Abstract
Proteolytic control of Caulobacter cell cycle proteins is primarily executed by ClpXP, a dynamically localized protease implicated in turnover of several factors critical for faithful cell cycle progression. Here, we show that the transient midcell localization of ClpXP that precedes cytokinesis requires the FtsZ component of the divisome. Although ClpAP does not exhibit subcellular localization, FtsZ is a substrate of both ClpXP and ClpAP in vivo and in vitro. A peptide containing the C-terminal portion of the FtsA divisome protein is a substrate of both ClpXP and ClpAP in vitro but is primarily degraded by ClpAP in vivo. Caulobacter carries out an asymmetric division in which FtsZ and FtsA are stable in stalked cells but degraded in the non-replicative swarmer cell where ClpAP alone degrades FtsA and both ClpAP and ClpXP degrade FtsZ. While asymmetric division in Caulobacter normally yields larger stalked and smaller swarmer daughters, we observe a loss of asymmetric size distribution among daughter cells when clpA is depleted from a strain in which FtsZ is constitutively produced. Taken together, these results suggest that the activity of both ClpXP and ClpAP on divisome substrates is differentially regulated in daughter cells.
Introduction
Caulobacter crescentus expresses multiple functions at precise times in the cell cycle, including chromosome replication and segregation, polar differentiation, and an asymmetric cell division yielding a non-replicative swarmer cell and a stalked cell that immediately enters the replicative cell cycle (Fig. 1A; Curtis and Brun, 2010). Cell cycle progression is dependent on temporally and spatially regulated proteolytic events catalysed by proteases such as ClpXP (Jenal, 2009; Bhat et al., 2013). ClpXP positioned at the cell pole during the swarmer-to-stalked cell transition is responsible for the proteolysis of the flagellar basal body components, the polar chemotaxis machinery (Alley et al., 1992; 1993; Tsai and Alley, 2001), and the CtrA master regulator (Domian et al., 1997; Jenal and Fuchs, 1998; McGrath et al., 2006). CtrA∼P in the swarmer cell binds to the origin of replication and inhibits DNA replication initiation (Quon et al., 1998). Following the proteolysis of CtrA in the stalked cell, chromosome replication is initiated and the ClpXP protease is released from the cell pole. At the time of predivisional cell cytokinesis, ClpXP accumulates at the cell division ring (Iniesta et al., 2006; McGrath et al., 2006), suggesting that ClpXP may play a role in the regulation of the divisome, the dynamic multicomponent structure that co-ordinates the invagination of the inner and outer membrane and remodelling of the cell wall during cell division. Consistent with this notion, the major divisome components FtsZ (a tubulin-like GTPase) and FtsA (an actin-like protein) are maintained at low levels in the stalked cell but cleared from the daughter swarmer cell (Kelly et al., 1998; Martin et al., 2004).
Fig 1.
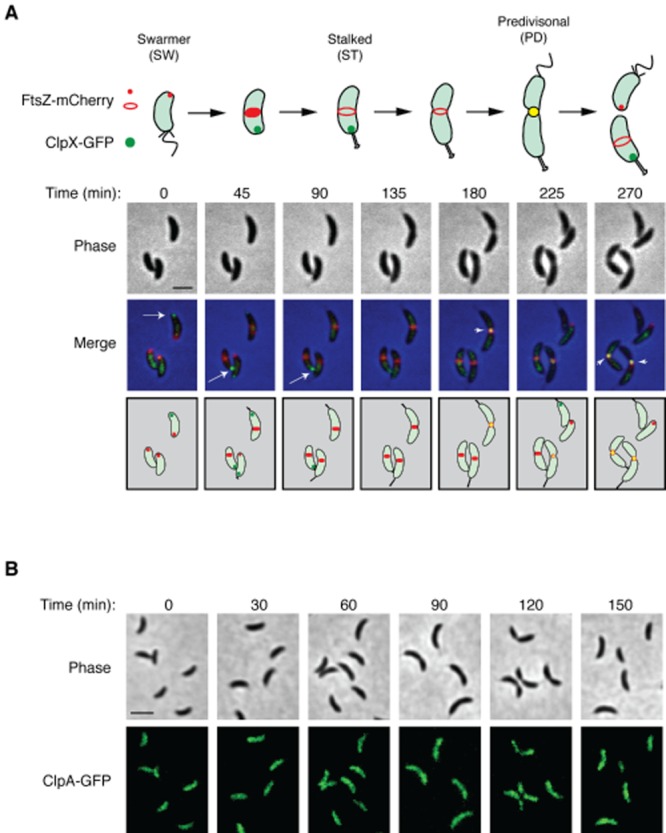
Dynamics of ClpX, ClpA and FtsZ subcellular localization.A. Cell cycle-dependent colocalization of ClpX–GFP and FtsZ–mCherry. Caulobacter bearing vanA::PvanA–clpX–egfp, xylX::PxylX–ftsZ–mCherry (LS5345) was induced with 0.5 mM vanillate for 2 h and 0.3% xylose for 1 h. Swarmer cells were isolated and allowed to proceed synchronously through the cell cycle on an agarose pad and imaged every 45 min using phase and fluorescence microscopy. A schematic of the cell cycle is shown below the panel. Scale bar represents 2.06 μm.B. Fluorescence microscopy of ClpA–GFP during the cell cycle. Caulobacter bearing pPvanA–clpA–egfp (LS5360) was induced with 0.5 mM vanillate for 2 h. Swarmer cells were isolated and allowed to proceed synchronously through the cell cycle. Aliquot of cells were sampled every 30 min, placed on an agarose pad and imaged using phase and fluorescence microscopy. Scale bar represents 2.6 μm.
Precise control of the levels of major cell division proteins is critical to the function of the Caulobacter divisome. In the absence of FtsZ, all other known divisome components fail to localize to the division site and cells fail to divide (Goley et al., 2011). Conversely, ftsZ overexpression results in gross cell division defects and cell death (Wang et al., 2001). Depletion of FtsA, which directly interacts with FtsZ, results in cell morphology defects consistent with incomplete envelope invagination and eventual cell death (Martin et al., 2004). The levels of these key cell division proteins change dramatically over the course of the cell cycle due in part to the cell-cycle-dependent control of ftsZ, ftsA and ftsQ synthesis (Sackett et al., 1998) mediated by the cell-cycle-regulated proteolysis of the transcription factors that regulate the expression of these proteins (Domian et al., 1997; Jenal and Fuchs, 1998; Gorbatyuk and Marczynski, 2005; McGrath et al., 2006).
Although regulated proteolysis of specific transcription factors plays a role in controlling the temporal availability of divisome-associated components, mounting evidence suggests a direct role for the ClpXP and ClpAP proteases in the cell-type specific degradation of the division proteins FtsZ and FtsA. In vitro studies suggest that E. coli FtsZ is a substrate of the ClpXP protease, and both FtsZ and FtsA are substrates of ClpP in Staphylococcus aureus and in Caulobacter (Bhat et al., 2013; Feng et al., 2013). In vivo, it has been shown that Caulobacter FtsZ is degraded in the predivisional cell just before cell division, but maintained at low levels specifically in the daughter stalked cell (Kelly et al., 1998). Although the CtrA and DnaA transcription factors contribute to the temporal regulation of divisome-associated components, FtsZ and FtsA levels vary as a function of the cell cycle even when they are constitutively produced (Wang et al., 2001; Martin et al., 2004), arguing that multiple factors contribute to the stability of divisome components in specific cell types.
Using in vivo and in vitro proteolysis assays of wild type and mutant FtsZ and FtsA proteins, we show that both proteins are substrates for ClpXP and ClpAP. We also show that while the presence of the Z-ring is required for ClpX localization to the division plane, ClpA is diffuse in the cytoplasm, suggesting that ClpX and ClpA may have different modes of interaction with Caulobacter divisome components. While FtsZ and FtsA are stable in stalked daughter cells, both ClpXP and ClpAP contribute to the removal of FtsZ in the daughter swarmer cell and only ClpAP is responsible for clearance of FtsA. A different mode of asymmetric activity of ClpXP is exhibited for the proteolysis of its CtrA substrate, where ClpXP localized to the stalked cell pole degrades CtrA in the daughter stalked cell and not in the daughter swarmer cell. Thus, differential proteolytic activity for specific substrates is a component of the asymmetric division that yields progeny of differing cell fate.
