Abstract
Background
Two recent technological advances dramatically reducing the rate of false-negatives in activity prediction by docking flexible 3D models of compounds include multi-conformational docking (mPockDock) and the docking of candidates to atomic property fields derived by co-crystallized ligands (mApfDock).
Results
The mApfDock and mPockDock provide the AUC of 90.4 and 83.8%, respectively. The mApfDock gave better performance when compounds required large induced-fit pocket changes unseen in crystallography, whereas the mPockDock is superior when the co-crystallized ligands do not represent sufficient chemical and binding location diversity.
Conclusion
Both approaches proved to be efficient for scaffold hopping; they are complementary when the coverage of the co-crystallized complexes is poor but become convergent when the complexes are diverse enough.
Background
Predicting biological activity of a compound directly from its 2D chemical sketch is one of the key challenges of computational biology and chemistry. Important applications of such a prediction include: identification of potential endocrine disruptors and environmental threats among 80,000 chemicals in the environment [1]; virtual ligand screening and finding de novo candidates with therapeutic activity [2–5]; repurposing a known drug for a different therapeutic target [6,7]; ‘scaffold hopping’ or replacement of a known active scaffold by a different chemotype with similar target activity; generation of focused libraries/derivatives for compound optimization; predicting poly-pharmacology of a compound [8], and so on. There are three principal method types that can be used to perform this task: the machine learning methods trained on many specific chemicals described by their 2D structure via derived properties and/or fingerprints (e.g., quantitative structure–activity relationship or chemical similarity) [9]; the 3D ligand-based methods that link the activity with a particular shape of 3D-property distribution and require one or a small number of ligands [10]; and the docking method, which derives the activity estimate from the predicted pose of a compound in the protein-binding pocket [11–13]. The pocket-docking method has the least (if any) dependence on prior knowledge of actives, and both (b) and (c) do not depend on a large training set and have the potential to capture the activity of an entirely new chemical structure never represented in a training set. For that reason we are focusing on improving the docking and scoring recognition methods using either the pockets or the known superimposed ligands.
The rapid growth of the protein crystallographic database, followed by the compilation of a comprehensive set of pockets, the Pocketome [14], provides a set of approximately 2000 flexible pocket ensembles with co-crystallized ligands. This set gives us a chance to compile a large and diverse recognition benchmark where either pockets or co-crystallized ligands may be used to recognize hundreds to thousands of known actives; use the benchmark to compare the improved versions of two main docking-based recognition methods, atomic property fields (APFs) docking and the multiple pocket conformation Internal Coordinates Mechanics (ICM) docking. The APF concept [10], a variation of Goodford’s GRID approach [15], is a continuous, multicomponent 3D potentials that represents preferences for key physicochemical atomic properties in various regions of 3D space occupied by the ligand [10]. In an independent comparative evaluation even a single ligand-generated APF-based molecular superposition outperformed several other methods in identifying correct alignment of bioactive conformations [16]. Our recent study also indicated that APFs offer an improvement in activity prediction compared with 2D fingerprint-based methods on a benchmark consisting of 320,000 molecular pairs [17]. Furthermore we evaluated and compared the pocket- and field-based models on a set of 13 G-protein-coupled receptors and 25 nuclear receptors [18]. However, that benchmark was somewhat limited and not designed to emphasize the ability of models to recognize new chemical scaffolds. Similarly, the Directory of Useful Decoys, one of the most popular benchmarks for molecular screening [19], has its limitations for the task at hand. In summary, the multipocket and cumulative field-based approaches have not been evaluated and optimized for the scaffold-hopping task on an unbiased and diverse benchmark set [16,18–22]. Here we explored the following questions: how to design a clean and unbiased and diverse benchmark explicitly for the scaffold-hopping task; can the docking/scoring to either multiple pockets (mPockDock) or multiple co-crystallized ligand fields (mApfDock) outperform the published shape or docking procedures [20]; for the field/shape docking, can cumulative fields from multiple ligands improve bioactivity prediction while reducing the bias to a specific ligand.
Key terms.
Virtual ligand screening
An in silico technique to screen a database of chemicals against activity models in order to identify new active candidates.
Scaffold hopping
An approach to discover structurally distinct compounds with the same activity as the original molecules.
Poly-pharmacology
An ability of a molecule to interact with multiple biological targets.
Atomic property fields
Continuous 3D potential maps for seven pharmacophore features.
Molecular docking
A method for predictions of mode of interaction of molecular ligands to binding pockets of macromolecules.
Here, we tested the improvements in both docking methods by considering multiple co-crystalized ligands and a selection of multiple binding pockets. The combined APFs were constructed from two to 76 co-crystalized ligands per model. A clean yet challenging benchmark suitable for evaluation of the scaffold-hopping ability was then carefully compiled for 37 protein targets, including 11 to 239 actives structurally dissimilar to the co-crystallized ‘seed’ compounds and 10-times larger number of decoys with matched properties. The recognition ability of the mApfDock was compared with the mPockDock [23] to demonstrate the complementarity of the two approaches for the scaffold hopping or activity prediction when both ligands and protein pockets are available.
Materials & methods
Scaffold-hopping benchmark preparation
All available bioactive drug-like small molecules were extracted from the ChEMBL database, which contained 2,787,240 published bioactivities of 578,715 distinct drug-like molecules for 7493 targets (ChEMBL version 5) [24,25]. Confidence levels assigned in ChEMBL to the links between molecular targets and published assays were used to filter out the unreliable data points. In addition, owing to the existence of complicated and inconsistent activity formats of the published assays, the normalization of bioactivities into a uniform unit, logarithm of molar concentration (M), was performed to allow further comparisons of bioactivities between different assays.
The active set (binders) and the decoy set (non-binders), we build with the following four steps. First, only molecules with bioactivities expressed as association/dissociation or inhibitory constant (Ka/Kd/Ki) or half maximal inhibitory/effective concentration (IC50/EC50) were included in the active set. IC50 was converted to the Ki constant with the Cheng–Prusoff equation [26]. Second, a criterion of collecting molecules with bioactivities <1 μM was set to define the biologically active binders. Good justification for setting 1 μM as a cutoff is that weaker binders have poorly defined molecular features favorable for binding and thus may not contribute constructively to the recognition; practically, these are compounds with potencies better than 1 μM that are typically considered as viable leads; and, activities in the range above 10 μM are often nonspecific/promiscuous upon secondary evaluation. Third, compounds were clustered based on 2D structural fingerprints and those with structural similarity closer than 0.3 linear fingerprint Tanimoto distance (TD) were excluded from active sets. TD is defined as TD = 1−T where T is the Tanimoto similarity coefficient calculated as the number of matching non-zero positions/bits between two fingerprints (m) divided by the total number of non-zero positions/bits (n):
Fourth, protonation state at pH 7.4 was assigned using ICM [27] and partial charges were assigned using MMFF94 [28,29].
