Short abstract
Pharmacogenomics and related genomic technologies may hold the potential to improve efficacy and safety in prescription, but complex factors affect their clinical success
Introduction
After bold claims and much anticipation, the impact of new genomic information and technologies is emerging and evolving in a complex fashion. This is certainly true of pharmacogenetics and pharmacogenomics, the study of the effect of genetic or genomic variation on drug response, which is purported to improve the safety and efficacy of prescription. The first products and services have reached the market in recent years, but their highly touted potential has not yet resulted in widespread clinical uptake, for a variety of reasons. This review explores some of the complex factors affecting the clinical success of pharmacogenetic and pharmacogenomic drugs and drug related tests. A series of criteria to inform clinicians, policy makers, and the public has already been established for predictive genetic tests1,2; given the importance of genetic testing for pharmacogenomics, the clinical uptake of pharmacogenomic drugs and strategies reflects similar factors.
Methods
The observations made in this paper draw on a broad literature review conducted during a research internship with the Nuffield Council on Bioethics' Working Party on Pharmacogenetics (my conclusions do not necessarily reflect the views of the council or its working party). I mined the Council's library, citations from key papers, and meetings of the working party. More recently, I searched Medline-PubMed and articles from the Wellcome Trust and the UK Human Genetics Commission, using the terms “pharmacogenetics or pharmacogenomics,” and variations of “economics,” “regulation,” “licensing,” and “ethics.”
Factors affecting the clinical uptake of pharmacogenomic products
Uncertainties in how genomic technologies will affect and be affected by the discovery, development, licensing, and postmarketing factors of drugs mean that the success of “candidates” is far from certain.3-6 Assessments of economic, regulatory, ethical, legal, and social considerations have already identified notable opportunities and challenges associated with pharmacogenomics.7-12
Summary points
Many factors will determine the success of genomics based technologies, products, and services designed to improve human health and disease
Pharmacogenetics and pharmacogenomics purport to increase safety and efficacy in prescription, but clinical uptake of their first few examples has not been widespread
Complex criteria are involved in the clinical success of drugs and drug related tests based on pharmacogenomics
The introduction of a conceptual framework and its application to recent case studies will help evaluate current pharmacogenomic strategies and will also help in guiding their future development, licensing, and prescription
Clinical factors such as the severity of the associated disease, availability of treatment, and effectiveness and accessibility of screening and surveillance methods will also have a direct impact on the uptake of pharmacogenomic strategies and have previously been outlined in the context of predictive genetic tests.1 They can be broadly categorised as technical accuracy (analytic validity); predictive power (clinical validity); potential for improving healthcare and health outcomes (clinical utility); and ethical, legal, and social implications.2 These criteria have also been related to pharmacogenomics,13 but additional ones should be considered in more depth since pharmacogenomics entails an intricate relation between genetic testing, diagnosis, and treatment. Although various definitions have been used, for the purpose of this paper a pharmacogenomic test can be defined as the correlation between a genetic anomaly and the efficacy or safety of a drug therapy, independent of whether the assay is at the level of genomics or proteomics. Industry leaders, clinicians, researchers, and policy makers have now begun to describe a series of factors that can define its clinical uptake and success.