Results
Subcellular localization of ClpX, ClpA and FtsZ as a function of the cell cycle
ClpX, ClpP and FtsZ have been shown to transiently localize to the Caulobacter cell division plane (McGrath et al., 2006; Thanbichler and Shapiro, 2006). To determine the relative temporal pattern of ClpX and FtsZ localization during the cell cycle, we performed time-lapse microscopy and imaged a synchronized population of cells expressing chromosomal Pvan–clpX–egfp and Pxyl–ftsZ–mcherry in a wild-type background (Fig. 1A). Swarmer cells were placed on an agarose pad and imaged every 45 min as they progressed through the cell cycle (Fig. 1A). During the swarmer-to-stalked cell transition (time points 0 and 45 min), ClpX–GFP localized to the flagellated pole while FtsZ–mCherry was faintly visible at the opposite pole (Fig. 1A). In early stalked cells, FtsZ–mCherry formed a prominent band at the incipient division plane (t = 45 and 90 min). A ClpX–GFP focus was not observed in early predivisional cells (t = 135 min) but appeared at the division site in late predivisional cells exhibiting deep constrictions. ClpX–GFP and FtsZ–mCherry colocalized (yellow focus) at the division site just before the completion of cell division.
ClpA, an AAA+ chaperone, interacts with ClpP to form the ClpAP protease (Thompson and Maurizi, 1994). To determine if ClpA is dynamically localized as a function of the cell cycle, we imaged samples taken from a synchronized population of cells harbouring a medium-copy plasmid expressing pPvan–clpA–egfp at 30-minute intervals over a 150-minute time-course (Fig. 1B). We found that throughout the cell cycle, ClpA–GFP was punctate and/or diffuse, with occasional randomly positioned foci. We confirmed that the ClpA–GFP fusion was functional in vivo by demonstrating that it recuses the cell cycle-dependent degradation of a known ClpA substrate, the FliF protein (Fig. S1A; Grunenfelder et al., 2004). Furthermore, we demonstrated that the GFP moiety remained fused to ClpA (Fig. S1B). These results suggests that our ClpA–GFP fusion is functional and that ClpA–GFP does not localize to distinct regions of the cell during cell cycle progression, in contrast to the observed dynamic localization of ClpX to the divisome.
ClpX localization to the division plane is dependent on FtsZ
The observed colocalization of ClpX–GFP and FtsZ–mCherry at the division plane raises the possibility that one or more divisome components are directly responsible for recruitment of ClpXP to the site of cell division. To test if the Z-ring is required for ClpX localization to the division plane, we introduced a chromosomal PvanA–clpX–egfp construct into the ftsZ depletion strain YB1585 to generate ftsZ::ftsZΔC xylX::PxylX–ftsZ, vanA::PvanA–clpX–egfp (Wang et al., 2001). In the resulting strain (LS5346), expression of the sole copy of ftsZ is controlled by the xylX promoter, thereby allowing depletion of FtsZ when cells are grown in the absence of xylose. LS5346 was grown in the presence of vanillate (to induce ClpX–GFP expression) and in the absence of xylose (to eliminate FtsZ from the cell) for 6 h (Fig. S2A) and imaged at 30-minute intervals. The FtsZ-depleted cells appeared elongated with no discernable constrictions and ClpX–GFP was punctate throughout the cell (Fig. 2A).
Fig 2.
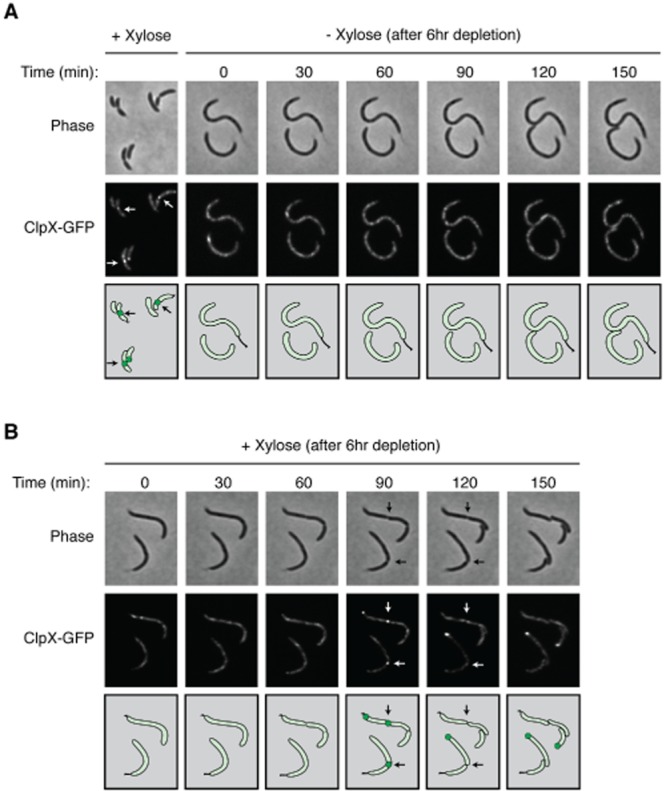
ClpX–GFP requires FtsZ for localization to the division plane.A. ClpX–GFP was imaged in a Caulobacter strain bearing ftsZ::ftsZΔC xylX::PxylX–ftsZ, vanA::PvanA–clpX–egfp (LS5346) grown in minimal M2G media in the presence of 0.5 mM vanillate and 0.3% xylose. This strain was then grown in the absence of xylose for 6 h to deplete FtsZ before being placed on an agarose pad (t = 0 min) lacking xylose. Cells were imaged at 30-minute intervals using phase and fluorescence microscopy.B. Recovery of ClpX–GFP at the division plane upon re-synthesis of FtsZ. Cells depleted of FtsZ were placed onto an agarose pad (t = 0 min) containing xylose and imaged at 30-minute intervals.
To determine if ClpX localization could be restored upon induction of FtsZ, strain LS5346 was grown for 6 h in the presence of vanillate to induce clpX–egfp and in the absence of xylose to deplete FtsZ. The FtsZ-depleted strain was then placed on a M2G agarose pad supplemented with xylose and imaged by fluorescence microscopy (Fig. 2B). By 30 min after FtsZ induction, FtsZ levels were similar to those observed prior to depletion (Fig. S2B), and cells displayed deep constrictions at 90 min, indicating restoration of FtsZ function. Completion of cell division occurred between 90 and 120 min post induction with ClpX–GFP properly localizing to the cell division site (Fig. 2B). Thus, ClpX localization to the division plane requires the presence of FtsZ. If FtsZ is the sole recruiter of ClpX, then FtsZ may stimulate its own destruction by concentrating ClpXP proteolytic activity at the Z-ring. Since multiple divisome components are recruited to the Z-ring, it is possible that one or more of these components are a target for ClpX recruitment to the incipient division site.
FtsZ is a substrate of ClpXP and ClpAP in vivo and in vitro
Previous studies have established a role for ClpP in the regulation of FtsZ and FtsA levels, although it has not yet been shown conclusively that these divisome components are direct substrates of ClpP, nor is it clear whether ClpP-dependent proteolysis of FtsZ/A also requires the ClpA and/or ClpX chaperones. To determine whether ClpAP-mediated proteolysis also contributes to the cell-cycle-dependent clearance of FtsZ, we used a clpA null mutant generated by replacing the chromosomal copy of clpA with an interposon (Grunenfelder et al., 2004). Because clpX is an essential gene in Caulobacter, we determined whether FtsZ is a substrate of the ClpXP protease by employing an inducible, catalytically dead, dominant-negative variant of ClpX, hereafter referred to as ClpX* (Potocka et al., 2002). We constructed a medium-copy plasmid bearing clpX* under the control of the vanillate-inducible PvanA promoter. To demonstrate the effect of ClpX* on ClpXP-mediated proteolysis in vivo, we measured the levels of a known ClpXP substrate, CtrA, in the presence and absence of ClpX*. As expected, we found that the levels of CtrA are increased when ClpX* is expressed (Fig. S3), confirming a significant reduction in ClpXP activity upon expression of clpX*. Thus, in the presence of vanillate, ClpP is effectively titrated by ClpX* and ClpXP substrates are stabilized.
