Abstract
Summary
Giardia duodenalis is a common intestinal parasite in humans and a wide range of livestock species. It is a genetically heterogeneous parasite that has been characterized in seven distinct genetic assemblages or cryptic species, and molecular markers can be used to differentiate both animal-specific and potentially zoonotic genotypes. Little is known about G. duodenalis and the range of assemblages occurring in domestic livestock species in the UK. Here, we present data on the occurrence and molecular diversity of G. duodenalis detected in the faeces or large intestinal contents of cattle, sheep, pigs, goats and camelids from farms in the north-west of England. Both healthy and clinically diseased animals were included in the survey. The presence of Giardia spp. and assemblages was determined by sequencing of the small-subunit ribosomal RNA gene. The potential association of infection with various clinical and epidemiological parameters was studied in cattle using both univariate and multivariate analyses. Giardia spp. were detected in 127 (34.3%) of the 370 animals tested. G. duodenalis assemblage E was found to be predominant in cattle and sheep, followed by assemblage A. Mixed infections with assemblages A and E were also detected. Interestingly, some cattle, sheep and pigs were found to be infected with more unexpected assemblages (C, D, F). Pre-weaned calves were more likely to test positive than adult animals, but no association between the occurrence of overt intestinal disease and G. duodenalis infection was detected. The common occurrence of assemblage A and the finding of unusual assemblages in atypical hosts suggest that in future, a multilocus analysis should be used to confirm the actual diversity of G. duodenalis in livestock and the presence of potentially zoonotic genotypes. These data also suggest that there is a need to re-evaluate the clinical significance of G. duodenalis infection in livestock.
Keywords: Giardia duodenalis, Giardia intestinalis, livestock, assemblages diversity, molecular typing, UK
Introduction
The genus Giardia includes some of the most common intestinal flagellates of vertebrates. These parasites have a wide host range, including mammals, birds and amphibians. Amongst the six currently accepted species of Giardia, G. duodenalis (syn. intestinalis/lamblia) has the broadest host range and is the species with the greatest public and animal health significance in terms of gastrointestinal disease. G. duodenalis is detected frequently in many mammals (Feng and Xiao, 2011) and is one of the most common intestinal parasites in pets like dogs (Batchelor et al., 2008) and in livestock (Thompson et al., 2008). The reported prevalence rates in production animals vary considerably, both geographically and according to the different diagnostic methods employed (Feng and Xiao, 2011). In cattle, a prevalence of between 6.6% in New Zealand (Learmonth et al., 2003) and up to 57.8% in Canada and Australia (O'Handley et al., 2000) has been reported. A cumulative parasite prevalence of 73% to 100% in both dairy and beef calves has been observed in North America (Xiao and Herd, 1994; Ralston et al., 2003), and also a farm prevalence as high as 96% has been recently reported in Canada (Dixon et al., 2011). Several studies have demonstrated that Giardia infections in cattle tend to occur more commonly towards the end of the neonatal period, and they are often chronic in nature (O'Handley and Olson, 2006). A higher prevalence of Giardia in pre (40%)- and post-weaned (52%) calves compared with adult cattle (∼27%) has been reported (Trout et al., 2005, 2007). In sheep and goats, Giardia infections are also frequently observed (Taylor et al., 1993; Geurden et al., 2008a; Robertson et al., 2010). On the other hand, few studies have assessed the prevalence of Giardia in pigs (Maddox-Hyttel et al., 2006; Armson et al., 2009).
Whilst G. duodenalis is recognized as a common cause of intestinal disease in humans, evidence that this parasite also causes significant disease in livestock is less convincing. It has been suggested that the multifactorial aetiology of diarrhoea complicates the interpretation of the relationship between Giardia infection and clinical disease (O'Handley and Olson, 2006). Nevertheless, Giardia infection alone or in combination with other enteric parasites has been associated with diarrhoea in young calves (O'Handley et al., 1999). The presence of morphological alterations in the intestinal epithelium of scouring calves with giardiasis has been shown (Barigye et al., 2008), and a significant improvement in weight gain was observed in calves that were treated for giardiasis with fenbendazole compared with untreated infected animals (Geurden et al., 2010a). Malabsorption, reduced feed efficiency and severe weight loss were also observed in infected lambs during a confirmed Giardia outbreak (Aloisio et al., 2006). These findings suggest that Giardia infection can have detrimental effects on livestock productivity; however, specific information relating to clinical outcomes and risk factors for giardiasis in livestock is lacking.
