ degradation pathway requires the mRNA to be first decapped by the decapping complex Dcp1/2 and then degraded by the Xrn1p exonuclease enzyme.
The poly(A) tail is added to an mRNA in the nucleus and processed to a length of approximately 60 nucleotides upon export, upon which it is subject to deadenylation by two different enzyme complexes.10–14 Deadenylation most likely accounts for the median mRNA tail length, which has been determined to be 27 nucleotides in yeast.15 The rate of deadenylation is central to defining the half-life of an mRNA. The Pan2/3 complex is thought to be responsible for the initial deadenylation of an mRNA. After initial poly(A) shortening, the multiprotein deadenylase complex, Ccr4/Pop2/Not primarily acts to shorten the poly(A) tail to approximately 10 nucleotides14,16,17 (for a recent review, see Refs 18–19). The oligoadenylated mRNA (approximately 10 adenosines) can be degraded in either the 5′→3′ or the 3′→5′ direction by Xrn1p (after decapping) or the exosome, respectively.9,20–23
After deadenylation, most mRNAs in yeast are degraded in the 5′→3′ direction. The process begins with the removal of the 5′ 7-methyl-guanosine cap by the decapping complex, consisting of Dcp1p, Dcp2p, and Edc3p (for a detailed discussion of decapping see Ref 24). Three steps are required for the mRNA to be decapped. First, the decapping complex on the mRNA must be assembled. Second, the 5′ cap must be exposed; therefore suggesting that the translation initiation factors eIF4G and eIF4E must be released from the mRNA.25 Finally, following the recruitment of the decapping complex, the phosphate bond that links the cap to the body of the mRNA is hydrolyzed, leaving a monophosphorylated 5′ end (Figure 1).21,26 Removal of the mRNA cap allows 5′→3′ degradation to proceed by the exonuclease Xrn1p.23
Alternatively, oligoadenylated mRNA can be degraded from the 3′→5′ direction by the exosome (Figure 1). The core exosome complex consists of a ring of six RNase PH-like proteins and three smaller RNA-binding proteins, which are required for its formation and stabilization. Rrp44p (Dis3p), an additional RNase R-like subunit, provides the exosome's exonuclease activity. The ring structure of the exosome serves as a binding site for the Ski2/3/8 complex and Ski7p, both of which are required for cytoplasmic degradation by the exosome.
Initiation of Translation
The initiation of translation is a complex process; however, for brevity, only the major steps of initiation will be outlined here, with an emphasis on the factors that are discussed in the text (Figure 2). For a more detailed discussion of translation initiation, see Ref 27. Several initiation factors are critical for mRNA translation, and of those that bind mRNA, the cap-binding complex eIF4F is central. The cap-binding complex (eIF4F) consists of the initiation factors eIF4E (Cdc33p in yeast), eIF4G (Tif4631p and Tif4632p), and eIF4A (Tif1p and Tif2p). This complex has three key functions that affect mRNA decay. First, it binds the 7-methyl-guanosine mRNA cap. Second, it links translation initiation factors that are assembled on the 5′ cap with the 3′ poly(A) tail via the poly(A) binding protein, Pab1p. Finally, it promotes translation.
Fig 2.
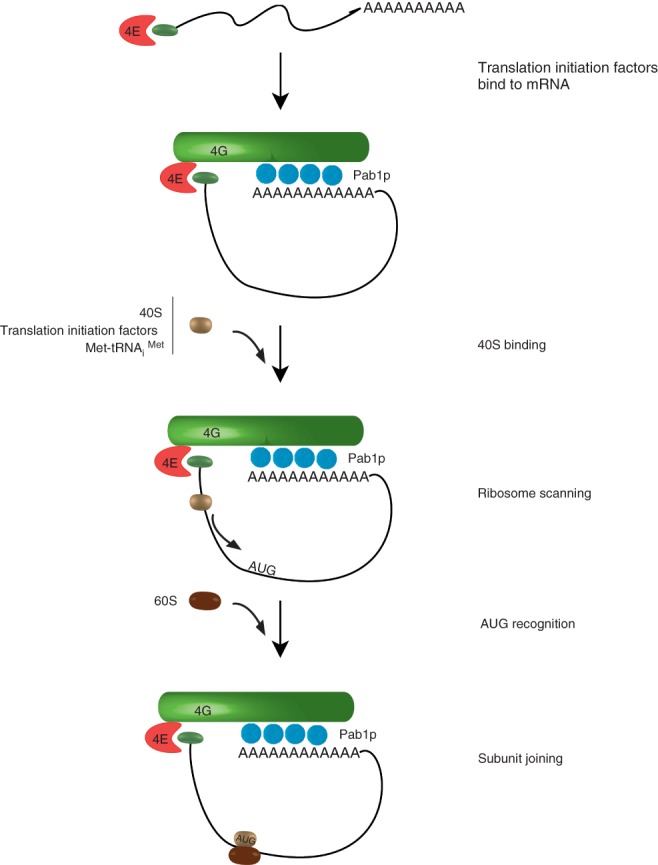
Initiation of translation. Messenger RNA (mRNA) translation begins after export, with association with translation initiation factors including eIF4E, eIF4G, and Pab1p. Translation initiation begins with the formation of the 43S preinitiation complex consisting of the small ribosomal subunit (40S), methionyl tRNA, and additional initiation factors. The preinitiation complex binds to the mRNA to form a 48S complex. The small ribosomal subunit then scans the message in the 5′→3′ direction until it recognizes an AUG codon. In association with other initiation factors, the large ribosomal subunit (60S) forms an 80S complex, which commences translation.
The small ribosomal subunit, along with methionyl-tRNAiMet and additional translation initiation factors, compose the 43S preinitiation complex. This complex binds to the mRNA and the cap-binding complex, forming the 48S initiation complex. The small subunit then scans the message from 5′→3′ direction and initiates translation predominantly at the first AUG codon. Initiation factors assist in the further joining of the large 60S subunit, forming an 80S complex, which then translates the mRNA.
TRANSLATION INITIATION AND mRNA DECAY
A reduction in mRNA translation has a strong effect on mRNA decay. However, this effect depends on the source of the reduction of translation, that is, a direct reduction in translation initiation or a global response to a stress. Sources of translational inhibition can be intrinsic or extrinsic to the mRNA (or cell). The intrinsic effect of translation inhibition provokes mRNA decay and results from mutations in initiation factors, cis-acting sequences of the mRNA and proteins that specifically inhibit mRNA translation initiation (Figure 3). In intrinsic inhibition of translation, the changes in translation are specific to the inhibited mRNA(s), with initiation factor harboring mutations, likely mimicking such an effect.
Fig 3.
