Abstract
The degradation of plant biomass by saprophytes is an ecologically important part of the global carbon cycle, which has also inspired a vast diversity of industrial enzyme applications. The xyloglucans (XyGs) constitute a family of ubiquitous and abundant plant cell wall polysaccharides, yet the enzymology of XyG saccharification is poorly studied. Here, we present the identification and molecular characterization of a complex genetic locus that is required for xyloglucan utilization by the model saprophyte Cellvibrio japonicus. In harness, transcriptomics, reverse genetics, enzyme kinetics, and structural biology indicate that the encoded cohort of an α-xylosidase, a β-galactosidase, and an α-l-fucosidase is specifically adapted for efficient, concerted saccharification of dicot (fucogalacto)xyloglucan oligosaccharides following import into the periplasm via an associated TonB-dependent receptor. The data support a biological model of xyloglucan degradation by C. japonicus with striking similarities – and notable differences – to the complex polysaccharide utilization loci of the Bacteroidetes.
Introduction
The saccharification of diverse types of plant biomass is both an ecologically important part of the global carbon cycle and a biotechnologically relevant aspect of the food, feed, biofuel, biomaterials, and cleaning-product industries. Hydrolysis of the complex plant cell wall to acquire sugars for growth presents a significant challenge for microorganisms, and requires a wide range of enzyme activities to address the tremendous diversity of glycosidic, peptide, polyphenolic, and ester linkages present (Carpita and McCann, 2000; Jovanovic et al., 2009). Whereas the complete hydrolysis of cellulose microfibres is difficult due to their semi-crystalline nature, the monosaccharide and linkage complexity of the hemicelluloses and pectins additionally confounds enzymatic attack. Moreover, the intertwining of plant cell wall biomolecules into a composite material provides an additional level of structural complexity mitigating biomass saccharification, and there is growing evidence that addition of hemicellulases to industrial enzyme cocktails can improve efficacy (Hu et al., 2013; Jabbour et al., 2013). As such, carbohydrate-active enzyme (CAZyme) discovery remains a vibrant area of research (Jovanovic et al., 2009; Mewis et al., 2013; Hemsworth et al., 2014; del Pulgar and Saadeddin, 2014).
Cellvibrio japonicus (previously Pseudomonas fluorescens subsp. cellulosa) is a Gram-negative bacterium that was first isolated from Japanese soil in the 1950s and has since become intensely studied due to its ability to degrade all common plant cell wall polysaccharides (Hazlewood and Gilbert, 1998; Deboy et al., 2008). As a model saprophytic organism, the complete genome sequence of C. japonicus has been determined and genetic tools have been established (Deboy et al., 2008; Gardner and Keating, 2012). C. japonicus is thus important both for CAZyme discovery and for fundamental molecular studies of polysaccharide degradation by Gram-negative environmental bacteria. Furthermore, recent metabolic engineering of a strain with the ability to produce ethanol has demonstrated the potential of C. japonicus for industrial chemical production in consolidated bioprocessing (Gardner and Keating, 2010). Whereas the mannan-, xylan-, and arabinan-degrading systems have been intensively studied (Cartmell et al., 2008; 2011,; Deboy et al., 2008; Emami et al., 2009 and references therein], the capacity of C. japonicus to degrade the ubiquitous hemicellulose family of xyloglucans has received little attention.
The xyloglucans (XyG) are a structurally complex family of polysaccharides found in all terrestrial plants (Carpita and McCann, 2000; Popper et al., 2011). In dicots alone, XyGs may account for 20–25% of primary cell wall dry weight (Scheller and Ulvskov, 2010) and, as such, constitute an important terrestrial carbon sink. XyGs are composed of a cellulose-like linear β(1→4)-glucan backbone appended with branching α(1→6)-xylosyl moieties, which can in turn be extended with additional glycosides (e.g. galacto-, fuco- and arabinosyl moieties) in a tissue- and species-dependent manner (Peña et al., 2008; Tuomivaara et al., 2014). Thus, the complete saccharification of XyGs necessitates a cohort of enzymes to address the linkage diversity present in individual XyG variants. Aligned with our continuing interest in xyloglucan-active enzymes (Gilbert et al., 2008; Ariza et al., 2011; Mark et al., 2011; Eklöf et al., 2012; 2013,; Larsbrink et al., 2014), we previously reported the biochemical and structural characterization of the main α-xylosidase of C. japonicus, CjXyl31A, which specifically hydrolyses terminal non-reducing-end xylose moieties of xyloglucan oligosaccharides (XyGOs) (Larsbrink et al., 2011). In the same study, we also identified that C. japonicus produces at least one extracellular endo-xyloglucanase capable of generating XyGOs by xyloglucan backbone cleavage. However, a key remaining question is, which are the other players in C. japonicus that might work in concert with these enzymes to fully deconstruct complex xyloglucans?
Here, we report the identification and molecular characterization of a unique xyloglucan utilization locus (XyGUL) in the genome of C. japonicus (Fig. 1). In addition to the α-xylosidase CjXyl31A, this locus contains a predicted β-galactosidase, a predicted α-l-fucosidase, and a predicted TonB-dependent receptor (Ferguson and Deisenhofer, 2002; Koebnik, 2005), whose collective presence indicates a role in dicot (fucogalacto)xyloglucan degradation and uptake. Using a combination of transcriptional analysis, reverse genetics, subcellular protein localization, recombinant enzyme kinetics, and structural biology, we were able to verify these predictions of glycoside function and extend a biological model of XyG utilization by this important saprophyte.
Figure 1.
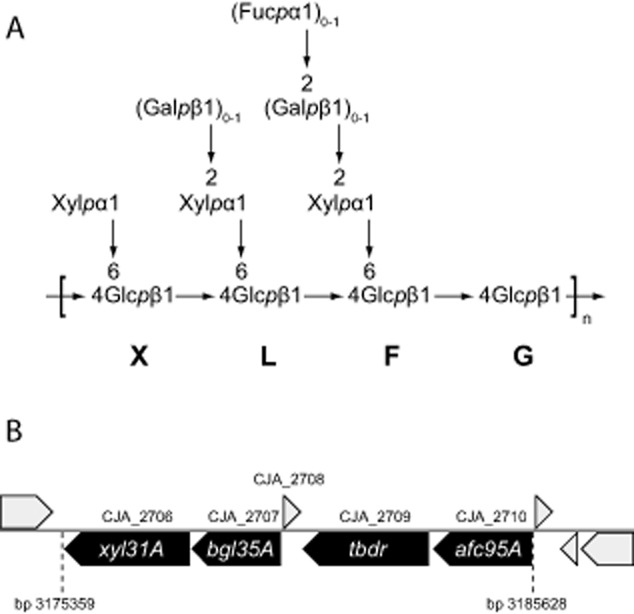
Xyloglucan structure and the arrangement of xyloglucan-degrading genes within the C. japonicus genome.
A. Typical (fucogalacto)xyloglucan structure, with abbreviated motif nomenclature according to Tuomivaara et al. (2014) indicated.
B. The xyloglucan utilization locus (XyGUL) in the genome of C. japonicus; hypothetical proteins are shown in grey.
Results
Genome walking reveals a putative xyloglucan utilization locus (XyGUL) in C. japonicus
Genome walking in silico revealed the presence of open reading frames encoding a predicted β-galactosidase from glycoside hydrolase (GH) family 35 (CjBgl35A, encoded by locus tag CJA_2707, bgl35A), a predicted TonB-dependent receptor (TBDR, encoded by CJA_2709), and a predicted α-l-fucosidase from glycoside hydrolase family 95 (CjAfc95A, encoded by CJA_2710, afc95A) upstream from the gene encoding the known α-xylosidase CjXyl31A (CJA_2706, xyl31A) (Fig. 1). Protein and gene names used here correspond to those proposed in Table S2 of Deboy et al. (2008). The predicted protein sequences of both CjBgl35A and CjAfc95A were compared to previously characterized enzymes in GH35 and GH95, respectively, using blast. Interestingly, CjBgl35A showed a high homology only to the Xanthomonas campestris pv. campestris GalD β-galactosidase, with a protein identity of 55% and similarity of 70% (over 536 residues), while similarity to other characterized GH35 enzymes catalogued in the CAZy database (Lombard et al., 2014; http://www.cazy.org/GH35_characterized.html) was poor. Moreover, the β-galactosidase activity of GalD has been indicated primarily through genetic studies and minimal specificity data exists for this enzyme (Yang et al., 2007), such that no reliable prediction of substrate specificity for CjBgl35A can be made. CjAfc95A, in contrast, had high similarity to all five characterized α(1→2)-l-fucosidases of GH95 (Lombard et al., 2014; http://www.cazy.org/GH95_characterized.html), with identities of 31–35% and similarities of 47–52%. In particular, the two plant enzymes (from Arabidopsis thaliana and Lilium longiflorum) and the fungal enzyme (from Aspergillus nidulans) have been shown to cleave the α(1→2)-fucosyl residues from XyGOs (Bauer et al., 2006; Ishimizu et al., 2007; Nagae et al., 2007; Leonard et al., 2008). On the basis of these bioinformatics analyses, we hypothesized that the GHs were involved in the cleavage of β(1→2)-galactosyl and α(1→2)-l-fucosyl residues of dicot (fucogalacto)xyloglucan, respectively, while the TBDR may function in carbohydrate import (Dejean et al., 2013).
CjBgl35A and CjAfc95A are XyG-specific glycosidases
To test predictions of the catalytic specificity of the GHs, we performed initial substrate screens on aryl glycosides (Glcp/Galp/Xylp-β-PNP, l-Araf-α-PNP and l-Fuc-α-CNP) with both enzymes produced recombinantly in Escherichia coli. To further investigate the potential roles of the enzymes in vivo, xyloglucan polysaccharides (XyG) and oligosaccharides (XyGOs) from tamarind and iceberg lettuce were examined as natural substrates, including product analysis by HPAEC-PAD and MALDI-TOF. Full kinetic data for both enzymes are listed in Table 1.