Giardia duodenalis is a complex of seven distinct genotypic assemblages (A to G) or cryptic species (Monis et al., 2003) that can only be distinguished by PCR amplification and sequencing of appropriate genes. These groupings show no morphological variation, but they differ in several other traits, including in vitro and in vivo growth rates, drug sensitivity, host infectivity and genome content (Thompson and Monis, 2012). Some assemblages show a rather wide host range, whereas others appear to be restricted to particular families or species of animals. Humans appear to be exclusively infected with the assemblages A and B, but these two groups are also widely reported in dogs, cats, livestock and also wild mammals (Feng and Xiao, 2011). Other groups are thought to be more host-specific, with assemblage E occurring in livestock, C and D in canids and F and G in cats and rodents, respectively (Cacciò and Ryan, 2008). In livestock, the ‘hoofed livestock’-specific assemblage E is the most common assemblage found in cattle, sheep, goats and pigs (Castro-Hermida et al., 2007; Trout et al., 2007; Geurden et al., 2008a; Armson et al., 2009), followed by genotypes belonging to assemblage A (Geurden et al., 2008b; Sprong et al., 2009). Assemblage B has also been reported to sporadically infect ruminants (Coklin et al., 2007; Dixon et al., 2011). The detection of genotypes belonging to the assemblages A and B in these species is potentially very important when evaluating the role of livestock in the possible zoonotic transmission of this parasite (Khan et al., 2011).
Given both the potential for G. duodenalis to exert a pathogenic effect on production animals and the potential for livestock to harbour zoonotic genotypes of this parasite, it is surprising that only one study has been published so far on the occurrence of G. duodenalis assemblages in livestock in the UK (Geurden et al., 2012). However, this study involved only a limited number of calves, and nothing is known about the occurrence of this parasite in farm animals other than cattle in the UK with the exception of a report on infection in lambs (Taylor et al., 1993). In this article, we present the results of a study of the occurrence and molecular diversity of Giardia spp. in faeces and large intestine content samples collected from 370 livestock from farms in north-west England. The assemblages of G. duodenalis were determined by PCR amplification and sequencing of the small-subunit ribosomal RNA (SSU rRNA) gene.
Materials and Methods
Samples and data collection
Samples from farm animals submitted to the Animal Health and Veterinary Laboratories Agency (AHVLA) Regional Laboratory in Preston, Lancashire, between September 2007 and June 2008 were included in this study. Samples tested consisted of surplus material (i.e. large intestine contents or faeces) submitted by private veterinary surgeons to AHVLA Preston either for disease diagnosis (the majority of samples) or for monitoring purposes and faeces collected during diagnostic post-mortem examinations at AHVLA Preston. All samples or animals submitted during the study period were eligible for the survey. The majority of specimens came from the area served by the Preston Regional Laboratory (i.e. counties of Lancashire and Cumbria) (n = 261, 70.5%) and Cheshire (n = 44, 11.9%). Samples submitted from other counties including Yorkshire (n = 27, 7.3%), Greater Manchester (n = 16, 4.3%), Isle of Man, Merseyside and Staffordshire (n = 15, 4%) and Derbyshire and Clwyd (n = 7, 1.9%) were also included. Where available, details of the animal gender, age, production system, housing and presenting clinical signs were recorded using a standard submission form completed by the private veterinary surgeon and/or by AHVLA veterinary investigation officers. The actual consistency of faeces (graded as normal, soft or diarrhoeic) was recorded at the time of sample examination. Also, in the case of samples/carcasses submitted for diagnostic purposes, the final disease diagnosis for each animal was recorded, based on examination protocols and diagnostic criteria adopted by AHVLA and as recorded in its VIDA database (Hall et al., 1980). Samples were not tested for G. duodenalis infection by AHVLA – Preston. For those samples where tests for other enteropathogens were carried out, the results of these other examinations were also recorded. Approximately 2 g of large intestine contents and/or faeces from each animal submission was collected into individual screw cap plastic vials, stored at 4°C and forwarded in iced parcels to the Department of Infection Biology, Institute of Infection and Global Health, University of Liverpool, on a monthly basis. Here, they were stored at 4°C before DNA extraction.