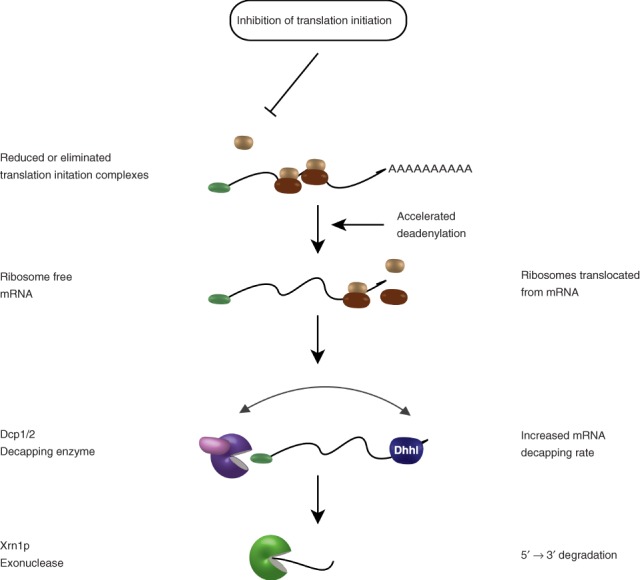
Intrinsic inhibition of translation and messenger RNA (mRNA) stability. Intrinsic inhibition of translation directly or indirectly inhibits initiation on specific messages. Subsequent to inhibition, mRNA is freed from ribosomes as they translocate off the mRNA. Decapping activators assemble on mRNA, predominantly at the 3′ UTR, and recruit the decapping enzyme. mRNA is decapped and degraded by Xrn1p in the 5′→3′ direction.
Intrinsic effects can be thought of as those that affect specific mRNAs or classes thereof. These mRNAs are specifically designated by the cell to be translationally repressed. The resulting mRNA degradation may reinforce the cellular program of inhibited translation. However, some examples of this form of inhibition globally affect many mRNAs, such as mutations in the cap-binding protein, which strongly inhibit translation initiation. The mechanism by which mutations globally affecting translation are often not physiological, but instead mimic translational repression of a specific mRNA.
On the other hand, extrinsic inhibition of initiation results from changes in the environment of the cell (Figure 4). Extrinsic inhibition feeds into stress response pathways and generally slows mRNA degradation by inhibiting poly(A) tail shortening. Extrinsic effects have two key distinctive consequences. First, extrinsic mRNA decay effects are insensitive to the intrinsic effects on translation initiation since deadenylation is inhibited. Deadenylation is the initial step in general mRNA decay, and is upstream of the decay steps affected by intrinsic repression. Second, extrinsic effects on decay are believed to occur as a consequence of the stress, such as glucose deprivation, heat shock and osmotic stress, rather than as a consequence of translational repression, leading it to be described as translation-independent.28
Fig 4.

Extrinsic inhibition of translation and messenger RNA (mRNA) stability. Stress external to the cell is sensed and initiates a host of responses, including the inhibition of translation initiation. The first step in the common mRNA decay pathway is deadenylation, which is inhibited by extrinsic effects on translation by an unknown mechanism. Once acclimated to the stress, the cell will either degrade mRNAs, re-commence translation or place mRNAs into non-translating storage.
In contrast to intrinsic effects, extrinsic translational repression globally affects mRNA translation. The suspension of decay can give the cell time to decide whether to maintain or alter its gene expression before destroying unnecessary mRNAs. In the case of temporary stress, mRNAs can return to translation without requiring the mRNAs to be transcribed again.29 However, if gene expression is altered, mRNAs that are no longer necessary can be destroyed once the inhibition of decay is lifted.
The mRNA Cap-Binding Complex Promotes mRNA Stability
Mutations in translation initiation factors reduce translation and increase mRNA deadenylation and decapping. The most central of the translation initiation factors in this process are those located at the 5′ and 3′ ends of the mRNA. At the 5′ end is the cap-binding complex (eIF4F), and at the 3′ end is the poly(A) binding protein (Pab1p). Both factors enhance translation and protect the mRNA from degradation.
Several studies have shown that mutating the cap-binding complex to reduce initiation rates increases mRNA degradation.25,30,31 A comprehensive study looked at changes in mRNA half-lives using mutations in multiple initiation factors, especially those in the cap-binding complex.25 This study examined two mRNAs that, broadly speaking, represent a stable (PGK1) and an unstable mRNA (MFA2).10,11,21,32 Shifting the cells to temperatures in which the various initiation factors are mostly inactive reduced the half-lives of both mRNAs by up to nearly seventy percent compared with wild-type, and these decreases in the half-lives were largely proportional to the degree to which the mutation inhibited translation.25,31,33 One observation was that an extremely poorly translating eIF4E mutant exhibited the most extreme decrease in half-life from 17 to 6 min for the PGK1 mRNA. A similar result was obtained using an eIF4E mutant that failed to interact with eIF4G, which similarly reduced the half-lives of specific mRNAs.31
Interestingly, the acceleration of the rate of decay of the short-lived MFA2 mRNA was much more refractory to the inhibition of translation initiation than PGK1.25 One reason for this discrepancy could be that the MFA2 mRNA decay rate is most likely near the maximum possible rate in yeast.34,35 Because the rate-limiting step of mRNA decay is deadenylation, only the most extreme perturbations may significantly alter the decay of such an unstable mRNA.
