Abstract
TLR signaling is essential to innate immunity against microbial invaders and must be tightly controlled. We have previously shown that TLR9 undergoes proteolytic cleavage processing by lysosomal proteases to generate two distinct fragments. The C-terminal cleavage product plays a critical role in activating TLR9 signaling; however, the precise role of the N-terminal fragment, which remains in lysosomes, in the TLR9 response is still unclear. In this article, we report that the N-terminal cleavage product negatively regulates TLR9 signaling. Notably, the N-terminal fragment promotes the aspartic protease-mediated degradation of the C-terminal fragment in endolysosomes. Furthermore, the N-terminal TLR9 fragment physically interacts with the C-terminal product, thereby inhibiting the formation of homodimers of the C-terminal fragment; this suggests that the monomeric C-terminal product is more susceptible to attack by aspartic proteases. Together, these results suggest that the N-terminal TLR9 proteolytic cleavage product is a negative self-regulator that prevents excessive TLR9 signaling activity.
Toll-like receptors are critical sensors for pathogen-associated molecular patterns, and they play key roles in provoking innate immune responses and enhancing adaptive immunity against microbial infection (1, 2). In resting myeloid cells, the predominant intracellular localization of TLR3, TLR7, TLR8, and TLR9 in the endoplasmic reticulum (ER) changes to an endolysosomal compartment, wherein they mediate the recognition of viral and bacterial nucleic acids (3–6). TLR ligation triggers recruitment of signaling adaptor molecules that leads to NF-κB activation and induces the expression of genes encoding immune and proinflammatory molecules (7, 8). Although TLR signaling is essential for the host’s immune response to pathogens, excessive activation of TLR signaling contributes to autoimmune and chronic inflammatory diseases (9). TLR signaling must thus be tightly controlled to maintain a proper immune balance.
Recent studies have reported that TLR9 undergoes proteolytic processing by endolysosomal proteases to generate the C-terminal cleavage fragment, which functions as an active receptor that is required for the binding of CpG-DNA and initiation of signal transduction (10–12), and the N-terminal half of the TLR9 ectodomain is required for DNA sensing (13). However, the precise functional role of the N-terminal fragment of TLR9, which remains with the C-terminal product in the endolysosome after proteolytic cleavage, is still not clearly understood. In this article, we report that the N-terminal cleavage product of TLR9 accelerates the dissociation of C-terminal fragment dimerization through physical interaction and promotes aspartic protease-mediated degradation of the C-terminal fragment, thus blocking TLR9 signal transduction. Our results collectively show an autoregulatory negative feedback mechanism of TLR9 activation by an N-terminal cleavage product in C-terminal–mediated signal transduction, suggesting that TLR9 is a self-regulatory protein. This is necessary to induce TLR tolerance capable of preventing fatal inflammatory conditions, which are associated with autoimmune diseases.
Materials and Methods
DNA constructs
All mouse TLR9-related constructs were fused at the C terminus to Myc or GFP. Wild-type TLR9-Myc, recombinant C-terminal TLR9 fragment (Cterm), and Unc93b-hemagglutinin (HA) have been described previously (10, 14). The recombinant N-terminal TLR9 fragment (Nterm-TM-GFP) was generated by overlap extension PCR with the primers 5′-GCTAGATCTGCCACCATGGTTCTCCGTCGAAGGACTCTG-3′ (XbaI-TLR9; forward), 5′-ACAGCCAAGAGTGAAAGGCCAAAGCACCTGTCCATGAAGTTCTTAGAAGCAGG-3′ (TM-470; reverse), 5′-CCTGCTTCTAAGAACTTCATGGACAGGTGCTTTGGCCTTTCACTCTTGGCTGT-3′ (470-TM; forward), and 5′-ATGCGTCGACCCGAGATGGTGCAGTATAGGCACCAC-3′ (SalI-TM-TLR9; reverse), and was finally fused at the C terminus with GFP (pEGFP-N1). TM-GFP encoding the TLR9 TM was fused at the N terminus with the H2-Kb signal sequence (MVPCTLLLLLAAALAPTQTRA). Nterm-Δ441-470-TM-GFP encoding the N-terminal TLR9 fragment and TM, but not the cleavage site, was generated by overlap extension PCR with the primers 5′-GTCAGAAGCCACCCCTGAAGAGTGCTTTGGCCTTTCACTCTTGGCTG-3′ (440-TM; forward) and 5′-CAGCCAAGAGTGAAAGGCCAAAGCACTCTTCAGGGGTGGCTTCTGAC-3′ (TM-440; reverse). Two different TLR9 ectodomain constructs tagged at the C terminus with Myc (Nterm-440-Myc [1–440] and Nterm-470-Myc [1–470]) were generated by PCR with the primers 5′-ATTAGATCTGCCACCATGGTTCTCCGTCGAAGGACTC-3′ (forward), 5′-CGTAGAATTCTTACAAGTCCTCTTCAGAAATGAGCTTTTGCTCCTCTTCAGGGGTGGCTTCTGACAG-3′ (Nterm-440-Myc [1–440]; reverse), and 5′-CGTAGAATTCTTACAAGTCCTCTTCAGAAATGAGCTTTTGCTCCCTGTCCATGAAGTTCTTAGAAGCAGG-3′ (Nterm-470-Myc [1–470]; reverse). All constructs were cloned into the retroviral pMSCV (puro) or pLHCX (hygro) vector (Clontech, Mountain View, CA) and were verified by sequencing.
Reagents
1826-CpG DNA and 1826-Biotin-CpG DNA (5′-Bio-TsCsCsAsTsg-sAsCsgsTsTsCsCsTsgsAsCsgsTsT-3′) were purchased from TIB Molbiol (Berlin, Germany). PNGase F and endoglycosidase H (Endo H) were purchased from New England Biolabs (Ipswich, MA). The monoclonal Myc Ab (catalog no. 2276) and GFP Ab (catalog no. ab290) were obtained from Cell Signaling Technology (Danvers, MA) and Abcam (Cambridge, U.K.), respectively. Streptavidin agarose beads and LPS (Escherichia coli 026: B6) were purchased from Pierce (Rockford, IL) and Sigma-Aldrich (St. Louis, MO). Proteasome inhibitor, MG132, and lysosomal proteases inhibitor, chloroquine, were purchased from Sigma-Aldrich. Aspartic protease inhibitor, pepstatin A (catalog no. 516481), was purchased from Calbiochem (Darmstadt, Germany).
Mice and cell lines
All animals were maintained under specific pathogen-free conditions according to guidelines set by the committee for animal care at Yonsei University. Immortalized wild-type and TLR9-deficient bone marrow–derived macrophages (BMDMs) were obtained from BEI Resources. Murine RAW 264.7 macrophages (ATCC TIB-71), human embryonic kidney (HEK) 293T cells (ATCC CRL-11268), and immortalized bone marrow macrophage cell lines were cultured in DME supplemented with 10% heat-inactivated FBS and penicillin/streptomycin. Cells were grown at 37°C in humidified air with 5% CO2.