DNA extraction and Giardia spp. genotyping
Faecal samples underwent molecular characterization to detect the presence of Giardia spp. and to determine the species and assemblage of the parasite. Total genomic DNA was extracted directly from samples using the QIAamp® DNA Stool Mini Kit (QIAGEN, Manchester, UK) as per manufacturer's instructions, with only minor modification. Specifically, purified genomic DNA was eluted in 100 μl of elution buffer (instead of 200 μl, as recommended by the manufacturer). A 292-bp fragment of the SSU rRNA gene was then amplified by PCR using the forward primer RH11 (5′-CATCCGGTCGATCCTGCC-3′) and the reverse primer RH4 (5′-AGTCGAACCCTGATTCTCCGCCAGG-3′) (Hopkins et al., 1997). PCRs were carried out in 20 μl volumes, with the final mix containing 1× Buffer (containing 18 mm Tris–HCl, 4.4 mm (NH4)2SO4, 1.8 mm MgCl2, 452 μg BSA and 1.76 μm EDTA), 400 μm of each dNTP, 10 pmol of each primer, 2.5 units of Taq DNA polymerase (QIAGEN) and 1–5 μl of purified genomic DNA. The reactions were performed in a DNA Engine Dyad® Peltier Thermal Cycler, with an initial denaturation step at 94°C for 3 min, a set of 35 cycles (each consisting of 20 s at 94°C, 20 s at 59°C and 30 s at 72°C) and a final extension at 72°C for 10 min. To determine the amplified fragments, 5 μl of PCR products was electrophoresed on a 1% agarose gel stained with ethidium bromide. A positive (G. duodenalis isolate WB C6 genomic DNA) and negative (PCR water) control samples were included in each PCR.
PCR products were purified using the QIAquick® PCR Purification kit (QIAGEN) as per the manufacturer's instructions and sent for sequencing to either Geneservice, Nottingham, or the DNA Sequencing Core service, Molecular Biology Support Unit at Cardiff University. All products were sequenced in both directions using the RH11 and RH4 primers, and obtained sequences were edited using BioEdit (version 7.0.9). To determine the species and/or the assemblage of Giardia, sequences were aligned with ClustalW with the following reference sequences from each assemblage: assemblage A [GenBank: AF199446], B [GenBank: AF199447], C [GenBank: AF199449], D [Genbank: AF199443], E [Genbank: AF199448] and F [Genbank: AF199444].
Statistical analyses
All statistical analyses were performed with PASW® Statistics 18 (IBM, Armonk, NY, USA), with the level of statistical significance set at 95% (P < 0.05). The potential association of each tested variable with Giardia infection was examined in cattle. Univariate analysis was used to examine potential associations of each variable with Giardia infection using a chi-squared test, and wherever appropriate, odds ratios (OR) with 95% confidence intervals (CI) were calculated using Giardia-negative animals as controls. Variables with P < 0.2 were then included into a multivariate logistic regression model to test the association of infection with the various risk factors. Interactions between risk factors were included as well. The final model was fitted using a stepwise method with variables significant at P < 0.05 retained in the final model. Model fit was assessed using the Hosmer–Lemeshow goodness-of-fit test.
Results
Giardia spp. prevalence and Giardia duodenalis assemblages
Of the 370 samples tested by PCR, 127 (34.3%) gave a positive result for Giardia spp. Of the 127 PCR-positive samples, 91 (71.6%) were successfully sequenced, confirming infection with G. duodenalis assemblages. No other species of Giardia were identified. The prevalence of Giardia infection in the six species surveyed and the distribution of the assemblages in the whole sample are reported in Table 1 and Fig. 1, respectively. Figure2 shows the occurrence of the assemblages in the two most represented species in our sample. In general, G. duodenalis assemblage E was the most common type found (57.1% of the samples), followed by assemblage A (22%) and mixed infections with both assemblages A and E (7.7%). The predominance of the assemblages A and E was observed in both cattle and sheep. Unexpectedly, the canine-specific assemblage C was found in a pig and two cattle and also mixed with assemblage A in the latter. Furthermore, assemblage D (alone or mixed with A) was detected in a few cattle and a ewe. Interestingly, two pigs were found harbouring the feline-specific assemblage F.
Table 1.
PCR prevalence of Giardia spp. in 370 farm animal faecal/large intestine content samples
| Animal species (no. samples tested) | No. PCR-positive for Giardia spp. (%) | No. with SSU rRNA sequence |
|---|---|---|
| Cattle (n = 283) | 93 (32.9) | 63 |
| Sheep (n = 64) | 28 (43.7) | 24 |
| Pig (n = 7) | 4 (57.1) | 3 |
| Goat (n = 9) | 1 (11.1) | – |
| Alpaca (n = 6) | 1 (16.7) | 1 |
| Llama (n = 1) | 0 (0) | – |
| Total | 127 | 91 |
Fig 1.