Reducing initiation rates using mutations in the cap-binding complex leads to enhanced mRNA decay by increasing the rates of deadenylation and decapping.25 For example, the PGK1 mRNA is not degraded until it is oligoadenylated and thus subject to decapping. The enhanced rate of decapping is facilitated by competition between eIF4E and Dcp1/2 for the mRNA cap, as eIF4E can block Dcp1/2 decapping activity.36 Supporting a role for eIF4E inhibiting the accessibility of the cap to the decapping enzyme, a dcp1 temperature-sensitive mutant with approximately 15% decapping activity alone and in combination with an eIF4E mutant was examined for its decapping ability. When combined with an eIF4E mutant, the dcp1 mutant was no longer defective in mRNA decapping and decay, suggesting that the disassembly of eIF4F in the eIF4E mutant promotes greater decapping.36 Similarly, in vitro, decapping can be stimulated by the addition of a cap analogue, which can compete with eIF4E for the mRNA. These and other experiments support a model where active release or passive destabilization of the entire cap-binding complex promotes decapping by allowing access to the mRNA cap.36,37
The Poly(A) Tail and Decay
The rate of removal of the poly(A) tail, or deadenylation, is usually the rate-limiting step in general mRNA decay. A long mRNA poly(A) tail serves to block entry to the 5′→3′ and 3′→5′ decay pathways. Once the tail has been shortened, the mRNA is then subject to decay by the exosome (3′→5′) or decapping and 5′→3′ Xrn1p digestion. Furthermore, similar to the cap-binding complex, the poly(A) tail and the poly(A) binding protein (Pab1p) serve to enhance translation and inhibit decay. The poly(A) tail is linked to the 5′ end and cap-binding complex (eIF4F) through the poly(A) binding protein (Pab1p), which directly associates with eIF4G.38 The cytoplasmic Pab1p binds a 3′ poly(A) tail of at least 10–12 nucleotides21,39 and inhibits the major deadenylation complex and mRNA decapping.9,40–42 Shortening of the poly(A) tail results in the loss of bound Pab1 protein(s), which further increases the rate of Ccr4/Pop2/Not deadenylation. Taken together, the consequence of a shorter tail is decreased translation and increased decay. However, the effect of the poly(A) tail on translation has been called into question. Contrary to previous studies, a recent report suggests that deadenylation may not affect translation but primarily enhances mRNA decay in yeast.15
Inhibition of Translation in cis Increases the mRNA Degradation Rate
Sequence variations in mRNAs can inhibit their translation and enhance the rate of their decay. By altering mRNA sequences, the effect of translational blocks on decay rates can be examined. Translation initiation can be reduced either by physically blocking ribosome scanning using RNA secondary structure or by altering the recognition of the start codon. These mutations accelerated the rate of decay of mRNAs, such as the stable PGK1 mRNA.32,33,43 Other naturally occurring, cis-acting sequences that promote decay and translational repression tend to be localized in the 3′ UTR and involve trans-acting factors; these are discussed later in this review and in more depth by others.9,44,45 However, inhibiting translation of short-lived mRNAs in cis does not appear to significantly affect their half-lives, similar to the effect of reducing translation using initiation factor mutations.33,46,47
The acceleration of mRNA decay by the intrinsic inhibition of translation initiation is primarily due to a several-fold increase in deadenylation and mRNA decapping rates, as observed for cis mutations.43 Given that Pab1p-associated mRNAs are inhibited in decapping, the decapping effect may be a consequence of deadenylation.
Trans-Acting Factors Repress Translation and Increase the mRNA Degradation Rate
Trans-acting factors are significant players in the linkage between mRNA translation and degradation. These factors largely cluster at two steps in the decay process decapping or deadenylation.
Decay factors may assemble on mRNAs and cause translational repression or, alternatively, assemble as a consequence of repression. Such a role was proposed when it was discovered that the decapping activators Dhh1p and Pat1p, working together, are required to facilitate translational repression in response to glucose starvation.48 The linkage with translational repression is consistent with the model suggesting that the replacement of translational initiation factors with mRNA decay proteins is an initial step in mRNA degradation.49,50 Several of these factors, including Dhh1p, Scd6p, and Sbp1p, have a decapping effect that results from their ability to repress translation.48,51–55 Of these, the DEAD box helicase Dhh1p has the strongest effect on the mRNA half-life.53,54 Pat1p is unique among decapping factors in that it functions to repress translation and directly activate the mRNA decapping enzyme, which is consistent with its strong effect on mRNA half-life.54,56
Factors that activate decapping can also assist in deadenylation. For example, the decapping activators Dhh1p and Pat1p are able to interact with and recruit deadenylases in yeast and other eukaryotes.57–61 Trans-acting factors can also target mRNA for degradation by increasing the rate of deadenylation through recruitment of deadenylases to an mRNA. These factors can either promote or cause translational repression directly or by accelerating the reduction of the poly(A) tail such as for the Ccr4/Pop2/Not deadenylase complex in Xenopus.62 Specific recruitment of deadenylases by trans-acting factors occurs in yeast and higher eukaryotes, and the PUF family of proteins is prominent among these.63,64
Trans-acting factors that act to repress translation also promote mRNA degradation as a consequence of repression. Similarly, a repressed mRNA may assemble additional trans-acting factors that reinforce repression and concomitantly stimulate decay.
Models for the Acceleration of mRNA Decay by Intrinsic Translational Repression
Intrinsic translational repression marks an individual or a class of mRNAs for specific repression (Figure 3). Such ‘marks’ can be deposited on nascent mRNA in the nucleus and affect cytoplasmic mRNA decay and translation. RNA polymerase II subunits Rpb4p and Rpb7p are such examples. They are hypothesized to be associated with transcribing mRNA in the nucleus and once in the cytoplasm affect both decay and translation.65,66 A ‘mark’ of a reduced translation can lead to increased decay of the target mRNA. Similarly, while mutations in the cap-binding complex and other translational initiation factors can affect an entire mRNA population, mutations in these factors may effectively mimic what occurs at a more local level, for example with cis RNA mutations.
mRNA decapping is an active process whereby translation initiation factors binding the cap are exchanged for a distinctly new set of proteins, with the decapping enzyme replacing eIF4E.36,49 Similarly, defects in eIF3, which interacts with eIF4F, also promote increased mRNA degradation.25,30 The cap-binding complex can play a central role in transitions to decay, thus, linking translation factors to the assembly of decapping enhancers.55
Binding of decapping enhancers can reinforce translational repression, while simultaneously promoting mRNA decapping.54 This process leads to the removal of the cap-binding complex itself. Coordination of this process may be accomplished by decapping enhancer proteins, such as Dhh1p and Pat1p, which can also interact and/or recruit the deadenylase complex to mRNA in yeast and other eukaryotes (see previous section).57–61
The inhibition of translation and deadenylation result in the formation of RNA granules, such as P bodies. P bodies are RNA-protein aggregates that accumulate non-translating mRNAs that are stored or degraded.29,67,68 Similar to mRNAs whose translation initiation is inhibited, these granules lack ribosomes and are enriched in proteins involved in 5′→3′ mRNA decay. Although P bodies are at the scene of action, their linkage with decay and translation remains unclear.69
Extrinsic Translation-Independent Effects on mRNA Decay
Extrinsic effects on translation are generated external to the cell (or mRNA) and include external environmental stresses such as glucose deprivation, heat shock, and osmotic stress. One of the first cellular responses to a stress in yeast is often translational repression.70–72 A key differentiating characteristic between intrinsic and extrinsic inhibition of translation is that stresses concomitantly induce a host of responses in the cell including inhibition of mRNA decay (Figure 4).73–77 In contrast, intrinsic effects simply act to inhibit translation and, as a consequence, enhance mRNA degradation (Figure 3). The inhibition of mRNA decay is thought to be a direct effect of stress, rather than a consequence of translational inhibition, as in the intrinsic case. As a result, this response may allow reprogramming and assessment of stress before committing to a new gene expression program (Figure 4).