Medical need
Pharmacogenomic approaches are especially valuable where they help to avoid acute outcomes: adverse reactions with grave clinical consequences, severe effects on quality of life, and high financial cost. For example, hypersensitivity reactions caused by abacavir (Ziagen), an antiretroviral drug, in the presence of the HLA-5701*B variant in white populations can have severe health consequences14—implying that genotyping before prescription can have an important positive effect on patients' quality of life. This is in addition to the serious and costly nature of HIV infection itself, which abacavir is intended to treat. Because of the substantial cost of treatment and potential for avoiding serious adverse events, disease management strategies are similarly under investigation for viral genotyping when treating hepatitis C patients with interferon and ribavirin.15
Where adverse reactions against existing drugs are mild in health and monetary terms, industry may conclude that large expenditure on regulatory issues and research and development will not justify the added benefit; alternately, clinicians may prefer to use traditional prescription by trial and error rather than incorporate molecular testing into their practice. From a broad health systems perspective, costs to consider include counselling, additional clinical visits to deal with complications, and other appropriate follow up.16
Clinical validity and utility of testing
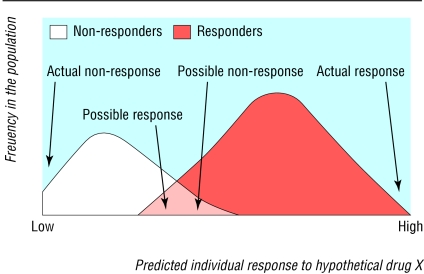
The importance of effective genetic testing for pharmacogenomics means that many factors influencing pharmacogenomics have already been identified by studies analysing genetic testing. For example, tests should be both sensitive and specific for the variant(s) being studied. However, inherent uncertainties can none the less “represent a major limitation”1 to clinical validity since test results represent probabilistic rather than definitive information, and health professionals must interpret tests with this in mind. In the context of pharmacogenomics, this means that, although the identification and characterisation of differing genetic or molecular profiles is likely to make the response to a drug more predictable than before, in some cases individuals will inevitably respond or not respond to a particular drug when testing predicts otherwise (figure).12
Figure 1.
Clinical validity of testing—the probabilistic nature of pharmacogenomics. Although pharmacogenomic test results will enable health professionals to predict drug response better, individual patients may still react in an unpredictable manner. Following testing, individuals classified as responders, for example, are likely to respond to a prescribed drug (far right), but may (rarely but significantly) be non-responders (middle area of overlap). Similarly, individuals classified as non-responders will probably not respond to a prescribed drug (far left), but may (rarely but significantly) be responders (middle area of overlap). A comparable situation will exist for the overlap between responders and patients predicted to experience adverse reactions. Adapted from reference 12
Strategies will thus be more successful when a drug prescribed on the basis of genotype has a low probability of false positive and false negative responses both within and across populations. In the case of abacavir, HLA-5701*B is neither 100% sensitive nor specific for hypersensitivity reactions: many people experiencing adverse reactions to drugs lack HLA-5701*B, whereas others who can tolerate abacavir possess the variant. The inconsistency in people without the variant experiencing adverse reactions was seen in white people (48-60% sensitivity) but was particularly pronounced in Hispanic patients (20-22% sensitivity) and even more so in black patients, for whom presence of the allele was not statistically associated with hypersensitivity,17 a finding that points to the existence of multiple parallel pathways and complex heterogeneity. As the field evolves further, drug response caused by gene variants whose prevalence differs across regions and populations will therefore require recording and sharing of information, trial results, and increased regulatory collaboration across jurisdictions. This will help to ensure that the safety of patients and the “first, do no harm” tenet of medicine are upheld rigorously.
At the level of clinical trials, because the number of patients required to develop a robust test increases drastically as the prevalence of a relevant variant decreases, the most clinically valuable compounds will be those for which adverse response or optimal efficacy genotypes occur at a notable frequency—not just in the trial population but in the patient population as a whole. With clinical utility in mind, developers will naturally focus on identifying gene variants or expression patterns that are predicted to affect response dramatically rather than marginally, based on a current understanding of molecular characteristics and disease pathogenesis. Both of these criteria are fulfilled by trastuzumab (Herceptin), a monoclonal antibody targeting tumours with overexpression of the ERBB2 (HER2) protein, which account for roughly 25-30% of cases of metastatic breast cancer. Pre-screening for HER2 expression levels reportedly resulted in one ninth the stage III trial size population that would have been required otherwise18; for tumours with overexpression, combination treatments including trastuzumab showed statistically significant improvements in median times to progression of disease, duration of response, failure of treatment, and survival time.19
A final consideration is the longer term utility of information derived from a pharmacogenomic test: the results of genotyping at one of the cytochrome P450 loci, which encode enzymes that metabolise many drugs (including codeine and clozapine, among others),20 represent a case where test results ordered for one prescription may well be relevant and revealing for other diseases and future prescriptions.