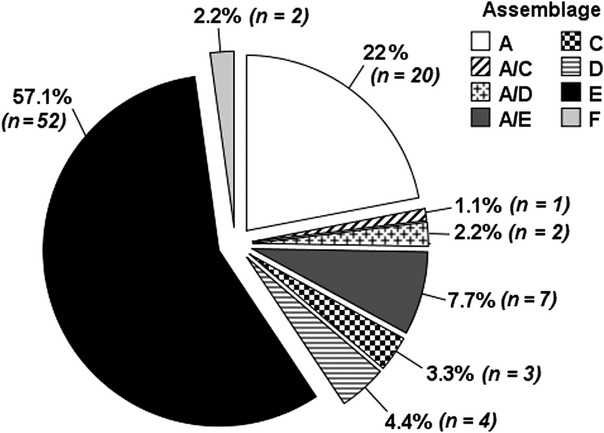
Distribution of G. duodenalis assemblages in 91 farm animal samples genotyped at the SSU rRNA locus.
Fig 2.
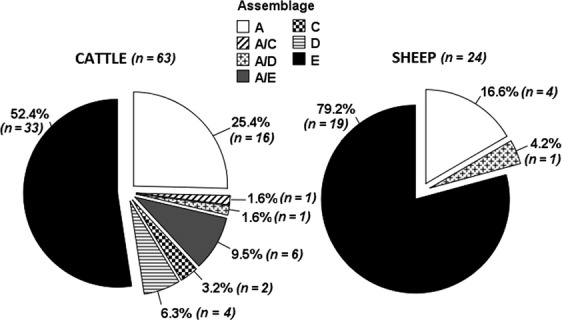
Distribution of G. duodenalis assemblages in cattle and sheep samples genotyped at the SSU rRNA locus.
Risk factors for infection in cattle
The potential association between G. duodenalis infection and parameters relating to age, housing, production type, clinical presentation and final disease diagnosis was examined specifically for cattle. The prevalence of infection varied according to the age group, with the highest prevalence of infection was seen in pre-weaned animals (43.6%), followed by neonatal calves (31.8%), post-weaned animals (26.1%) and adult cattle (23%). The risk of G. duodenalis infection was significantly higher only in pre-weaned calves (univariate: OR 2.59, 1.42–4.73 95% CI, P = 0.002).
Prevalence of G. duodenalis infection did not vary significantly according to production system, with an infection prevalence of 39% recorded in cattle belonging to suckler systems, and an infection prevalence of 31.1% in cattle from non-suckler (dairy, calf rearer or beef fattener) enterprises. Although the prevalence of infection in housed animals (32.9%, 46 positives of 140) was higher than that in cattle that were kept outdoors or in mixed accommodation (22%, 11 positives of 50), this difference was not statistically significant (P = 0.150).
There was no significant difference in the prevalence of infection between cattle submissions with a reported clinical history of diarrhoea (infection prevalence 32.9%, 54 positives of 164) and cattle submissions that had no clinical history of diarrhoea reported (26.8%, 26 positives of 97). The consistency of the faeces, determined in the laboratory as normal, soft or diarrhoeic, was also not associated with an increased risk of G. duodenalis infection (P > 0.05). Finally, both undiagnosed digestive disease and the diagnosis of another enteric infection showed no statistical association with infection (P > 0.05).
Age group was the only significant variable, which could be retained in the final multivariate model for factors associated with G. duodenalis occurrence in cattle (Table 2). To control for the large variation in weaning age between suckler cattle and dairy, calf rearer and beef finisher enterprises, a variable relating to management systems was also forced into the model, but inclusion of this production-related variable had little effect on odds ratios or statistical association recorded in univariate analysis. Pre-weaned animals were significantly more likely to be infected with the parasite.
Table 2.
Multivariate analysis of variables significantly associated with the detection of Giardia in cattle from north-west England
| No. positive/total (prevalence) (%) | Odds ratio | 95% CI | P-value | |
|---|---|---|---|---|
| Age group | ||||
| Adult | 32/139 (23) | Baseline | ||
| Post-weaned | 6/23 (26.1) | 1.18 | 0.43–3.25 | 0.754 |
| Pre-weaned | 34/78 (43.6) | 2.59 | 1.42–4.73 | 0.002 |
| Neonatal | 7/22 (31.8) | 1.54 | 0.56–4.30 | 0.404 |
| Purpose | ||||
| Dairy, calf rearer or beef fattener | 69/222 (31.1) | Baseline | ||
| Suckler | 23/59 (39) | 1.34 | 0.67–2.65 | 0.407 |
Hosmer–Lemeshow goodness-of-fit test P = 0.2671.