Retroviral transduction
HEK 293T cells were transfected with plasmids encoding VSV-G and Gag-Pol, as well as pMSCV-TLR9-Myc, pLHCX-Cterm-Myc, pMSCV-Cterm-GFP, pMSCV-Nterm-TM-GFP, pMSCV-Nterm-Δ441-470-TM-GFP, or pMSCV-TM-GFP. Forty-eight hours after transfection, media containing viral particles were collected, filtered through a 0.45-μm membrane, and incubated with RAW macrophages, immortalized macrophages, or bone marrow–derived dendritic cells (BMDCs) for 24 h. Cells were selected with puromycin or hygromycin for 48 h or 4 d.
Preparation of BMDMs
BMDCs were prepared as described previously (15). In brief, BMDM cells were generated from wild-type C57BL/6 mice (Orient Bio, Gyeonggi-do, South Korea), in medium containing 25 ng/ml M-CSF (Biolegend, San Diego, CA). Femurs and tibiae were collected from 4-wk-old mice. After removing bone-adjacent muscles, we extracted marrow cells by flushing with a 25-gauge needle. Bone marrow cells were then resuspended in DMEM (10% FBS and 1% antibiotics) with M-CSF (5 ng/ml). Fresh medium was replenished on days 2 and 4. BMDMs were generated after 6–8 d of culture.
Pulse-chase analysis, immunoprecipitation, and Endo F/H assay
Cells were “starved” for 1 h in medium without methionine and cysteine, then were labeled with [35S]methionine/cysteine and chased for the indicated times. Cells were lysed with 1% Nonidet P-40 (NP-40) in PBS supplemented with protease inhibitors for 1 h at 4°C. After pre-clearing of lysates with protein G-Sepharose (Sigma-Aldrich), primary Abs and then protein G-Sepharose were added to the supernatants and incubated at 4°C. The protein G-Sepharose beads were washed five times with 0.1% NP-40/PBS. Proteins were eluted from the beads by boiling in 1% SDS. Digestion with PNGase F or Endo H (New England Biolabs) was performed at 37°C for 3 h. For coimmunoprecipitation experiments, cells were lysed in 1% digitonin (Calbiochem) in buffer containing 25 mM HEPES, 100 mM NaCl, 10 mM CaCl2, and 5 mM MgCl2 (pH 7.6) supplemented with protease inhibitors. Buffers containing 0.1% digitonin were used for all subsequent steps. Bound proteins were eluted by boiling in SDS sample buffer or 1% SDS. Proteins were separated by SDS-PAGE, transferred onto a nitrocellulose membrane, blocked with 5% skim milk in PBS with 0.1% Tween 20 (Sigma-Aldrich) for 2 h, and probed with the appropriate Abs for 4 h at room temperature. Membranes were washed three times with PBS with 0.1% Tween 20 and incubated with HRP-conjugated streptavidin for 1 h. The immunoblots were visualized with ECL detection reagent (Pierce).
ELISA
Cells were treated with 1 μM CpG-DNA or 80 ng/ml LPS for 12 h. The media were collected and analyzed by ELISA using hamster anti-mouse/rat IL-6 (BD Biosciences, San Jose, CA) as a capture Ab and biotin-labeled rabbit anti-mouse as a secondary Ab (BD Biosciences).
Intracellular staining for IL-6
BMDCs were cultured for 5 d with GM-CSF and were incubated with normal DMEM. Cells were then stimulated with agonists for 6 h in the presence of 10 μg/ml brefeldin A. The cells were fixed with 4% formaldehyde for 10 min at room temperature and permeabilized with 0.5% saponin in FACS buffer (PBS with 2% BSA and 0.05% sodium azide) for 10 min. Cells were stained with PE-conjugated anti–IL-6 (Biolegend, San Diego, CA) for 30 min. Fluorescence intensity was measured on a flow cytometer (BD Biosciences).
Immunofluorescence assay
For immunofluorescent staining, cells were fixed in 3.7% formaldehyde and permeabilized with 0.1% Triton X-100 before incubation with TAMRA-CpG-DNA (Life Technologies, Carlsbad, CA) or LysoTracker (Life Technologies). Cells were imaged using a fluorescence microscope.
Luciferase assay
HEK293T cells in 12-well plate were transfected with 150 ng NF-κB firefly luciferase, 10 ng Renilla luciferase, 100 ng Unc93B1, and 150 ng full-length TLR9 or C-term of TLR9 in combination with 100 ng TM-GFP, Nterm-TM-GFP, Nterm 440-Myc, or Nterm 470-Myc. After 24 h, cells were stimulated with 1 μM CpG-DNA for 12 h; luciferase assays were performed using Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and Victor X5 (Perkin Elmer, Waltham, MA). Firefly luciferase activity was normalized to Renilla luciferase activity.
Ethics statement
All animal experiments were performed in accordance with the Korean Food and Drug Administration guidelines. Protocols were reviewed and approved by the Institutional Animal Care and Use Committee of Yonsei Laboratory Animal Research Center. All mice were maintained in the specific pathogen-free facility of the Yonsei Laboratory Animal Research Center.
Results
Generation of lysosomal-targeted N-terminal TLR9 recombinant fragments
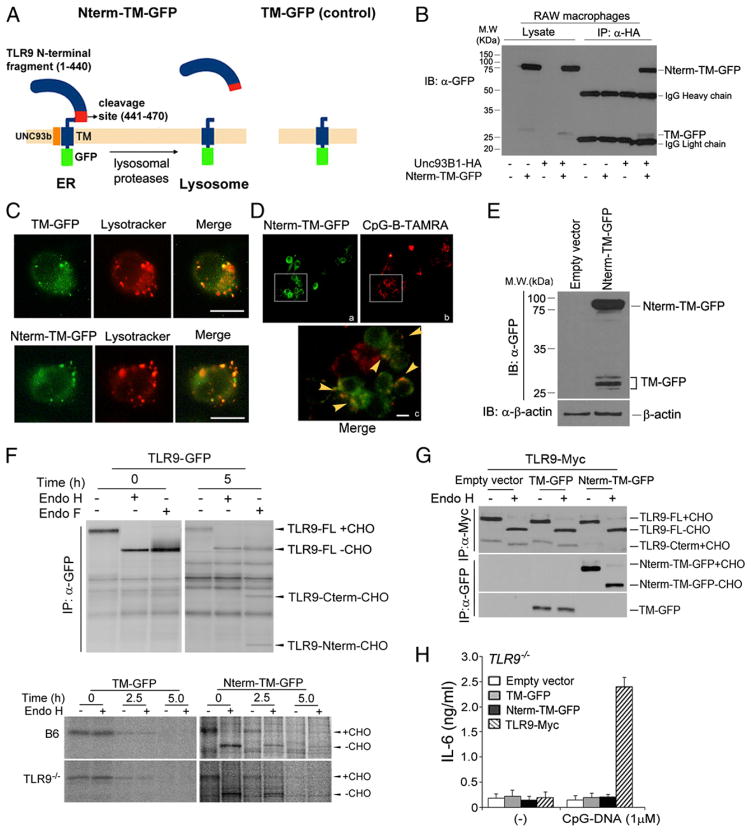
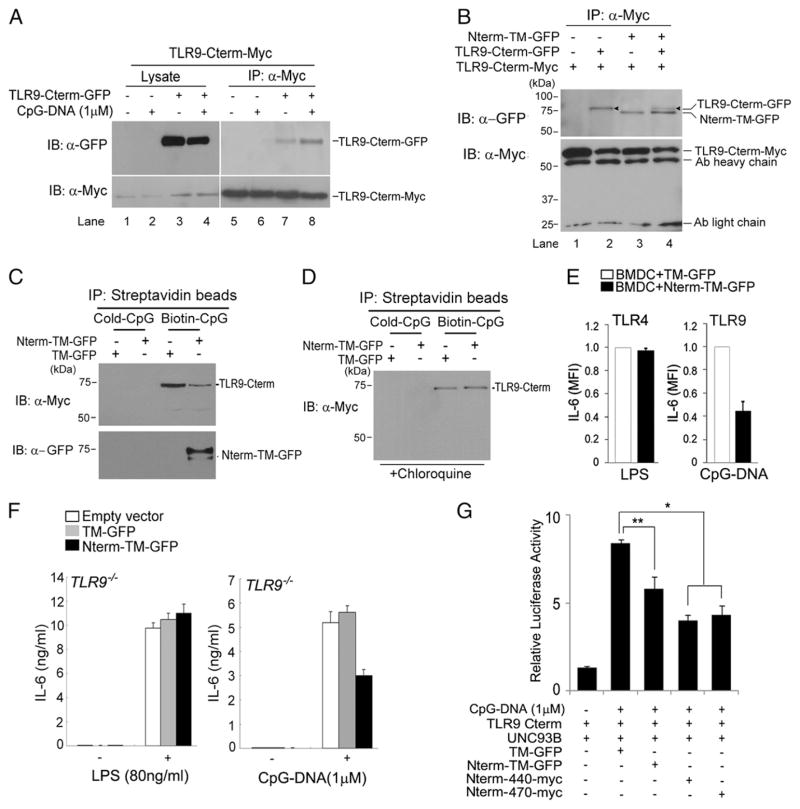