The decoy sets were generated from the ChemBridge catalog with over 700,000 available compounds. High-quality decoy sets should resemble the physical properties of the active ligands; therefore, six physical properties were used to match the active sets: molecular weight, the number of hydrogen bond acceptors and donors, the number of rotatable bonds, the number of atoms, and LogP. For each of 37 targets, the actives were clustered at 0.3 TD and the average properties were calculated for each cluster. Decoy compounds that resemble these physical properties within a range of ±1 SD of the mean for each parameter were collected. In addition the decoys closer than 0.3 TD to each other were excluded from the decoy sets to avoid redundancy. In the absence of any information about activity of decoy molecules against a given target, we removed the similar-to-actives (TD < 0.5) decoy molecules to reduce the chance of choosing an ‘active’ decoy by accident. Finally, the decoys were assigned the protonation state and partial charges the same way as for the actives. All molecules were assigned to 3D coordinates by the Molsoft convert 2D to 3D procedure.
Protein target selection
Protein targets were selected from the original Pocketome [14] set compiled from experimental co-crystal structures in the Protein Data Bank [30]. The selection criteria included the following: the target should have more than two distinct Protein Data Bank noncovalent co-crystallized small-molecule ligands (referred to as ‘seeds’ for the mApfDock models) and more than 10 known actives with relevant activity values from ChEMBL activity database. Targets with prodrug actives or peptide-binding sites were excluded as well and low-resolution structures.
3D APF models, docking & scoring
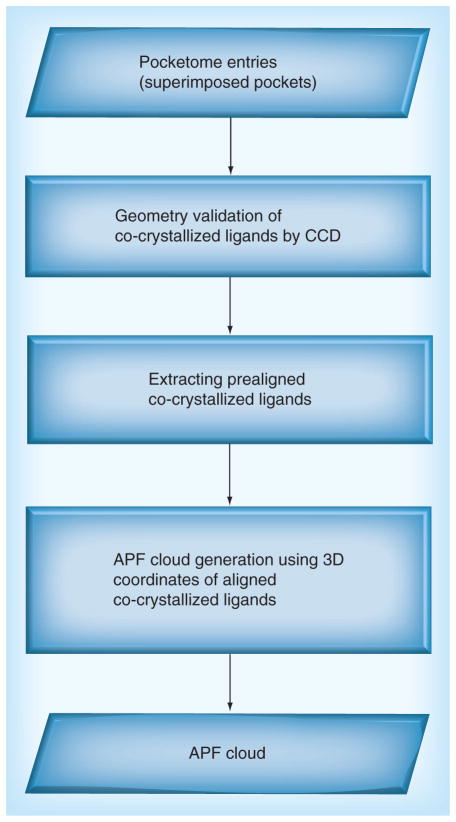
APFs represent seven physicochemical properties of the observed atoms of co-crystalized ligands inside the pocket space as continuous 3D potentials represented as a set of discrete values on a grid. The fields include hydrogen bond acceptor, hydrogen bond donor, charged, lipophilic, sp2 hybridized, size and electronegative/electropositive [10]. In this study, the mApfDock model was generated from multiple co-crystallized ligands extracted from an ensemble of superimposed pockets from Pocketome [14]. The outline of the APF map generation is shown in Figure 1. The superposition of co-crystallized ligand structures was the consequence of the iterative protein pocket superposition. The algorithm evolves a set of Gaussian weights gradually to find the better superimposable core of atom pairs between two proteins [31]. The geometry and connectivity of crystallographic ligand structures in the Pocketome was validated using the Chemical Component Dictionary database [32].
Figure 1. Flowchart of atomic property field cloud generation.
APF: Atomic property field; CCD: Chemical Component Dictionary.
For each target the actives and decoys were docked into the mApfDock grids using the ICM stochastic global optimization procedure [33]. Molecules were sampled by random torsion modifications or translation/rotation moves followed by local gradient minimization and acceptance/rejection of the new conformation according to Metropolis criterion [10]. The history-based feedback mechanism and temporary annealing schedules improve the sampling efficiency. The docking performance of the mApfDock models was also evaluated by self-docking the seed ligands onto the mApfDock grids using the aforementioned procedure.
ICM grid docking
For each of 37 targets all pockets containing the seed ligands were refined around their cognate ligands. The refinement procedure introduced on the basis of our experience in the OpenEye docking competition [21] attempts to improve the placement of hydrogens, as well as the tips of interacting side chains around the bound ligands. The ICM Biased Probability Monte Carlo algorithm was used to sample the conformation of side chains of residues within 5 Å from the seed ligand [11]. The low-energy conformations with distinct geometry were saved in a conformational stack. The number of alternative conformations generated during this procedure was largely dependent on the pocket plasticity. The maximal number of structures in the conformational stack was set to 300 by adjusting the maximum angular root-mean-square deviation (RMSD) per variable when two structures are still considered as belonging to the same cluster. The binding score was then compared between the unrefined and refined models and the better model is picked.
Grid potential maps including van der Waals, electrostatic, hydrogen bonding and hydrophobicity were calculated for each new conformation [34]. The ICM molecular docking first sampled ligand conformations then created starting poses for the conformers in the binding pocket and then optimized the ligand-binding geometry in grid potential maps of each pocket conformer using the Biased Probability Monte Carlo simulation. Three independent runs of the ICM molecular docking were performed to ensure convergence of the Monte Carlo optimization and the top three best quality complexes were rescored [35]. The top-scoring ligand poses were merged into the full atom model of each protein conformer and evaluated with all-atom ICM ligand binding score [36,37]. More details can be found in [12,13,21].
Evaluating the prediction performance by normalized square root receiver operating characteristic AUC
The performance of the two algorithms at distinguishing active ligands from a large decoy set was evaluated as the area (AUC*) under the receiver operating characteristic (ROC) curve with the following axes: true positive rate (y-axis) versus the square root of total number of data points (x-axis). Since the AUC fails to address the early recognition problems in some cases [38], the NSQAUC described recently was used in the assessment of the screening power [39]. The NSQAUC is calculated as:
NSQAUC is more sensitive to early compound recognition than the commonly used linear AUC [40]. A model giving perfect separation of signals from noise returns the NSQAUC of 100, whereas a model giving a random guess returns the NSQAUC of 0.
Results & discussion
Generation of high-quality superimposed protein pockets
We collected a library of 37 high quality protein pockets from the Pocketome. The detailed target information is shown in Table 1. Each protein pocket was represented by an ensemble of superimposed pocket structures in order to capture the binding conformations of the corresponding complex ligands. The number of residues surrounding the protein pockets for pocket superposition process and the average of pairwise backbone RMSDs between the superimposed pockets are shown in Table 2. This RMSD reveals the degree of conformational variability of the protein pocket and its steric compatibility with various ligands in the ensemble. The smaller value of RMSD of superimposed pockets ensures the quality of molecular alignment. The 3D coordinates of the corresponding ligands from superimposed pockets were used as a template to construct the mApfDock model rather than using molecular alignment that overlaps common substructures or pharmacophores in neighboring areas in space. The assumption of common substructural fragments leading to similar biological activity and binding conformation is not appropriate to all cases, because small structural changes in molecules sometimes lead to the complete different biological activities and binding conformations, so-called discontinuities in protein–ligand interaction. Therefore, using the 3D coordinates of co-crystallized ligands from the superimposed pockets provides an advantage of preserving the near-real and unbiased ligand-binding conformations that reveal the actual protein–ligand interactions.