To assay for FtsZ proteolysis in vivo, we generated an inducible gene fusion comprising a tandem-affinity purification (TAP) tag fused to the N-terminus of FtsZ (TAP–FtsZ) that was then integrated at the chromosomal xylX locus, thereby placing TAP–FtsZ expression under the control of a xylose-inducible promoter. This construct was then introduced into a wild-type, clpX*, or ΔclpA strain. Each resulting strain was grown in the presence of xylose (and, in the case of clpX*, vanillate) for 2 h, washed, and resuspended in media lacking xylose. Samples were then taken every 15 min and cell lysates were subjected to SDS-PAGE and western immunoblotting using TAP-specific antisera. In the presence of ClpX*, the stability of TAP–FtsZ increased more than twofold, whereas the stability of TAP–FtsZ in the absence of a functional clpA allele increased roughly fivefold compared to TAP–FtsZ in a wild-type background (Fig. 3A). As control, we also show that the stability of another cell-cycle-regulated divisome component, FtsQ, does not change in the presence of the ClpX* or a clpA null allele (Fig. S4A and B). These data suggest that FtsZ may be a direct target of both ClpAP and ClpXP proteolysis in vivo.
Fig 3.
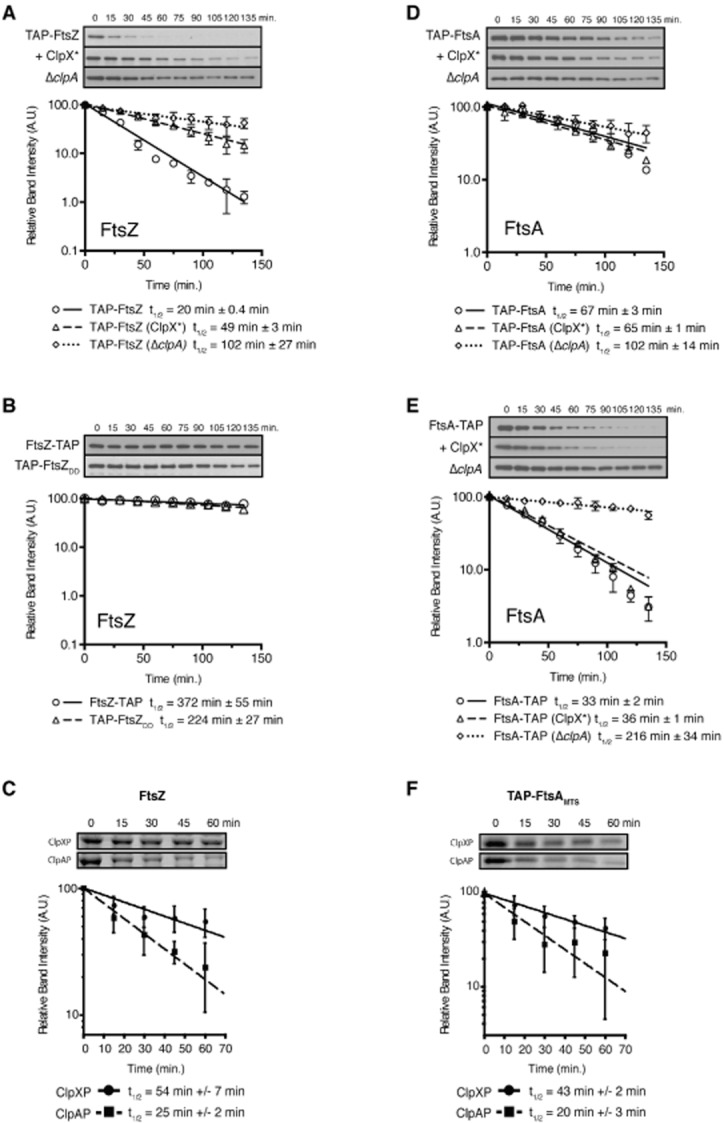
FtsZ and FtsA are substrates of ClpXP and ClpAP.A. TAP–FtsZ is stabilized in vivo in clpX* and ΔclpA strains. Caulobacter strains bearing xylX::PxylX–tap–ftsZ (LS5347), xylX::PxylX–tap–ftsZ pPvanA-clpX* (LS5350) or ΔclpA::Ω xylX::PxylX–tap–ftsZ (LS5356) were grown in PYE media in the presence of xylose 0.3% (and 0.5 mM vanillate when applicable) for 2 h. Cells were then washed and suspended in media lacking the inducers. Aliquots taken at 15-minute intervals were subjected to immunoblot analysis using TAP-specific antiserum to detect TAP–FtsZ.B. C-terminal mutants of FtsZ are stabilized in vivo. Caulobacter strains bearing xylX::PxylX–ftsZ–tap (LS5353) or xylX::PxylX–tap–ftsZ-DD (LS5354) were grown in PYE media the presence of the xylose (0.3%) inducer for 2 h, washed, and then resuspended in media lacking xylose. Aliquots were then taken and assayed at 15-minute intervals as described in A.C. FtsZ is degraded in vitro by ClpXP and ClpAP in the presence of ATP. Reaction conditions consist of 1 μM ClpX6 or 1 μM ClpA*6, 1 μM ClpP14, 1 μM FtsZ.D. N-terminal-blocked TAP–FtsA is relatively stable in vivo. Caulobacter strains bearing xylX::PxylX–tap–ftsA (LS5348), xylX::PxylX–tap–ftsA pPvanA-clpX* (LS5351) or ΔclpA::Ω xylX::PxylX–tap–ftsA (LS5357) and assayed as described in A.E. C-terminal blocked FtsA–TAP is degraded in vivo and specifically stabilized in a ΔclpA strain. Caulobacter strains bearing xylX::PxylX–ftsA–tap (LS5355), xylX::PxylX–ftsA–tap pPvanA-clpX* (LS5366) or ΔclpA::Ω xylX::PxylX–ftsA–tap (LS5367).F. FtsA bearing an N-terminal fusion of the TAP epitope tag to the membrane targeting sequence (MTS) is degraded in vitro by ClpAP and ClpXP in the presence of ATP. Reaction conditions consist of 1 μM ClpX6 or 1 μM ClpA*6, 1 μM ClpP14, 1 μM TAP–FtsAMTS. Quantification of triplicate in vitro reactions is shown below the images of each reaction. Each panel in this figure shows a graph of the relative band intensity as a function of time of the respective immunoblot. Data from each experiment is plotted on a semi-log scale, with fitted lines representing single-exponential decays with the half-life shown below. Error is shown as the standard error of the mean of three experiments.
The carboxy-terminal sequence of ClpXP substrates is important for their recognition and proteolysis (Flynn et al., 2003). Additionally, the C-terminus of FtsZ is known to be critical for ClpX recognition in E. coli (Camberg et al., 2009). We therefore reasoned that if FtsZ is a direct substrate of ClpXP or ClpAP, then the C-terminus of FtsZ should be critical for proteolysis in vivo. To test this, we generated C-terminal mutations in FtsZ and assayed the stability of each mutant in vivo (Fig. 3B). Whereas TAP–FtsZ was no longer detectable after 75 min following the removal of inducer, we found that FtsZ–TAP (in which the TAP tag is present at the very C-terminus) and TAP–FtsZDD (a variant in which the alanine and asparagine at the very C-terminus of FtsZ are changed to two aspartic acids) were completely stabilized, exhibiting in vivo half lives that were longer than that of TAP–FtsZ by more than an order of magnitude (compare Fig. 3A and B). These findings are consistent with the notion that FtsZ is a direct substrate of both ClpXP and ClpAP and reveal a putative degradation motif at the C-terminus of FtsZ.
To determine conclusively whether FtsZ is a direct target of ClpXP and/or ClpAP, we tested the ability of purified ClpAP and ClpXP to degrade FtsZ by incubating purified Caulobacter FtsZ with ClpAP or ClpXP in vitro. In order to prevent auto-degradation of ClpA by ClpAP, we employed a ClpA variant that lacks the intrinsic C-terminal degradation tag (ClpA*; Maglica et al., 2008). Using this in vitro proteolysis assay, we found that FtsZ is degraded in the presence of either ClpA*P or ClpXP, albeit with differential efficiency; in our assay, FtsZ exhibited an approximate half-life of 25 min in the presence of ClpA*P and 54 min when incubated with ClpXP (Fig. 3C). Additionally, we found that proteolysis of FtsZ by ClpXP requires the N-terminal zinc-binding domain of ClpX, as a truncated variant lacking the first 61 amino acids corresponding to the N-terminal domain (ClpXΔN61) was not capable of degrading FtsZ in vitro (Fig. S5). This is consistent with previous studies in which the N-terminus of E. coli ClpX was shown to be important for degradation of E. coli FtsZ (Camberg et al., 2009; Sugimoto et al., 2010). Collectively, these results provide evidence that FtsZ is a direct substrate of the ClpAP and ClpXP proteases, and that this activity requires both the C-terminus of FtsZ and, in the case of ClpXP, the N-terminal domain of ClpX.