In general, inhibition of deadenylation has been identified as the source of increased stability under stress. For example, such stabilization has been observed for glucose deprivation,28,35 osmotic stress28,78–80 and, in certain cases, with heat shock depending on, among other factors, the intensity of the heat shock.28,81 It should be noted that the moderate heat shock used in temperature-sensitive translation initiation alleles was not found alter the mRNA stability.28
Certain mRNAs may avoid stabilization under these conditions. For example, under glucose deprivation, classes of mRNAs, such as mRNAs encoding ribosomal proteins, undergo enhanced decay or have reduced abundance.35,82 Similarly, under osmotic stress certain groups of mRNAs also resist the trend of stabilization.78,79 Instead, these mRNAs evade the overall trend by recruiting specific decay factors to initiate more rapid deadenylation or by channeling into an alternative decay pathway. Such results suggest that increased decapping and destabilization may also be occurring during a stress that otherwise generally stabilizes mRNA. When mRNA is examined for changes in stability due to stress, timing can be important, as the greatest effects on mRNA abundance are observed after 20 min of stress. Observations after this time period may represent a shifting from the preservation of mRNA to commitment to degradation.
Taken together, extrinsic inhibition of translation due to stress appears to generally stabilize mRNA. This apparent paradox is reconciled by inhibition of deadenylation by stress. Since deadenylation is the first and rate-limiting step of mRNA degradation, this limits the effective rate of mRNA decay (Figure 4).11
At least three points suggest that stabilization of mRNA resulting from extrinsic inhibition of initiation is not due to effects on inhibition of translation initiation. First, inhibition of deadenylation occurs when translation is halted by the addition of the drug cycloheximide.28 Second, within 2 min of stress, deadenylation is inhibited, which is faster than the translational repression response.28,77 Third, inhibition of deadenylation occurs in an eIF3 translation initiation mutant that exhibits increased decay in the absence of stress.25,28 These data suggest that the stress-mediated inhibition of the deadenylase is upstream to the intrinsic effect of increased decay. Furthermore, the effects on deadenylation appear to be conserved since similar observations were made under stress conditions in mammalian cells.83,84
These data suggest that the observed alterations in mRNA decay during intrinsic and extrinsic inhibition of translation are fundamentally from different source(s). The extrinsic effects allow the cell to reprogram and assess environmental conditions. These effects may originate by being imprinted on mRNA during transcription during stress, such as for Rpb4p and Rpb7p.85 Conversely, intrinsic inhibition may have evolved to target individual groups of mRNAs for repression, rapid degradation, and elimination.
TRANSLATIONAL ELONGATION AND mRNA DEGRADATION
The reciprocal relationship between translational initiation and mRNA degradation promoted the idea that the primary mechanism of 5′→3′ decay is to first exit translation, then decap and degrade the mRNA.48 Ribosome association was thought to inhibit eukaryotic mRNA degradation, as suggested, in part, by numerous studies showing reduced mRNA decay when using the drug cycloheximide.86–90 Cycloheximide affects the elongation of ribosomes on mRNA91 and therefore supports a model that elongating ribosomes inhibit mRNA degradation.
Cycloheximide affects mRNA decay by indirectly inhibiting mRNA decapping, resulting in the accumulation of deadenylated mRNA.46 Evidence that the cycloheximide effect is indirect comes from mRNA stabilization that was observed in a poorly translated mRNA variant with a stem loop in the 5′ un-translated region.46 Furthermore, cycloheximide slows decapping of a translating MFA2 mRNA and arrests ribosomes, which block further 5′→3′ mRNA degradation.46
Consistent with the inhibition of degradation by pausing of elongation, a temperature-sensitive mutant of the tRNA CCA-adding enzyme similarly stabilizes mRNA.92 The mutant results in tRNAs lacking the proper CCA sequence at their 3′ ends, resulting in uncharged tRNAs and increased polysomes, consistent with the slowing of elongation.92,93 Similarly, a mutation in threonyl-tRNA synthetase produces stabilization of other mRNAs, presumably by the same mechanism.94
Further support for mRNA decay during translation comes from the biochemical localization of mRNA decay factors and decay intermediates to translating fractions using sucrose gradient ultracentrifugation (or polysome profiles). The 5′→3′ exonuclease Xrn1p is one decay factor that is present in polysomes.95 While the majority of Xrn1p was found in the non-translating RNP fractions or in the pellet, an mRNA decay intermediate was similarly enriched in polysomes and the RNP fractions. The human decapping enzyme was likewise found in a salt-sensitive polysomal fraction using differential ultracentrifugation.96
Co-Translational mRNA Degradation
Recently, mRNA decay has been proposed to occur co-translationally.97–99 A key observation was the localization of decapped mRNA with translating ribosomes in a sucrose gradient.97 Because decapped mRNAs are rapidly degraded, the authors used an xrn1 mutant in which decapped mRNA cannot be degraded in the 5′→3′ direction. To further examine if the association is translation-dependent, the investigators performed three additional experiments. First, they treated the extract with EDTA to collapse the polysomes into 40S and 60S ribosomal subunits. This treatment eliminated translating ribosomes from the gradient and shifted the decapped mRNAs to the non-translating fractions. Second, the sedimentation of uncapped mRNA in the polysomes was ORF length dependent suggesting ribosomal association. The shorter RPL25 mRNA had decapped mRNAs predominantly in the lighter polysomes, consistent with an ORF length that can accommodate only a limited number of ribosomes. Finally, the addition of a stem loop structure in the 5′ UTR similarly shifted the PGK1 mRNA into lighter, less translating fractions. These data suggest that mRNA can be decapped while remaining associated with ribosomes (Figure 5).
Fig 5.
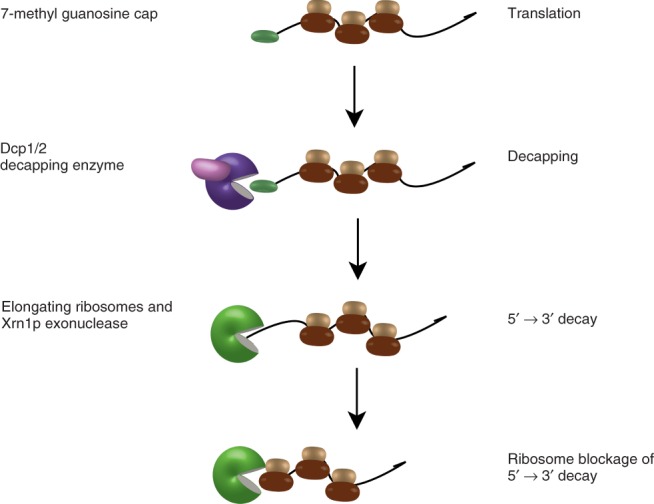
Co-translational messenger RNA (mRNA) decay. A translating mRNA is decapped by the Dcp1/2 decapping complex. Once the 5′ 7-methyl-guanosine cap is removed by the decapping enzyme, 5′→3′ exonucleolytic decay mediated by the exonuclease Xrn1p proceeds. During decay, the ribosomes block the exonuclease until they translocate off the mRNA.