Table 1.
Information of protein targets and associated benchmarks.
| ChEMBL ID | Swiss ID | Target | Organism | Domain | Seeds | Actives | Decoys |
|---|---|---|---|---|---|---|---|
| 275 | P19793 | RXRA | Human | 221–462 | 6 | 14 | 207 |
| 10090 | P0A6K3 | DEF | Escherichia coli | 2–169 | 4 | 14 | 417 |
| 10460 | P16184 | DYR | Pneca | 1–206 | 11 | 25 | 440 |
| 11 | P00734 | THRB | Human | 364–622 | 20 | 200 | 2832 |
| 157 | Q02127 | PYRD | Human | 28–395 | 4 | 19 | 561 |
| 12536 | P47811 | MK14 | Mouse | 2–359 | 35 | 23 | 3359 |
| 11473 | P78536 | ADA17 | Human | 216–478 | 5 | 52 | 1122 |
| 133 | P37231 | PPARG | Human | 232–505 | 15 | 35 | 1745 |
| 11678 | P24941 | CDK2 | Human | 1–298 | 76 | 24 | 516 |
| 242 | P15121 | ALDR | Human | 1–316 | 13 | 21 | 1046 |
| 11110 | P22894 | MMP8 | Human | 101–262 | 16 | 67 | 1837 |
| 19607 | P00800 | THER | Bacth | 233–548 | 10 | 14 | 1739 |
| 19 | P03372 | ESR1 | Human | 302–551 | 32 | 45 | 1097 |
| 20073 | P11509 | CP2A6 | Human | 24–494 | 6 | 11 | 205 |
| 11359 | Q08499 | PDE4D | Human | 380–722 | 12 | 24 | 364 |
| 174 | Q92731 | ESR2 | Human | 258–505 | 10 | 48 | 755 |
| 11871 | P39900 | MMP12 | Human | 105–264 | 8 | 14 | 174 |
| 11109 | P08254 | MMP3 | Human | 101–270 | 18 | 115 | 1397 |
| 10185 | P55263 | ADK | Human | 18–362 | 3 | 18 | 481 |
| 10140 | P06239 | LCK | Human | 228–505 | 7 | 60 | 1295 |
| 11408 | P53779 | MK10 | Human | 40–403 | 8 | 21 | 673 |
| 13000 | P03956 | MMP1 | Human | 102–267 | 4 | 146 | 2425 |
| 9 | P00533 | EGFR | Human | 695–1022 | 4 | 104 | 2667 |
| 163 | Q07869 | PPARA | Human | 197–468 | 3 | 31 | 340 |
| 10599 | Q07343 | PDE4B | Human | 321–700 | 12 | 26 | 608 |
| 10980 | P35968 | VGFR2 | Human | 810–1171 | 4 | 103 | 2669 |
| 10074 | P23946 | CMA1 | Human | 22–247 | 2 | 12 | 186 |
| 36 | P06401 | PRGR | Human | 676–933 | 5 | 43 | 1689 |
| 25 | P04150 | GCR | Human | 504–777 | 2 | 17 | 1229 |
| 10193 | P00915 | CAH1 | Human | 2–261 | 6 | 167 | 5315 |
| 3 | O76074 | PDE5A | Human | 533–863 | 7 | 39 | 1693 |
| 236 | P08473 | NEP | Human | 55–750 | 5 | 38 | 1152 |
| 11140 | P27487 | DPP4 | Human | 38–766 | 12 | 70 | 1215 |
| 11442 | P06737 | PYGL | Human | 1–847 | 2 | 13 | 2960 |
| 10702 | P50579 | AMPM2 | Human | 110–478 | 11 | 14 | 2214 |
| 15 | P00918 | CAH2 | Human | 2–260 | 38 | 239 | 3144 |
| 17047 | P05979 | PGH1 | Sheep | 23–594 | 2 | 13 | 459 |
| Mean | 12 | 52 | 1412 | ||||
| SD | 14 | 56 | 1147 |
Table 2.
Pairwise backbone root-mean-square deviations between superimposed pockets and the average of root-mean-square deviations between crystallographic and atomic property field predicted ligand conformation.
| Target | Seeds | Residues (n) | RMSD (Å)
|
|
|---|---|---|---|---|
| Pockets | APF self-docking | |||
| RXRA | 6 | 45 | 0.68 | 1.13 |
|
| ||||
| DEF | 4 | 24 | 0.60 | 0.84 |
|
| ||||
| DYR | 11 | 22 | 0.61 | 1.44 |
|
| ||||
| THRB | 20 | 30 | 0.31 | 1.53 |
|
| ||||
| PYRD | 4 | 24 | 0.19 | 0.99 |
|
| ||||
| MK14 | 35 | 57 | 2.22 | 2.69 |
|
| ||||
| ADA17 | 5 | 26 | 0.49 | 0.48 |
|
| ||||
| PPARG | 15 | 59 | 2.28 | 3.98 |
|
| ||||
| CDK2 | 76 | 49 | 3.24 | 1.96 |
|
| ||||
| ALDR | 13 | 32 | 0.58 | 2.21 |
|
| ||||
| MMP8 | 16 | 27 | 0.37 | 2.82 |
|
| ||||
| THER | 10 | 19 | 0.18 | 2.42 |
|
| ||||
| ESR1 | 32 | 49 | 2.88 | 1.38 |
|
| ||||
| CP2A6 | 6 | 14 | 0.22 | 1.33 |
|
| ||||
| PDE4D | 12 | 23 | 0.18 | 1.87 |
|
| ||||
| ESR2 | 10 | 47 | 6.25 | 1.79 |
|
| ||||
| MMP12 | 8 | 21 | 0.46 | 2.52 |
|
| ||||
| MMP3 | 18 | 36 | 1.13 | 3.36 |
|
| ||||
| ADK | 3 | 25 | 2.34 | 0.78 |
|
| ||||
| LCK | 7 | 39 | 2.70 | 0.85 |
|
| ||||
| MK10 | 8 | 44 | 0.95 | 1.77 |
|
| ||||
| MMP1 | 4 | 26 | 0.45 | 0.80 |
|
| ||||
| EGFR | 4 | 40 | 4.48 | 0.89 |
|
| ||||
| PPARA | 3 | 33 | 0.82 | 0.76 |
|
| ||||
| PDE4B | 12 | 26 | 0.28 | 2.15 |
|
| ||||
| VGFR2 | 4 | 34 | 2.35 | 0.79 |
|
| ||||
| CMA1 | 2 | 26 | 0.33 | 0.77 |
|
| ||||
| PRGR | 5 | 27 | 2.04 | 0.44 |
|
| ||||
| GCR | 2 | 29 | 0.24 | 0.64 |
|
| ||||
| CAH1 | 6 | 22 | 0.11 | 1.96 |
|
| ||||
| PDE5A | 7 | 44 | 2.71 | 2.18 |
|
| ||||
| NEP | 5 | 23 | 0.33 | 1.22 |
|
| ||||
| DPP4 | 12 | 26 | 0.21 | 0.84 |
|
| ||||
| PYGL | 2 | 23 | 0.03 | 0.48 |
|
| ||||
| AMPM2 | 11 | 26 | 0.21 | 1.42 |
|
| ||||
| CAH2 | 38 | 34 | 0.25 | 1.62 |
|
| ||||
| PGH1 | 2 | 16 | 0.09 | 0.81 |
Pocket RMSD is described by averaging across pairwise backbone RMSDs of residues that are used for pocket superimposition and APF self-docking RMSD is described by averaging across all RMSDs of crystallographic seeds.