Table 1.
Activity of CjBgl35A and CjAfc95A on various substrates
| Enzyme | Substrate | kcat (s−1) | Km (mM) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| CjBgl35A | Gal-β-PNP | 10.1 ± 0.2 | 0.81 ± 0.05 | 12.5 |
| XLLG | 3.8 ± 0.08 | |||
| Tamarind xyloglucana | 0.0031 ± (6.5 × 10−5) | |||
| CjAfc95A | l-Fuc-α-CNP | 26.5 ± 1.1 | 2.1 ± 0.2 | 12.9 |
| XLFG | 19.2 ± 0.5 | 0.21 ± 0.017 | 89.8 | |
| Lettuce xyloglucanb | 1.85 ± 0.084 | 0.26 ± 0.03 | 7.1 |
Kinetic parameters calculated from the available galactosyl moieties of the substrate (16%) as reported by the manufacturer.
Kinetic parameters calculated from the available fucosyl moieties of the substrate, as determined by end-point hydrolysis by CjAfc95A.
CjBgl35A
CjBgl35A was able to hydrolyse Gal-β-PNP but did not act upon any of the other substrates in the initial screen. The pH optimum of the enzyme was determined to be pH 5.5 in acetate buffer (Supplemental Fig. S1), but the pH-rate profile deviated from the bell-shaped profile expected for an enzyme mechanism involving two ionizing residues. The kcat/Km value for the reaction on Gal-β-PNP was 12.5 s−1 mM−1, which is in the same range as another bacterial GH35 β-galactosidase, BgaC from Streptococcus pneumonia (49.3 s−1 mM−1) and the A. thaliana BGAL4 (19.1 s−1 mM−1) (Ahn et al., 2007; Cheng et al., 2012). Hydrolysis of the substrate lactose, which is the preferred substrate for many enzymes in GH35, could not be observed; CjBgl35A is thus likely not able to generally address β(1→4)-linkages.
In contrast, HPAEC-PAD indicated that the enzyme released galactose from both tamarind seed (galacto)xyloglucan and the doubly galactosylated XLLG nonasaccharide (see Fig. 1 for XyGO nomenclature). Enzyme saturation could not be reached for either substrate (Supplemental Fig. S2). Notably, a nasturtium (Tropaeolum majus) β-galactosidase also exhibited a strictly linear dependence of rate on substrate concentration using either nasturtium or tamarind xyloglucan as substrates (Edwards et al., 1988) (it is likely, but not conclusive, that this is a GH35 enzyme: GenBank CAW88932). Apparent kcat/Km values were nonetheless calculated by linear regression of the initial velocities versus substrate concentration plot. Notably, the apparent kcat/Km value for the degalactosylation of tamarind xyloglucan is c. 1000-fold lower than that for XLLG (Table 1). This difference, estimated from the total galactose content of the polysaccharide, is likely to represent a lower limit, given the possibility of contaminating galactosylated oligosaccharides (acting as alternate substrates) below our limits of detection and the clear preference of CjBgl35A for one of the two galactosyl residues (vide infra). Regardless, the data indicate that the galactosidase is most likely to act subsequent to the hydrolysis of xyloglucan polysaccharide chains into component oligosaccharides.
Product analysis by MALDI-TOF MS showed that CjBgl35A was able to hydrolyse both galactosyl moieties from XLLG to form XXXG (Fig. 2A). A time-course study using HPAEC-PAD revealed an order of preference for the galactosyl residues on the two L (Gal–Xyl–Glc) moieties: XXLG was produced essentially exclusively during the initial stages of the reaction, with XXXG appearing only when a majority of the XLLG had been converted into XXLG. Given this preference, the initial-rate the kinetic data (Table 1, Supplemental Fig. S2) likely reflect the hydrolysis of the galactosyl moiety most distal from the reducing-end (underlined: XLLG). To assess whether the enzyme could cleave galactosyl moieties underlying a terminal fucosyl moiety, a 1:2 mixture of the oligosaccharides XXFG and XLFG (Fig. 1A) was prepared from fucosylated XyG extracted from iceberg lettuce by digestion with a Bacteroides ovatus GH5 endo-xyloglucanase (Larsbrink et al., 2014) and size-exclusion chromatography. Extended incubation of a high concentration of CjBgl35A with this mixture (10 μM enzyme, c. 0.5 mM substrate, 16 h, 25°C) followed by MALDI-TOF MS demonstrated that only XLFG was hydrolysed, yielding XXFG, which was not further transformed (Fig. 2C). The data indicate that CjBgl35A acts strictly as an exo-galactosidase that requires terminal galactosyl moieties on XyGOs.
Figure 2.
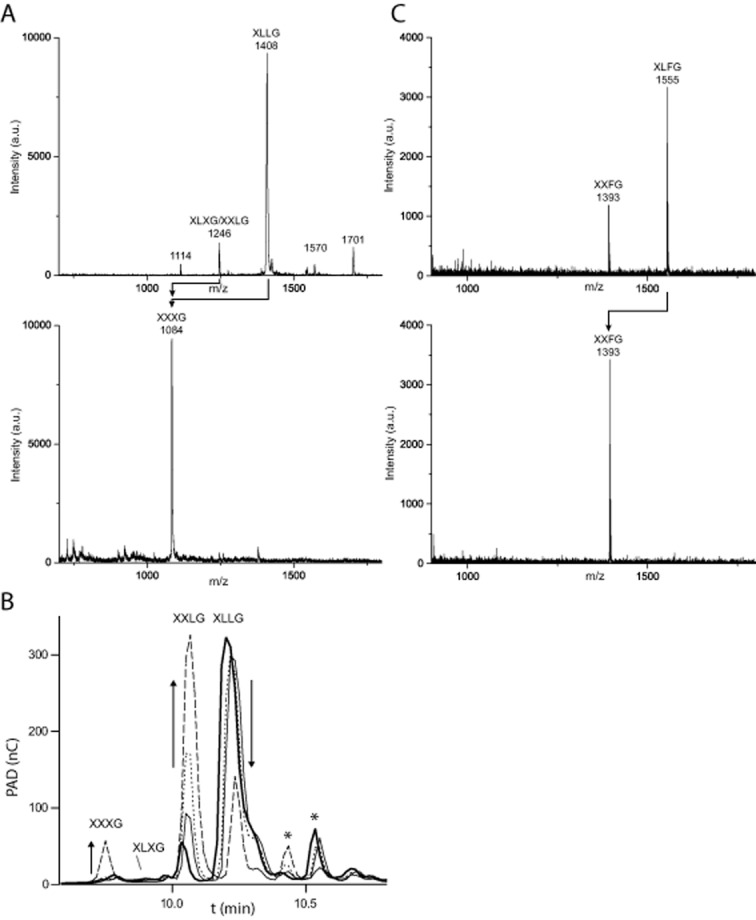
Analysis of products from CjBgl35A activity on galactosylated xyloglucan oligosaccharides.
A. MALDI-TOF spectra of a preparation of XLLG containing a minor amount of XXLG/XLXG before and after incubation with CjBgl35A, indicating complete hydrolysis of both galactosyl residues. Observed m/z values are consistent with [M + Na]+ adducts. Minor peaks at m/z 1570 and 1701 correspond to unidentified Hex7Pen3 and Hex7Pen4 oligosaccharides (Tuomivaara et al., 2014).
B. HPAEC-PAD chromatograms of a time-course study of the conversion of XLLG into XXXG via XXLG as a primary product. Arrows indicate trends in peak intensity changes for the starting material and major products. XLLG (bold line) was incubated with CjBgl35A and samples were analysed after 5 min (thin line), 20 min (dotted line) and 2 h (dashed line). Formation of XLXG is not observed, while XXXG appears only after the majority of the XLLG has been converted into XXLG, revealing that the galactosyl moiety closest to the non-reducing end (cf. Fig. 1) is the preferred substrate for the enzyme. Asterisks indicate minor components, which may correspond to unidentified oligosaccharides observed by MALDI-TOF MS.
C. MALDI-TOF spectra of the hydrolysis of a preparation containing XLFG and XXFG by CjBgl35A. XLFG is completely converted into XXFG while the ‘F’ unit is unchanged, demonstrating that fucosylation blocks the strictly exo-acting galactosidase. Observed m/z values are consistent with [M+Na]+ adducts.
At present, no other bacterial enzymes from GH35 have been assayed for XyG specificity, to our knowledge, so direct comparisons with CjBgl35A are difficult to make. However, many of these are specific for lactose, a substrate not hydrolysed by CjBgl35A, which suggests distinct activity profiles. Rather, the specificity of CjBgl35A is similar to plant β-galactosidases with a general ability to remove side-chain galactosyl units from xyloglucan polysaccharides; the GH35 member from A. thaliana, Bgal10 (At5g63810) produces XXLG and XXXG sequentially due to a preference for the underlined side-chain in XLLG (Sampedro et al., 2012 and references therein).
CjAfc95A
CjAfc95A did not cleave any of the four PNP glycosides tested (vide supra), but readily hydrolysed l-Fuc-α-CNP. Using this substrate, the pH-rate profile was classically bell-shaped with a pH optimum of pH 6.5 in citrate buffer (Supplemental Fig. S3). The observed kcat/Km value for CjAfc95A (12.9 mM−1 s−1; Table 1 and Fig. S4) compares favourably with that of a Bifidobacterium longum subsp. infantis GH95 member (0.12 mM−1 s−1) (Sela et al., 2012). On natural substrates, product analysis by HPAEC-PAD and MALDI-TOF (Fig. 3) indicated that XLFG and XXFG were converted to XLLG and XXLG respectively. This specificity is similar to characterized GH95 α-l-fucosidases from the plants A. thaliana and L. longiflorum, both of which are active on fucosylated XyGOs (Ishimizu et al., 2007; Leonard et al., 2008).
Figure 3.