The previous experiments were performed in a genetic background where decapped mRNAs are not degraded. This strain has a deletion for the cytoplasmic 5′→3′ exonuclease Xrn1p, which normally rapidly degrades uncapped mRNA. To explore whether this occurs in wild-type yeast, a pause site composed of 10 rare mostly arginine codons was inserted into an mRNA.97 This sequence was intended to stall ribosomes, which would then block 5′→3′ decay and generate mRNA decay intermediates. One possibility is that the decay product could be generated by No-Go decay due to the stalling in the region of the rare codons.6,100 However, the decay product was still observed in a mutant where No-Go decay was inhibited, suggesting it is independent of No-Go decay. Furthermore, a NMD decay mutant did not affect the appearance of the decay product. Taken together, these results suggest that the mRNA decay intermediate generated by stalled ribosomes degrades through the general 5′→3′ mRNA degradation pathway.
In summary, the authors propose that mRNA decapping in vivo occurs predominantly on polyribosomes, with the ribosomes blocking the 5′→3′ degradation of Xrn1p, as previously observed in cycloheximide-treated cells.46
These data appear to contradict evidence that mRNA decapping is greatly enhanced when translation initiation is inhibited. These data could be reconciled, if one considers a model in which mRNAs are deadenylated while still in translation.41,101,102 Deadenylated mRNA is generally a prerequisite for mRNA decay. As such, the assembly of a decapping complex on deadenylated mRNA could begin before ribosomes have fully run off. These mRNAs may have already begun the process of exchanging their translation initiation factors for mRNA decapping and decay proteins.
If this occurs, some portion of the mRNA that is marked for decay will decap mRNAs that are no longer initiating new translation but still elongating. If so, then the large majority of mRNAs may be translating when the poly(A) tail is shortened such that the tail no longer inhibits decapping. While previous experiments suggest that decapping on translating mRNAs is slower, the large amount of mRNA that is engaged in translation could account for co-translational mRNA decapping and decay. The state of an mRNA as it is being decapped while undergoing elongation is, therefore, an important area for further study to understand the mechanism of mRNA decay.
The Role of Trans-Acting Factors in Elongation and mRNA Degradation: Dhh1p and Stm1p
It is currently unclear how mRNAs can be recognized for general decay on translating polysomes. A promising candidate for this function is the DEAD box helicase Dhh1p. Dhh1p was previously shown to affect mRNA stability, decapping and to inhibit translational initiation.48,51,58,103 Recently published studies further examined the role of Dhh1p using tethering assays.51,99 Both studies observed that translation inhibition occurs for Dhh1p tethered mRNA in mutants that are inactive in the 5′→3′ decay pathway (i.e., dhh1 and xrn1), and the mRNA levels are not decreased, suggesting that these functions are separable.
Experiments from the Coller laboratory went further to suggest that translation inhibition is at least partially due to the inhibition of elongation. They provide evidence that Dhh1p is found in translating polysomes, mRNA tethered to Dhh1p localizes to polysomes and that Dhh1p affects decay of mRNA having sequences that stall ribosomes.
Their work proposed that Dhh1p slows translational elongation, while earlier experiments failed to find Dhh1p association with translating mRNA.48 However, using the reasoning that Dhh1p association with translating mRNA may be transient, crosslinking was used to stabilize the potential interactions. Using crosslinking, Dhh1p was found within the translating portion of the polysome gradient, consistent with ribosomal association.99 Furthermore, this result was confirmed by affinity purifying ribosomes and subjecting them to sucrose gradient ultracentrifugation. Similarly, the association was also independently observed without crosslinking after the diauxic shift, suggesting a possible regulation of Dhh1p.104 The transience of the interaction may be linked to the ATPase function of Dhh1p. While it is not required for translational repression, this function affects the decay of short-lived mRNA as well as the localization of Dhh1p in P bodies.99
Likewise, mRNA tethered to Dhh1p localized to polysomes, consistent with an increased load of ribosomes and suggestive of translational stalling (Figure 6).99 The results of sedimentation after affinity purification of ribosomes suggest that these rapidly sedimenting and apparently highly translating mRNAs are ribosome associated.
Fig 6.
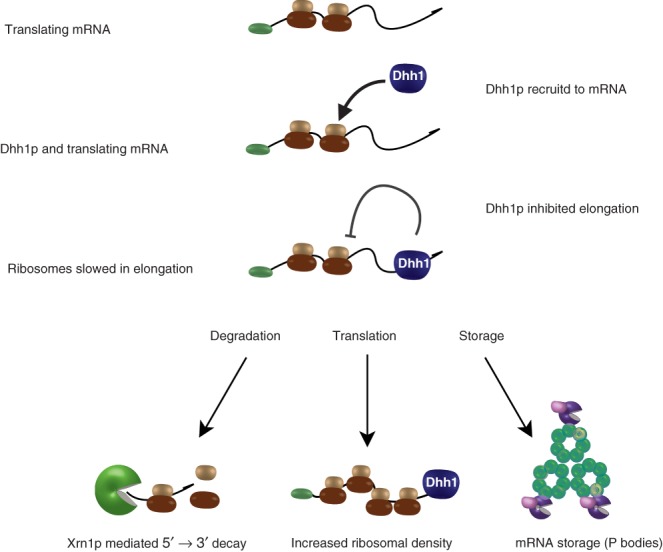
Dhh1p-mediated inhibition of ribosomal translocation. A translating messenger RNA (mRNA) directly or indirectly recruits Dhh1p. Once associated with an mRNA, Dhh1p acts to inhibit elongation of the message by the ribosomes. The affected mRNA can then undergo one of three fate choices: 5′→3′ degradation by decapping and exonucleolytic decay, storage in P bodies, or other structures or accumulation of translationally slowed ribosomes.