FtsA is a substrate of ClpAP in vivo but a substrate of ClpAP and ClpXP in vitro
Like FtsZ, the actin-like divisome protein FtsA is a putative substrate of the ClpP protease in Caulobacter (Bhat et al., 2013). In light of the observations described in the preceding sections, we wished to determine whether FtsA is also a direct substrate of ClpXP and/or ClpAP. To that end, we measured the stability of TAP–FtsA in wild-type Caulobacter and in strains harbouring a clpX* or ΔclpA mutant allele as described above. In contrast to TAP–FtsZ, we found that the stability of TAP–FtsA was relatively unchanged in the presence of ClpX* and only modestly affected by the absence of functional ClpA (Fig. 3D). This result indicates that the ClpXP and ClpAP proteases exhibit differential activity toward FtsZ and FtsA in vivo.
For some ClpXP substrates, an exposed amino-terminus is important for recognition (Flynn et al., 2003). We reasoned that any ClpXP activity against FtsA might be masked in the assay described above due to the presence of an N-terminal TAP tag. Therefore, we examined the stability of FtsA bearing a TAP tag at the C-terminus (FtsA–TAP). We found that FtsA–TAP was less stable than TAP–FtsA (Fig. 3E; compare to Fig. 3D), suggesting that the FtsA N-terminus may be important for regulation of FtsA levels in vivo. To test if ClpA or ClpX specifically recognizes the N-terminus of FtsA, we assayed the stability of FtsA–TAP in strains harbouring ΔclpA or clpX* mutant alleles. We found that the stability of FtsA–TAP was relatively unaffected by the presence of ClpX* while the stability of FtsA–TAP in a ΔclpA background increased dramatically (Fig. 3E). These data imply that ClpA is likely the primary degradation factor for FtsA and that N-terminal recognition motifs exist within the FtsA peptide.
Next, we addressed the possibility that ClpAP, and not ClpXP, is directly responsible for proteolysis of FtsA by examining the ability of purified ClpAP or ClpXP to degrade FtsA in vitro. However, the insolubility of Caulobacter FtsA upon overproduction in E. coli precluded any biochemical assays that require purified FtsA protein. As an alternative, we performed in vitro proteolysis experiments using purified TAP–FtsAMTS, a hybrid protein containing the membrane targeting sequence of FtsA (comprising the last 15 residues of the mature protein) fused to an N-terminal TAP tag. We found that in vivo, the stability of TAP–FtsAMTS was unaltered when expressed in the presence of ClpX* (data not shown). Surprisingly though, we found that in the presence of ATP, both ClpA*P and ClpXP degraded TAP–FtsAMTS in vitro (Fig. 3F) despite the fact that this protein lacks the exposed N-terminus of FtsA. We quantified the band intensities from each reaction and found that TAP–FtsAMTS had a half-life of approximately 20 min in the presence of ClpAP and approximately 43 min in reactions containing ClpXP. We also show that both ClpXP and ClpAP fail to degrade purified FtsA lacking the N-terminal 18 amino acids and the C-terminal 15 amino acids of the peptide (Fig. S6A–D) arguing that both the N-terminus and the C-terminus of FtsA serve as recognition motifs for ClpAP and ClpXP. Although the contribution of ClpAP to FtsA processing can be detected in vivo and in vitro, the proteolytic activity of ClpXP against FtsA was only observed in vitro, suggesting that FtsA is protected from degradation by ClpXP in vivo.
FtsZ and FtsA are differentially degraded in progeny swarmer cells following asymmetric cell division
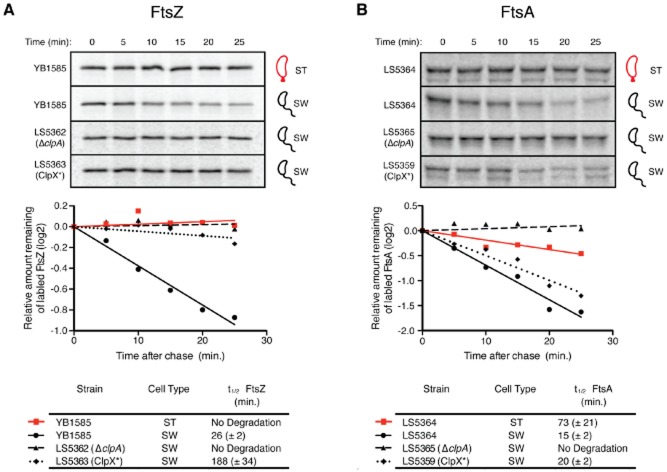
It was demonstrated previously that FtsZ and FtsA are stabilized in the progeny stalked cells, while in the progeny swarmer cells the levels of both divisome components rapidly decreased below the detection threshold (Kelly et al., 1998; Martin et al., 2004). We have shown that both FtsZ and FtsA are substrates of ClpAP and ClpXP in vitro but that FtsA is not a substrate of ClpXP in vivo. To specifically assess whether both proteases are active in progeny swarmer cells, we bypassed transcriptional regulation of ftsZ and constitutively expressed ftsZ from the xylX promoter in strains bearing ΔclpA or clpX* alleles and compared the half-life of FtsZ in stalked cells to that observed in swarmer cells. Strains were incubated in the presence of xylose for 5 min and then labelled with [35S]-methionine for 5 min. Cells were then chased with unlabelled methionine, after which, samples were taken at 5-minute intervals over a 25-minute time-course. FtsZ was then immunoprecipitated, subjected to SDS-PAGE, and quantified using a phosphorimager. Under these assay conditions we found that FtsZ was rapidly degraded in clpA+ clpX+ (YB1585) swarmer cells with a half-life of roughly 26 min, whereas we detected little or no degradation in either ΔclpA (LS5362) or clpX* (LS5363) swarmer cells (Fig. 4A), demonstrating that both ClpA and ClpX contribute to the proteolysis of FtsZ in the swarmer cell.
Fig 4.
Asymmetric protease function.A. Pulse chase analysis of [35S]-Methionine labelled FtsZ in strain ftsZ::ftsZΔC xylX::PxylX–ftsZ (YB1585) stalked cells and swarmer cells and in strains bearing ftsZ::ftsZΔC xylX::PxylX–ftsZ ΔclpA::Ω (LS5362) or ftsZ::ftsZΔC ΔxylX::PxylX–ftsZ pPvanA-clpX* (LS5363).B. Pulse chase analysis of [35S]-Methionine labelled FtsA in Caulobacter bearing xylX::PxylX–ftsA (LS5364) stalked cells and swarmer cells, and swarmers in strains bearing xylX::PxylX–ftsA ΔclpA::Ω (LS5365) or xylX::PxylX–ftsA pPvanA-clpX* (LS5359). Labelled FtsZ or FtsA was immunoprecipitated with antibodies to either FtsZ or FtsA and the relative protein levels were plotted on a semi-log graph versus time. t ½ values for each time point after the chase. All strains were grown in the presence of 0.3% xylose for 2 h prior to synchrony to induce FtsZ or FtsA and that both LS5363 and LS5359 were grown in the presence 0.5 mM vanillate for 2 h prior to synchrony to induce ClpX*.
The same experimental conditions were used to determine the half-life of FtsA in swarmer cells. Like FtsZ, FtsA was barely detectable in wild type swarmer cells (Martin et al., 2004). In order to increase the concentration of FtsA for efficient immunoprecipitation detection, we generated a strain that expresses ftsA from the xylX promoter (LS5364) along with derivatives containing ΔclpA (LS5365) or pPvanA-clpX* (LS5359). For each strain, a synchronized population of cells was grown in media supplemented with xylose (and vanillate, in the case of LS5359) and utilized in pulse-chase assays as described above. We found that FtsA had a half-life of approximately 15 min in clpA+ clpX+ (LS5364) swarmer cells and a marginally longer half-life of approximately 20 min in the presence of ClpX*; however, in the absence of ClpA, we observed no degradation of FtsA in vivo (Fig. 4B). Collectively, these results suggest that proteolysis of FtsA in progeny swarmer cells is primarily mediated through ClpAP, while the proteolysis of FtsZ in swarmer cells is mediated by both ClpXP and ClpAP.