Discussion
Giardia spp. was detected by PCR in 34% of animals sampled in this survey, and G. duodenalis was the sole Giardia species identified using molecular sequencing. The infection prevalence varied between different host species, with the highest observed in sheep (43.7% of samples positive) and pigs (57.1% positive), although relatively small numbers of samples from these two species were examined. In cattle, which represented the majority of samples examined, the prevalence of G. duodenalis infection was nearly 33%. Our study was not a formally structured random epidemiological survey, and many of the samples came from diseased animals submitted for diagnostic purposes (although not specifically for gastrointestinal disease), so our figures may therefore not reflect the true population prevalence. Nevertheless, these results show that G. duodenalis is a common intestinal parasite of cattle of all ages and also of other farm animals in the area under study. In cattle in this survey, the occurrence of infection with G. duodenalis was higher in immature cattle than in adult animals. The highest risk of infection was seen in the pre-weaned age group, and this was independent of the production system (dairy or suckler enterprise). In the UK, the age of calves at weaning varies widely between production systems; whilst it is usual practice to wean dairy calves from 6 to 10 weeks of age, suckler calves are commonly weaned much later at around 6–10 months of age. The age-related prevalence of infection is in accordance with the work of others, who have also demonstrated significantly higher infection prevalence in young calves compared with older animals (Trout et al., 2005; O'Handley and Olson, 2006; Santín et al., 2009).
Differences in management, including differences in animals stocking density, hygiene regimes or water supply, could have an important effect on the exposure of cattle to intestinal parasites like Giardia. The prevalence of Cryptosporidium infection was reported to vary according to production system in England and Wales (Featherstone et al., 2010), with suckler calves at lower risk of infection than those of dairy, calf-rearing or beef-fattening enterprises. Although it was not possible to test the effect of housing in their study, animal management practices such as stocking density of similar age animals were proposed as possible explanations for the variation in prevalence. However, no statistical association between G. duodenalis infection and housing was detected in our study. There is relatively little information about the effect of farm management on the occurrence of G. duodenalis, and the results are not consistent. A 3-fold increase in the risk of infection was reported in housed cattle in Canada (Ruest et al., 1998), and the authors attributed this to the fact that the parasite cysts survival and infectivity may be reduced outdoors compared with a closed environment. In a study of cattle in Denmark, the type of calf housing did not influence the prevalence of Giardia and Cryptosporidium, but maternity pens were proposed as an indirect source of infection (Maddox-Hyttel et al., 2006). In a large multicentre study across Europe, calves that were kept in contact with the dam were found to be significantly more at risk of infection (Geurden et al., 2012). Conversely, in the same study, disinfection of the calves housing emerged as a protective factor against G. duodenalis infection.
There was no association between G. duodenalis and digestive disease in cattle in our sample. Neither unformed faeces at the testing stage nor presenting with diarrhoea as one of the clinical signs were found to be significantly associated with the presence of the parasite. It is certainly plausible that soft/diarrhoeic faeces and diarrhoea are associated with G. duodenalis infection, and this has been demonstrated in cattle and sheep in both natural and experimental infections (Geurden et al., 2010b). The lack of significance in our survey could be related to the correlation with other variables, most notably the occurrence of co-infections with other enteropathogens causing similar symptoms. Despite reports of the pathogenic potential of G. duodenalis in livestock (Castro-Hermida et al., 2005; Aloisio et al., 2006; Geurden et al., 2010b), many laboratories do not routinely test livestock samples for evidence of this particular infection. However, despite the lack of clinical significance of giardiasis shown in our study, the possibility of this infection should perhaps be considered as part of the differential diagnosis in livestock with digestive system disease.
Genotyping at the SSU rRNA locus confirmed the predominance of G. duodenalis assemblage E genotypes in cattle and sheep. Also assemblage A genotypes were frequently found in this survey, and mixed infections with assemblages A and E were reported in cattle and an alpaca. These findings are in accordance with other reports on G. duodenalis genetic diversity in cattle across the world (Geurden et al., 2008b; Sprong et al., 2009), including the UK (Geurden et al., 2012). The common occurrence of assemblage A isolates observed in our study warrants further investigation due to the zoonotic potential of genotypes within this assemblage. The SSU rRNA gene is particularly suitable for screening large numbers of samples as it is both easy to amplify because of its multicopy nature and shows no cross-amplification with the DNA of other organisms (Hopkins et al., 1997). However, this locus is too conserved to detect variability at the subassemblage level (Wielinga and Thompson, 2007), and the use of more variable markers (such as beta-giardin, triose phosphate isomerase or glutamate dehydrogenase genes) is needed to identify the presence of homologous human and animal, and therefore potentially zoonotic, genotypes (Sprong et al., 2009).