In the endolysosomal compartment, TLR9 is proteolytically cleaved into a C-terminal fragment, which is capable of immune signaling (10, 11), and an N-terminal cleavage fragment, the functional role of which is still unclear. Onji et al. (13) showed that the N-terminal half of the TLR9 ectodomain is required for DNA sensing; however, the effect of the TLR9 ectodomain construct lacking the transmembrane region might be different from the original properties of the N-terminal fragment of TLR9, which remains with the C-terminal product in the endolysosome after proteolytic cleavage. To address the precise role of the N-terminal cleavage product in TLR9 signaling, we first needed to express only the TLR9 N-terminal region in the endolysosomal compartment. We generated the TLR9 chimeras TLR9 Nterm-TM-GFP and TM-GFP (Fig. 1A). TLR9 Nterm-TM-GFP contains the TLR9 N-terminal fragment (residues 1–440), the cleavage site comprising the region corresponding to aa 441–470, and the TM that is critical for binding Unc93B1, which is responsible for facilitating transport to endolysosomes from the ER (14, 16). The use of the GFP tag at the C terminus allowed us to determine whether the chimera was cleaved at the cleavage site in the endolysosome, as the chimera proteins can be detected using an anti-GFP Ab. TM-GFP encodes the TLR9 transmembrane region and the GFP tag at the C terminus, and is used as a negative control. We next established RAW macrophage derivatives that stably expressed either Nterm-TM-GFP or TM-GFP. We examined whether Nterm-TM-GFP is capable of interacting with Unc93B1. We found that Nterm-TM-GFP interacted with Unc93B1 (Fig. 1B). To further evaluate the ability of the chimeric proteins to relocate to the lysosome, we used LysoTracker and examined their colocalization after exposing cells to CpG-DNA. Although the relocation ability of these chimeras was not as high as wild-type TLR9, ~40% of chimeric proteins were able to translocate to the lysosome where they colocalized with CpG-DNA (Fig. 1C, 1D). Furthermore, based on detection of a band corresponding to TM-GFP, we concluded that Nterm-TM-GFP is proteolytically cleaved in lysosomes. These results suggest that the N-terminal cleavage fragment is indeed overexpressed in lysosomes (Fig. 1E).
FIGURE 1.
Generation of lysosomal-targeted TLR9 N-terminal cleavage fragments. (A) Schematic showing generation of lysosomal-targeted TLR9 N-terminal chimeric constructs. Nterm-TM-GFP contains the N-terminal TLR9 proteolytic cleavage product, the cleavage sites (residues 441–470), TM, and GFP at the C terminus; TM-GFP encodes only the TLR9 TM and GFP. (B) RAW cells expressing TLR9 Nterm-TM-GFP in the presence or absence of Unc93B1-HA were lysed with 1% digitonin lysis buffer. Unc93B1 proteins were immunoprecipitated with the anti-HA Ab. Immunoprecipitated samples were resolved by SDS-PAGE and analyzed using anti-GFP Western blot analyses. (C) RAW cells expressing TM-GFP or TLR9 Nterm-TM-GFP were stimulated with 1 μM CpG-DNA for 1 h and washed. Cells were treated with LysoTracker and imaged using immunofluorescence assay. Scale bars, 5 μm. (D) RAW macrophages stably expressing TLR9 Nterm-TM-GFP were stimulated with 1 μM TAMRA-labeled CpG-DNA for 30 min, washed, and imaged after an additional 3-h incubation. Arrowheads indicate colocalization between Nterm-TM-GFP with CpG-B-TAMRA. Scale bars, 5 μm. (E) Lysates from RAW macrophages stably expressing Nterm-TM-GFP were analyzed by Western blot analyses using anti-GFP or anti–β-actin Ab. (F, upper panels) Wild-type TLR9-GFP–expressing macrophages were labeled with [35S]methionine/cysteine and chased for 5 h. Cell lysates were immunoprecipitated with the anti-Myc Ab, and immunoprecipitates were treated with Endo H or Endo F. (F, bottom panels) B6 or TLR9−/− immortalized macrophages stably expressing TM-GFP or Nterm-TM-GFP were labeled with [35S]methionine/cysteine for 30 min and chased for the indicated times. Each chimeric protein was recovered using the anti-GFP Ab, followed by treatment with Endo H. (G) Nterm-TM-GFP does not affect trafficking of wild-type TLR9. RAW cells expressing empty vector, TM-GFP, or TLR9 Nterm-TM-GFP in the presence of full-length TLR9-Myc were lysed with 1% NP-40 lysis buffer. Lysates were immunoprecipitated with the anti-Myc or anti-GFP Ab. Immunoprecipitated proteins were resolved by SDS-PAGE and analyzed using anti-Myc or anti-GFP Western blot analyses. (H) The N-terminal TLR9 product does not activate TLR9 signaling. ELISA showing IL-6 production by TLR9−/− immortalized macrophages stably expressing empty vector, TM-GFP, Nterm-TM-GFP, or wild-type TLR9, stimulated for 12 h with CpG-DNA (1 μM). Data are representative of at least three independent experiments (average ± SD).