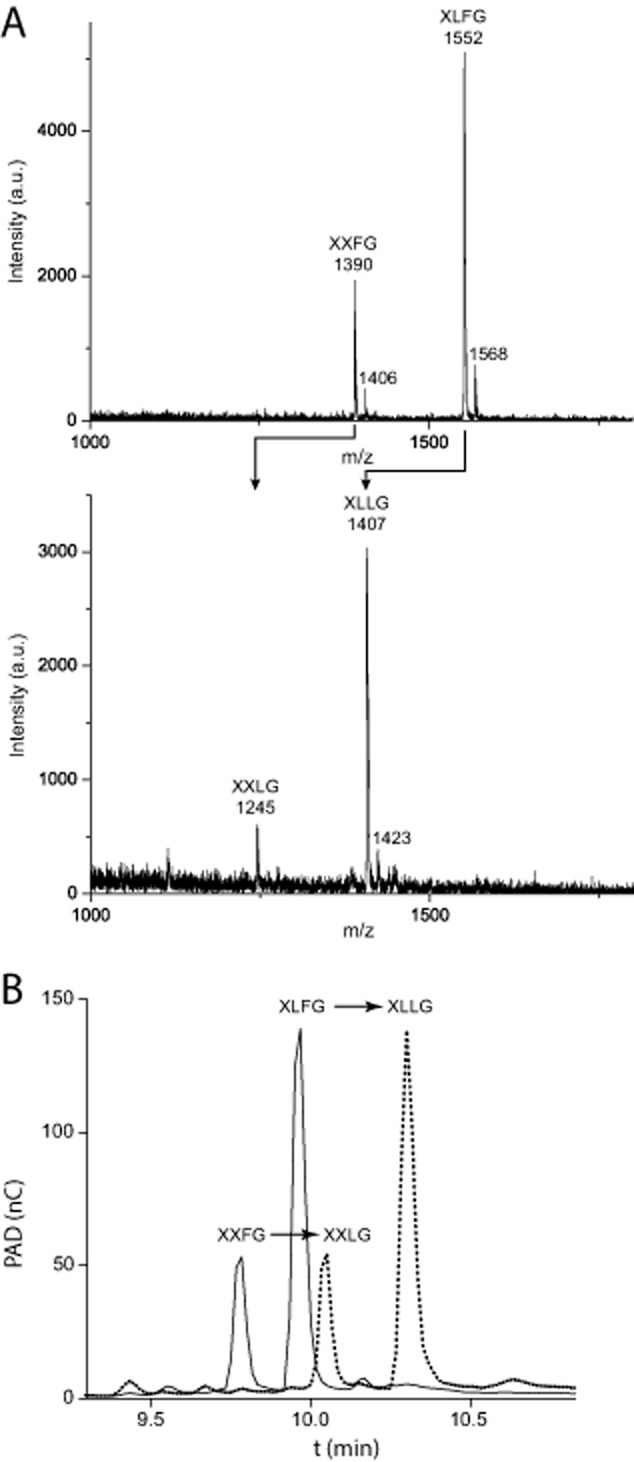
Analysis of products from CjAfc95A activity on fucosylated xyloglucan oligosaccharides.
A. MALDI-TOF spectra of the hydrolysis of a preparation containing XLFG and XXFG into XLLG and XXLG, respectively, after incubation with CjAfc95A.
B. HPAEC-PAD chromatograms of the same conversion. Observed m/z values are consistent with [M+Na]+ adducts; minor amounts of [M+K]+ adducts are also observed.
The release of fucose from both the polysaccharide and the mixture of fucosylated XyGOs was also quantified by HPAEC-PAD (Table 1). Although the kinetics are confounded by the two-component XyGO mixture, the relative apparent kcat/Km value for the oligosaccharides was sevenfold higher than that for l-Fuc-α-CNP, due to a 10-fold lower apparent Km value. This suggests that carbohydrate-binding interactions in the positive enzyme subsites significantly enhance catalysis (for subsite nomenclature see Davies et al., 1997). Indeed, the intrinsic leaving-group ability of 2-chloro-4-nitrophenol [pKa 5.45 (Ibatullin et al., 2008)] is potentially 10 orders of magnitude greater than a sugar hydroxyl group [pKa c. 16 (Damude et al., 1996)]. Similar strong effects of extended XyGO binding in positive enzyme subsites are exhibited by the α-xylosidase in the locus (Larsbrink et al., 2011; Silipo et al., 2012).
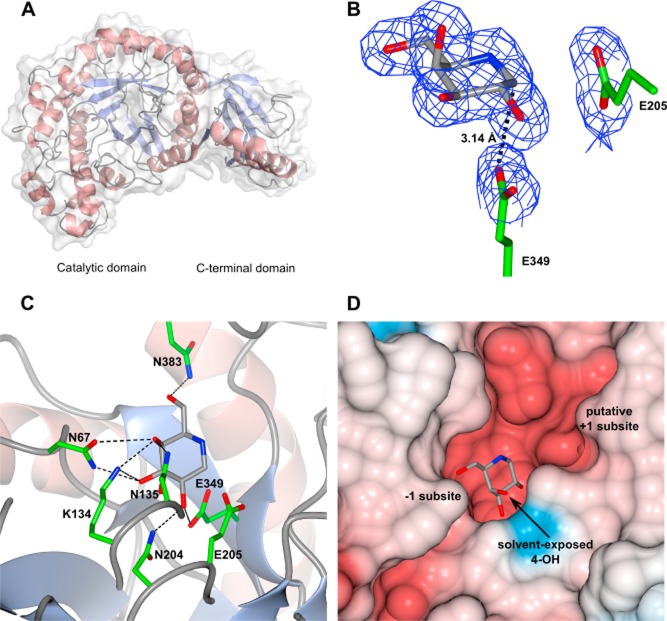
The tertiary structure of CjBgl35A reveals the molecular basis for substrate recognition
To complement our structure–function analysis of CjXyl31A (Larsbrink et al., 2011; Silipo et al., 2012), we wished to complete the tertiary structural characterization of the remaining GHs encoded by the putative C. japonicus XyGUL to reveal the determinants of xyloglucan recognition. Whereas CjBgl35A has proven amenable to crystallography, CjAfc95A has thus far resisted our attempts, despite extensive effort.
The three-dimensional structure of CjBgl35A reveals a relatively unusual two-domain architecture comprising an N-terminal catalytic (β/α)8 (TIM) barrel domain (residues 37–419), appended to a smaller, C-terminal, mixed α/β structure (residues 420–575) consisting of two short α helices and nine β strands (Fig. 4 and Supplemental Table S2; PDB ID 4D1I). Given its poor sequence similarity to other GH35 enzymes (vide supra), this two-domain fold exhibits low homology to other currently known protein structures; only one significant structural match to another bacterial GH35 β-galactosidase, from Caulobacter crescentus strain CB15, was found. Analysis using the Dali server (Holm and Rosenström, 2010) shows Bgl35A and the C. crescentus enzyme (PDB accession code 3U7V) to have a Z-score of 61.5, an r.m.s.d. value of 1.2 Å mapped across 508 Cα positions, and a sequence identity of 56%. The next most-similar protein (GH5 endo-β-mannanase, PDB code 1QNO) has a Z-score of 25.1, an r.m.s.d. value of 3.0 Å across 289 Cα positions, and a sequence identity of just 12%.
Figure 4.
Crystallography of CjBgl35A.
A. The tertiary structure viewed along the barrel axis of the N-terminal catalytic domain. The mixed α/β motif of the C-terminal domain is indicated, while a transparent surface overlay illustrates the rigid nature of the domain interface.
B. 1-Deoxygalactonojirimycin (DGJ) bound in the active site (carbon atoms in grey); the proposed catalytic residues, E349 and E205, are also illustrated (carbon atoms in green). The pseudoanomeric position of the ligand can be observed approximately 3.14 Å from the carboxylate group of E349, the likely catalytic nucleophile. Electron density is depicted as a maximum-likelihood weighted 2Fo−Fc synthesis contoured at 0.29 e Å−3 (1.0 σ).
C. Additional residues in the −1 subsite interacting with GDJ.
D. Surface representation, coloured according to electrostatic potential, showing the position of DGJ within the −1 subsite. A further cavity immediately adjacent to the pseudoanomeric position of DGJ is suggested to be the +1 subsite, accommodating the branched oligosaccharide substrates within the active-site cleft.
Panel A was assembled using PyMOL version 1.3r1 (Schrödinger, LLC), while panels B–D were created using CCP4Mg (McNicholas et al., 2011).
The (β/α)8 structure of the CjBgl35A N-terminal catalytic domain reveals a centrally positioned cleft running laterally across the open end of the barrel. This topology appears to be consistent with the requirement for binding of extended, branched oligo/polysaccharide substrates such as XyGOs and/or XyG and thus the cleft was anticipated to contain the catalytic active site. Subsequent soaking of native Bgl35A crystals with the iminosugar 1-deoxygalactonojirimycin [DGJ; 1,5-Dideoxy-1,5-imino-d-galactitol (Paulsen et al., 1980)] confirmed both the location of the active centre and, importantly, unveiled crucial ligand-protein interactions including the identity of the likely catalytic amino acids (Fig. 4B and C, and Supplemental Table S2; PDB ID 4D1J). The Kd for this interaction in solution was determined to be 485 ± 42 nM by ITC (Fig. S5).
In the complex structure, DGJ is located within a deep cavity, approximately at the centre of the proposed active-site cleft. DGJ is co-ordinated in this −1 subsite by a complex hydrogen-bonding network mediated by the side-chain groups of residues N67 (interacting with O3), K134 (O3 and O4), N135 (O4), N204 (O2), and N383 (O6) (Fig. 4C). The side-chains of E205 and E349 occupy classical positions expected for the catalytic pair, comprising a proton donor-acceptor and nucleophile consistent with the retaining mechanism expected for all GH35 members (Fig. 4B). Furthermore, the position of the catalytic proton donor-acceptor residue is consistent with an anti-protonation trajectory (Heightman and Vasella, 1999).