ClpA is required for efficient regulation of FtsZ cell cycle-timing and positioning of the cytokinetic ring
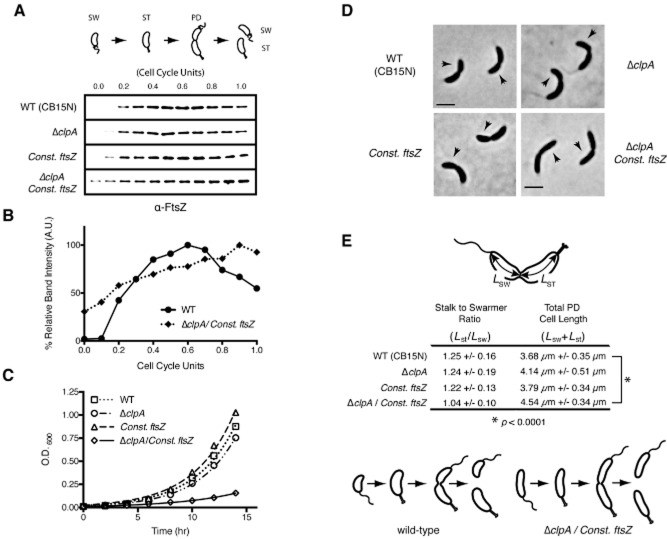
We analysed the relative levels of FtsZ in synchronized populations of wild-type (LS101), single mutant (UJ838 or YB1585) and double mutant (LS5362) strains using antibodies directed against FtsZ. In both the clpA deletion strain and the wild-type strain, the dynamic pattern of FtsZ accumulation were comparable: FtsZ levels were undetectable in swarmer cells (t = 0.0 cell cycle units), peaked around 0.45 cell cycle units, and then decreased to about 50% of the peak value as division plane constriction was initiated at 0.80 cell cycle units (Fig. 5A). In cells constitutively expressing ftsZ, FtsZ levels were fairly comparable to that observed in wild-type and ΔclpA strains, except that there was a twofold increase in protein levels at 0.2 cell cycle units. However, in the double mutant constitutively expressing ftsZ and lacking clpA, we detected increased levels of FtsZ in the swarmer cell (0 and 0.1 cell cycle units) and only a minimal decrease in FtsZ protein levels at 1.0 cell cycle units, suggesting that ClpA directly or indirectly contributes to the degradation of FtsZ at the time of cell division (Fig. 5A and B).
Fig 5.
ClpA is required for regulation of FtsZ cell cycle accumulation and the asymmetry of cell division.A. Relative levels of FtsZ as a function of the cell cycle. Swarmer cells from strains wild-type (LS101), ΔclpA::Ω (UJ838), ftsZ::ftsZΔC xylX::PxylX–ftsZ (YB1585 – Const. ftsZ) and ΔclpA::Ω ftsZ::ftsZΔC xylX::PxylX–ftsZ (LS5362 – double mutant), grown in M2G media supplemented with 0.3% xylose when appropriate, were isolated and allowed to progress synchronously through the cell cycle. Aliquots were taken at the intervals shown and subjected to immunoblot analysis using antibody to FtsZ. One cell cycle unit is equal to one doubling time. Each lane reflects the total amount of protein from a normalized density of culture (0.3 OD600) at the indicated cell cycle unit. SW, swarmer cells; ST, stalked cells; PD, predivisional cells.B. Relative band intensities of the FtsZ immunoblot assays as a function of cell cycle units of the WT and the constitutive ftsZ/ΔclpA double mutant strains. Values were normalized so that the maximum value of each immunoblot equals 1.C. Growth curves of LS101, UJ838, YB1585 and LS5362. The optical density (OD600) of each culture grown in M2G minimal media was monitored every 2 h for 14 h. The plot represents the average measured optical density from three independent experiments.D. Phase contrast microscopy of wild-type (LS101), UJ838, YB1585, LS5362. An aliquot of cells from each strain was placed on a M2G agarose pad and imaged at 1.25 cell cycle units (predivisional cells are shown). Black arrows point to the incipient swarmer cell. Scale bars represent 2.62 μm.E. Measured cell length (Lst + Lsw) and the relative position of the incipient division site (Lst/Lsw) for wild-type (n = 100), UJ838 (n = 102), YB1585 (n = 100) and the double mutant LS5362 (n = 105) predivisional cells from images represented in D. Error is represented as SD from the mean. The incipient swarmer cell length (Lsw) and incipient stalked cell length (Lst) are defined as the distance along the center line from either the stalked pole or the incipient flagellated pole to the site of invagination. The schematic illustrates the cell shape of wild-type and ΔclpA/Const. ftsZ double mutant.
A clpA null mutant is viable and exhibits minor cell morphology defects (Jenal and Fuchs, 1998; Grunenfelder et al., 2004). To investigate the role of ClpA with respect to the cell cycle-regulated accumulation of FtsZ, we first measured the doubling times of wild-type (LS101), ftsZ::ftsZΔC xylX::PxylX–ftsZ (YB1585), ΔclpA (UJ838), and ΔclpA ftsZ::ftsZΔC xylX::PxylX–ftsZ (LS5362) strains. The LS5362 double mutant had a 3.9-hour doubling time as compared to approximately 2.5 h for the other three strains (Fig. 5C), suggesting that simultaneous disruptrion of ftsZ transcriptional control and the ClpA protease is detrimental to Caulobacter cells.
We imaged strains LS101, UJ838, YB1585 and the double mutant strain LS5362 by phase microscopy (Fig. 5D). We measured the average lengths of predivisional cells for each strain and found that the double mutant LS5362 was roughly 23% longer than wild type (4.54 versus 3.68 μm) and roughly 10% and 20% longer than UJ838 and YB1585 (4.14 and 3.79 μm) respectively (Fig. 5D and E). To determine the relative position of the incipient division site within each predivisional cell, we calculated the ratio of the incipient stalked cell length to the incipient swarmer cell length (Lst/Lsw) for each strain and found that while the region of invagination for wild-type, UJ838 and YB1585 was biased asymmetrically towards the incipient swarmer pole (Lst/Lsw = 1.25, 1.24 and 1.22 respectively), the site of invagination for LS5362 was located centrally along the long axis of the cell (Lst/Lsw = 1.04, P < 0.0001) (Fig. 5D and E). Collectively, these observations suggest that regulation of FtsZ levels by ClpA contributes to the precise timing of FtsZ accumulation and the position of the cell division plane.
Discussion
During cytokinesis in C. crescentus, the contraction of the Z-ring complex culminates in the generation of two inner-membrane-bounded intracellular compartments followed by the subsequent invagination and closure of the outer membrane to complete cell division (Wang et al., 2001; Judd et al., 2003). Each step of this process is highly regulated at multiple levels, including temporal control of transcription of the genes encoding the divisome components (Kelly et al., 1998; Sackett et al., 1998), spatial control of division site placement through a protein gradient formed by the FtsZ polymerization inhibitor MipZ (Thanbichler and Shapiro, 2006), and the temporally ordered septal recruitment and incorporation of divisome components (Goley et al., 2011). It was previously shown that residual FtsZ and FtsA are stable in progeny stalked cells following completion of cell division but are cleared from the non-replicative daughter swarmer cell (Kelly et al., 1998; Martin et al., 2004). Here, we show that this clearance is catalysed by the AAA+ ATPases ClpX and ClpA in conjunction with the ClpP peptidase, and that FtsZ and FtsA are differentially regulated by ClpXP and ClpAP in the incipient swarmer cell upon asymmetric cell division (Fig. 6).
Fig 6.