We next examined the rates of TM-GFP or Nterm-TM-GFP biosynthesis and maturation by performing pulse-chase experiments and analyzing Endo H or Endo F digestion. We metabolically labeled RAW macrophages expressing wild-type TLR9-GFP, TM-GFP, or Nterm-TM-GFP, chased for the indicated times, and recovered the proteins from lysates by immunoprecipitating with the anti-GFP Ab. Immunoprecipitates were digested with Endo H and Endo F. As shown in previous studies in BMDMs or RAW macrophages (10, 11, 17), wild-type TLR9 trafficked through the Golgi compartment, whereas full-length, Endo H–resistant TLR9-GFP was barely detectable (Fig. 1F, upper panels). Although both chimeras were less stable than wild-type TLR9, the biosynthesis and maturation rates of Nterm-TM-GFP and TM-GFP occurred with similar kinetics as wild-type TLR9 (Fig. 1F, bottom panels). Because the TM-GFP chimera does not have the extracellular domain containing multiple glycosylation sites, it was not cleaved by Endo H. We did not observe differences in kinetics or stability between TM-GFP and Nterm-TM-GFP. In addition, the kinetics of both chimeric proteins was not affected by the presence or absence of endogenous TLR9 (Fig. 1F, bottom panels). We examined that trafficking of wild-type TLR9 was not affected by Nterm-TM-GFP or TM-GFP (Fig. 1G). Because Nterm-TM-GFP and TM-GFP do not have the C-terminal active region that is required for subsequent TLR9 signal transduction (10–12), TLR9−/− immortalized macrophages that expressed the chimeric constructs were unable to activate TLR9 signaling after treatment with CpG-DNA (Fig. 1H).
The N-terminal proteolytic cleavage product of TLR9 negatively regulates its signaling
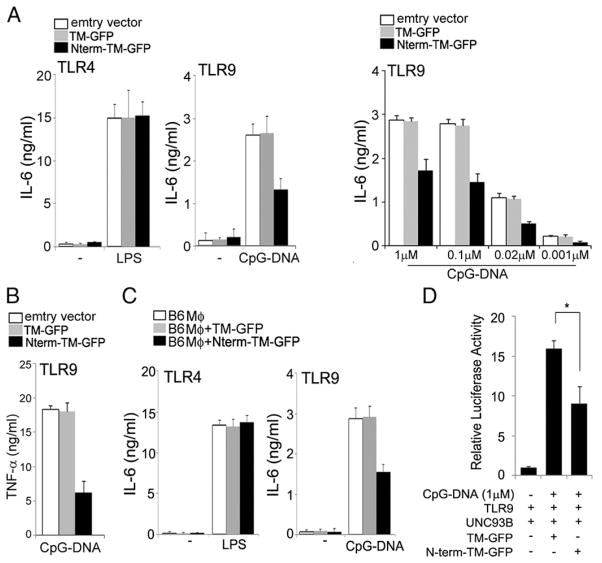
We next assessed the specific functional effects of N-terminal cleavage fragments on TLR9 stability and activation. We exposed RAW macrophages that stably expressed either TLR9 Nterm-TM-GFP or TM-GFP to TLR4 (LPS) and TLR9 (CpG-DNA) agonists and measured IL-6 production. We found that overexpression of the N-terminal TLR9 cleavage product in endolysosomes inhibited IL-6 production by macrophages stimulated with CpG-DNA, but did not influence IL-6 release after LPS exposure (Fig. 2A). We also found that induction of TNF-α production by CpG-DNA was impaired upon TLR9-Nterm-TM-GFP overexpression in macrophages (Fig. 2B). Similarly, CpG-DNA–stimulated, immortalized macrophages that stably expressed Nterm-TM-GFP showed reduced IL-6 production. In contrast, CpG-DNA stimulation of immortalized macrophages expressing TM-GFP did not affect IL-6 synthesis (Fig. 2C). To confirm these findings, we used an NF-κB–luciferase reporter gene to examine whether the N-terminal fragment inhibits TLR9 signaling activation. Indeed, cells transfected with the N-terminal cleavage product inhibited TLR9 signaling (Fig. 2D). We thus conclude that the N-terminal TLR9 proteolytic cleavage product that is normally generated along with the C-terminal fragment by lysosomal proteases is an important negative regulator of TLR9 signaling.
FIGURE 2.
The N-terminal TLR9 proteolytic cleavage product negatively regulates its signaling. (A) The influence of the N-terminal cleavage product on TLR9 signaling. IL-6 production was measured in RAW macrophages expressing TM-GFP or Nterm-TM-GFP after stimulation for 12 h with LPS (80 ng/ml), CpG-DNA (1 μM), or titration with the indicated concentrations of CpG-DNA. IL-6 production was measured by ELISA. (B) TNF-α production by macrophages stably expressing TM-GFP or Nterm-TM-GFP exposed for 12 h to CpG-DNA (1 μM). (C) IL-6 production by wild-type immortalized macrophages stably expressing TM-GFP or Nterm-TM-GFP after stimulation with LPS (80 ng/ml) or CpG-DNA (1 μM) for 12 h. (D) NF-κB luciferase reporter assays of HEK293T cells expressing full-length TLR9 in the presence of the indicated expression plasmids were performed 12 h after stimulation with 1 μM CpG-DNA. *p < 0.05 (Student t test). Data are representative of at least three independent experiments (average ± SD).
The N-terminal TLR9 cleavage product targets a C-terminal active receptor for degradation in the endolysosome
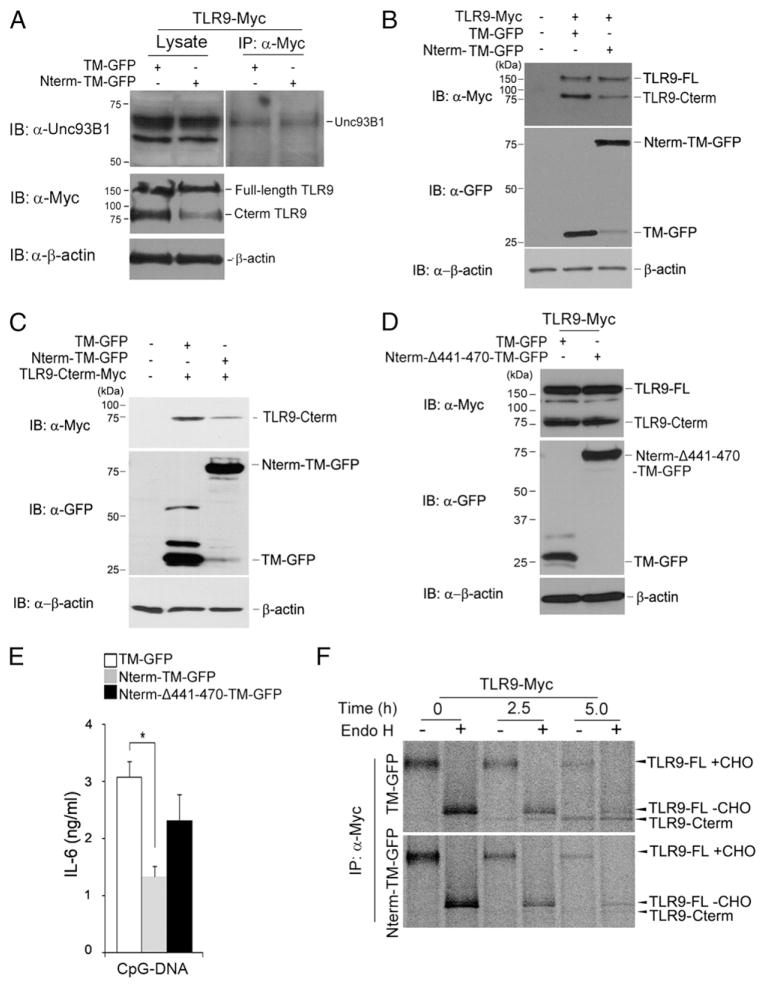
To establish how the N-terminal fragment negatively controls TLR9 responses, we investigated whether the N-terminal fragment affects the interaction of full-length TLR9 with Unc93B1. However, we observed no differences in this interaction between TM-GFP– and Nterm-TM-GFP–expressing cells (Fig. 3A). We next determined whether the N-terminal TLR9 product influences proteolytic cleavage processing and generation of a C-terminal fragment. In RAW macrophages expressing both full-length TLR9 tagged at the C terminus with Myc (TLR9-Myc) and Nterm-TM-GFP, we observed a significant reduction in C-terminal TLR9 cleavage products, but not in full-length TLR9. Unlike Nterm-TM-GFP–expressing macrophages, the introduction of TM-GFP did not affect the C-terminal fragment (Fig. 3B). Next, we sought to determine whether the reduced yield of C-terminal fragments was due to decreased stability of the C-terminal products. We generated RAW macrophages that stably expressed the recombinant C-terminal TLR9 fragment tagged at the C terminus with Myc (TLR9-Cterm-Myc), and transduced them with a retrovirus encoding Nterm-TM-GFP or TM-GFP. We found that the stability of the C-terminal fragment was reduced in Nterm-TM-GFP–expressing cells (Fig. 3C).