A pocket positioned directly adjacent to the pseudoanomeric position of DGJ is proposed to be the +1 catalytic subsite that is responsible for binding side-chain xylosyl moieties present on XyG/XyGOs. Curiously, the apparent solvent-exposed nature of the non-reducing end of the ligand suggests the possibility to accommodate extended substrates, since the axial 4-OH position of the six-membered ring points directly out into the large cleft (Fig. 4D). However, in fucosylated XyGOs such as XXFG and XLFG, the pendant fucosyl residue is α(1→2)-linked to the galactosyl residue (Fig. 1). Steric considerations (Fig. 4D) therefore explain the requirement for CjAfc95A to first remove the terminal Fuc from such XyGOs before the underlying Gal can be addressed by CjBgl35A (Fig. 2).
Despite the observation of separate protein domains (Fig. 4A), it seems likely that these together solely constitute a catalytic module, i.e. a distinct biochemical/biological function is not indicated. Structure/sequence homology searching of the C-terminal region in isolation produced no significant results that would indicate similarity to known regulatory or carbohydrate-binding modules (CBMs), despite the observation of a trough-like surface somewhat reminiscent of a substrate recognition cleft (Gilbert et al., 2013). In addition, the observation that the early part of the C-terminal domain is formed from a distortion of the final helix of the barrel domain, and that the initial β-strand of this domain stacks directly against the penultimate barrel helix, strongly suggests a rigid, monolithic structure.
Genetic analysis defines the biological importance of the XyGUL
With a firm understanding of the determinants of substrate recognition by the glycoside hydrolases CjXyl31A (Larsbrink et al., 2011), CjBgl35A, and CjAfc95A, we sought to confirm that the putative XyGUL indeed constituted a co-ordinately regulated locus. Further, we were interested to determine whether this multi-gene locus was exclusively responsible for XyG saccharification by C. japonicus.
Transcriptional analysis
Quantitative PCR (qPCR) was performed on all four predicted XyGUL genes (Fig. 1; xyl31A, bgl35A, tbdr, afc95A) to monitor possible upregulation during growth on minimal medium containing tamarind XyGOs versus a glucose control. Additional predicted β-galactosidases and α-l-fucosidases found in the C. japonicus genome (Deboy et al., 2008) (bgl2A, bgl2B, bgl2C and afc95B), but not in the putative XyGUL, were also included to determine their potential involvement in XyG deconstruction. Of these, the predicted β-galactosidase gene, bgl2B, and the other predicted α-l-fucosidase gene, afc95B, showed the same level of expression in both growth conditions, and were therefore used as reference genes. The four XyGUL genes were all clearly upregulated 12- to 45-fold when C. japonicus was grown in M9-XyGO medium (Fig. 5A).
Figure 5.
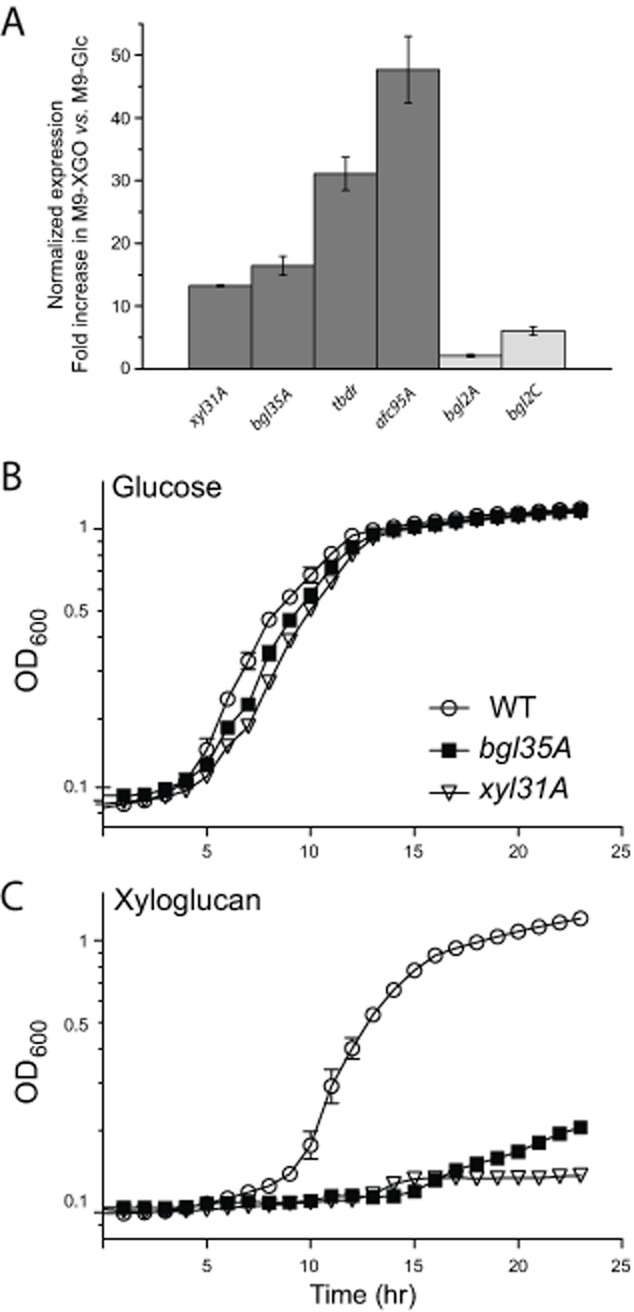
Transcriptomic and reverse genetic analysis of the C. japonicus xyloglucan utilization locus (XyGUL).
A. Normalized increase in expression of the locus genes (cf. Fig. 1) in cells grown on M9-XyGO medium versus to M9-Glc medium, compared with other predicted galactosidase-encoding (bgl2A, bgl2B, bgl2C) and fucosidase-encoding (afc95B) genes (Deboy et al., 2008). bgl2B and afc95B had identical expression in both growth conditions and therefore served as reference genes. Error bars indicate the standard error of the mean.
B and C. Growth analysis of C. japonicus wild-type (WT) and bgl35A and xyl31A knockout mutant strains in MOPS minimal medium with either 0.25% glucose (B) or 0.25% xyloglucan (C) as sole carbon sources. All growth experiments were performed in biological triplicate; error bars represent the standard deviation of the mean (in many cases the error bars are smaller than the data point marker).
The predicted β-galactosidase genes bgl2A and bgl2C, which are not in the putative XyGUL were also slightly (< 5-fold) upregulated in the M9-XyGO cultures (Fig. 5A). However, genome analysis suggests that these genes are unlikely to be purposely involved in XyG catabolism and may instead be responding to a general sensing signal due to galactose released by the XyGUL enzymes. For example, bgl2A (CJA_0496) is located together with a predicted endo-1,4-galactanase, gal53A-2 (CJA_0497) and a TonB-dependent receptor (CJA_0498), and might instead be primarily involved in (arabino)galactan metabolism. bgl2C (CJA_2610) is co-located with a predicted chitinase, chi18D (CJA_2611), which may point towards a principal function related to fungal cell wall or insect carapace degradation.
To assay whether any of the locus genes were transcribed as a polycistronic mRNA, PCR was performed in an attempt to amplify the regions between each of the genes in the cDNA, using the primers from the qPCR study. The xyl31A and bgl35A genes were found to be co-transcribed on one bicistronic mRNA strand (data not shown). This observation is supported by the similar expression levels of these two genes indicated by qPCR (Fig. 5A) and the distance between the genes (41 bases), which is unlikely to accommodate a promoter region. Similar analyses of the bgl35A/tbdr and tbdr/afc95A pairs suggested that the TonB-dependent receptor gene and afc95A are transcribed individually. The non-coding regions directly upstream of these genes are 88 and 199 bases in length respectively, and are therefore large enough to contain individual promoters.
Reverse genetics of xyl31A and bgl35A
To assess physiological function, xyl31A and bgl35A gene-disruption mutants were generated, confirmed by PCR (Fig. S6), and assayed for growth on tamarind xyloglucan. Inactivation of xyl31A mutation results in a severe growth defect, with a growth rate of 0.06 h−1 (peak OD600 = 0.13) on xyloglucan, compared to 0.41 h−1 (peak OD600 = 1.2) of the wild-type strain (Fig. 5B and C). As expected from its bicistronic relationship with xyl31A, disruption of bgl35A likewise results in a severe growth defect (growth rate 0.07 h−1, peak OD600 = 0.21). Both mutants grow in a wild-type manner on the polysaccharides xylan (from beechwood), carboxymethyl cellulose, and pectin (from apple).
Subcellular localization of XyGUL function
We previously demonstrated that CjXyl31A is cell membrane-bound, with attachment likely mediated through N-terminal lipidation (Larsbrink et al., 2011). In contrast, LipoP (Juncker et al., 2003) predicts that neither CjBgl35A nor CjAfc95A are N-terminally lipidated. Furthermore, SignalP (Petersen et al., 2011) fails to identify a signal peptide in CjAfc95A. To further refine our spatial model of XyG utilization in C. japonicus, the subcellular locations of CjBgl35A, and CjAfc95A were investigated by specific chromogenic assays of cell fractions. Notably, no significant α-l-fucosidase activity could be detected in any of the protein fractions from the M9-Glc cultures (Table 2). Likewise, β-galactosidase activity was absent in the secreted protein fraction from M9-Glc cultures, while only very weak activity was observed in the periplasmic and cytoplasmic protein fractions. In contrast, growth on M9 medium containing XyGOs to induce XyGUL expression resulted in a significant increase in both activities in all three fractions. The specific activity levels were however clearly highest in the periplasmic fractions versus the secreted and cytoplasmic fractions (activity in these fractions, which is at least eightfold lower in all cases, may represent some degree of cross-contamination).
Table 2.