Relative ease of use
Rapid, reliable, inexpensive, and easily interpretable molecular tests improve the value of pharmacogenomic approaches for clinicians, especially where current non-genetic methods of monitoring drug response are inefficient or unavailable.21 Genomic technologies used to predict drug response must also be robust and widely accessible in diverse settings in clinical practice. Widespread clinical uptake of trastuzumab, for example, was enabled by the approval of an immunohistochemistry test that uses well established methods and readily available equipment to determine HER2 expression levels and select patients who are most likely to benefit from treatment, thereby avoiding some unnecessary prescription. Conversely, although evidence points to a major genetic role in essential hypertension, existing techniques to monitor blood pressure already allow doctors to adjust drug selection and dosage quickly when necessary, making new strategies involving pharmacogenomic testing less appealing.
In the case of patients with acute lymphocytic leukaemia and a high risk thiopurine methyl transferase (TPMT) genotype, which produces a thiopurine metabolising enzyme associated with extreme toxicity to the drug 6-mercaptopurine, some researchers have argued against molecular testing and in favour of the current standard, regular red blood cell counts.22 They believe that moving to molecular testing could delay important treatment and would entail additional cost and complexity. There are also fears that its interpretation might confound doctors, although an ongoing debate has prompted the US Food and Drug Administration to undertake a formal review of the genetic risk of 6-mercaptopurine.
The importance of easy to use molecular testing to inform prescription is underlined by the statement that regulators would be willing to assess diagnostic tests alongside their linked, pharmacogenomics based drug candidates,23 as was done with trastuzumab in the United States. It also meshes well with the argument that predictive genetic testing has increased utility when traditional screening and surveillance methods are inefficient or expensive.
Existing choice of treatments
As knowledge of biological pathways grows and the mechanisms of drug action and metabolism become clearer, pharmacogenomic opportunities and strategies should be evaluated in the context of the existing choice of treatments. This context will be fed into by other criteria already established. Where many drugs are already available and are known to target a variety of pathways, the availability of correlation studies—along with robust, rapid, and reliable molecular profiling in the clinical setting—will assume greater importance. Where few drugs exist for an illness but often result in severe and costly adverse events, clinical pharmacogenomic testing will be critical to avoiding adverse reactions to drugs, and a more urgent need arises to develop medicines designed for a particular underserved molecular profile or population.
The suggestion that drug development resources should be concentrated on cases where fewer treatment options exist none the less leaves the door open for clinical diagnostic testing to refine prescription where only a limited set of drugs is available: in particular, modulation of dosage is an already existing and powerful tool, enabling doctors to improve treatment response or minimise adverse effects. Variants of CYP2D6, a cytochrome P450 enzyme found in the liver,20 can result in a spectrum of activity depending on the medicine in question, from people with a slow metabolism who experience adverse effects because of an accumulation of drugs in the body, to people with an ultrarapid metabolism who only experience a therapeutic effect when prescribed drugs at a high concentration. Patients with acute lymphocytic leukaemia can similarly be treated by modulating the dosage on the basis of their TPMT risk profile. One consultant argues that pharmacogenomics will reap its largest revenue gains by expanding the clinical uptake of existing drugs (by greater flexibility in the adjustment of dosages)18 rather than by stratifying existing markets through a greater selection of drugs.