FIGURE 3.
The N-terminal cleavage product degrades C-terminal active receptor. (A) RAW macrophages expressing either TM-GFP or TLR9 Nterm-TM-GFP were lysed with 1% digitonin lysis buffer. TLR9-Myc proteins were immunoprecipitated with the anti-Myc Ab. Immunoprecipitated samples were resolved by SDS-PAGE and analyzed by Western blot analyses using the Unc93B1, Myc, or β-actin Ab. (B) TLR9 stability in RAW macrophages expressing either TM-GFP or Nterm-TM-GFP was monitored by immunoblotting with anti-Myc, anti-GFP, or anti–β-actin. Full-length TLR9 was tagged at the C terminus with Myc. TLR9-FL, full-length TLR9; TLR9 Cterm, C-terminal fragment of TLR9. (C) Immunoblot analyses of the recombinant Myc-tagged C-terminal TLR9 fragment (residues 471–1032) in RAW macrophages expressing either TM-GFP or Nterm-TM-GFP. Lysates were immunoblotted with anti-Myc, anti-GFP, or anti–β-actin. (D) Immunoblot analyses of the recombinant Myc-tagged C-terminal TLR9 fragment in RAW macrophages expressing either TM-GFP or Nterm-Δ441-470-TM-GFP. (E) IL-6 production by macrophages stably expressing TM-GFP or Nterm-Δ441-470-TM-GFP exposed for 12 h to CpG-DNA (1 μM). *p < 0.01 (Student t test). (F) Wild-type TLR9-Myc–expressing macrophages stably expressing either TM-GFP or Nterm-TM-GFP were labeled with [35S]methionine/cysteine for 1 h and chased for the indicated times. Cell lysates were immunoprecipitated with the anti-Myc Ab, and immunoprecipitates were treated with Endo H. Data are representative of three independent experiments. Data are representative of at least three independent experiments (average ± SD).
To test whether the N-terminal region that includes a TM affects the stability of the C-terminal product, we generated an additional construct, Nterm-Δ441-470-TM-GFP, which contains the N-terminal TLR9 fragment and TM, but not the cleavage site. Unlike Nterm-TM-GFP, cells transfected with Nterm-Δ441-470-TM-GFP did not degrade the C-terminal fragment (Fig. 3D). Moreover, cells transfected with Nterm-Δ441-470-TM-GFP did not efficiently inhibit the CpG-DNA–driven TLR9 response, suggesting that the endolysosomal cleavage process that generates the soluble N-terminal fragment is important for inhibition of TLR9 signaling (Fig. 3E). To confirm these results, we performed pulse-chase experiments. We found the N-terminal TLR9 fragment degraded the C-terminal fragment (Fig. 3F). Thus, we conclude that the N-terminal TLR9 fragment generated after proteolytic cleavage in the lysosome facilitates C-terminal fragment degradation.
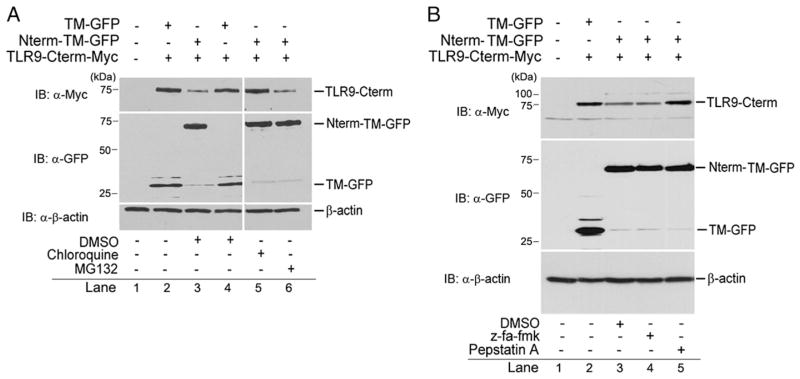
Aspartic protease is necessary for degradation of the C-terminal TLR9 fragment by N-terminal product
To investigate whether the degradation of the C-terminal fragment by the N-terminal TLR9 cleavage product was dependent on either proteasomal or lysosomal proteases, we monitored the protein levels of a recombinant C-terminal fragment in N-terminal fragment-expressing RAW macrophages in the presence of MG132 or chloroquine, an inhibitor of proteasome or lysosomal proteases. We found that the degradation of the C-terminal fragment by the N-terminal product was considerably inhibited by RAW macrophages treated with chloroquine but was unaffected by exposure to MG132 (Fig. 4A, lanes 3, 5, 6). In addition, to explore whether cathepsin activity was essential for TLR9 activation and is also involved in degradation of the C-terminal fragment in the N-terminal–mediated negative regulation, we exposed the RAW macrophages to z-FA-fmk, a cysteine protease inhibitor, or to pepstatin A, an inhibitor of aspartic proteases. Notably, z-FA-fmk had no effect on the degradation of the C-terminal fragment by the N-terminal TLR9 cleavage product, but the degradation was affected by pepstatin A (Fig. 4B, lanes 3–5). We concluded that the degradation of C-terminal product by the N-terminal TLR9 fragment is induced by aspartic proteases in the lysosome. In contrast, cathepsin shows no signs of such degradation, unlike generation of the functional C-terminal proteolytic cleavage product.
FIGURE 4.