APF: Atomic property field; RMSD: Root-mean-square deviation.
Analysis of the benchmark
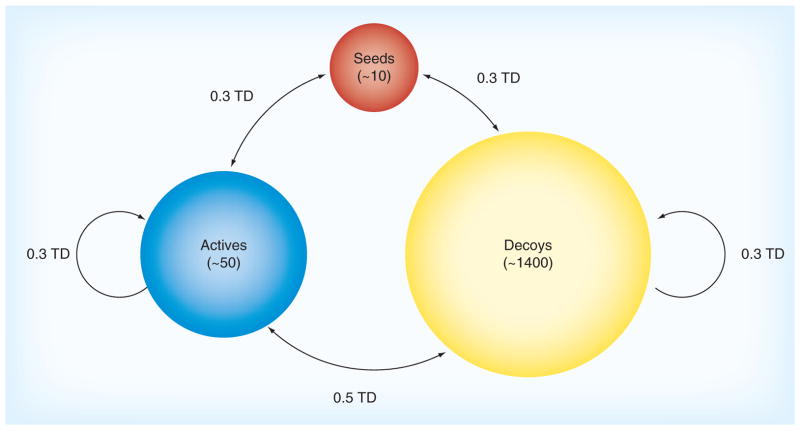
The discriminating power of mApfDock model is evaluated by its capacity to recognize a small number of active molecules, binders, from a much greater number of decoy molecules, non-binders. We expect that the requirement of the benchmark sets composed of at least 10 active ligands and more than 10-times larger number of decoy ligands should lend statistical meaningfulness to the evaluation of the prediction power of the model (Table 1). On the other hand, the relationship of the decoy molecules to active ligands is critical in assessing enrichment factors of mApf-Dock models. The outline for structural relationships between active, decoy and seed ligands are shown in Figure 2. The average number of seeds, actives and decoys is 10, 50 and 1400, respectively. According to Verdonk, Rognan, Jain and Irwin’s study, the artificially good enrichments always result from a biased decoy set when the following two issues are not carefully handled: the worry of overfitting and massively incomplete sampling of chemical space [41–44]. The unbiased decoy sets should resemble physical properties of the actives well enough, while being topologically dissimilar to active sets. If the misleadingly biased decoy sets are used, the discriminating power of the model will be simply a separation of trivial physical properties between active and decoy.
Figure 2. Outline of the structural relationships between seed, active and decoy sets.
The average number of seeds, actives and decoys is approximately 10, 50 and 1400, respectively.
Redundant benchmark molecules with similar structures can statistically lead to the biased assessment. In the Directory of Useful Decoys, the selected decoys are above 0.1 TD from active ligand [42]. It is explicitly designed to present physically similar but chemically dissimilar decoys. Therefore, it might lead to biased assessment and is not suitable for benchmarking the ‘scaffold hopping’. The distribution of chemical distance in ChEMBL has a mean of 0.554 and a SD of 0.148 [22], resulting in a distance of 0.258, a lower bound of the distribution, with 2 SD below the mean. We believe that the criteria of topological dissimilarity above 0.3 TD between active, decoy and seed ligand should result in unbiased benchmarking for the scaffold hopping.
Prediction of ligand-binding geometry by APF self-docking
The composition of templates in structural and geometric/coordinate aspects plays a critical role in the robustness of the mApfDock models. The prediction accuracy of the mApfDock models was initially evaluated by self-docking the seed molecules that were used to construct the mApfDock models onto the APF clouds, calculating the RMSD between the predicted and crystallographic ligand conformation. For most (28 out of 37) of the APF clouds, seed molecules were re-docked with the average RMSD smaller than 2.0 Å, indicating the high power of the APF cloud in predicting bound ligand conformations (Table 2). In addition, the result of self-docking shows that using more seed ligands in the construction of the APF cloud might lead to the larger RMSD values for the prediction of ligand conformations. Several factors might be involved in this trend: the structural similarity, the consistency of molecular properties, the size and flexibility of the seed ligands. The re-docking for a small set of highly similar ligands is trivial because they share similar properties mostly. However, APF clouds for larger sets of more diverse ligands may become dithered as different binding modes get blended, resulting in more ambiguity.
Comparison of molecular recognition between mApfDock & mPockDock
Our previous study showed a good performance of mApfDock in measuring spatial chemical distance between two chemicals as compared with the 2D Tanimoto and Shape Tanimoto measures [17]. In this study, we evaluate the ability of APF target-specific and pocket-specific models generated using multiple crystal-graphic ligands to accommodate experimentally determined active ligands and recognize them in a comparatively large set of random drug-like decoys. This further development of the APF approach aims at scaffold hopping and expansion of the candidate pool in lead generation. The screening performance of mApfDock was tested on the benchmark containing 37 protein targets. Each target-specific benchmark included at least two seeds and 10 active ligands and decoy sets that were at least 10-times larger than the active set. Protein pocket information for each target and the corresponding Protein Data Bank seeds and benchmark are shown in Table 1.