Specific galactosidase and fucosidase activities for proteins produced under different growth conditions
| Growth medium | Substrate | Specific activity in cell fractions (μM released product·s−1·μg protein−1) | ||
|---|---|---|---|---|
| Secreted | Periplasmic | Intracellular | ||
| M9-XyGO | Gal-β-PNP | 1.25 ± 0.14 | 18.3 ± 3 | 1.0 ± 0.3 |
| l-Fuc-α-CNP | 160 ± 47 | 1400 ± 160 | 28.8 ± 6.6 | |
| M9-Glc | Gal-β-PNP | n/d | 0.86 ± 0.05 | 0.058 ± 0.003 |
| l-Fuc-α-CNP | n/d | n/d | n/d | |
Discussion
Transcript and reverse genetics analyses allow us to conclude that xyl31A, bgl35A, tbdr, and afc95A (Fig. 1) constitute a xyloglucan utilization locus (XyGUL), which is the primary genetic determinant conferring C. japonicus with the ability to saccharify this ubiquitous plant cell wall polysaccharide. Together with substrate specificity and structural analysis of the encoded glycoside hydrolases, subcellular localization data allow us to propose an updated model of XyG degradation in C. japonicus (Fig. 6). In this model, XyG polysaccharide is hydrolysed into component oligosaccharides by one or more endo-xyloglucanase(s). The liberated XyGOs are then imported into the periplasm via the TBDR (Ferguson and Deisenhofer, 2002; Koebnik, 2005; Dejean et al., 2013), where the exo-glycosidases CjXyl31A, CjBgl35A, and CjAfc95A work in concert, together with a currently unidentified β-glucosidase(s), to yield monosaccharides for further catabolism.
Figure 6.
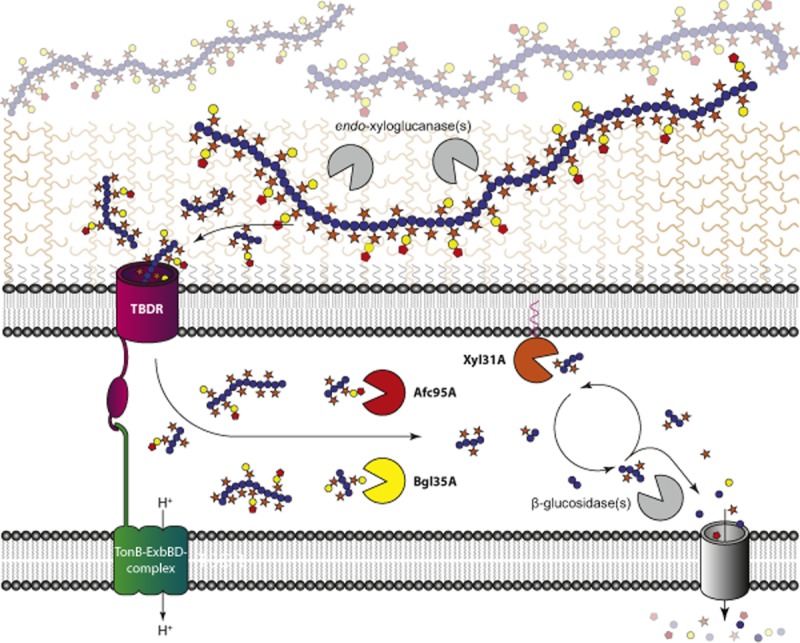
Proposed pathway of (fucogalacto)xyloglucan degradation by C. japonicus. Sugar symbols are as follows: Glc – blue circles, Xyl – orange stars, Gal – yellow circles, Fuc – red pentagons. Secreted enzymes with endo-xyloglucanase activity depolymerize the polysaccharides into xyloglucan oligosaccharides which are imported into the periplasm by the TonB-dependent receptor of the locus. In the periplasm, CjBgl35A and CjAfc95A strip off galactose and fucose from the oligosaccharides respectively. In concert, CjXyl31A and an unknown β-glucosidase cleave off terminal xylose and glucose residues from the non-reducing end in an iterative manner.
Notably, structural enzymology provides some of the strongest support for this model. The α-xylosidase, CjXyl31A, has a strict non-reducing-end specificity due to a deep, pocket-shaped active site and thus cannot access xylosyl residues positioned along the intact polysaccharide chain (Larsbrink et al., 2011). Likewise, the apparent kcat/Km value of CjBgl35A for the oligosaccharide XLLG is three orders of magnitude higher than for tamarind XyG polysaccharide (Table 1), which, together with the observed pocket topology of the active-site (Fig. 4), suggests that cleavage of the polysaccharide to XyGOs also occurs prior to the action of this enzyme. As highlighted above, steric limitations of the CjBgl35A active-site also indicate that CjAfc95A must remove terminal fucosyl residues prior to fully enable degalactosylation by CjBgl35A.
It is particularly interesting to note the obvious differences between the XyGUL system of C. japonicus vis-à-vis the polysaccharide utilization loci (PULs) of Bacteroidetes (Koropatkin et al., 2012). A recently characterized XyGUL from B. ovatus comprises a complete cohort of GHs sufficient to address each linkage in solanaceous (arabinogalacto)xyloglucan, including both a keystone endo-xyloglucanase and six diverse exo-glycosidases, in addition to a TBDR, a hybrid two-component sensor/regulator, and two cell-surface carbohydrate-binding proteins (Larsbrink et al., 2014; Terrapon and Henrissat, 2014). In contrast, the C. japonicus XyGUL – in addition to being alternatively directed towards dicot (fucogalacto)xyloglucan – is significantly less complete: It does not encode the requisite endo-xyloglucanase(s) necessary to initiate the saccharification pathway by polysaccharide backbone cleavage, yet C. japonicus does secrete significant endo-xyloglucanase activity into the medium under XyGO-induction (Larsbrink et al., 2011). Likewise, the C. japonicus XyGUL does not encode a β-glucosidase, which would be required to digest the β(1→4)-glucan backbone of the XyGOs. Finally, the XyGUL appears to lack a substrate sensor/transcriptional regulator analogous to that which regulates xylan-specific genes in C. japonicus (Emami et al., 2009). Thus, although the C. japonicus XyGUL shares a theme of genetic colocalization and co-regulation with the Bacteroidetes PULs, it is notably less evolved with respect to complexity and completeness. It is also notable that the TBDRs of the C. japonicus and B. ovatus XyGULs do not share any significant sequence similarity, despite predicted functional homology based on their association with a similar complement of GHs.
In general, the colocalization and co-ordinated regulation of CAZyme-encoding genes in C. japonicus has not been widely studied. Indeed, annotation of the C. japonicus genome revealed that many of these genes are not clustered on the chromosome, although interesting exceptions can be observed (Deboy et al., 2008). For example, early work by Gilbert, Hazlewood and co-workers revealed that endo-xylanase B of GH10 (CJA_3280) and α-l-arabinofuranosidase C of GH62 (CJA_3281) are encoded by adjacent genes in C. japonicus. Post-genomic re-examination by McClendon et al. has revealed that these are likely to constitute part of a Xylan Utilization Locus that additionally encodes a predicted endo-xylanase of GH30 (CJA_3279), feruloyl esterase D (Fee1B) of CE1 (CJA_3282) (Ferreira et al., 1993), a predicted β-galactosidase of GH98 (CJA_3286), and a second predicted feruloyl esterase (Fee1A) of CE1 (CJA_3287) (McClendon et al., 2011). Likewise, the endo-xylanase Xyn10D (CJA_2888) (Emami et al., 2009) is located between the α-glucuronidase GlcA67A (CJA_2887) (Nagy et al., 2003) and the acetylxylan esterase Axe2B (CJA_2889) (Zhang et al., 2014). On the other hand, the reason for colocalization of other genes is sometimes less obvious: the broad-specificity xylan-active α-l-arabinofuranosidase Abf51A (CJA_2769) (Beylot et al., 2001) is found in tandem with the endo-mannanase Man26A (CJA_2770) (Hogg et al., 2001), which perhaps suggests a greater diversity of mannan substructures than previously appreciated. Regardless, we posit that increased consideration of CAZyme colocalization within genomes can significantly facilitate understanding the basic biology of C. japonicus and other bacteria, as well as advance the discovery of novel enzyme cohorts for biotechnological applications (Martens et al., 2014). For example, a recent study of a mannan-degrading locus of Bacteroides fragilis has identified a syntenic locus in C. japonicus (CJA_0241–0246) putatively comprising a sugar/cation symporter, a mannosyl-glucose phosphorylase, a mannobiose 2-epimerase, a GH5 endo-mannanase and a GH27 α-galactosidase (Deboy et al., 2008; Senoura et al., 2011).
The data presented here shed new light on the genetic, biochemical, and structural basis of hemicellulose utilization by C. japonicus and, moreover, reveal a novel, matched set of glycosidases for biotechnological applications. Indeed, the importance of xyloglucan saccharification has been arguably under-appreciated in the context of biofuel production (Gilbert et al., 2008), although recent studies are beginning to address this issue (Hu et al., 2013; Jabbour et al., 2013), buoyed by an increased understanding of specific, xyloglucan-active enzymes (Gilbert et al., 2008; Ariza et al., 2011; Larsbrink et al., 2011; Eklöf et al., 2012). Interestingly, the xyloglucan utilization locus described here encodes neither the endo-xyloglucanase(s) nor the β-glucosidase(s) required for full degradation of XyG. The proteins necessary for sensing XyG in the environment are also presently unknown. Future studies from our collaboration will focus on the identification of the corresponding genes elsewhere in the genome, in addition to functional studies on the TonB-dependent receptor of the locus.
Experimental procedures
Ultrapure water, purified on a Milli-Q system (Millipore) to a resistivity of ρ > 18.2 MΩ cm, was used in all experiments. Galactose, lactose, Gal-β-PNP, Glc-β-PNP, Xyl-β-PNP and l-Araf-α-PNP were purchased from Sigma. l-Fuc-α-CNP and fucose were purchased from Carbosynth. Tamarind xyloglucan (XyG) was purchased from Megazyme. XLLG was prepared from tamarind XyG as described previously (Greffe et al., 2005). Lettuce XyG and XLFG were isolated as described below. C. japonicus Ueda107 was obtained from the National Collections of Industrial, Marine, and Food Bacteria (Aberdeen, Scotland).