The role of slowing or pausing elongation by Dhh1p is not yet clear, as it may perform several functions. Three of these possibilities include promoting the storage of translationally silent mRNA, inhibiting translation or directly promoting decay (Figure 6). First, it is possible that elongation slowing is an important element to prepare mRNAs for storage. While P bodies have not been shown to contain elongationally stalled messages, these mRNAs may be stored elsewhere or be in a transitory state. Second, elongation slowing may be a contributor to the overall translation rate, as observed for long stretches of rare codons, which can result in feedback inhibition of initiation.105 Third, elongation slowing may simply be used to promote decay, such as for elongation-slowed mRNAs. An example is the PGK1 mRNA, which is a transcript whose decay is normally relatively insensitive to the presence of Dhh1p. PGK1 can be rendered sensitive to Dhh1p by the addition of rare codons, which slows translation, suggesting a possible role of Dhh1p in quality control surveillance (Figure 7).99 This process could be in competition with No-Go decay.6,100 However, Dhh1p may represent an independent quality control system consistent with a recent report suggesting that the No-Go dependent decay does not occur on multiple proline codon sequences that stall ribosomes.106 However, the nature or extent of stalling that targets an mRNA to either a Dhh1p or No-Go dependent decay pathway has not yet been determined.
Fig 7.
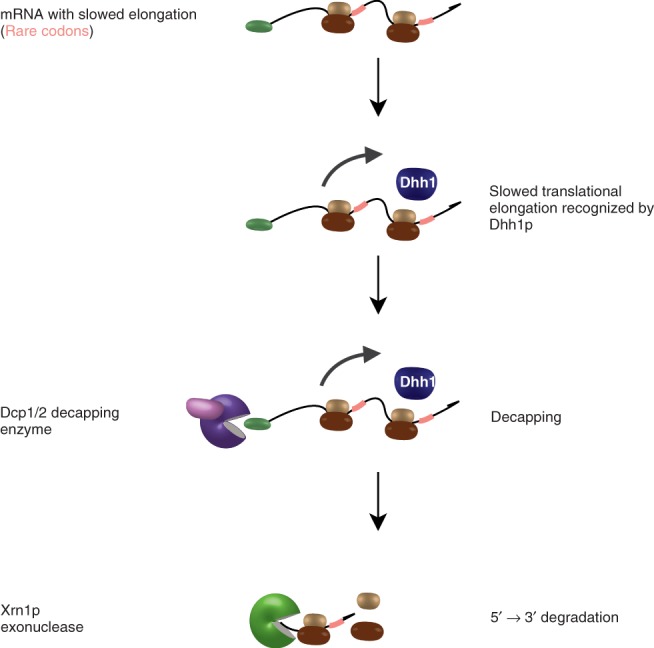
Detection of slowed elongation by Dhh1p. Slowed ribosomal elongation of a translating messenger RNA (mRNA) containing rare codons or other slow stretches of mRNA (indicated in pink) is detected by Dhh1p, resulting in accelerated decapping by the decapping complex Dcp1/2 and consequent degradation of the mRNA by the exonuclease Xrn1p.
Although Dhh1p can inhibit initiation and elongation, it is unclear what determines its specific function or what prompts Dhh1p to promote storage or decay. Dhh1p may act through additional proteins since Dhh1p has no consensus RNA-binding site based on a recent CLIP study.107 One prominent candidate is Stm1p, with which it has a genetic interaction.108 Consistent with this model, Stm1p is ribosome associated and implicated in translational slowing.109–111 These proteins share similarities, such as sensitivity to translation elongation-inhibiting drugs.99,110 Additionally, Stm1p promotes the decay of an mRNA that has been shown to be degraded in a Dhh1p-dependent fashion.108,112 Interestingly, the decay rates of some transcripts are altered by Dhh1p but not by Stm1p, suggesting that Dhh1p also targets a set of transcripts independent of Stm1p.58,103,108
Another possibility is that multiple protein partners could modulate Dhh1p function. This possibility was recently investigated through biochemical and crystallographic experiments by the Conti laboratory.113 The authors demonstrated that Dhh1p can tightly bind Edc3p, Pat1p, and RNA, but these binding relationships are exclusive, with Edc3p and Pat1p interacting with the same region of Dhh1p. Given this exclusivity, these data suggest a model whereby the alteration of protein binding partners could allow Dhh1p to possess alternative functions. Furthermore, the Dhh1p-binding regions of Edc3p and Pat1p only occupy a small portion of the surface of Dhh1p, leading to possible combinatorial effects with additional proteins.
These alterations in binding partners may be linked to localization in P bodies. For example, the Weis laboratory observed that Dhh1p-tethered mRNA is localized to P bodies in an xrn1 deletion mutant, while tethering Dhh1p in the dcp2 deletion caused localization to highly translating polysomes.51 Whether these effects are due to the altered abundance of P bodies in these mutants114 or have implications for elongation-stalled mRNAs in P bodies remains to be examined.
CONCLUSION
Fundamentally, the processes of translation and the general degradation of mRNA are tightly linked. The origin of the perturbation of translation has a major impact on the subsequent effects on degradation. Translation can impact the rates of mRNA decay in three major ways.
First, intrinsic translation repression of an mRNA provides insight into the mechanism of how an individual mRNA exits translation and begins the process of decay (Figure 3). As a consequence of reduced translation, mRNAs appear to be driven toward a fate of destruction due to the assembly of decay factors that replace the translation initiation machinery and reinforce the initial repression and enhance decay rates. This type of mechanism ensures that mRNAs that have limited translatability will be directed toward decay by default. Second, extrinsic translational repression of mRNA by stress conditions targets the bulk of the mRNA that is present at the time (Figure 4). By inhibiting mRNA decay upstream at deadenylation, the inhibition of translation is uncoupled from the increased decay that is observed with intrinsic translational inhibition. To avoid entry into the common mRNA degradation pathways, mRNA stability is increased by inhibiting deadenylation, which is the first step of general mRNA decay. The slowing of mRNA decay allows time for the cell to assess its commitment to its current genetic program and, after adaptation, to commence degradation of unnecessary mRNAs (Figure 4). Third, mRNA decay may occur co-translationally, with mRNA containing elongation pauses being targeted for enhanced decay.
Acknowledgments
We thank Mridula Muppavarapu and Marcus Johansson for comments on the manuscript. The research of the authors is supported by grants from Umeå University and Vetenskapsrådet (The Swedish Research Council) 621-2010-4602. Funding for open access from the Bernhard och Signe Bäckströms Fond. We apologize to those authors whose work we could not cite owing to space limitations.