Unexpectedly, the diversity of G. duodenalis appeared to be greater than that anticipated in our livestock samples, with the supposedly canine-specific assemblages C and/or D being found in some cattle, a pig and sheep. These two assemblages have never been reported from cattle or sheep before. Similarly to our study, assemblage D was also found in two pigs from Denmark (Langkjaer et al., 2007), and a goat isolate was identified as assemblage C in Australia as well (Ng et al., 2011). Furthermore, in our survey, two pig isolates were also typed as the feline-specific assemblage F, similar to a report of infection in two animals in Australia (Armson et al., 2009). In all the above reports, as in this study, molecular typing involved solely the use of the SSU rRNA locus. The detection of unusual and animal-specific assemblages in human faecal samples has sometimes been associated with the use of this marker (Feng and Xiao, 2011). Because in our survey it was not possible to confirm the assemblage at other genetic loci, the occurrence of these unusual assemblages must be interpreted with caution. An alternative possible explanation for the finding of canine and feline assemblages in atypical hosts is that these animals may simply act as mechanical vectors by ingesting parasite cysts of dog or cat origin. The use of a multilocus sequence typing approach is recommended in future studies to confirm the assignment of an isolate to a particular assemblage and to detect potentially zoonotic subgenotypes of G. duodenalis in the area. Nevertheless, recent reports showed the occurrence of supposedly host-specific assemblages in a variety of different hosts, including mixed infections with the E and C assemblages in pet chinchillas (Levecke et al., 2011), and also assemblage C was isolated from a human (Soliman et al., 2011). Our results along with the aforementioned reports suggest that the actual host range of G. duodenalis assemblages should be re-evaluated in future studies involving farm animals using a multilocus approach.
Acknowledgments
The authors thank colleagues at AHVLA Preston for their assistance with this survey, and in particular, R Cutler for retrieval of epidemiological and VIDA data. CM is supported by a studentship from the UK Health Protection Agency; WT is grateful to the Government of Thailand for funding of his studentship; SML was supported by ESRC; JMW is grateful to the support of Novartis Animal Health for funding NR.
References
- Aloisio F, Filippini G, Antenucci P, Lepri E, Pezzotti G, Cacciò SM. Pozio E. Severe weight loss in lambs infected with Giardia duodenalis assemblage B. Vet. Parasitol. 2006;142:154–158. doi: 10.1016/j.vetpar.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armson A, Yang R, Thompson J, Johnson J, Reid S. Ryan UM. Giardia genotypes in pigs in Western Australia: prevalence and association with diarrhoea. Exp. Parasitol. 2009;121:381–383. doi: 10.1016/j.exppara.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Barigye R, Dyer NW, Newell TK, Khaitsa ML, Trout JM, Santín M. Fayer R. Molecular and immunohistochemical detection of assemblage E, Giardia duodenalis in scouring North Dakota calves. Vet. Parasitol. 2008;157:196–202. doi: 10.1016/j.vetpar.2008.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor DJ, Tzannes S, Graham PA, Wastling JM, Pinchbeck GL. German AJ. Detection of endoparasites with zoonotic potential in dogs with gastrointestinal disease in the UK. Transbound. Emerg. Dis. 2008;55:99–104. doi: 10.1111/j.1865-1682.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- Cacciò SM. Ryan U. Molecular epidemiology of giardiasis. Mol. Biochem. Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Castro-Hermida JA, Delafosse A, Pors I, Ares-Mazas E. Chartier C. Giardia duodenalis and Cryptosporidium parvum infections in adult goats and their implications for neonatal kids. Vet. Rec. 2005;157:623–627. doi: 10.1136/vr.157.20.623. [DOI] [PubMed] [Google Scholar]