Cloning of CjBgl35A and CjAfc95A
The open reading frames encoding CjBgl35A and CjAfc95A (GenBank Accession No. ACE85180.1 and ACE83895.1 respectively) were amplified by PCR from genomic DNA of C. japonicus Ueda107 using Phusion polymerase (Finnzymes), using forward primers incorporating a CACC overhang to enable TOPO cloning, and reverse primers excluding stop codons (Eurofins MWG Operon; Supplemental Table S1). The forward primer for CjBgl35A was designed to exclude the predicted native signal peptide (residues 1–36), while no signal peptide was predicted for CjAfc95A (Petersen et al., 2011). The PCR products were cloned into the pENTR/SD/D-TOPO entry vectors (Invitrogen) and recombined into pET-DEST42 destination vectors (Invitrogen) following the manufacturer's instructions.
Initial protein crystals produced from the pET-DEST42 construct, featuring a larger C-terminal tag, yielded only relatively weak diffraction to approximately 4.0 Å (data not shown) and proved difficult to optimize further. As such, primers were designed to allow recloning of the gene fragment encoding CjBgl35A into a pET28a vector modified for Ligation Independent Cloning (LIC), and featuring a shorter, N-terminal hexahistidine tag (Eurofins MWG Operon; Supplemental Table S1). The PCR product was cloned into a pre-prepared linear vector stock using the InFusion-HD cloning kit (Clontech) according to the manufacturer's instructions.
Recombinant gene expression and protein purification
pET-DEST42 constructs were transformed into E. coli BL21(DE3) by electroporation, and proteins were produced and purified by immobilized metal affinity chromatography (IMAC) following an established protocol (Larsbrink et al., 2011). The modified pET28a-Bgl35A construct was transformed into E. coli TUNER(DE3) cells via the heat-shock method, with recombinant protein purified by IMAC and size-exclusion chromatography. Protein purity was verified by SDS-PAGE. Protein concentration was determined from A280 values; the extinction coefficients used for CjBgl35A and CjAfc95A were 118260 and 151845 M−1 cm−1 respectively [ProtParam server (Gasteiger et al., 2005)].
Crystallization, data collection and structure solution of CjBgl35A
Crystals of CjBgl35A suitable for full X-ray data collection were grown using hanging drop vapour diffusion at 19°C, with equal volumes of pure protein and reservoir solution (2.6 M sodium acetate pH 7.2). Ligand complex formation with GDJ was achieved by soaking native crystals in 10 mM GDJ (final) for a period of approximately 1 h. Since concentrated sodium acetate present within the mother-liquor solution proved an adequate cryo-protectant, no additional solvents were added to crystals prior to flash cooling in liquid N2. Full diffraction data for both native and ligand complex CjBgl35A crystals were collected at beamline I03 of the Diamond Light Source (Didcot, Oxfordshire, UK). Measured reflection intensities were indexed, integrated and scaled using XDS (Kabsch, 2010a, b) and the CCP4 suite (Winn et al., 2011) implementation of Aimless. The structure of CjBgl35A was solved by molecular replacement with PHASER (McCoy et al., 2007), employing the co-ordinates of the C. crescentus GH35 (PDB code 3U7V) as a phasing model. An initial atomic model was constructed using the CCP4 (Winn et al., 2011) implementation of Buccaneer and refined via the maximum-likelihood method using numerous cycles of REFMAC (Murshudov et al., 2011; Winn et al., 2011), with additional manual correction using COOT (Emsley et al., 2010). The structure of CjBgl35A in complex with GDJ was produced by refining the scaled data (processed as above) against the finalized model of the native protein and visually inspecting calculated electron density maps for evidence of ligand binding. The final atomic model of the complex was subsequently refined and corrected as above. Models and structure factors for both native CjBgl35A and the GDJ complex have been deposited into the PDB with respective accession codes 4D1I and 4D1J.
High-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD)
Oligo- and monosaccharides were analysed on a Dionex ICS-3000 HPLC system operated by Chromelion software version 7 (Dionex) using a Dionex CarboPac PA200 column for Gradient A and a Dionex CarboPac PA1 column for Gradient B. Solvent A was water, solvent B was 1 M sodium hydroxide and solvent C was 1 M sodium acetate. The gradients used were as follows: Gradient A: 0 to 5 min, 10% B, 2% C; 5 to 12 min, 10% B and a linear gradient from 2% to 30% C; 12 to 12.1 min, 50% B, 50% C; 12.1 to 13 min, an exponential gradient of B and C back to initial conditions; 13 to 17 min, initial conditions. Gradient B: column pre-conditioned prior to injection by −13 to −3 min, 12% B, 6.8% C; −3 to 0 min, 100% A; 0 to 25 min, 100% A.
Matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) analysis of oligosaccharides
MALDI-TOF was performed on oligosaccharide samples using a Voyager-DE STR instrument (Applied Biosystems) in positive linear mode with an acceleration of 20 kV and an extraction delay time of 150 ns. The samples were prepared by mixing equal amount of sample (1 μl) with the matrix (10 mg ml−1 2,5 dihydroxybenzoic acid in 1:1 acetonitrile : H2O, containing 0.05% trifluoroacetic acid).
Xyloglucan extraction from iceberg lettuce
Using a modified and scaled-up version of a published protocol (Hsieh and Harris, 2009), XyG polysaccharide was extracted from approximately 500 g of fresh iceberg lettuce leaves, obtained from a local grocery store. The plant material was homogenized in 70% aqueous ethanol using a high-speed blender, collected by filtration using Miracloth (Millipore) and ground to a fine powder in liquid nitrogen using a ceramic mortar and pestle. Alcohol-soluble polysaccharides and smaller sugars were removed by repeated 70% ethanol wash and filtration steps. Non-cellulosic polysaccharides were extracted in 6 M sodium hydroxide, containing 1% sodium borohydride to prevent alkaline peeling, followed by neutralization with acetic acid.
Ethanol was added to the neutralized solution to a final concentration of 70% to precipitate polysaccharides, followed by centrifugation at 24 000 g for 15 min. The supernatant was discarded and the pellet washed three times with 70% ethanol. The washed pellet was dissolved in water and loaded onto a Q Sepharose column (GE Lifesciences), pre-equilibrated with 10 mM imidazole (pH 7.0) to bind charged polysaccharides (e.g. pectins) to the matrix. Neutral hemicellulosic polysaccharides were eluted in three column volumes of the same buffer. The resulting hemicellulose-containing fraction was incubated with 150 units of the xylanase CjCBM22-GH10 (Xyn10A) and 150 units of mannanase 26A (both purchased from NZYtech) for 16 h at 37°C, to hydrolyse contaminating xylan and mannan respectively. The resulting oligosaccharides were subsequently removed in the supernatant following ethanol precipitation of the lettuce XyG (final yield 180 mg after lyophilization from water). Prior to analysis by HPAEC-PAD (gradient B), 1 mg of lettuce XyG was hydrolysed by incubation with 2 M trifluoroacetic acid (TFA) for 3 h at 120°C (1 ml total volume). Hydrolysis products were vacuum-dried and re-suspended in deionized water, followed by filtration (0.2 μm). The lettuce xyloglucan comprised Glc, Xyl, Gal and Fuc, with pectin derived contaminations of approximately 5%.
In a second extraction, fucosylated xylogluco-oligosaccharides (XyGOs) were obtained via a similar procedure lacking the ion exchange step. Seven hundred and fifty milligrams of lyophilized crude lettuce XyG was dissolved in 100 ml 50 mM ammonium acetate, pH 5.5, to which was added 1.28 mg B. ovatus BoGH5A endo-xyloglucanase (Larsbrink et al., 2014). The reaction was incubated overnight at 35°C followed by lyophilization (dry weight 0.3 g). XyGOs were dissolved in 1 ml water and loaded onto a 100 cm XK16 column packed with 200 ml Bio-Gel P-2 Gel (Bio-Rad), which had been equilibrated with water. Eluted fractions were analysed by MALDI-TOF and fractions containing predominantly XXXG, XXFG and XLFG, respectively, were pooled and lyophilized.
Enzyme assays
All assays were carried out at 25°C, at the pH optimum of the respective enzyme. Curve fitting and processing of kinetic data were performed using Origin 8 software (OriginLab).
pH dependence
Measurements of the pH-dependence of CjBgl35A and CjAfc95A were performed with Gal-β-PNP and l-Fuc-α-CNP as substrates, respectively, using the assays described below. Buffers (50 mM) are indicated in Figs S1 and S3.
Chromogenic assays
Activities on PNP glycosides were analysed by a stopped assay as previously described (Larsbrink et al., 2011), using enzyme concentrations in the μM range for initial screens and several hours incubation. For initial rate kinetics on Gal-β-PNP for CjBgl35A, 42 nM enzyme was used and reactions were stopped after 10 min. Assays utilizing l-Fuc-α-CNP were monitored continuously for the release of 2-chloro-4-nitrophenolate using a Cary 300 spectrophotometer (Agilent Technologies). For the determination of the kinetic parameters of CjAfc95A, 22 nM enzyme was used. An extinction coefficient of 12 936 M−1 cm−1, determined from a standard curve, was used to calculate product concentration from A405 values.
HPAEC-PAD-based assays
Enzymatic reactions on XyG polysaccharides and XyGOs were performed in 50 μl reactions, containing 50 mM buffer at the pH optimum of the assayed enzyme, and were stopped by addition of 2 μl of 5 M sodium hydroxide prior to HPAEC-PAD analysis. For the reaction of CjBgl35A on XLLG, 2.7 nM enzyme was used, the reaction was terminated after 10 min. For the reaction of CjBgl35A on tamarind XyG, 0.16 μM enzyme was used, the reaction was terminated after 130 min. For the reaction of CjAfc95A on XLFG, 0.64 nM enzyme was used, and the reaction was terminated after 15 min. For the reaction of CjAfc95A on lettuce XyG, 270 nM enzyme was used, and the reaction was terminated after 20 min. To quantify the release of galactose or fucose from the reactions, the respective commercial monosaccharides were used as standards.