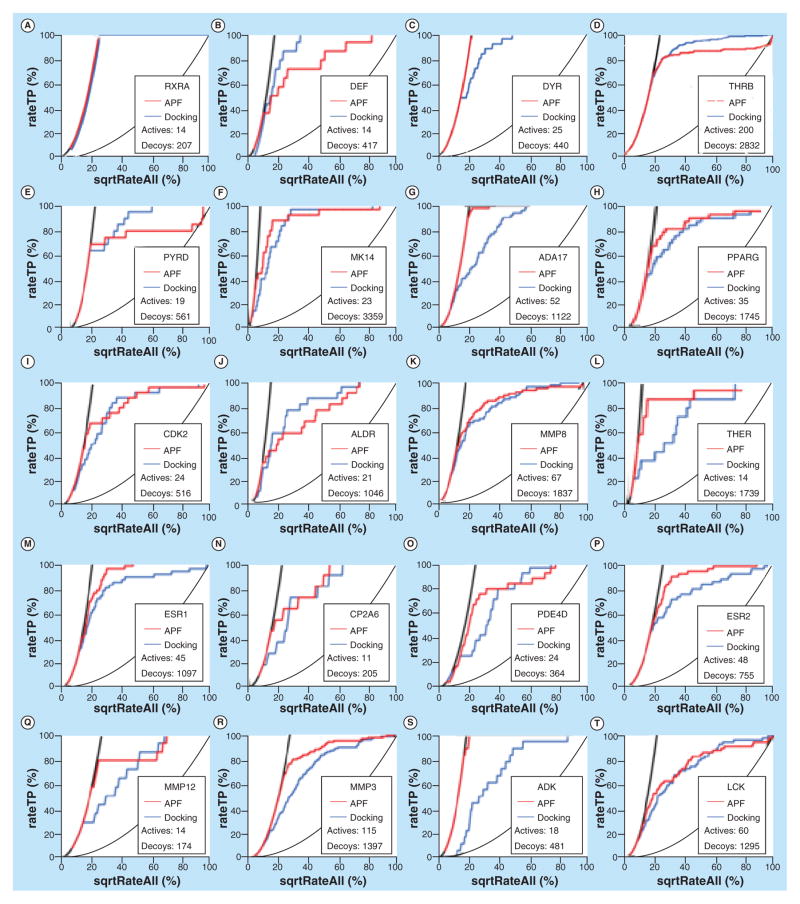
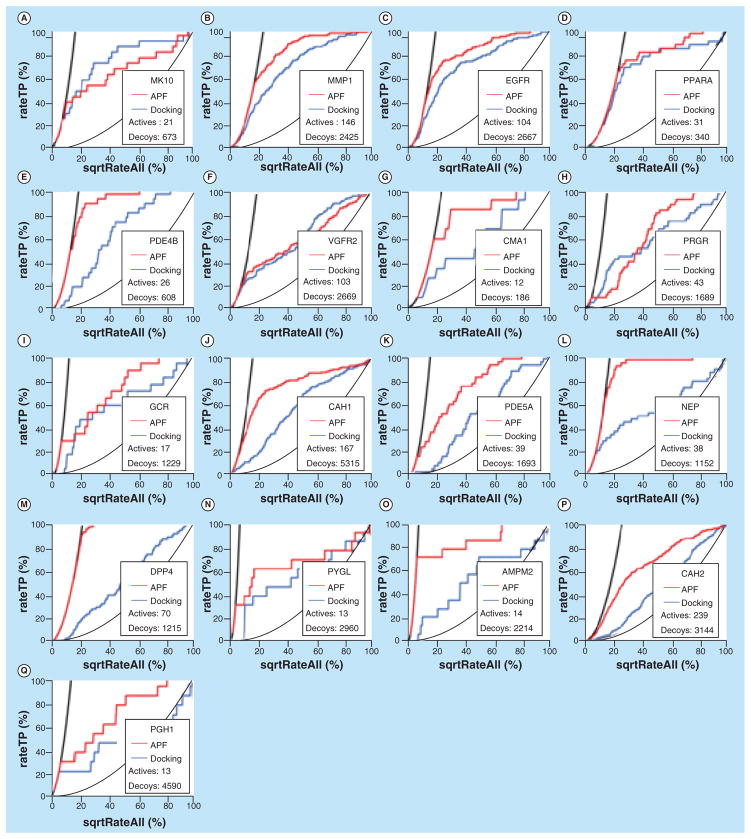
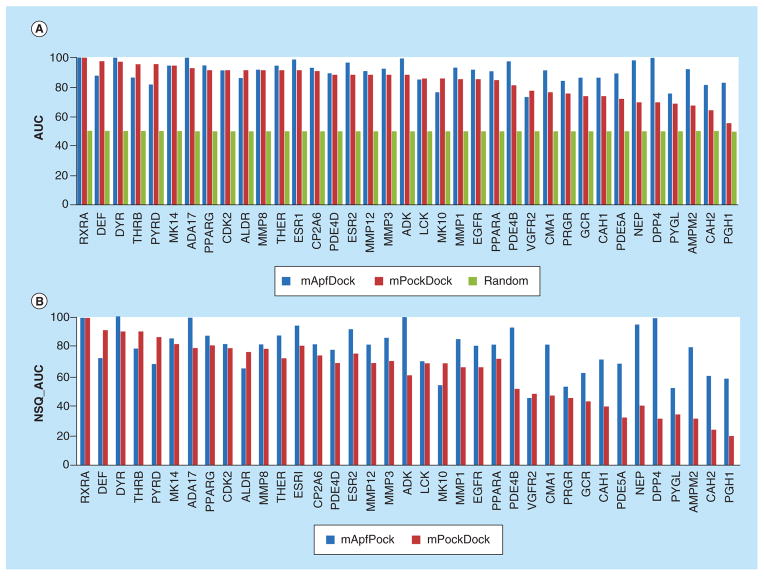
As the crystal structures of protein pockets are available, the performance in molecular recognition using mPockDock is then compared with mApfDock models. The benchmark was docked against all binding pockets of seed ligands and further optimized by induced-fit simulation. The comparison of the performance in molecular recognition between mApfDock and mPockDock, as measured by NSQAUC curve, is shown in Figures 3 & 4 & Table 3. The NSQAUC and corresponding AUC values of mApfDock and mPock-Dock are shown in the Figure 5. The power of mApf-Dock in recognizing distinct structures is much better than mPockDock in most cases (30/37 = 81%). Even better, the molecular recognition using mApfDock gave the improvement of more than the NSQAUC of 10 in 20 out of 37 cases compared with the mPock-Dock. The 19% of cases where the mPockDock gave better performance show that both methods are relevant, in particular when the absence of consistency in ligand-binding determinants results in poorly defined property fields. In general, the mApfDock gave the mean NAQ_AUC of 78.4% compared with 62.9% generated by the mPockDock.
Figure 3. Comparison of the performance in molecular recognition between mApfDock and mPockDock, as measured by the normalized square root of the area under the receiver operating characteristic curves for each target.
The normalized false-positive rate (x-axis) is plotted against the true-positive rate (y-axis). Receiver operating characteristic curves of mApfDock and mPockDock measurements are shown in red and blue, respectively. (A) RXRA, (B) DEF, (C) DYR, (D) THRB, (E) PYRD, (F) MK14, (G) ADA17, (H) PPARG, (I) CDK2, (J) ALDR, (K) MMP8, (L) THER, (M) ESR1, (N) CP2A6, (O) PDE4D, (P) ESR2, (Q) MMP12, (R) MMP3, (S) ADK and (T) LCK.