Aspartic protease is required for N-terminal product-mediated C-terminal product degradation. RAW macrophages expressing recombinant Myc-tagged C-terminal fragment transduced a retrovirus expressing TM-GFP or Nterm-TM-GFP, incubated for 6 h with DMSO, chloroquine (5 μM), or MG132 (10 μM) (A), or for 12 h with z-FA-fmk (10 μM) or pepstatin A (1 μM) (B); proteins were analyzed with anti-Myc, anti-GFP, or anti–β-actin by immunoblot. Data are representative of three independent experiments.
The N-terminal cleavage product blocks TLR9 C-terminal fragment dimerization by physically interacting with the C-terminal fragment
Full-length TLR9 forms homodimers in the absence of CpG-DNA, suggesting that TLR9 dimers induce a conformational change capable of ligand binding and bringing the TIR domains in close proximity to recruit adaptor proteins to initiate signal transduction (18). In addition, Li et al. (19) reported that the cleaved C-terminal fragment usually forms a monomer in solution, and that CpG-DNA enhances dimerization. To explore why the C-terminal product is vulnerable to aspartic proteases in macrophages expressing the N-terminal fragment, we analyzed the effect of the N-terminal TLR9 fragment on C-terminal cleavage product dimerization. We first examined whether recombinant C-terminal products form homodimers in RAW macrophages. We used RAW cells that stably coexpressed recombinant C-terminal TLR9 fragments tagged with either Myc (TLR9 Cterm-Myc) or GFP and exposed them to CpG-DNA. We immunoprecipitated TLR9 Cterm-Myc using the anti-Myc Ab and examined dimerization by immunoblotting with the anti-GFP Ab. We found that cleaved C-terminal TLR9 products formed homodimers even in the absence of CpG-DNA, and that homodimer formation was increased by CpG-DNA (Fig. 5A, lanes 7, 8). We next investigated whether the N-terminal TLR9 product disrupts homodimerization of the C-terminal fragment. We used RAW macrophages that expressed Myc- or GFP-tagged C-terminal TLR9 products with either Nterm-TM-GFP or TM-GFP and exposed them to an aspartic protease inhibitor to detect the C-terminal fragment, which is degraded by N-terminal product expression (Fig. 4). In the anti-Myc immunoprecipitates, we immunoblotted with the anti-GFP Ab and detected that the C-terminal product strongly interacted with the N-terminal fragment (Fig. 5B, lane 3). We also found that C-terminal product dimer formation was significantly reduced by N-terminal TLR9 product expression, suggesting that disruption of C-terminal product dimerization by the N-terminal fragment sensitizes the C-terminal functional receptor to aspartic protease degradation in the lysosome (Fig. 5B, lanes 2, 4).
FIGURE 5.
The N-terminal cleavage product blocks TLR9 C-terminal fragment dimerization by physically interacting with the C-terminal fragment. (A) Immunoblot analyses of RAW macrophages expressing the Myc-tagged C-terminal fragment with or without the GFP-tagged C-terminal fragment and incubated with CpG-DNA (1 μM) for 6 h at 37°C. Anti-Myc was used for immunoprecipitation, and anti-GFP or anti-Myc were used for immunoblot analyses. (B) Analyses of immunoprecipitates of RAW macrophages stably expressing the Myc-tagged C-terminal fragment and GFP-tagged C-terminal fragment in the presence or absence of Nterm-TM-GFP. TLR9 C-terminal fragments recovered by immunoprecipitation with anti-Myc were treated with Endo F and immunoblotted with anti-GFP. Arrowheads indicate C-terminal TLR9 fragments dimers. (C and D) Immunoblot analyses of RAW cells stably expressing TLR9 Cterm-Myc with TM-GFP or TLR9 Nterm-TM-GFP, and incubated for 3 h at 37°C with biotinylated CpG-DNA (3 μM) in the absence (C) or presence of chloroquine (5 μM) (D); CpG-DNA and bond materials were precipitated with streptavidin-agarose and analyzed with anti-Myc or anti-GFP. (E) Flow cytometric analyses of intracellular IL-6 in BMDCs from wild-type transduced with either TM-GFP or Nterm-TM-GFP, stimulated for 6 h with LPS (80 ng/ml) or CpG-DNA (1 μM) in the presence of brefeldin A (day 6 of culture). Cells were fixed and stained with the anti–IL-6 Ab. (F) IL-6 production by immortalized TLR9−/− cells expressing Myc-tagged TLR9 with either TM-GFP or Nterm-TM-GFP induced by incubation with LPS (80 ng/ml) or CpG-DNA (1 μM). (G) NF-κB luciferase reporter assays of HEK293T cells expressing the C-terminal fragment of TLR9 in the presence of the indicated expression plasmids were performed 12 h after stimulation with 1 μM CpG-DNA. *p < 0.05, **p < 0.01 (Student t test). Data are representative of two (A–D) or three (E–G) independent experiments (average ± SD).
We next analyzed the interaction between CpG-DNA and the C-terminal TLR9 fragment in the presence and absence of the N-terminal product. As expected, given the degradation of the C-terminal TLR9 fragment by aspartic proteases in the lysates of cells expressing the N-terminal cleavage product, the association between the C-terminal TLR9 product and biotin-CpG in Nterm-TM-GFP–expressing macrophages was much weaker than that in TM-GFP–expressing cells (Fig. 5C, upper panel).
Because a previous study suggested that the N-terminal region of the TLR9 ectodomain participates in ligand binding (20), we examined whether the N-terminal product directly binds CpG-DNA (Fig. 5C, bottom). We found that the N-terminal fragment binds CpG-DNA, implying that it could compete with the C-terminal fragment for DNA binding in the lysosome. To determine whether this is the case, we examined the influence of the N-terminal product on the binding affinity of the C-terminal product for CpG-DNA in the presence of chloroquine because the lysosomal inhibitor blocks the ability of the N-terminal product to degrade the C-terminal fragment. Unlike control conditions, we did not observe a decreased interaction of biotin-CpG-DNA with the C-terminal product in Nterm-TM-GFP–expressing macrophages treated with chloroquine (Fig. 5D). Therefore, we conclude that although the N-terminal product can bind CpG-DNA, it does not directly compete for CpG-DNA binding. Rather, the N-terminal product selectively degrades the C-terminal product, thereby leading to reduced CpG-DNA binding.
To confirm the negative self-regulatory effect of the N-terminal product on TLR9 signaling in primary cells, we purified BMDCs and transduced them with a retrovirus encoding the N-terminal fragment. After exposing these cells to LPS or CpG-DNA, we measured cytokine production. BMDCs expressing Nterm-TM-GFP inhibited responsiveness to CpG-DNA stimulation, as indicated by IL-6 production. In contrast, IL-6 production was not affected by the CpG-DNA stimulation of BMDCs expressing TM-GFP (Fig. 5E). In addition, introduction of the N-terminal TLR9 product into TLR9−/− immortalized macrophages expressing wild-type TLR9 inhibited the CpG DNA-driven production of IL-6. In contrast, IL-6 production elicited by the LPS stimulation of BMDCs or TLR9−/− immortalized cells expressing Nterm-TM-GFP was not affected (Fig. 5F). We also observed that the N-terminal fragment inhibited signaling responses by the C-terminal cleavage fragment of TLR9 when stimulated with CpG-DNA, as monitored by an NF-κB–luciferase reporter gene (Fig. 5G). Although Onji et al. (13) have reported that the expression of the N-terminal ectodomain of TLR9 is involved in the full activation of TLR9 signaling, we failed to observe the full activation of TLR9 by a combination of the N-terminal and C-terminal fragments (Fig. 5G). Two different TLR9 ectodomain constructs tagged at the C terminus with Myc (Nterm-440-Myc [1–440] and Nterm-470-Myc [1–470]) were seen to block the signaling activation of the C-terminal TLR9 cleavage product, as with the inhibition of the lysosomal N-terminal fragment. These results collectively established that the N-terminal TLR9 cleavage product inhibits the homodimerization of the C-terminal fragment through competitive inhibition by physical interaction, and thus facilitates the aspartic protease-dependent degradation of monomeric C-terminal active receptor. We therefore conclude that the N-terminal TLR9 cleavage product is a negative self-regulator targeting the C-terminal functional receptor, a role that is crucial for the homeostasis of the TLR9 response.