REFERENCES
- 1.Jonas S, Izaurralde E. The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev. 2013;27:2628–2641. doi: 10.1101/gad.227843.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang Y-F, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 3.Kervestin S, Jacobson A. NMD: a multifaceted response to premature translational termination. Nat Rev Mol Cell Biol. 2012;13:700–712. doi: 10.1038/nrm3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- 5.Klauer AA, van Hoof A. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip Rev RNA. 2012;3:649–660. doi: 10.1002/wrna.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harigaya Y, Parker RR. No-go decay: a quality control mechanism for RNA in translation. Wiley Interdiscip Rev RNA. 2010;1:132–141. doi: 10.1002/wrna.17. [DOI] [PubMed] [Google Scholar]
- 7.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 8.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 9.Parker RR. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhlrad D, Parker RR. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 11.Decker CJ, Parker RR. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 12.Hsu CL, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CE, Tarun SZ, Boeck R, Sachs AB. PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5744–5753. doi: 10.1128/mcb.16.10.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker RR. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 15.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denis CL, Chen J. The CCR4-NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acid Res Mol Biol. 2003;73:221–250. doi: 10.1016/s0079-6603(03)01007-9. [DOI] [PubMed] [Google Scholar]
- 17.Thore S, Mauxion F, Seraphin B, Suck D. X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep. 2003;4:1150–1155. doi: 10.1038/sj.embor.7400020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C-YA, Shyu A-B. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA. 2011;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker RR. Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J. 2002;21:1427–1436. doi: 10.1093/emboj/21.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hoof A, Frischmeyer PA, Dietz HC, Parker RR. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 21.Muhlrad D, Decker CJ, Parker RR. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5″→3″ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 22.Chang JH, Xiang S, Xiang K, Manley JL, Tong L. Structural and biochemical studies of the 5″→3″ exoribonuclease Xrn1. Nat Struct Mol Biol. 2011;18:270–276. doi: 10.1038/nsmb.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jinek M, Coyle SM, Doudna JA. Coupled 5′ nucleotide recognition and processivity in xrn1-mediated mRNA decay. Mol Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1:253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz DC, Parker RR. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.She M, Decker CJ, Chen N, Tumati S, Parker RR, Song H. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat Struct Mol Biol. 2006;13:63–70. doi: 10.1038/nsmb1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilgers V, Teixeira D, Parker RR. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae. RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brengues M, Teixeira D, Parker RR. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnes CA. Upf1 and Upf2 proteins mediate normal yeast mRNA degradation when translation initiation is limited. Nucleic Acids Res. 1998;26:2433–2441. doi: 10.1093/nar/26.10.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown JT, Yang X, Johnson AW. Inhibition of mRNA turnover in yeast by an xrn1 mutation enhances the requirement for eIF4E binding to eIF4G and for proper capping of transcripts by Ceg1p. Genetics. 2000;155:31–42. doi: 10.1093/genetics/155.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaGrandeur T, Parker RR. The cis acting sequences responsible for the differential decay of the unstable MFA2 and stable PGK1 transcripts in yeast include the context of the translational start codon. RNA. 1999;5:420–433. doi: 10.1017/s1355838299981748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linz B, Koloteva N, Vasilescu S, McCarthy JE. Disruption of ribosomal scanning on the 5′-untranslated region, and not restriction of translational initiation per se, modulates the stability of nonaberrant mRNAs in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:9131–9140. doi: 10.1074/jbc.272.14.9131. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munchel SE, Shultzaberger RK, Takizawa N, Weis K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol Biol Cell. 2011;22:2787–2795. doi: 10.1091/mbc.E11-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwartz DC, Parker RR. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol. 2000;20:7933–7942. doi: 10.1128/mcb.20.21.7933-7942.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilela C, Velasco C, Ptushkina M, McCarthy JE. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the eIF4F translation initiation complex. EMBO J. 2000;19:4372–4382. doi: 10.1093/emboj/19.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarun SZ, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz DC, Parker RR. Interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiae. Cold Spring Harb Monogr Arch. 2000;39:807–826. [Google Scholar]
- 41.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 42.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muhlrad D, Decker CJ, Parker RR. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghosh S, Jacobson A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdiscip Rev RNA. 2010;1:351–361. doi: 10.1002/wrna.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta. 2012;1819:593–603. doi: 10.1016/j.bbagrm.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Beelman CA, Parker RR. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. [PubMed] [Google Scholar]
- 47.Coller J, Gray N, Wickens M. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coller J, Parker RR. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tharun S, Parker RR. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 50.Coller J, Parker RR. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 51.Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J Cell Biol. 2011;194:527–537. doi: 10.1083/jcb.201007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal SP, Dunckley T, Parker RR. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:5120–5130. doi: 10.1128/MCB.01913-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle J-C, Fromont-Racine M, Jacobson A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc Natl Acad Sci USA. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nissan T, Rajyaguru P, She M, Song H, Parker RR. Decapping activators in Saccharomyces cerevisiae act by multiple mechanisms. Mol Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajyaguru P, She M, Parker RR. Scd6 targets eIF4G to repress translation: RGG motif proteins as a class of eIF4G-binding proteins. Mol Cell. 2012;45:244–254. doi: 10.1016/j.molcel.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker RR. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 57.Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A. Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics. 1998;148:571–579. doi: 10.1093/genetics/148.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker RR. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maillet L, Collart MA. Interaction between Not1p, a component of the Ccr4-Not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem. 2002;277:2835–2842. doi: 10.1074/jbc.M107979200. [DOI] [PubMed] [Google Scholar]
- 60.Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol Cell Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 64.Miller MA, Olivas WM. Roles of Puf proteins in mRNA degradation and translation. Wiley Interdiscip Rev RNA. 2011;2:471–492. doi: 10.1002/wrna.69. [DOI] [PubMed] [Google Scholar]
- 65.Harel-Sharvit L, Eldad N, Haimovich G, Barkai O, Duek L, Choder M. RNA polymerase II subunits link transcription and mRNA decay to translation. Cell. 2010;143:552–563. doi: 10.1016/j.cell.2010.10.033. [DOI] [PubMed] [Google Scholar]