APF: Atomic property field.
Figure 4. Comparison of the performance in molecular recognition between mApfDock and mPockDock, as measured by the normalized square root of the area under the receiver operating characteristic curves for each target.
The normalized false-positive rate (x-axis) is plotted against the true-positive rate (y-axis). Receiver operating characteristic curves of mApfDock and mPockDock measurements are shown in red and blue, respectively. (A) MK10, (B) MMP1, (C) EGFR, (D) PPARA, (E) PDE4B, (F) VGFR2, (G) CMA1, (H) PRGR, (I) GCR, (J) CAH1, (K) PDE5A, (L) NEP, (M) DPP4, (N) PYGL, (O) AMPM2, (P) CAH2 and (Q) PGH1.
APF: Atomic property field.
Table 3.
The NSQAUC and corresponding receiver operating characteristic values generated by mApfDock and mPockDock measurement.
| Target | mApfDock
|
mPockDock
|
||
|---|---|---|---|---|
| ROC (%) | NSQAUC (%) | ROC (%) | NSQAUC (%) | |
| RXRA | 100.0 | 100.0 | 100.0 | 100.0 |
|
| ||||
| DEF | 88.4 | 72.3 | 98.0 | 90.8 |
|
| ||||
| DYR | 100.0 | 100.0 | 97.1 | 90.5 |
|
| ||||
| THRB | 87.2 | 78.6 | 95.7 | 90.3 |
|
| ||||
| PYRD | 81.2 | 68.0 | 95.5 | 86.4 |
|
| ||||
| MK14 | 94.7 | 85.6 | 94.8 | 81.8 |
|
| ||||
| ADA17 | 99.8 | 99.3 | 93.4 | 79.4 |
|
| ||||
| PPARG | 94.8 | 87.3 | 92.0 | 80.7 |
|
| ||||
| CDK2 | 91.8 | 81.3 | 91.6 | 79.1 |
|
| ||||
| ALDR | 85.8 | 65.0 | 91.4 | 75.9 |
|
| ||||
| MMP8 | 91.3 | 81.2 | 91.3 | 78.3 |
|
| ||||
| THER | 94.8 | 87.3 | 91.1 | 71.6 |
|
| ||||
| ESR1 | 98.4 | 93.9 | 90.9 | 80.6 |
|
| ||||
| CP2A6 | 93.1 | 81.1 | 90.8 | 74.3 |
|
| ||||
| PDE4D | 89.8 | 77.2 | 88.6 | 68.7 |
|
| ||||
| ESR2 | 96.3 | 91.2 | 88.0 | 74.9 |
|
| ||||
| MMP12 | 91.0 | 81.5 | 87.6 | 68.6 |
|
| ||||
| MMP3 | 92.9 | 85.7 | 86.9 | 70.1 |
|
| ||||
| ADK | 99.9 | 99.5 | 86.7 | 60.7 |
|
| ||||
| LCK | 85.8 | 69.9 | 86.5 | 68.3 |
|
| ||||
| MK10 | 76.6 | 53.0 | 85.9 | 68.4 |
|
| ||||
| MMP1 | 93.6 | 85.0 | 85.6 | 66.2 |
|
| ||||
| EGFR | 92.0 | 80.9 | 85.0 | 65.7 |
|
| ||||
| PPARA | 90.7 | 81.1 | 84.3 | 71.2 |
|
| ||||
| PDE4B | 97.3 | 92.5 | 81.3 | 51.2 |
|
| ||||
| VGFR2 | 73.5 | 44.9 | 77.6 | 48.2 |
|
| ||||
| CMA1 | 91.4 | 81.1 | 76.0 | 47.2 |
|
| ||||
| PRGR | 83.4 | 52.2 | 75.2 | 45.4 |
|
| ||||
| GCR | 86.3 | 61.7 | 73.8 | 43.0 |
|
| ||||
| CAH1 | 86.1 | 71.0 | 73.5 | 39.2 |
|
| ||||
| PDE5A | 89.1 | 68.4 | 71.4 | 31.4 |
|
| ||||
| NEP | 98.0 | 94.8 | 69.4 | 40.2 |
|
| ||||
| DPP4 | 99.7 | 98.9 | 69.2 | 31.2 |
|
| ||||
| PYGL | 75.3 | 51.5 | 68.2 | 33.7 |
|
| ||||
| AMPM2 | 92.1 | 80.2 | 67.7 | 31.2 |
|
| ||||
| CAH2 | 81.6 | 60.0 | 64.2 | 23.0 |
|
| ||||
| PGH1 | 82.8 | 58.5 | 55.4 | 19.9 |
|
| ||||
| Mean | 90.4 | 78.4 | 83.8 | 62.9 |
ROC: Receiver operating characteristic.
Figure 5. Comparison of the recognition power between mApfDock and mPockDock over 37 targets.
The performance is measured by (A) AUC and the corresponding (B) NSQAUC.
In the most extreme case (CMA1) the mApfDock model that was constructed using only two distinct seed ligands with a dissimilarity of 0.5 TD gave the NAQ_ AUC of 81.1% in contrast to 47.2% obtained using the mPockDock. The predicted docking conformations of representative active ligands with the highest APF score are shown in Figure 6. The selected targets are the top nine datasets that gave the most significant difference in NSQAUC between the mApfDock and the mPock-Dock. Remarkably, the best scoring ligand of CMA1 is also the most active inhibitor (pKi of 8.9) found in ChEMBL database (Figure 6I). In addition, the rest of the representative ligands, except PGH1, in Figure 6 not only generate the highest APF score but also have bioactivity below 10 nM. The most active ligand of PGH1 from ChEMBL has the pKi of 8.4, which generates the fourth highest APF score and the representative ligand (Figure 6F) has the pKi of 7.0, which is still bioactively significant. Overall, the atomic properties of docking conformations are well superimposed onto the corresponding properties of seed ligands.
Figure 6. The predicted docking conformation of active ligands superimposed on the atomic property field cloud for nine representative targets.
(A) DPP4, (B) NEP, (C) AMPM2, (D) PDE4B, (E) ADK, (F) PGH1, (G) CAH2, (H) PDE5A, (I) CMA1. These targets give the top nine well-performed mApfDock models compared with mPockDock measurements. The atomic property field cloud is shown in blue wire by hydrogen bond donor, red wire by hydrogen bond acceptor, green wire by aromaticity, yellow wire by lipophilicity, aquamarine contour by positive charge and pink contour by negative charge. Each representative active ligand generated the highest atomic property field score in each target set.