Factors that could influence the selection of drug candidates by pharmaceutical companies
Medical need
Clinical and economic consequences of forgoing available treatment
Clinical and economic consequences of the adverse reactions that could be prevented
Improvements in quality of life due to avoiding adverse reactions
Clinical validity and utility
Technical accuracy (analytic validity) of the diagnostic test
Clinical validity of pharmacogenomic tests for drug response
Prevalence of the variants under consideration in the population
Degree of impact of variants on drug response
Dominant versus recessive modes of inheritance for associated variants
Long term utility of information from pharmacogenomic tests
Ease of use
Speed, reliability, cost, robustness, easy interpretation, and accessibility of diagnostic tests
Availability and effectiveness of existing (non-genomic) screening methods
Choice of treatments
Availability and selection of existing drugs for a particular disease
Existence of pharmacogenomic association studies
Potential for modulating dosages to optimise the response to a drug
Case studies
The criteria and framework outlined here (box) can help to describe and explain the clinical utility and uptake of certain pharmacogenomic strategies and the relative lack of success of others. In the case of abacavir, for example, while one group has implemented genotyping of all patients who are prescribed the drug,14 GlaxoSmithKline's lack of recommendation on testing reflects the substantial number of people with false positive or false negative test results and the company's ongoing attempts to identify a set of markers with greater sensitivity and specificity in diverse clinical settings and across populations.17 Despite the substantial medical and financial cost of hypersensitivity reactions, this highlights the importance of clinical validity in the framework outlined here.
For breast cancer, the emergence of trastuzumab and its rapid and successful clinical uptake is due to its high impact on patients' quality of life and life expectancy (medical need); the resistance of HER2 positive cancers to conventional chemotherapy (lack of effective alternative treatments); and the accurate, straightforward, and easily interpreted tests that are available along with the drug (clinical validity and utility, ease of use). This combination of attributes makes Herceptin a prototypical example of the application of genomic technologies to health care.
In contrast to these examples, TPMT and CYP2D6 illustrate the ambiguity that clinicians face when considering whether to adopt pharmacogenomic strategies. For TPMT, prospective genotyping for children with acute lymphocytic leukaemia has become accepted in certain medical centres in the United States; individuals with a high risk genotype (producing a low activity enzyme) are prescribed a lower dosage of 6-mercaptopurine, which would theoretically eliminate the need for vigilant monitoring of patients' blood cell counts. Despite this medical need, however, several factors have prevented more widespread uptake of TPMT genotyping.22 Firstly, the test's ease of use has been called into question: its high cost ($100-300; £55-164; €82-245) and turnaround time may delay urgently needed treatment, whereas erythrocyte monitoring can be done by oncologists cheaply, effectively, and alongside treatment. Secondly, the clinical validity and utility of genotyping have been criticised: most studies have focused on four alleles that are prevalent in white populations, but variants more often found in Asian populations are not commonly included. Data on allelic frequencies for all populations should be documented better, and relevant tests must be both sensitive and specific within and across these groups.24 In contrast, blood cell monitoring is universally valid if handled quickly and as long as the patient has not had a recent blood transfusion.25 Finally, and also relating to ease of use, confusion over the interpretation of genetic test results could lead to underdosing of heterozygotes, for example, and imperfect understanding by doctors may result in inappropriate treatment.
As with TPMT, pharmacogenomic strategies including CYP2D6 have not been used much in clinical practice (with exceptions such as in Scandinavia) despite longstanding evidence of its role in the metabolism of 20-25% of medicines.20 Adverse reactions associated with the more than 70 known CYP2D6 variants are undesirable but rarely life threatening, and alternative treatments are usually available. Although CYP2D6 genotyping could be useful for a range of prescriptions, the large number of alleles and the differences in allele prevalence between populations can make the interpretation of molecular test results complex for health professionals. In this case, a combination of difficulty in interpreting test results, mild adverse reactions, multiple treatment alternatives, and low clinical utility has kept CYP2D6 testing out of widespread clinical use thus far. The impact of a new technology introduced in mid-2003, a diagnostic chip that can test for several cytochrome P-450 variants, could alter this equation by improving speed and efficiency and by simplifying the interpretation of test results while lowering costs.26
Additional educational resources
For health professionals, researchers, and policy makers
Nuffield Council on Bioethics report. Pharmacogenetics: ethical issues (www.nuffieldfoundation.org/pharmacogenetics). Covers research and development issues, regulation and public policy, and ethical implications for clinical practice.
University of Cambridge Epidemiology for Policy Group. My Very Own Medicine: What Must I Know? Information Policy for Pharmacogenetics (www.phpc.cam.ac.uk/epg/IPP.html). Summarises a range of current issues and makes policy recommendations.