Isothermal titration calorimetry (ITC)
Isothermal titration calorimetry (ITC) was performed with a MicroCal Auto-iTC200 system (GE Healthcare). Assays were conducted in triplicate at 25°C, with GDJ (390 μM) titrated into the ITC cell containing pure CjBgl35A (38.5 μM). Dissociation constants (Kd) for each titration were subsequently calculated and averaged using the Origin 7 software package (MicroCal).
Transcription analysis
Total RNA was extracted from cultures of C. japonicus grown in liquid M9 minimal media supplemented with either a mixture of tamarind XyGOs (4 g l−1) or glucose (10 g l−1). Five millilitres overnight cultures were used to inoculate 200 ml cultures of the same media, and the OD600 of the cultures were monitored. When the OD600 had reached 1, corresponding to a late exponential growth phase (Gardner and Keating, 2010), 2 ml culture was added to 4 ml RNAprotect Bacteria Reagent (Qiagen) and the cells were harvested by centrifugation at 4300 g for 5 min. Total RNA was extracted using the RNeasy mini kit (Qiagen), following the manufacturer's instructions, including in-solution DNase treatment and subsequent clean-up. The final yield of total RNA was on average 42 μg for XyGO cultures and 21 μg for Glc cultures. One microgram of each biological sample was reverse transcribed into cDNA by the iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed on a CFX96 Touch instrument using Hard-Shell white well plates (Bio-Rad). Primers for the locus genes (Eurofins MWG Operon; Table S1), as well as the predicted β-galactosidase encoding genes CjBgl2A, CjBgl2B, CjBgl2C and the predicted α-l-fucosidase encoding gene CjAfc95B were used to analyse the respective genes for each growth condition in biological duplicates and technical triplicates. The protocol used for amplification consisted of: 95°C 3 min, 40 cycles of 95°C, 15 s; 58°C, 15 s; 72°C, 15 s, followed by a melt curve (65°C to 95°C read in 0.5°C steps). The qPCR results were analysed using the Bio-Rad CFX Manager 2.0 software (Bio-Rad). To analyse whether the genes were transcribed as an operon in a single polycistronic mRNA strand, the primers from the qPCR study were used to amplify the regions between adjacent genes using Phusion polymerase (Finnzymes) and the constructed cDNA as template.
Enzyme localization studies
The remaining cells from the minimal media cultures, above, were harvested by centrifugation at 4300 g for 10 min, and subjected to osmotic shock as described previously (Larsbrink et al., 2011). The secreted proteins in the supernatant media were collected, concentrated and washed with 50 mM sodium phosphate, pH 7.0, using Amicon Ultra filter units (10 kDa cut-off; Millipore). The remaining cells were re-suspended in 5 ml sodium phosphate, pH 7.0, and lysed by sonication. The lysate was centrifuged at 25 000 g for 45 min and the resulting supernatant liquid was collected and concentrated with the same filter units and buffer as above to obtain soluble cytoplasmic proteins. The three fractions containing secreted, periplasmic, and cytoplasmic proteins were each assayed for β-galactosidase and α-l-fucosidase activities using the chromogenic assays described above; protein concentrations were measured by the Bradford method.
Construction of targeted gene disruptions and strain growth analysis
Directed gene disruptions from integration of suicide plasmid pK18mobsacB (Schafer et al., 1994) into the genome were facilitated by tri-parental mating as done previously (Gardner and Keating, 2010; 2012,). Confirmation of correct gene disruptions was performed by PCR using primers listed in Table S1. C. japonicus strains were grown on MOPS minimal medium (Neidhardt et al., 1974) with glucose [0.25% (wt/vol)] or tamarind xyloglucan [0.25% (wt/vol)] as sole carbon sources. Growth assays were performed in a TECAN M200Pro at 30°C with vigorous shaking. Growth measurements were taken via optical density (OD) at 600 nm. Growth experiments were performed in biological triplicates.
Acknowledgments
We are grateful to Dr Lauren S. McKee for assistance in manuscript preparation. Work in the Brumer lab was supported by the Mizutani Foundation for Glycoscience, and The Swedish Research Council Formas (via CarboMat – the KTH Advanced Carbohydrate Materials Centre), faculty funding from the Michael Smith Laboratories/UBC, the Natural Sciences and Engineering Research Council of Canada (Discovery Grant), the Canada Foundation for Innovation and the British Columbia Knowledge Development Fund. Work in the Davies lab was supported by the Biotechnology and Biological Science Research Council (BBSRC; Grant BB/I014802/1). The staff of the Diamond Light Source is thanked for provision of data collection facilities. Work in the Gardner lab was supported by start-up funds provided by the Provost's Office at UMBC. We thank Prof. Harry J. Gilbert for inspiration.
Conflicts of interest
The authors declare no competing interests.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
Supporting Information
References
- Ahn YO, Zheng M, Bevan DR, Esen A, Shiu SH, Benson J, et al. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 35. Phytochemistry. 2007;68:1510–1520. doi: 10.1016/j.phytochem.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Ariza A, Eklöf JM, Spadiut O, Offen WA, Roberts SM, Besenmatter W, et al. Structure and activity of Paenibacillus polymyxa xyloglucanase from glycoside hydrolase family 44. J Biol Chem. 2011;286:33890–33900. doi: 10.1074/jbc.M111.262345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Vasu P, Persson S, Mort AJ. Somerville CR. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc Natl Acad Sci USA. 2006;103:11417–11422. doi: 10.1073/pnas.0604632103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beylot MH, Emami K, McKie VA, Gilbert HJ. Pell G. Pseudomonas cellulosa expresses a single membrane-bound glycoside hydrolase family 51 arabinofuranosidase. Biochem J. 2001;358:599–605. doi: 10.1042/0264-6021:3580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N. McCann M. The cell wall. In: Jones RL, editor; Biochemistry and Molecular Biology of Plants. Somerset, NJ: John Wiley & Sons; 2000. pp. 55–108. Buchanan, B.B., Gruissem, W., [Google Scholar]
- Cartmell A, Topakas E, Ducros VMA, Suits MDL, Davies GJ. Gilbert HJ. The Cellvibrio japonicus mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J Biol Chem. 2008;283:34403–34413. doi: 10.1074/jbc.M804053200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartmell A, McKee LS, Peña MJ, Larsbrink J, Brumer H, Kaneko S, et al. The Structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem. 2011;286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Wang L, Jiang YL, Bai XH, Chu J, Li Q, et al. Structural insights into the substrate specificity of Streptococcus pneumoniae beta(1,3)-galactosidase BgaC. J Biol Chem. 2012;287:22910–22918. doi: 10.1074/jbc.M112.367128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damude HG, Ferro V, Withers SG. Warren RAJ. Substrate specificity of endoglucanase A from Cellulomonas fimi: fundamental differences between endoglucanases and exoglucanases from family 6. Biochem J. 1996;315:467–472. doi: 10.1042/bj3150467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies GJ, Wilson KS. Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem J. 1997;321:557–559. doi: 10.1042/bj3210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deboy RT, Mongodin EF, Fouts DE, Tailford LE, Khouri H, Emerson JB, et al. Insights into plant cell wall degradation from the genome sequence of the soil bacterium Cellvibrio japonicus. J Bacteriol. 2008;190:5455–5463. doi: 10.1128/JB.01701-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean G, Blanvillain-Baufume S, Boulanger A, Darrasse A, de Bernonville TD, Girard AL, et al. The xylan utilization system of the plant pathogen Xanthomonas campestris pv campestris controls epiphytic life and reveals common features with oligotrophic bacteria and animal gut symbionts. New Phytol. 2013;198:899–915. doi: 10.1111/nph.12187. [DOI] [PubMed] [Google Scholar]
- Edwards M, Bowman YJL, Dea ICM. Reid JSG. A beta-d-galactosidase from nasturtium (Tropaeolum majus L.) cotyledons – purification, properties, and demonstration that xyloglucan is the natural substrate. J Biol Chem. 1988;263:4333–4337. [PubMed] [Google Scholar]
- Eklöf JM, Ruda MC. Brumer H. Distinguishing xyloglucanase activity in endo-beta(1-4)glucanases. In: Gilbert HJ, editor; Methods in Enzymology: Cellulases. Amsterdam: Elsevier; 2012. pp. 97–120. Vol. 510, Ch. 6. [DOI] [PubMed] [Google Scholar]
- Eklöf JM, Shojania S, Okon M, McIntosh LP. Brumer H. Structure–function analysis of a broad specificity Populus trichocarpa endo-beta-glucanase reveals an evolutionary link between bacterial licheninases and plant XTH gene products. J Biol Chem. 2013;288:15786–15799. doi: 10.1074/jbc.M113.462887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami K, Topakas E, Nagy T, Henshaw J, Jackson KA, Nelson KE, et al. Regulation of the xylan-degrading apparatus of Cellvibrio japonicus by a novel two-component system. J Biol Chem. 2009;284:1086–1096. doi: 10.1074/jbc.M805100200. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG. Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson AD. Deisenhofer J. TonB-dependent receptors – structural perspectives. Biochim Biophys Acta. 2002;1565:318–332. doi: 10.1016/s0005-2736(02)00578-3. [DOI] [PubMed] [Google Scholar]
- Ferreira LMA, Wood TM, Williamson G, Faulds C, Hazlewood GP, Black GW. Gilbert HJ. A modular esterase from Pseudomonas fluorescens subsp. cellulosa contains a noncatalytic cellulose-binding domain. Biochem J. 1993;294:349–355. doi: 10.1042/bj2940349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG. Keating DH. Requirement of the Type II secretion system for utilization of cellulosic substrates by Cellvibrio japonicus. Appl Environ Microbiol. 2010;76:5079–5087. doi: 10.1128/AEM.00454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JG. Keating DH. Genetic and functional genomic approaches for the study of plant cell wall degradation in Cellvibrio japonicus. In: Gilbert HJ, editor; Methods in Enzymology: Cellulases. Amsterdam: Elsevier; 2012. pp. 331–347. Vol. 510, Ch. 18. [DOI] [PubMed] [Google Scholar]