An intrinsic complexity in the structure-based molecular recognition is related to the flexibility of the protein-binding pockets. Co-crystal structures of identical ligand-binding domains bound to different chemotypes reveal more or less pronounced conformational changes to accommodate binding of ligands. The induced-fit optimization gave the average improvement of NSQAUC of 13.7% in the mPockDock. This trend toward the improvement in the mPockDock conformed to our published paper [21]. However, because different chemotypes can induce alternative conformational changes to the ligand-binding pocket, such as rotation of side chains and small loop rearrangements, each of them represents only a fraction of the total molecular chemical recognition properties and has somewhat limited potential for virtual screening of novel chemotypes. By contrast, the mApfDock represents the virtual binding site by multiproperty potential and does not rely on the geometry of the binding site. The conformational changes of the ligand binding site, therefore, have less impact on the performance of mApfDock. However, the absence of consistency in the ligand-binding determinants results in poorly defined property fields and is unfavorable to the mApf-Dock. As the results of this study, there is no single perfect approach for molecular recognition. As both approaches are complementary to each other, a hybrid model could be designed to improve the screening, activity prediction and scaffold hopping.
Conclusion
In this study, we demonstrated and evaluated the utility of two physical models of ligand binding based on specific experimental binding pocket geometry or bound ligand geometry for the recognition of new bioactives with minimal historical bias. The multiple-crystallographic ligand field approach, mApfDock, was compared with the docking approach with multiple crystallographic pocket conformations for a set of 37 proteins. We showed that the cumulative APF fields derived from an ensemble of spatially overlaid co-crystallized ligands reveal the conserved binding determinants and result in stronger pharmacophoric signals. A carefully assembled benchmark ensures the unbiased assessment for the recognition power and provides an evaluation for scaffold-hopping capability. On average, the mApf-Dock and mPockDock approach generates the NSQAUC of 78.4 and 62.9%, respectively (note that NSQAUC ranges from −100 to 100%, in contrast to the regular AUC that ranges from 0 to 100%). The cases with poor mApfDock performances were found to result from the unusual or rare binding modes captured by the crystallography. The cases where mPockDock gave worse performance might indicate insufficient conformational sampling, unexpected induced fit or deficiencies in the scoring function. However, the two approaches converge when the number and diversity of input crystallographic complexes is sufficiently high.
Future perspective
Predicting all essential biological, physiological and disease-related functions and effects of a chemical compound directly from its chemical structure is one of the greatest challenges. Here we tested 3D models for 37 activities and the approach can now be extended into approximately 700 human pockets. However, in the future, when all essential proteins, complexes and biological assemblies are defined at the level of 3D structures, and co-crystal structures with some ligands, it should be possible to make comprehensive and accurate predictions by docking flexible, and potentially chemically reactive, 3D models of chemicals to the entire human pocketome of tens of thousands of essential pockets. Alternatively, a growing body of experimental data on chemical activities can be used to generate hypotheses just based on chemically similar compounds. The chemical features that discriminate pharmacological actions are not always distinguishable in agreement with the structural basis of the ligand-binding pocket. Furthermore, each structural model will gain in accuracy and ability to discriminate between specific types of functional modulation (e.g., agonist, antagonist, allosteric etc.). These type of activity prediction panels may be used for identifying or repurposing candidates with desired pharmacological actions and minimal drug-associated adverse reactions, designing guidelines and alerts for the chemical and nutritional industries to avoid unintended consequences and better understanding molecular biochemistry and chemical biology of biological metabolites and their receptors.
Executive summary.
Cumulative 3D pharmacophoric fields derived from an ensemble of spatially overlaid co-crystallized ligands reveal the conserved binding determinants and result in strong models to recognize activity of any chemical compound with much less historical bias than the chemical descriptor-based machine learning methods.
The multi-pocket docking approach is complementary to the atomic property fields approach, in particular when the co-crystallized ligands do not represent sufficient chemical and binding location diversity.
The two approaches converge when the number and diversity of complexes used in a model is sufficiently high; the ‘field’ approach is more permissive to compounds requiring large induced-fit pocket changes previously unseen in crystallography.
Acknowledgments
The authors thank I Kufareva, M Neves and M Rueda for valuable discussion, C Edwards and E Raush for support and K Wright for help with manuscript preparation.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Financial & competing interests disclosure
This work was partially supported by NIH grants R01 GM071872, R01 HL048908, and RC2 LM010994. R Abagyan and M Totrov have financial interest in Molsoft, LLC, the source of the software used in the project. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.McRobb FM, Sahagun V, Kufareva I, Abagyan R. In silico analysis of the conservation of human toxicity and endocrine disruption targets in aquatic species. Environ Sci Technol. 2014;48(3):1964–1972. doi: 10.1021/es404568a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma DL, Chan DSH, Leung CH. Molecular docking for virtual screening of natural product databases. Chem Sci. 2011;2(9):1656–1665. [Google Scholar]
- 3.Leung CH, Chan DS, Yang H, et al. A natural product-like inhibitor of NEDD8-activating enzyme. Chem Commun. 2011;47(9):2511–2513. doi: 10.1039/c0cc04927a. [DOI] [PubMed] [Google Scholar]
- 4.Chan DS, Lee HM, Yang F, et al. Structure-based discovery of natural-product-like TNF-alpha inhibitors. Angew Chem Int Ed Engl. 2010;49(16):2860–2864. doi: 10.1002/anie.200907360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HM, Chan DSH, Yang F, et al. Identification of natural product Fonsecin B as a stabilizing ligand of c-myc G-quadruplex DNA by high-throughput virtual screening. Chem Commun. 2010;46(26):4680–4682. doi: 10.1039/b926359d. [DOI] [PubMed] [Google Scholar]
- 6.Pollastri MP, Campbell RK. Target repurposing for neglected diseases. Future Med Chem. 2011;3(10):1307–1315. doi: 10.4155/fmc.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma DL, Chan DS, Leung CH. Drug repositioning by structure-based virtual screening. Chem Soc Rev. 2013;42(5):2130–2141. doi: 10.1039/c2cs35357a. [DOI] [PubMed] [Google Scholar]
- 8.Kharkar PS, Warrier S, Gaud RS. Reverse docking: a powerful tool for drug repositioning and drug rescue. Future Med Chem. 2014;6(3):333–342. doi: 10.4155/fmc.13.207. [DOI] [PubMed] [Google Scholar]