FDA Draft Guidance for Industry: Pharmacogenomic Data Submissions (www.fda.gov/cder/guidance/5900dft.pdf). Provides guidance on when pharmacogenomic data should be submitted and how it will be assessed by regulatory authorities.
UK Department of Health Genetics white paper. Our inheritance, our future—realizing the potential of genetics in the NHS (www.dh.gov.uk/PolicyAndGuidance/HealthAndSocialCareTopics /Genetics/GeneticsGeneralInformation/GeneticsGeneral Article/fs/en?CONTENT_ID=4016430&chk=RnGBgL). Outlines the potential for genetics in health care, and proposes investments in training, manpower, research, and education to prepare the NHS.
PharmGKB, Pharmacogenetics and Pharmacogenomics Knowledge Base (www.pharmgkb.org). Database that contains genomic, phenotypic, and clinical information collected from pharmacogenetic research studies.
For patients
National Institute of General Medical Sciences. Medicines for you. www.nigms.nih.gov/funding/medforyou.html
Human Genome Project Information. Pharmacogenomics. www.ornl.gov/sci/techresources/Human_Genome/medicine/pharma.shtml
The Wellcome Trust. Pharmacogenetics. www.wellcome.ac.uk/en/genome/genesandbody/hg07b001.html
All provide a basic introduction to pharmacogenetics for laypeople, and links to further reading.
Conclusion
Pharmacogenomics and related genomic technologies have been widely reported to hold the potential to improve efficacy and safety in prescription. To that end, an initial set of factors describing the clinical uptake of drugs developed by using pharmacogenomic strategies might be categorised in terms of medical need, clinical validity and utility, relative ease of use, and the nature of currently available treatments. These criteria are explored and illustrated here in several examples. They should stimulate debate on the impact of pharmacogenomics on the clinical environment and, conversely, on the effect of clinical factors on the development and implementation of pharmacogenomics.
Other criteria and examples may well emerge in the course of such a discussion, and the clinical validity, utility, and uptake of these strategies may change along with advances in technology or revisions to how health professionals (particularly doctors and pharmacists) are trained. Pharmacogenomics and related genomic advances are clearly placing a unique lens on the multiple actors participating in the development, regulation, and prescription of drugs, as well as the complex interactions within our health systems. Finally, the ethical, legal, social, economic, and regulatory implications of such a framework require further investigation, including considerations of equity, distributive justice, and the particular opportunities and challenges presented by various health systems and their organisation.
I thank Tor Lezemore for critical reading of this manuscript, and the secretariat of the Nuffield Council on Bioethics for their hospitality during my research internship.
Funding: Research for this paper was made possible by a Commonwealth scholarship held in the Department of Social Policy, London School of Economics and Political Science.
Competing interests: None declared.
References
- 1.Evans JP, Skrzynia C, Burke W. The complexities of predictive genetic testing. BMJ 2001;322: 1052-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burke W, Atkins D, Gwinn M, Guttmacher A, Haddow J, Lau J, et al. Genetic test evaluation: Information needs of clinicians, policy makers, and the public. Am J Epidemiol 2002;156: 311-8. [DOI] [PubMed] [Google Scholar]
- 3.Birmingham K. Experts predict bleak post-genomic era for drug R&D. Nature Med 2001;7: 262. [DOI] [PubMed] [Google Scholar]
- 4.Norton RM. Clinical pharmacogenetics: Applications in pharmaceutical R&D. Drug Discov Today 2001;6: 180-5. [DOI] [PubMed] [Google Scholar]