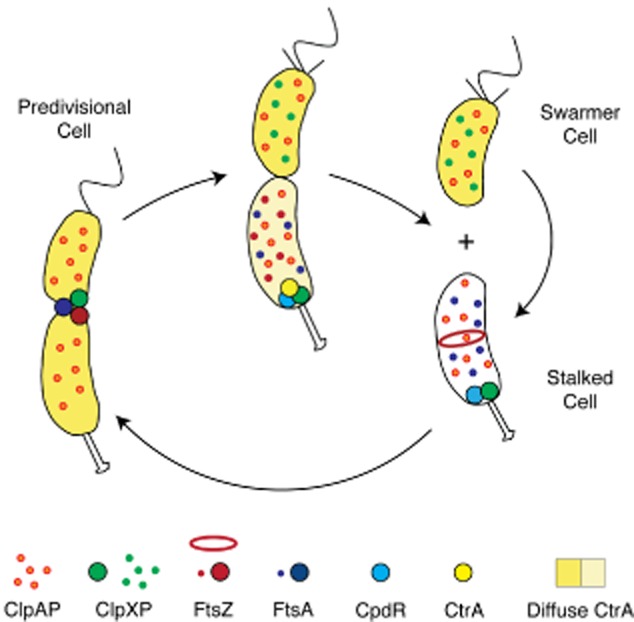
Schematic of the dynamic deployment of the ClpXP and ClpAP proteases and the FtsZ and FtsA divisome components during Caulobacter cell cycle progression. ClpXP and ClpAP are diffuse in the swarmer cell. Upon transition from a swarmer to stalked cell, the CpdR single domain response regulator localizes to the incipient stalked pole where it functions to recruit ClpXP to the cell pole. There, ClpXP degrades the polar flagellar basal body, components of the chemotaxis complex, and the CtrA inhibitor of DNA replication (Iniesta et al., 2006). As the stalked cell transitions to a predivisional cell, CpdR is degraded and ClpXP is freed from the cell pole (Iniesta and Shapiro, 2008). Near the end of the division process, ClpXP is recruited to the FtsZ ring (McGrath et al., 2006), while ClpAP remains diffuse throughout the cell cycle. Following cell division, ClpAP degrades FtsA while both ClpXP and ClpAP degrade FtsZ in the swarmer cell, but not in the daughter stalked cell.
Data presented here show that FtsZ is a substrate of both ClpAP and ClpXP in vivo and in vitro (Fig. 3). C-terminal mutations in FtsZ have a stabilizing effect in vivo, suggesting that the motif that underlies recognition of FtsZ by Clp is contained within the C-terminus of Caulobacter FtsZ; indeed, such a mechanism has been established for ClpXP-directed FtsZ degradation in E. coli (Camberg et al., 2009). In addition, we found that a fusion protein containing the membrane targeting sequence (i.e. the C-terminal 15 amino acids) of FtsA is degraded both by ClpAP and by ClpXP in vitro (Fig. 3F). This demonstrates the sufficiency of the FtsA C-terminus for protease susceptibility and raises the possibility that FtsA is also a direct substrate of ClpXP and ClpAP in the cell. However, we find that ClpXP exhibits minimal activity toward FtsA in vivo (even in the absence of ClpA), and we observe a significant contribution of the FtsA N-terminus to ClpAP-mediated proteolysis in Caulobacter (Fig. 3E). The observation that in vitro proteolysis of FtsZ and TAP–FtsAMTS does not require adaptor proteins clearly demonstrates that either protease directly interacts with both FtsZ and FtsA. However in vivo, adaptor proteins might in fact mediate the stability of FtsZ and FtsA during the course of the cell cycle. Collectively, these observations indicate distinct yet partially overlapping proteolytic regulatory programs for FtsA and FtsZ. Based on our findings, we propose that FtsZ proteolysis in vivo occurs through the combined activity of the ClpAP and ClpXP proteases via a C-terminal substrate recognition motif, and that degradation of FtsA is primarily dependent on the ClpAP protease, which recognizes both termini of the substrate.
Precise control of the levels of key cell division proteins is critical to the function of the Caulobacter divisome. In the absence of FtsZ, all other known divisome components fail to form midcell foci and division does not occur (Goley et al., 2011). Conversely, ftsZ overexpression results in gross cell division defects and cell death (Wang et al., 2001). Similarly, depleting or overexpressing FtsA (which interacts with FtsZ) causes cell morphology defects, inhibition of cell division, incomplete envelope invagination, and cell death (Martin et al., 2004). In this study, we have shown that FtsZ is still subject to cell cycle control when it is constitutively transcribed or when ClpA is absent from the cell; however, we also found that constitutive expression of ftsZ in a ΔclpA background results in the progressive accumulation of FtsZ over the cell cycle and causes impaired cell growth and a failure to divide asymmetrically. Taken together, we interpret these results to mean that robust temporal regulation of divisome dynamics is (i) essential for normal growth and division, and (ii) achieved through transcriptional and post-translational control of FtsZ. Furthermore, since the intracellular ratio of FtsZ to FtsA is tightly regulated over the course of the cell cycle (Kelly et al., 1998; Sackett et al., 1998; Quardokus et al., 2001; Martin et al., 2004), the defects in cell growth and division that we observe upon FtsZ accumulation in the ΔclpA/ftsZ-constitutive background imply a loss of optimal FtsZ/FtsA stoichiometry, which is presumably required for normal positioning of the division machinery. Therefore, the dynamic variation in the relative levels of these divisome components is not only critical for cell viability but also appears to mediate division site selection.
Although FtsZ and FtsA are both susceptible to proteolysis by ClpAP and ClpXP in vitro, our findings imply that ClpAP alone is responsible for the turnover of residual FtsA in the swarmer cell while both ClpXP and ClpAP degrade residual FtsZ in the same compartment (Fig. 4A and B). The apparent protection of FtsA from ClpXP-mediated proteolysis in vivo argues that protease activity in Caulobacter is subject to exquisite regulation. The differential activity of ClpXP in the swarmer and stalked cells stands in stark contrast to the activity of ClpXP on its CtrA substrate (Fig. 6). The CtrA master cell cycle regulator is stable in the daughter swarmer cell but rapidly degraded by the ClpXP protease in the daughter stalked cell (Jenal and Fuchs, 1998; McGrath et al., 2006). While CtrA is degraded in the daughter stalked cell, the opposite holds true for the FtsZ and FtsA divisome components. We find it noteworthy that divergent proteolytic control paradigms could be orchestrated by a single protease, and we suggest that our results establish differential control of protease activity as a fundamental determinant of asymmetric cell division in Caulobacter.
The observation that essential ClpXP substrates involved in cell cycle progression (e.g. CtrA, FtsZ and FtsA) exhibit differential stability in the daughter swarmer and stalked cells begs the question as to what regulatory mechanisms govern substrate-specific proteolytic schemes in a specific cell type. With respect to CtrA degradation, a complex pathway involving multiple polar proteins co-ordinates the localization of both ClpXP and its CtrA substrate to the incipient stalked pole (Domian et al., 1996; Quon et al., 1996; Iniesta et al., 2006; McGrath et al., 2006; Iniesta and Shapiro, 2008; Duerig et al., 2009). This pathway is governed by transcriptional regulation, by spatially controlled phospho-transfer reactions, and even by proteolysis of the localization factors to ensure the timely clearance of CtrA during the cell cycle. The FtsZ-dependent recruitment of ClpXP to the division plane (Fig. 2A and B) suggests that dynamic localization of protease activity may also underlie regulated proteolysis of FtsZ and FtsA in vivo. Although ClpX–GFP and FtsZ–mCherry appear to colocalize at the division plane in the late predivisional cell (Figs 1A and 2B), it remains formally possible that the observed colocalization in fact occurs at the swarmer side of the division plane following compartmentalization. This hypothesis is attractive insofar as it explains the cell type-specific degradation of FtsZ, and because it is supported by a previous study in which FtsZ turnover was shown to increase rapidly just prior to the separation of the two daughter cells (Kelly et al., 1998). A second mechanism driving differential proteolysis may involve other components of the division machinery, which actively compete for binding sites of ClpX substrates. In E. coli, ZipA has been shown to bind directly to the globular C-terminal region of FtsZ (Pazos et al., 2013) thereby preventing proteolysis by ClpXP. Although Caulobacter is thought to lack a ZipA homologue, FtsA has also been shown to bind to the C-terminus of FtsZ in E. coli (Ma and Margolin, 1999; Pichoff and Lutkenhaus, 2002). Since the mechanism of interaction between FtsZ and FtsA is likely to be conserved, it is possible that FtsA functions in a similar manner to ZipA, protecting FtsZ, or at least a pool of FtsZ from proteolysis until the completion of cell division.