- Castro-Hermida JA, Mezo M. González-Warleta M. Natural infection by Cryptosporidium parvum and Giardia duodenalis in sheep and goats in Galicia (NW Spain) Small Ruminant Res. 2007;72:96–100. [Google Scholar]
- Coklin T, Dixon B, Parrington L. Farber J. Prevalence and molecular characterization of Giardia duodenalis and Cryptosporidium spp. in dairy cattle in Ontario, Canada. Vet. Parasitol. 2007;150:297–305. doi: 10.1016/j.vetpar.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Dixon B, Parrington L, Cook A, Pintar K, Pollari F, Kelton D. Farber J. The potential for zoonotic transmission of Giardia duodenalis and Cryptosporidium spp. from beef and dairy cattle in Ontario, Canada. Vet. Parasitol. 2011;175:20–26. doi: 10.1016/j.vetpar.2010.09.032. [DOI] [PubMed] [Google Scholar]
- Featherstone CA, Giles M, Marshall JA, Mawhinney IC, Holliman A. Pritchard GC. Cryptosporidium species in calves submitted for post-mortem examination in England and Wales. Vet. Rec. 2010;167:979–980. doi: 10.1136/vr.c3948. [DOI] [PubMed] [Google Scholar]
- Feng Y. Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurden T, Thomas P, Casaert S, Vercruysse J. Claerebout E. Prevalence and molecular characterisation of Cryptosporidium and Giardia in lambs and goat kids in Belgium. Vet. Parasitol. 2008a;155:142–145. doi: 10.1016/j.vetpar.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Geurden T, Geldhof P, Levecke B, Martens C, Berkvens D, Casaert S, Vercruysse J. Claerebout E. Mixed Giardia duodenalis assemblage A and E infections in calves. Int. J. Parasitol. 2008b;38:259–264. doi: 10.1016/j.ijpara.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Geurden T, Vandenhoute E, Pohle H, Casaert S, De Wilde N, Vercruysse J. Claerebout E. The effect of a fenbendazole treatment on cyst excretion and weight gain in calves experimentally infected with Giardia duodenalis. Vet. Parasitol. 2010a;169:18–23. doi: 10.1016/j.vetpar.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Geurden T, Vercruysse J. Claerebout E. Is Giardia a significant pathogen in production animals? Exp. Parasitol. 2010b;124:98–106. doi: 10.1016/j.exppara.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Geurden T, Vanderstichel R, Pohle H, Ehsan A, von Samson-Himmelstjerna G, Morgan ER, Camuset P, Capelli G, Vercruysse J. Claerebout E. A multicentre prevalence study in Europe on Giardia duodenalis in calves, with molecular identification and risk factor analysis. Vet. Parasitol. 2012;190(3–4):383–390. doi: 10.1016/j.vetpar.2012.06.039. [DOI] [PubMed] [Google Scholar]
- Hall SA, Dawson PS. Davies G. VIDA II: a computerised diagnostic recording system for veterinary investigation centres in Great Britain. Vet. Rec. 1980;106:260–264. doi: 10.1136/vr.106.12.260. [DOI] [PubMed] [Google Scholar]
- Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA. Thompson RC. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J. Parasitol. 1997;83:44–51. [PubMed] [Google Scholar]
- Khan SM, Debnath C, Pramanik AK, Xiao L, Nozaki T. Ganguly S. Molecular evidence for zoonotic transmission of Giardia duodenalis among dairy farm workers in West Bengal, India. Vet. Parasitol. 2011;178:342–345. doi: 10.1016/j.vetpar.2011.01.029. [DOI] [PubMed] [Google Scholar]
- Langkjaer RB, Vigre H, Enemark HL. Maddox-Hyttel C. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology. 2007;134:339–350. doi: 10.1017/S0031182006001533. [DOI] [PubMed] [Google Scholar]
- Learmonth JJ, Ionas G, Pita AB. Cowie RS. Identification and genetic characterisation of Giardia and Cryptosporidium strains in humans and dairy cattle in the Waikato Region of New Zealand. Water Sci. Technol. 2003;47:21–26. [PubMed] [Google Scholar]
- Levecke B, Meulemans L, Dalemans T, Casaert S, Claerebout E. Geurden T. Mixed Giardia duodenalis assemblage A, B, C and E infections in pet chinchillas (Chinchilla lanigera) in Flanders (Belgium) Vet. Parasitol. 2011;177:166–170. doi: 10.1016/j.vetpar.2010.11.027. [DOI] [PubMed] [Google Scholar]
- Maddox-Hyttel C, Langkjær RB, Enemark HL. Vigre H. Cryptosporidium and Giardia in different age groups of Danish cattle and pigs - Occurrence and management associated risk factors. Vet. Parasitol. 2006;141:48–59. doi: 10.1016/j.vetpar.2006.04.032. [DOI] [PubMed] [Google Scholar]