- 5.Owens J, Ramster B, Lawrence RN. Impact of SNP genotyping could save millions by 2010. Drug Discov Today 2001;6: 450. [Google Scholar]
- 6.Ward SJ. Impact of genomics in drug discovery. BioTechniques (Euro Edition) 2001: 64-9. [DOI] [PubMed]
- 7.Phillips KA, Veenstra DL, Van Bebber S, Sakowski J. An introduction to cost-effectiveness and cost-benefit analysis of pharmacogenomics. Pharmacogenomics 2003;4: 231-9. [DOI] [PubMed] [Google Scholar]
- 8.Bristol L. A regulatory protocol for pharmacogenomics services. Pharmacogenomics J 2002;2: 83-6. [DOI] [PubMed] [Google Scholar]
- 9.Issa AM. Ethical perspectives on pharmacogenomic profiling in the drug development process. Nature Rev Drug Discov 2002;1: 300-8. [DOI] [PubMed] [Google Scholar]
- 10.Rothstein MA, Epps PG. Ethical and legal implications of pharmacogenomics. Nature Rev Genet 2001;2: 228-31. [DOI] [PubMed] [Google Scholar]
- 11.Robertson JA, Brody B, Buchanan A, Kahn J, McPherson E. Pharmacogenetic challenges for the health care system. Health Aff 2002;21: 155-67. [DOI] [PubMed] [Google Scholar]
- 12.Shah J. Economic and regulatory considerations in pharmacogenomics for drug licensing and healthcare. Nature Biotechnol 2003;21: 747-53. [DOI] [PubMed] [Google Scholar]
- 13.Melzer D, Raven A, Detmer DE, Ling T, Zimmern RL. My very own medicine: What must I know? Cambridge: Department of Public Health and Primary Care, University of Cambridge, 2003.
- 14.Lindpaintner K. The importance of being modest: Reflections on the pharmacogenetics of abacavir. Pharmacogenomics 2002;3: 835-8. [DOI] [PubMed] [Google Scholar]
- 15.Moriguchi H, Uemura T, Kobayashi M, Chung RT, Sato C. Management strategies using pharmacogenomics in patients with severe HCV-1b infection: A decision analysis. Hepatology 2002;36: 177-85. [DOI] [PubMed] [Google Scholar]
- 16.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: A systematic review. JAMA 2001;286: 2270-9. [DOI] [PubMed] [Google Scholar]
- 17.Hughes AR, Mosteller M, Bansal AT, Davies K, Haneline SA, Lai EH, et al. Association of genetic variations in HLA-B region with hypersensitivity to abacavir in some, but not all, populations. Pharmacogenomics 2004;5: 203-11. [DOI] [PubMed] [Google Scholar]
- 18.Tollman P, Guy P, Altshuler J, Flanagan A, Steiner M. A revolution in R&D: how genomics and genetics are transforming the biopharmaceutical industry. Boston, MA: Boston Consulting Group, 2001.
- 19.National Institute for Clinical Excellence. Technology Appraisal Guidance No.34. Guidance on the use of trastuzumab for the treatment of advanced breast cancer. London: National Institute for Clinical Excellence, 2002.
- 20.Wolf CR, Smith G. Pharmacogenetics. Br Med Bull 1999;55: 366-86. [DOI] [PubMed] [Google Scholar]
- 21.Veenstra DL, Higashi MK, Phillips KA. Assessing the cost-effectiveness of pharmacogenetics. AAPS PharmSci 2000;2:article 29. [DOI] [PMC free article] [PubMed]
- 22.Marshall E. Preventing toxicity with a gene test. Science 2003;302: 588-90. [DOI] [PubMed] [Google Scholar]
- 23.Branca M. FDA fosters pharmacogenomics, Bio IT World 2002. www.bioitworld.com/archive/061202/horizons_lesko.html (accessed 18 May 2004).
- 24.Van Aken J, Schmedders M, Feuerstein G, Kollek R. Prospects and limits of pharmacogenetics: the thiopurine methyl transferase (TPMT) experience. Am J Pharmacogenomics 2003;3: 149-55. [DOI] [PubMed] [Google Scholar]
- 25.Krynetski E, Evans W. Drug methylation in cancer therapy: lessons from the TPMT polymorphism. Oncogene 2003;22: 7403-13. [DOI] [PubMed] [Google Scholar]
- 26.Roche Diagnostics. Roche diagnostics launches the AmpliChip CYP450 in the US, the world's first pharmacogenomic microarray for clinical applications: Roche Diagnostics, 2003. www.roche.com/med-cor-2003-06-25 (accessed 7 Jun 2004).