While the underlying cause of the observed late recruitment of ClpX to the Z-ring remains unclear, it is possible that the interaction between ClpXP and FtsZ is highly sensitive to the geometry of multimeric FtsZ at the division septum. This may explain why ClpX fails to localize to the incipient division site in the absence of FtsZ and why ClpX is not immediately localized to the division plane when FtsZ synthesis is induced following several hours of depletion (Fig. 2A and B; Fig. S2A and B). Although ClpXP is believed to function as a negative regulator of FtsZ polymerization dynamics in other organisms such as B. subtilis, E. coli, and M. tuberculosis (Weart et al., 2005; Camberg et al., 2009; Dziedzic et al., 2010; Sugimoto et al., 2010), the protease does not localize to the division septum in these systems under normal growth conditions (Kain et al., 2008; Kirstein et al., 2008; Simmons et al., 2008; Dziedzic et al., 2010). Furthermore, the levels of FtsZ and FtsA in B. subtilis, E. coli and M. tuberculosis do not exhibit cell cycle variability, whereas these divisome components are subject to multiple levels of cell-cycle-dependent regulation in Caulobacter. We therefore hypothesize that the FtsZ-dependent concentration of ClpXP activity at the division plane in Caulobacter represents a novel mechanism for the destabilization and disassembly of the divisome. This model of regulated proteolysis appears to be intrinsic to the progression of the Caulobacter cell cycle and likely contributes to the spatial and temporal regulation of divisome components following cell division.
Experimental procedures
Bacterial strains, plasmids, synchronization, growth conditions
All strains and plasmids used are detailed in Tables S1 and S2. Details of strain and plasmid construction are available upon request. C. crescentus strains were grown at 28°C in peptone yeast extract (PYE) medium or M2-glucose minimal media salts (M2G). When appropriate, the growth medium was supplemented with (liquid/solid) 02% dextrose, 0.3% d-xylose, 0.5 mM vanillate (pH 7.5), 1 (5) μg ml−1 gentamicin, 5 (25) μg ml−1 kanamycin, 1 (2) μg ml−1 oxytetracyclin, 25 (100) μg ml−1 spectinomycin, and/or 5 (5) μg ml−1 streptomycin. All experiments were performed in PYE or M2G with cells kept in logarithmic phase. For small-scale synchrony, cells were grown in 20 ml of M2G with antibiotics to OD600 ∼0.3, harvested by pelleting at 6000 g, resuspended in 650 μl of ice-cold M2 salts and combined with 750 μl of Percoll (Sigma-Aldrich). The cell suspension was centrifuged for 20 min at 4°C and 10 000 g. The bottom swarmer band was isolated, washed three times in 1 ml of ice-cold M2 salts and resuspended in warm M2G media for growth at 28°C. For large-scale synchrony, cells were grown in 1 l of M2G with antibiotics to OD600 ∼0.3, harvested by pelleting at 7000 g, resuspended in 180 ml of ice-cold M2 salts and combined with 60 ml of Ludox (Sigma-Aldrich). The cell suspension was centrifuged for 30 min at 4°C and 9000 g. The bottom swarmer band was isolated, washed three times in 20 ml of ice-cold M2 salts and resuspended in warm M2G media for growth at 28°C. For the depletion strains, cells were grown in PYE medium containing 0.3% xylose, washed with plain PYE medium three times, and then resuspended in M2G medium containing 0.2% glucose or 0.3% xylose. Samples were analysed by phase-contrast and fluorescence microscopy.
Microscopy
For localization studies, 0.3% xylose or 0.5 mM vanillic acid (pH 7.5) was added to induce expression of protein fusions when indicated. For time-course experiments, cells were grown in M2G medium, synchronized, and viewed on 1.5% agarose in M2G media. Phase-contrast and fluorescence microscopy images were obtained using a Leica DM 6000 B microscope with a HCX PL APO 100×/1.40 Oil PH3 CS objective, Hamamatsu EM-CCD C9100 camera and Metamorph microscopy automation and image analysis software. Images were processed with Adobe Photoshop. To determine length ratios for nascent swarmer and stalked cell compartments of predivisional cells (Lst/Lsw), phase contrast images were segmented automatically and analysed using Microbetracker (Garner, 2011; Sliusarenko et al., 2011) and cell-length distributions were then plotted using MATLAB (ver. 2011b).
Immunoblotting and analysis
For a given experiment, equivalent OD600 units of cell lysate were separated on a 10% SDS polyacrylamide gel. Western immunoblotting was performed with standard procedures using the following primary antibody dilutions: FtsZ 1:10 000 (Mohl et al., 2001), TAP (Figge et al., 2004) 1:10 000 (Sigma Aldrich). Secondary horseradish peroxidase-conjugated goat anti-rabbit and chemiluminescent substrate was applied prior to exposure to film. Relative band intensities were quantified by densitometry using the pro Fit or ImageJ Software program. Prism was used for graphical representation of each data set. To determine the rate of turnover for each TAP fusion, the slope was obtained from a time versus relative protein level (%) semi-log plot and fit to an exponential regression line. The half-life was then calculated using ln(2)/-slope. Error is shown as ± SE for each plot.
Protein purification and in vitro degradation reactions
C. crescentus ClpX and ClpP were purified as previously described (Rood et al., 2012). ClpA* is a variant of C. crescentus ClpA lacking the final 9 residues that was shown with the E. coli ClpA orthologue to be fully active, but less prone to autodegradation (Maglica et al., 2008). Briefly, untagged ClpA* was purified via ammonium precipitation, followed by phenyl-sepharose (Sigma-Aldrich), anion-exchange (Q Fast Flow and Mono-Q; GE Healthcare), and size-exclusion (Sephacryl-200; GE Healthcare) chromatography. FtsZ was purified as previously described (Thanbichler and Shapiro, 2006). TAP–FtsA-MTS was purified using standard Ni-NTA purification (Thermo-Fisher). ClpXP and ClpAP degradation reactions were performed as previously described (Maglica et al., 2008; Rood et al., 2012). Detailed purification and reaction protocols are available upon request.
Pulse-chase experiments
Pulse-chase analysis was used to determine the stability of FtsZ and FtsA as previously described with minor modifications (Martin et al., 2004). A synchronized population of swarmer cells was supplemented with 15 μCi ml−1 l-[35S]-methionine (PerkinElmer) for 5 min and chased with 1 mM unlabelled l-methionine and 0.2 mg ml−1 casamino acids. A one ml aliquot of cells was collected every 5 min, pelleted by centrifugation at 13 000 g, frozen in liquid nitrogen and stored at −80°C. Stalked cells were obtained by allowing a pure population of swarmer cells to proceed synchronously till they differentiated into stalked cells at 50 min into the cell cycle. Then l-[35S]-methionine was added to the pure stalked cell population. For immunoprecipitation of FtsZ, a cell pellet was thawed and resuspended in 50 μl of SDS lysis buffer (10 mM Tris-HCl pH 8.0, 1% SDS, 1 mM EDTA) and boiled for 5 min at 90°C for 5 min. Cell lysate from each sample was then resuspended in 0.8 ml of immunoprecipitation wash buffer (50 mM Tris-HCl pH 8.0, 450 mM NaCl and 1% Triton X-100). To precipitate proteins that bind non-specifically, each sample was incubated with 20 μl of protein A-agarose (Gottesman et al., 1998) for 1 h at room temperature and equivalent counts of radiolabelled proteins were then used for immunoprecipitation. Each sample was incubated with 5 μl of anti-FtsZ serum at room temperature for 1 h and then with 20 μl of protein A-agarose at room temperature for 1 h after which, beads were collected by centrifugation, washed and vortexed three times with IP was buffer with one final wash in PBS buffer (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4). Beads were then resuspended in 25 μl of SDS sample buffer and boiled for 5 min at 90°C. For immunoprecipitation of FtsA, a cell pellet was thawed and resuspended in 100 μl of FtsA lysis buffer (immunoprecipitation wash buffer, 4 mg ml−1 lysozyme (Sigma-Aldrich), protease inhibitor cocktail (Complete Mini, EDTA-free protease inhibitor cocktail tablet, Roche) and placed on ice for 30 min. Cell lysate from each sample was then resuspended in 0.9 ml of immunoprecipitation wash buffer (50 mM Tris-HCl pH 8.2, 450 mM NaCl and 1% Triton X-100). To precipitate proteins that bind non-specifically, each sample was incubated with 20 μl of protein A-agarose (Gottesman et al., 1998) for 1 h at room temperature and equivalent counts of radiolabelled proteins were then used for immunoprecipitation. Each sample was incubated with 5 μl of anti-FtsA serum at 4°C for 2 h and then with 20 μl of protein A-agarose at 4°C for 30 min after which, beads were collected by centrifugation, washed and vortexed three times with IP wash buffer with one final wash in PBS buffer (137 mM NaCl, 10 mM phosphate, 2.7 mM KCl, pH 7.4). Beads were then resuspended in 30 μl of SDS sample buffer and boiled for 5 min at 90°C. The resulting samples from both FtsZ and FtsA pulses were resolved by SDS-PAGE and dried gels were exposed against a Phosphor Screen (Molecular Dynamics) for at least 20 h. Labelled bands were scanned with a Phosphorimager and quantified using ImageJ. Half-life values were calculated as mentioned above.