- Monis PT, Andrews RH, Mayrhofer G. Ey PL. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect Genet. Evol. 2003;3:29–38. doi: 10.1016/s1567-1348(02)00149-1. [DOI] [PubMed] [Google Scholar]
- Ng J, Yang R, Vicky V, Cox P. Ryan U. Identification of zoonotic Cryptosporidium and Giardia genotypes infecting animals in Sydney's water catchments. Exp. Parasitol. 2011;128:138–144. doi: 10.1016/j.exppara.2011.02.013. [DOI] [PubMed] [Google Scholar]
- O'Handley RM. Olson ME. Giardiasis and cryptosporidiosis in ruminants. Vet. Clin. North. Am. Food. Anim. Pract. 2006;22:623–643. doi: 10.1016/j.cvfa.2006.07.002. [DOI] [PubMed] [Google Scholar]
- O'Handley RM, Cockwill C, McAllister TA, Jelinski M, Morck DW. Olson ME. Duration of naturally acquired giardiosis and cryptosporidiosis in dairy calves and their association with diarrhoea. J. Am. Vet. Med. Assoc. 1999;214:391–396. [PubMed] [Google Scholar]
- O'Handley RM, Olson ME, Fraser D, Adams P. Thompson RC. Prevalence and genotypic characterisation of Giardia in dairy calves from Western Australia and Western Canada. Vet. Parasitol. 2000;90:193–200. doi: 10.1016/s0304-4017(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Ralston BJ, McAllister TA. Olson ME. Prevalence and infection pattern of naturally acquired giardiasis and cryptosporidiosis in range beef calves and their dams. Vet. Parasitol. 2003;114:113–122. doi: 10.1016/s0304-4017(03)00134-1. [DOI] [PubMed] [Google Scholar]
- Robertson LJ, Gjerde BK. Hansen EF. The zoonotic potential of Giardia and Cryptosporidium in Norwegian sheep: a longitudinal investigation of 6 flocks of lambs. Vet. Parasitol. 2010;171:140–145. doi: 10.1016/j.vetpar.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Ruest N, Faubert GM. Couture Y. Prevalence and geographical distribution of Giardia spp. and Cryptosporidium spp. in dairy farms in Quebec. Can. Vet. J. 1998;39:697–700. [PMC free article] [PubMed] [Google Scholar]
- Santín M, Trout JM. Fayer R. A longitudinal study of Giardia duodenalis genotypes in dairy cows from birth to 2 years of age. Vet. Parasitol. 2009;162:40–45. doi: 10.1016/j.vetpar.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Soliman RH, Fuentes I. Rubio JM. Identification of a novel assemblage B subgenotype and a zoonotic assemblage in human isolates of Giardia intestinalis in Egypt. Parasitol. Int. 2011;60:507–511. doi: 10.1016/j.parint.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Sprong H, Cacciò SM. van der Giessen JWB. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl. Trop. Dis. 2009;3:1–12. doi: 10.1371/journal.pntd.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MA, Catchpole J, Marshall RN. Green J. Giardiasis in lambs at pasture. Vet. Rec. 1993;133:131–133. doi: 10.1136/vr.133.6.131. [DOI] [PubMed] [Google Scholar]
- Thompson RCA. Monis PT. Giardia – from genome to proteome. Adv. Parasitol. 2012;78:57–95. doi: 10.1016/B978-0-12-394303-3.00003-7. [DOI] [PubMed] [Google Scholar]
- Thompson RCA, O'Handley R. Palmer CS. The public health and clinical significance of Giardia and Cryptosporidium in domestic animals. Vet. J. 2008;177:18–25. doi: 10.1016/j.tvjl.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trout JM, Santín M, Greiner E. Fayer R. Prevalence and genotypes of Giardia duodenalis in post-weaned dairy calves. Vet. Parasitol. 2005;130:177–183. doi: 10.1016/j.vetpar.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Trout JM, Fayer R. Santín M. Prevalence of Giardia duodenalis genotypes in adult dairy cows. Vet. Parasitol. 2007;147:205–209. doi: 10.1016/j.vetpar.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Wielinga CM. Thompson RCA. Comparative evaluation of Giardia duodenalis sequence data. Parasitology. 2007;134:1795–1821. doi: 10.1017/S0031182007003071. [DOI] [PubMed] [Google Scholar]
- Xiao LH. Herd RP. Infection patterns of Cryptosporidium and Giardia in calves. Vet. Parasitol. 1994;155:257–262. doi: 10.1016/0304-4017(93)00645-f. [DOI] [PubMed] [Google Scholar]