- 9.Tropsha A. Best practices for QSAR model development, validation, and exploitation. Mol Informatics. 2010;29(6–7):476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 10.Totrov M. Atomic property fields: generalized 3D pharmacophoric potential for automated ligand superposition, pharmacophore elucidation and 3D QSAR. Chem Biol Drug Des. 2008;71(1):15–27. doi: 10.1111/j.1747-0285.2007.00605.x. [DOI] [PubMed] [Google Scholar]
- 11.Totrov M, Abagyan R. Flexible protein–ligand docking by global energy optimization in internal coordinates. Proteins. 1997;(Suppl 1):215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 12.Park SJ, Kufareva I, Abagyan R. Improved docking, screening and selectivity prediction for small molecule nuclear receptor modulators using conformational ensembles. J Comput Aided Mol Des. 2010;24(5):459–471. doi: 10.1007/s10822-010-9362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13•.Bottegoni G, Kufareva I, Totrov M, Abagyan R. Four-dimensional docking. a fast and accurate account of discrete receptor flexibility in ligand docking. J Med Chem. 2009;52(2):397–406. doi: 10.1021/jm8009958. Develops a new type of molecular docking method to improve the activity prediction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Kufareva I, Ilatovskiy AV, Abagyan R. Pocketome: an encyclopedia of small-molecule binding sites in 4D. Nucleic Acids Res. 2011;40:D535–D540. doi: 10.1093/nar/gkr825. Introduces a collection of druggable binding site ensembles enabling multiconformational pocket-based docking and cumulative 3D field-based docking. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodford PJ. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem. 1985;28(7):849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 16.Giganti D, Guillemain H, Spadoni JL, Nilges M, Zagury JF, Montes M. Comparative evaluation of 3D virtual ligand screening methods: impact of the molecular alignment on enrichment. J Chem Inf Model. 2010;50(6):992–1004. doi: 10.1021/ci900507g. [DOI] [PubMed] [Google Scholar]
- 17•.Grigoryan AV, Kufareva I, Totrov M, Abagyan RA. Spatial chemical distance based on atomic property fields. J Comput Aided Mol Des. 2010;24(3):173–182. doi: 10.1007/s10822-009-9316-x. Benchmarks the activity prediction of fingerprint-based and field-based methods on a set of 320,000 molecular pairs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Kufareva I, Chen YC, Ilatovskiy AV, Abagyan R. Compound activity prediction using models of binding pockets or ligand properties in 3D. Curr Top Med Chem. 2012;12(17):1869–1882. doi: 10.2174/156802612804547335. Compares the performance of pocket-based and field-based models in molecular recognition for G-protein-coupled receptors and nuclear receptors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J Med Chem. 2006;49(23):6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swann SL, Brown SP, Muchmore SW, et al. A unified, probabilistic framework for structure- and ligand-based virtual screening. J Med Chem. 2011;54(5):1223–1232. doi: 10.1021/jm1013677. [DOI] [PubMed] [Google Scholar]
- 21•.Neves MA, Totrov M, Abagyan R. Docking and scoring with ICM: the benchmarking results and strategies for improvement. J Comput Aided Mol Des. 2012;26(6):675–686. doi: 10.1007/s10822-012-9547-0. Evaluates the pose and activity prediction of the Internal Coordinates Mechanics docking method using Directory of Useful Decoys. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, Bajorath J. Systematic assessment of scaffold distances in ChEMBL: prioritization of compound data sets for scaffold hopping analysis in virtual screening. J Comput Aided Mol Des. 2012;26(10):1101–1109. doi: 10.1007/s10822-012-9603-9. [DOI] [PubMed] [Google Scholar]
- 23.Rueda M, Bottegoni G, Abagyan R. Consistent improvement of cross-docking results using binding site ensembles generated with elastic network normal modes. J Chem Inf Model. 2009;49(3):716–725. doi: 10.1021/ci8003732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaulton A, Bellis LJ, Bento AP, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40(Database issue):D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadatos G, Overington JP. The ChEMBL database: a taster for medicinal chemists. Future Med Chem. 2014;6(4):361–364. doi: 10.4155/fmc.14.8. [DOI] [PubMed] [Google Scholar]
- 26.Cheng Y, Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22(23):3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 27.Abagyan R, Totrov M, Kuznetsov D. ICM – a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem. 1994;15(5):488–506. [Google Scholar]
- 28.Halgren TA. Merck molecular force field. I Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17(5–6):490–519. [Google Scholar]
- 29.Halgren TA. MMFF VI. MMFF94s option for energy minimization studies. J Comput Chem. 1999;20(7):720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Rose PW, Beran B, Bi C, et al. The RCSB Protein Data Bank. redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abagyan R, Kufareva I. The flexible pocketome engine for structural chemogenomics. Methods Mol Biol. 2009;575:249–279. doi: 10.1007/978-1-60761-274-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Z, Chen L, Maddula H, et al. Ligand Depot: a data warehouse for ligands bound to macromolecules. Bioinformatics. 2004;20(13):2153–2155. doi: 10.1093/bioinformatics/bth214. [DOI] [PubMed] [Google Scholar]
- 33.Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J Mol Biol. 1994;235(3):983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 34.Totrov M, Abagyan R. Derivation of sensitive discrimination potential for virtual ligand screening. Presented at: RECOMB99 3rd International Conference on Computational Molecular Biology; Lyon, France. 11–14 April (1999).pp. 312–320. [Google Scholar]
- 35.Katritch V, Kufareva I, Abagyan R. Structure based prediction of subtype-selectivity for adenosine receptor antagonists. Neuropharmacology. 2011;60(1):108–115. doi: 10.1016/j.neuropharm.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schapira M, Totrov M, Abagyan R. Prediction of the binding energy for small molecules, peptides and proteins. J Mol Recognit. 1999;12(3):177–190. doi: 10.1002/(SICI)1099-1352(199905/06)12:3<177::AID-JMR451>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 37.Bursulaya BD, Totrov M, Abagyan R, Brooks CL. 3rd: comparative study of several algorithms for flexible ligand docking. J Comput Aided Mol Des. 2003;17(11):755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 38.Truchon JF, Bayly CI. Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. J Chem Inf Model. 2007;47(2):488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 39.Katritch V, Rueda M, Abagyan R. Ligand-guided receptor optimization. Methods Mol Biol. 2012;857:189–205. doi: 10.1007/978-1-61779-588-6_8. [DOI] [PubMed] [Google Scholar]
- 40.Katritch V, Rueda M, Lam PC, Yeager M, Abagyan R. GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex. Proteins. 2010;78(1):197–211. doi: 10.1002/prot.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bissantz C, Folkers G, Rognan D. Protein-based virtual screening of chemical databases. 1 Evaluation of different docking/scoring combinations. J Med Chem. 2000;43(25):4759–4767. doi: 10.1021/jm001044l. [DOI] [PubMed] [Google Scholar]
- 42.Irwin JJ. Community benchmarks for virtual screening. J Comput Aided Mol Des. 2008;22(3–4):193–199. doi: 10.1007/s10822-008-9189-4. [DOI] [PubMed] [Google Scholar]
- 43.Pham TA, Jain AN. Parameter estimation for scoring protein–ligand interactions using negative training data. J Med Chem. 2006;49(20):5856–5868. doi: 10.1021/jm050040j. [DOI] [PubMed] [Google Scholar]
- 44.Verdonk ML, Berdini V, Hartshorn MJ, et al. Virtual screening using protein–ligand docking: avoiding artificial enrichment. J Chem Inf Comput Sci. 2004;44(3):793–806. doi: 10.1021/ci034289q. [DOI] [PubMed] [Google Scholar]