The N-terminal portion of TLR9 has distinct functions in the ER or lysosome and a prolonged half-life compared with the C-terminal product
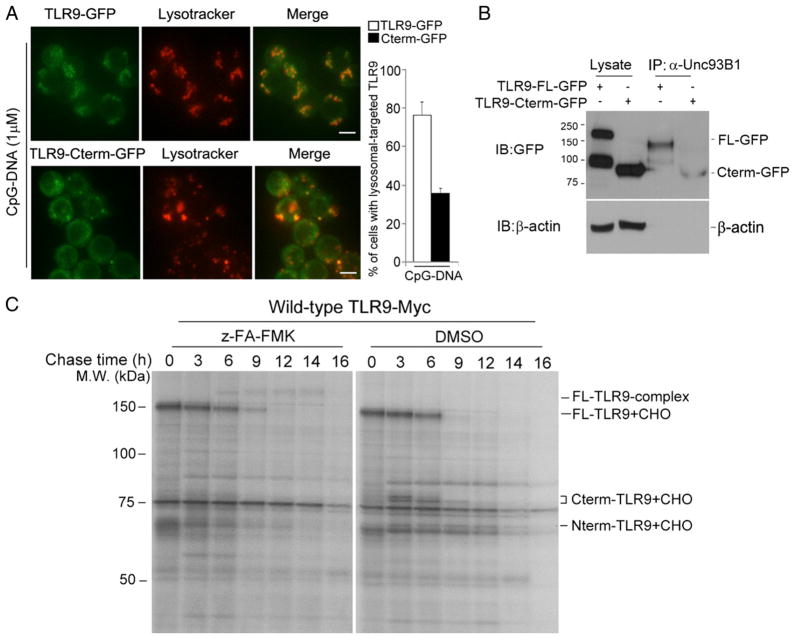
The recombinant C-terminal cleavage fragment activates NF-κB signaling and CpG-DNA–driven TNF-α production; however, as shown previously, the levels of the activation appear lower than those observed with full-length TLR9 (10, 19). Thus, we hypothesized that the N-terminal portion of TLR9 has distinct functions in the ER and lysosome. We first examined whether the N-terminal portion in the ER helps full-length TLR9 translocate efficiently to the lysosome (Fig. 6A). Indeed, full-length TLR9 was more efficiently trafficked to the lysosome compared with the recombinant C-terminal product, suggesting the N-terminal portion supports effective full-length protein relocation from the ER to the lysosome. To further test the possibility that the N-terminal portion provides strong affinity for Unc93B1, we examined the binding affinity of full-length TLR9 or the recombinant TLR9 C-terminal product with Unc93B1. The binding affinity of the C-terminal product for Unc93B1 was very slightly reduced when compared with full-length TLR9, but we did not observe a significant difference in their binding affinity (Fig. 6B). This result suggests that the N-terminal region of the TLR9 ectodomain is capable of binding unknown adaptor proteins in the ER, which supports full-length TLR9 relocation to the lysosome.
FIGURE 6.
The N-terminal region of TLR9 has distinct functions based on cellular localization. (A) RAW macrophages stably expressing either wild-type TLR9-GFP or C-terminal TLR9 fragments tagged with GFP (TLR9 Cterm-GFP) were stimulated with 1 μM CpG-DNA for 2 h, washed, and imaged after an additional 3 h with LysoTracker. Quantitative analysis of lysosomal-targeted TLR9 (right graph). Values represent the average percentage of cells with lysosomal-targeted TLR9 (n = 40–60 cells). Scale bars, 5 μm. (B) RAW macrophages expressing either wild-type TLR9-GFP or recombinant Cterm-GFP were lysed with 1% digitonin lysis buffer. Unc93B1 proteins were immunoprecipitated using the anti-Unc93B1 Ab. Immunoprecipitated samples were resolved by SDS-PAGE and analyzed using the anti-GFP Ab. (C) Wild-type TLR9-Myc–expressing macrophages were labeled with [35S]methionine/cysteine for 1 h and chased for the indicated times. Metabolically labeled cells were lysed with 1% digitonin lysis buffer, and lysates were immunoprecipitated using the anti-Myc Ab. Data are representative of three independent experiments.
To evaluate the stability profile of a cohort of newly synthesized full-length TLR9 or its N-terminal or C-terminal fragments over time, we pulse-labeled TLR9-Myc–expressing macrophages with [35S]methionine/cysteine for 30 min and chased for the indicated times (Fig. 6C). Because the C-terminal fragment physically interacts with the N-terminal product, we evaluated the kinetics and stability of full-length TLR9 and its N- or C-terminal fragments at each time point by immunoprecipitating with the anti-Myc Ab. As reported previously (10, 11, 21), we observed that TLR9 is proteolytically processed in a time-dependent manner via a multistep process, and its processing was blocked upon treatment with z-FA-FMK, a cathepsin inhibitor (Fig. 6C). Interestingly, the stability of the C-terminal fragment was reduced by ~60 and 25% after 6 and 9 h, respectively, whereas the majority of the N-terminal fragments were highly stable throughout the chase period. These results suggest that the N-terminal fragment is more stable over time than the C-terminal fragment in endolysosomes. This results in a switch in the negative feedback mechanism of TLR9 signaling by C-terminal product degradation.
Discussion
TLR signaling stimulates expression of inflammatory cytokines and antimicrobial proteins. However, prolonged or uncontrolled TLR activation leads to the development of septic shock and inflammatory diseases (22, 23). Therefore, signal-specific regulators responsible for repressing proinflammatory cytokines or blocking signal transduction pathways must negatively control TLR activation after the elimination of microbial populations. Proteolytic cleavage events are crucial for producing functional TLR9 receptors that bind CpG-DNA and initiate NF-κB activation (10–12). However, the role of the N-terminal TLR9 cleavage product generated after lysosomal protease–mediated cleavage was not elucidated in TLR9 signaling. We have now demonstrated that the N-terminal TLR9 cleavage product facilitates dimer dissociation of C-terminal cleavage products through physical interactions between N- and C-terminal fragments, thereby allowing the C-terminal TLR9 product to be more susceptible to aspartic protease-mediated degradation in the endolysosomal compartment. The broad specificity inhibitor pepstatin A most effectively blocked this degradation and, accordingly, inhibited C-terminal product degradation in cells expressing the N-terminal TLR9 fragment. CpG-DNA binding with TLR9 induces dimer formation to allow TIR domains to recruit adaptor proteins (18, 19). Our data show that blocking C-terminal product dimers via physical interaction with the N-terminal fragment promotes aspartic protease-mediated degradation of the C-terminal fragment, indicating that C-terminal product homodimer formation is necessary to prevent proteolytic degradation in the endolysosomal condition.