- 66.Bellofatto V, Wilusz J. Transcription and mRNA stability: parental guidance suggested. Cell. 2011;147:1438–1439. doi: 10.1016/j.cell.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 67.Sheth U, Parker RR. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker RR. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Decker CJ, Parker RR. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Simpson CE, Ashe MP. Adaptation to stress in yeast: to translate or not? Biochem Soc Trans. 2012;40:794–799. doi: 10.1042/BST20120078. [DOI] [PubMed] [Google Scholar]
- 71.Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuhn KM, DeRisi JL, Brown PO, Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol Cell Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
- 74.Bond U. Stressed out! Effects of environmental stress on mRNA metabolism. FEMS Yeast Res. 2006;6:160–170. doi: 10.1111/j.1567-1364.2006.00032.x. [DOI] [PubMed] [Google Scholar]
- 75.Grousl T, Ivanov P, Frýdlová I, Vasicová P, Janda F, Vojtová J, Malínská K, Malcová I, Nováková L, Janosková D, et al. Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J Cell Sci. 2009;122:2078–2088. doi: 10.1242/jcs.045104. [DOI] [PubMed] [Google Scholar]
- 76.Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192:289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Melamed D, Pnueli L, Arava Y. Yeast translational response to high salinity: global analysis reveals regulation at multiple levels. RNA. 2008;14:1337–1351. doi: 10.1261/rna.864908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Romero-Santacreu L, Moreno J, Pérez-Ortín JE, Alepuz P. Specific and global regulation of mRNA stability during osmotic stress in Saccharomyces cerevisiae. RNA. 2009;15:1110–1120. doi: 10.1261/rna.1435709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Molin C, Jauhiainen A, Warringer J, Nerman O, Sunnerhagen P. mRNA stability changes precede changes in steady-state mRNA amounts during hyperosmotic stress. RNA. 2009;15:600–614. doi: 10.1261/rna.1403509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller C, Schwalb B, Maier K, Schulz D, Dümcke S, Zacher B, Mayer A, Sydow J, Marcinowski L, Dölken L, et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Mol Syst Biol. 2011;7:458. doi: 10.1038/msb.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Castells-Roca L, García-Martínez J, Moreno J, Herrero E, Bellí G, Pérez-Ortín JE. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS One. 2011;6:e17272. doi: 10.1371/journal.pone.0017272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arribere JA, Doudna JA, Gilbert WV. Reconsidering movement of eukaryotic mRNAs between polysomes and P bodies. Mol Cell. 2011;44:745–758. doi: 10.1016/j.molcel.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gowrishankar G, Winzen R, Bollig F, Ghebremedhin B, Redich N, Ritter B, Resch K, Kracht M, Holtmann H. Inhibition of mRNA deadenylation and degradation by ultraviolet light. Biol Chem. 2005;386:1287–1293. doi: 10.1515/BC.2005.146. [DOI] [PubMed] [Google Scholar]
- 84.Gowrishankar G, Winzen R, Dittrich-Breiholz O, Redich N, Kracht M, Holtmann H. Inhibition of mRNA deadenylation and degradation by different types of cell stress. Biol Chem. 2006;387:323–327. doi: 10.1515/BC.2006.043. [DOI] [PubMed] [Google Scholar]
- 85.Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol Syst Biol. 2008;4:223. doi: 10.1038/msb.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raj NB, Pitha PM. Analysis of interferon mRNA in human fibroblast cells induced to produce interferon. Proc Natl Acad Sci USA. 1981;78:7426–7430. doi: 10.1073/pnas.78.12.7426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 88.Stimac E, Groppi VE, Coffino P. Increased histone mRNA levels during inhibition of protein synthesis. Biochem Biophys Res Commun. 1983;114:131–137. doi: 10.1016/0006-291x(83)91604-2. [DOI] [PubMed] [Google Scholar]
- 89.Gay DA, Sisodia SS, Cleveland DW. Autoregulatory control of β-tubulin mRNA stability is linked to translation elongation. Proc Natl Acad Sci USA. 1989;86:5763–5767. doi: 10.1073/pnas.86.15.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herrick D, Parker RR, Jacobson A. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2269–2284. doi: 10.1128/mcb.10.5.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peltz SW, Donahue JL, Jacobson A. A mutation in the tRNA nucleotidyltransferase gene promotes stabilization of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:5778–5784. doi: 10.1128/mcb.12.12.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Aebi M, Kirchner G, Chen JY, Vijayraghavan U, Jacobson A, Martin NC, Abelson J. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae. J Biol Chem. 1990;265:16216–16220. [PubMed] [Google Scholar]
- 94.Zuk D, Belk JP, Jacobson A. Temperature-sensitive mutations in the Saccharomyces cerevisiae MRT4, GRC5, SLA2 and THS1 genes result in defects in mRNA turnover. Genetics. 1999;153:35–47. doi: 10.1093/genetics/153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mangus DA, Jacobson A. Linking mRNA turnover and translation: assessing the polyribosomal association of mRNA decay factors and degradative intermediates. Methods. 1999;17:28–37. doi: 10.1006/meth.1998.0704. [DOI] [PubMed] [Google Scholar]
- 96.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu W, Petzold C, Coller J, Baker KE. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:244–247. doi: 10.1038/nsmb.1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sweet T, Kovalak C, Coller J. The DEAD-Box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuroha K, Akamatsu M, Dimitrova L, Ito T, Kato Y, Shirahige K, Inada T. Receptor for activated C kinase 1 stimulates nascent polypeptide-dependent translation arrest. EMBO Rep. 2010;11:956–961. doi: 10.1038/embor.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Long RM, McNally MT. mRNA decay: x (XRN1) marks the spot. Mol Cell. 2003;11:1126–1128. doi: 10.1016/s1097-2765(03)00198-9. [DOI] [PubMed] [Google Scholar]
- 102.Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013;29:691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Drummond SP, Hildyard J, Firczuk H, Reamtong O, Li N, Kannambath S, Claydon AJ, Beynon RJ, Eyers CE, McCarthy JE. Diauxic shift-dependent relocalization of decapping activators Dhh1 and Pat1 to polysomal complexes. Nucleic Acids Res. 2011;39:7764–7774. doi: 10.1093/nar/gkr474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, Haar von der T. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J. 2014;33:21–34. doi: 10.1002/embj.201385651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mitchell SF, Jain S, She M, Parker RR. Global analysis of yeast mRNPs. Nat Struct Mol Biol. 2013;20:127–133. doi: 10.1038/nsmb.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Balagopal V, Parker RR. Stm1 modulates mRNA decay and Dhh1 function in Saccharomyces cerevisiae. Genetics. 2009;181:93–103. doi: 10.1534/genetics.108.092601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Dyke MW, Nelson LD, Weilbaecher RG, Mehta DV. Stm1p, a G4 quadruplex and purine motif triplex nucleic acid-binding protein, interacts with ribosomes and subtelomeric Y′ DNA in Saccharomyces cerevisiae. J Biol Chem. 2004;279:24323–24333. doi: 10.1074/jbc.M401981200. [DOI] [PubMed] [Google Scholar]
- 110.Van Dyke N, Pickering BF, Van Dyke MW. Stm1p alters the ribosome association of eukaryotic elongation factor 3 and affects translation elongation. Nucleic Acids Res. 2009;37:6116–6125. doi: 10.1093/nar/gkp645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Balagopal V, Parker RR. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA. 2011;17:835–842. doi: 10.1261/rna.2677311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Muhlrad D, Parker RR. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sharif H, Ozgur S, Sharma K, Basquin C, Urlaub H, Conti E. Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res. 2013;41:8377–8390. doi: 10.1093/nar/gkt600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teixeira D, Parker RR. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]