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD. Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor; The Proteomics Protocol Handbook. New York: Humana Press; 2005. pp. 571–607. [Google Scholar]
- Gilbert HJ, Stålbrand H. Brumer H. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol. 2008;11:338–348. doi: 10.1016/j.pbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Gilbert HJ, Knox JP. Boraston AB. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol. 2013;23:669–677. doi: 10.1016/j.sbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Greffe L, Bessueille L, Bulone V. Brumer H. Synthesis, preliminary characterization, and application of novel surfactants from highly branched xyloglucan oligosaccharides. Glycobiology. 2005;15:437–445. doi: 10.1093/glycob/cwi013. [DOI] [PubMed] [Google Scholar]
- Hazlewood GP. Gilbert HJ. Structure and function analysis of Pseudomonas plant cell wall hydrolases. Prog Nucleic Acid Res Mol Biol. 1998;61:211–241. doi: 10.1016/s0079-6603(08)60828-4. [DOI] [PubMed] [Google Scholar]
- Heightman TD. Vasella AT. Recent insights into inhibition, structure, and mechanism of configuration-retaining glycosidases. Angew Chem Int Ed Engl. 1999;38:750–770. doi: 10.1002/(SICI)1521-3773(19990315)38:6<750::AID-ANIE750>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hemsworth GR, Henrissat B, Davies GJ. Walton PH. Discovery and characterization of a new family of lytic polysaccharide monooxygenases. Nat Chem Biol. 2014;10:122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg D, Woo EJ, Bolam DN, McKie VA, Gilbert HJ. Pickersgill RW. Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding. J Biol Chem. 2001;276:31186–31192. doi: 10.1074/jbc.M010290200. [DOI] [PubMed] [Google Scholar]
- Holm L. Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YSY. Harris PJ. Xyloglucans of monocotyledons have diverse structures. Mol Plant. 2009;2:943–965. doi: 10.1093/mp/ssp061. [DOI] [PubMed] [Google Scholar]
- Hu JG, Arantes V, Pribowo A. Saddler JN. The synergistic action of accessory enzymes enhances the hydrolytic potential of a ‘cellulase mixture’ but is highly substrate specific. Biotechnol Biofuels. 2013;6:112. doi: 10.1186/1754-6834-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibatullin FM, Baumann MJ, Greffe L. Brumer H. Kinetic analyses of retaining endo-(xylo)glucanases from plant and microbial sources using new chromogenic xylogluco-oligosaccharide aryl glycosides. Biochemistry. 2008;47:7762–7769. doi: 10.1021/bi8009168. [DOI] [PubMed] [Google Scholar]
- Ishimizu T, Hashimoto C, Takeda R, Fujii K. Hase S. A novel alpha 1,2-L-fucosidase acting on xyloglucan oligosaccharides is associated with endo-beta-mannosidase. J Biochem (Tokyo) 2007;142:721–729. doi: 10.1093/jb/mvm186. [DOI] [PubMed] [Google Scholar]
- Jabbour D, Borrusch MS, Banerjee G. Walton JD. Enhancement of fermentable sugar yields by alpha-xylosidase supplementation of commercial cellulases. Biotechnol Biofuels. 2013;6:58. doi: 10.1186/1754-6834-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic I, Magnuson JK, Collart F, Robbertse B, Adney WS, Himmel ME. Baker SE. Fungal glycoside hydrolases for saccharification of lignocellulose: outlook for new discoveries fueled by genomics and functional studies. Cellulose. 2009;16:687–697. [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H. Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010a;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010b;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koebnik R. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 2005;13:343–347. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA. Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsbrink J, Izumi A, Ibatullin FM, Nakhai A, Gilbert HJ, Davies GJ. Brumer H. Structural and enzymatic characterization of a glycoside hydrolase family 31 alpha-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochem J. 2011;436:567–580. doi: 10.1042/BJ20110299. [DOI] [PubMed] [Google Scholar]
- Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, et al. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard R, Pabst M, Bondili JS, Chambat G, Veit C, Strasser R. Altmann F. Identification of an Arabidopsis gene encoding a GH95 alpha1,2-fucosidase active on xyloglucan oligo- and polysaccharides. Phytochemistry. 2008;69:1983–1988. doi: 10.1016/j.phytochem.2008.03.024. [DOI] [PubMed] [Google Scholar]
- Lombard V, Ramulu HG, Drula E, Coutinho PM. Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon SD, Shin HD. Chen RR. Novel bacterial ferulic acid esterase from Cellvibrio japonicus and its application in ferulic acid release and xylan hydrolysis. Biotechnol Lett. 2011;33:47–54. doi: 10.1007/s10529-010-0394-6. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC. Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNicholas S, Potterton E, Wilson KS. Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark P, Zhang Q, Czjzek M, Brumer H. Ågren H. Molecular dynamics simulations of a branched tetradecasaccharide substrate in the active site of a xyloglucan endo-transglycosylase. Mol Simul. 2011;37:1001–1013. [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS. Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014 doi: 10.1016/j.jmb.2014.06.022. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mewis K, Armstrong Z, Song YC, Baldwin SA, Withers SG. Hallam SJ. Biomining active cellulases from a mining bioremediation system. J Biotechnol. 2013;167:462–471. doi: 10.1016/j.jbiotec.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagae M, Tsuchiya A, Katayama T, Yamamoto K, Wakatsuki S. Kato R. Structural basis of the catalytic reaction mechanism of novel 1,2-alpha-L-fucosidase from Bifidobacterium bifidum. J Biol Chem. 2007;282:18497–18509. doi: 10.1074/jbc.M702246200. [DOI] [PubMed] [Google Scholar]
- Nagy T, Nurizzo D, Davies GJ, Biely P, Lakey JH, Bolam DN. Gilbert HJ. The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of aldobiouronic acid as important substrate recognition determinants. J Biol Chem. 2003;278:20286–20292. doi: 10.1074/jbc.M302205200. [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL. Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen H, Hayauchi Y. Sinnwell V. Monosaccharides containing nitrogen in the ring, XXXVII. Synthesis of 1,5-dideoxy-1,5-imino-D-galactitol. Chem Ber. 1980;113:2601–2608. [Google Scholar]
- Peña MJ, Darvill AG, Eberhard S, York WS. O'Neill MA. Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology. 2008;18:891–904. doi: 10.1093/glycob/cwn078. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G. Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Herve C, Domozych DS, Willats WGT, Tuohy MG. Evolution and diversity of plant cell walls: from algae to flowering plants. In: Ort D, et al., editors. Annual Review of Plant Biology. Vol. 62. Palo Alto: Annual Reviews; 2011. pp. 567–588. Merchant, S.S., Briggs, W.R., [DOI] [PubMed] [Google Scholar]
- del Pulgar EMG. Saadeddin A. The cellulolytic system of Thermobifida fusca. Crit Rev Microbiol. 2014;40:236–247. doi: 10.3109/1040841X.2013.776512. [DOI] [PubMed] [Google Scholar]
- Sampedro J, Gianzo C, Iglesias N, Guitian E, Revilla G. Zarra I. AtBGAL10 is the main xyloglucan beta-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol. 2012;158:1146–1157. doi: 10.1104/pp.111.192195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G. Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Scheller HV. Ulvskov P. Hemicelluloses. Annu Rev Plant Biol. 2010;61:263–289. doi: 10.1146/annurev-arplant-042809-112315. [DOI] [PubMed] [Google Scholar]
- Sela DA, Garrido D, Lerno L, Wu SA, Tan KM, Eom HJ, et al. Bifidobacterium longum subsp. infantis ATCC 15697 alpha-fucosidases are active on fucosylated human milk oligosaccharides. Appl Environ Microbiol. 2012;78:795–803. doi: 10.1128/AEM.06762-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senoura T, Ito S, Taguchi H, Higa M, Hamada S, Matsui H, et al. New microbial mannan catabolic pathway that involves a novel mannosylglucose phosphorylase. Biochem Biophys Res Commun. 2011;408:701–706. doi: 10.1016/j.bbrc.2011.04.095. [DOI] [PubMed] [Google Scholar]
- Silipo A, Larsbrink J, Marchetti R, Lanzetta R, Brumer H. Molinaro A. NMR spectroscopic analysis reveals extensive binding interactions of complex xyloglucan oligosaccharides with the Cellvibrio japonicus glycoside hydrolase family 31 alpha-xylosidase. Chem Eur J. 2012;18:13395–13404. doi: 10.1002/chem.201200488. [DOI] [PubMed] [Google Scholar]
- Terrapon N. Henrissat B. How do gut microbes break down dietary fiber? Trends Biochem Sci. 2014;39:156–158. doi: 10.1016/j.tibs.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Tuomivaara ST, Yaoi K, O'Neill MA. York WS. Generation and structural validation of a library of diverse xyloglucan-derived oligosaccharides, including an update on xyloglucan nomenclature. Carbohydr Res. 2014 doi: 10.1016/j.carres.2014.06.031. doi: 10.1016/j.carres.2014.06.031. [DOI] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TC, Hu RM, Weng SF. Tseng YH. Identification of a hypothetical protein of plant pathogenic Xanthomonas campestris as a novel beta-galactosidase. J Mol Microbiol Biotechnol. 2007;13:172–180. doi: 10.1159/000103609. [DOI] [PubMed] [Google Scholar]
- Zhang XY, Rogowski A, Zhao L, Hahn MG, Avci U, Knox JP. Gilbert HJ. Understanding how the complex molecular architecture of mannan-degrading hydrolases contributes to plant cell wall degradation. J Biol Chem. 2014;289:2002–2012. doi: 10.1074/jbc.M113.527770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information