Acknowledgments
We thank Erin Goley and Shapiro lab members for helpful discussions and we thank Dante Ricci for critical reading of the manuscript. We thank Erin Goley and Yi-Chen Yeh for strains and Tao Long for support with microbeTracker. This work is supported in part by NIH RO1GM 32506 (Shapiro), by NIH R00GM084157 (Bhat/Chien) and by NIH Training Grant GM 07790 (Williams).
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher*s web-site.
Supporting information
References
- Alley MR, Maddock JR. Shapiro L. Polar localization of a bacterial chemoreceptor. Genes Dev. 1992;6:825–836. doi: 10.1101/gad.6.5.825. [DOI] [PubMed] [Google Scholar]
- Alley MR, Maddock JR. Shapiro L. Requirement of the carboxyl terminus of a bacterial chemoreceptor for its targeted proteolysis. Science. 1993;259:1754–1757. doi: 10.1126/science.8456303. [DOI] [PubMed] [Google Scholar]
- Bhat NH, Vass RH, Stoddard PR, Shin DK. Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol Microbiol. 2013;88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberg JL, Hoskins JR. Wickner S. ClpXP protease degrades the cytoskeletal protein, FtsZ, and modulates FtsZ polymer dynamics. Proc Natl Acad Sci USA. 2009;106:10614–10619. doi: 10.1073/pnas.0904886106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis PD. Brun YV. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol Mol Biol Rev. 2010;74:13–41. doi: 10.1128/MMBR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC. Shapiro L. The control of temporal and spatial organization during the Caulobacter cell cycle. Curr Opin Genet Dev. 1996;6:538–544. doi: 10.1016/s0959-437x(96)80081-5. [DOI] [PubMed] [Google Scholar]
- Domian IJ, Quon KC. Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, et al. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 2009;23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziedzic R, Kiran M, Plocinski P, Ziolkiewicz M, Brzostek A, Moomey M, et al. Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PLoS ONE. 2010;5:e11058. doi: 10.1371/journal.pone.0011058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Michalik S, Varming AN, Andersen JH, Albrecht D, Jelsbak L, et al. Trapping and proteomic identification of cellular substrates of the ClpP protease in Staphylococcus aureus. J Proteome Res. 2013;12:547–558. doi: 10.1021/pr300394r. [DOI] [PubMed] [Google Scholar]
- Figge RM, Divakaruni AV. Gober JW. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol Microbiol. 2004;51:1321–1332. doi: 10.1111/j.1365-2958.2003.03936.x. [DOI] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT. Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Garner EC. MicrobeTracker: quantitative image analysis designed for the smallest organisms. Mol Microbiol. 2011;80:577–579. doi: 10.1111/j.1365-2958.2011.07580.x. [DOI] [PubMed] [Google Scholar]
- Goley ED, Yeh YC, Hong SH, Fero MJ, Abeliuk E, McAdams HH. Shapiro L. Assembly of the Caulobacter cell division machine. Mol Microbiol. 2011;80:1680–1698. doi: 10.1111/j.1365-2958.2011.07677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk B. Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou Y. Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunenfelder B, Tawfilis S, Gehrig S, ØSterås M, Eglin D. Jenal U. Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF motor protein. J Bacteriol. 2004;186:4960–4971. doi: 10.1128/JB.186.15.4960-4971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA. Shapiro L. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc Natl Acad Sci USA. 2008;105:16602–16607. doi: 10.1073/pnas.0808807105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniesta AA, McGrath PT, Reisenauer A, McAdams HH. Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci USA. 2006;103:10935–10940. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U. The role of proteolysis in the Caulobacter crescentus cell cycle and development. Res Microbiol. 2009;160:687–695. doi: 10.1016/j.resmic.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Jenal U. Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd EM, Ryan KR, Moerner WE, Shapiro L. McAdams HH. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc Natl Acad Sci USA. 2003;100:8235–8240. doi: 10.1073/pnas.1433105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kain J, He GG. Losick R. Polar localization and compartmentalization of ClpP proteases during growth and sporulation in Bacillus subtilis. J Bacteriol. 2008;190:6749–6757. doi: 10.1128/JB.00589-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AJ, Sackett MJ, Din N, Quardokus E. Brun YV. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 1998;12:880–893. doi: 10.1101/gad.12.6.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirstein J, Strahl H, Moliere N, Hamoen LW. Turgay K. Localization of general and regulatory proteolysis in Bacillus subtilis cells. Mol Microbiol. 2008;70:682–694. doi: 10.1111/j.1365-2958.2008.06438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. Margolin W. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol. 1999;181:7531–7544. doi: 10.1128/jb.181.24.7531-7544.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Iniesta AA, Ryan KR, Shapiro L. McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Maglica Z, Striebel F. Weber-Ban E. An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity. J Mol Biol. 2008;384:503–511. doi: 10.1016/j.jmb.2008.09.046. [DOI] [PubMed] [Google Scholar]
- Martin ME, Trimble MJ. Brun YV. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol Microbiol. 2004;54:60–74. doi: 10.1111/j.1365-2958.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- Mohl DA, Easter J., Jr Gober JW. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol Microbiol. 2001;42:741–755. doi: 10.1046/j.1365-2958.2001.02643.x. [DOI] [PubMed] [Google Scholar]
- Pazos M, Natale P. Vicente M. A specific role for the ZipA protein in cell division: stabilization of the FtsZ protein. J Biol Chem. 2013;288:3219–3226. doi: 10.1074/jbc.M112.434944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S. Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocka I, Thein M, ØSterås M, Jenal U. Alley MR. Degradation of a Caulobacter soluble cytoplasmic chemoreceptor is ClpX dependent. J Bacteriol. 2002;184:6635–6641. doi: 10.1128/JB.184.23.6635-6641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quardokus EM, Din N. Brun YV. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol Microbiol. 2001;39:949–959. doi: 10.1046/j.1365-2958.2001.02287.x. [DOI] [PubMed] [Google Scholar]
- Quon KC, Marczynski GT. Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L. Marczynski GT. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood KL, Clark NE, Stoddard PR, Garman SC. Chien P. Adaptor-dependent degradation of a cell-cycle regulator uses a unique substrate architecture. Structure. 2012;20:1223–1232. doi: 10.1016/j.str.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackett MJ, Kelly AJ. Brun YV. Ordered expression of ftsQA and ftsZ during the Caulobacter crescentus cell cycle. Mol Microbiol. 1998;28:421–434. doi: 10.1046/j.1365-2958.1998.00753.x. [DOI] [PubMed] [Google Scholar]
- Simmons LA, Grossman AD. Walker GC. Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis. J Bacteriol. 2008;190:6758–6768. doi: 10.1128/JB.00590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T. Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto S, Yamanaka K, Nishikori S, Miyagi A, Ando T. Ogura T. AAA+ chaperone ClpX regulates dynamics of prokaryotic cytoskeletal protein FtsZ. J Biol Chem. 2010;285:6648–6657. doi: 10.1074/jbc.M109.080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M. Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Thompson MW. Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- Tsai JW. Alley MR. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jones BD. Brun YV. A set of ftsZ mutants blocked at different stages of cell division in Caulobacter. Mol Microbiol. 2001;40:347–360. doi: 10.1046/j.1365-2958.2001.02395.x. [DOI] [PubMed] [Google Scholar]
- Weart RB, Nakano S, Lane BE, Zuber P. Levin PA. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol Microbiol. 2005;57:238–249. doi: 10.1111/j.1365-2958.2005.04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information