Various mutations within the N-terminal region of TLR9 cause its inactivation, and the N-terminal half of the TLR9 ectodomain acts as a DNA sensor (19, 20), implying that the N-terminal region might be required for full activation. In addition, although the C-terminal cleavage fragment activates cytokine production elicited by CpG-DNA or NF-κB signaling, the levels of TNF-α production or NF-κB activation seem to be lower than those seen with full-length TLR9 (10, 19). Our results showed that in the ER, the N-terminal portion of the TLR9 ectodomain supports effective relocation of full-length TLR9 to endolysosomes. Once trafficked to the lysosome, the N-terminal cleavage product generated by lysosomal proteases plays a key role in negative regulation of the TLR9 response in late stages of the immune response. It remains possible that the N-terminal region may contribute to enhancement of full-length TLR9 maturation in the ER or proteolytic cleavage processing in lysosomes and that the N-terminal product plays different roles in the TLR9 response to infection in a time-dependent manner.
Soluble TLR2 (sTLR2), sTLR4, and sTLR9 can modulate their responses, thereby reducing inflammation and infection (24–27). Chockalingam et al. (25) previously showed that N-terminally tagged sTLR9 is generated by proteolysis and observed in the lysosome. It is also detectable by Western blot analyses in HEK293T cells. However, as reported previously (11), we failed to detect N-terminal HA-tagged sTLR9 in RAW macrophages (Supplemental Fig. 1). Secondary-structure prediction programs show another small extended loop in TLR9 (10) situated between LRR1 and LRR2 that could be another lysosomal protease cleavage site(s). It may be hard to detect N-terminally tagged fragments derived from N-terminally tagged full-length TLR9 because of their small size. Recent study has shown that the crystal structure of the C-terminal product of mouse TLR9 exhibits similarities to the structures of other nucleic acid–binding TLRs, particularly TLR8 C-terminal domain (28). However, more detailed structural analyses of TLR9 are necessary to understand the proteolytic cleavage events of TLR9.
In conclusion, our data demonstrate that remnant N-terminal cleavage products of TLR9 generated by lysosomal proteases have a critical role as negative self-regulators of TLR9 activation induced by C-terminal active receptors. In this manner, they maintain appropriate cytokine and chemokine production involved in TLR homeostasis.
Supplementary Material
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea supported by the Ministry of Education, Science and Technology (Grants 2011-0015372 and 2010-0009203) and by the National Research and Development Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea.
Abbreviations used in this article
- BMDC
bone marrow–derived dendritic cell
- BMDM
bone marrow–derived macrophage
- Endo
endoglycosidase
- ER
endoplasmic reticulum
- HA
hemagglutinin
- HEK
human embryonic kidney
- Nterm
N-terminal TLR9 fragment
- sTLR
soluble TLR
- TLR9 Cterm
C-terminal TLR9 fragment
- TM
transmembrane domain
Footnotes
Disclosures
The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 3.Latz E, Schoenemeyer A, Visintin A, Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T, Golenbock DT. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 4.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 5.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 6.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 7.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 8.Lee CC, Avalos AM, Ploegh HL. Accessory molecules for Toll-like receptors and their function. Nat Rev Immunol. 2012;12:168–179. doi: 10.1038/nri3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 10.Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, Sakka E, Lennon-Duménil AM, Amigorena S, Cabanie L, Manoury B. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Onji M, Kanno A, Saitoh S, Fukui R, Motoi Y, Shibata T, Matsumoto F, Lamichhane A, Sato S, Kiyono H, et al. An essential role for the N-terminal fragment of Toll-like receptor 9 in DNA sensing. Nat Commun. 2013;4:1949. doi: 10.1038/ncomms2949. [DOI] [PubMed] [Google Scholar]
- 14.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maehr R, Hang HC, Mintern JD, Kim YM, Cuvillier A, Nishimura M, Yamada K, Shirahama-Noda K, Hara-Nishimura I, Ploegh HL. Asparagine endopeptidase is not essential for class II MHC antigen presentation but is required for processing of cathepsin L in mice. J Immunol. 2005;174:7066–7074. doi: 10.4049/jimmunol.174.11.7066. [DOI] [PubMed] [Google Scholar]
- 16.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 17.Avalos AM, Kirak O, Oelkers JM, Pils MC, Kim YM, Ottinger M, Jaenisch R, Ploegh HL, Brinkmann MM. Cell-specific TLR9 trafficking in primary APCs of transgenic TLR9-GFP mice. J Immunol. 2013;190:695–702. doi: 10.4049/jimmunol.1202342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DC, Monks BG, McKnight CJ, Lamphier MS, Duprex WP, et al. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Berke IC, Modis Y. DNA binding to proteolytically activated TLR9 is sequence-independent and enhanced by DNA curvature. EMBO J. 2012;31:919–931. doi: 10.1038/emboj.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peter ME, Kubarenko AV, Weber AN, Dalpke AH. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J Immunol. 2009;182:7690–7697. doi: 10.4049/jimmunol.0900819. [DOI] [PubMed] [Google Scholar]
- 21.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tak PP, Firestein GS. NF-kappaB: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto Y, Gaynor RB. Role of the NF-kappaB pathway in the pathogenesis of human disease states. Curr Mol Med. 2001;1:287–296. doi: 10.2174/1566524013363816. [DOI] [PubMed] [Google Scholar]
- 24.Dulay AT, Buhimschi CS, Zhao G, Oliver EA, Mbele A, Jing S, Buhimschi IA. Soluble TLR2 is present in human amniotic fluid and modulates the intraamniotic inflammatory response to infection. J Immunol. 2009;182:7244–7253. doi: 10.4049/jimmunol.0803517. [DOI] [PubMed] [Google Scholar]
- 25.Chockalingam A, Cameron JL, Brooks JC, Leifer CA. Negative regulation of signaling by a soluble form of toll-like receptor 9. Eur J Immunol. 2011;41:2176–2184. doi: 10.1002/eji.201041034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zunt SL, Burton LV, Goldblatt LI, Dobbins EE, Srinivasan M. Soluble forms of Toll-like receptor 4 are present in human saliva and modulate tumour necrosis factor-alpha secretion by macrophage-like cells. Clin Exp Immunol. 2009;156:285–293. doi: 10.1111/j.1365-2249.2009.03854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raby AC, Le Bouder E, Colmont C, Davies J, Richards P, Coles B, George CH, Jones SA, Brennan P, Topley N, Labéta MO. Soluble TLR2 reduces inflammation without compromising bacterial clearance by disrupting TLR2 triggering. J Immunol. 2009;183:506–517. doi: 10.4049/jimmunol.0802909. [DOI] [PubMed] [Google Scholar]
- 28.Collins B, Wilson IA. Crystal structure of the C-terminal domain of mouse TLR9. Proteins. 2014 doi: 10.1002/prot.24616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.