Abstract
Coral diseases are characterized by microbial community shifts in coral mucus and tissue, but causes and consequences of these changes are vaguely understood due to the complexity and dynamics of coral-associated bacteria. We used 16S rRNA gene microarrays to assay differences in bacterial assemblages of healthy and diseased colonies displaying White Plague Disease (WPD) signs from two closely related Caribbean coral species, Orbicella faveolata and Orbicella franksi. Analysis of differentially abundant operational taxonomic units (OTUs) revealed strong differences between healthy and diseased specimens, but not between coral species. A subsequent comparison to data from two Indo-Pacific coral species (Pavona duerdeni and Porites lutea) revealed distinct microbial community patterns associated with ocean basin, coral species and health state. Coral species were clearly separated by site, but also, the relatedness of the underlying bacterial community structures resembled the phylogenetic relationship of the coral hosts. In diseased samples, bacterial richness increased and putatively opportunistic bacteria were consistently more abundant highlighting the role of opportunistic conditions in structuring microbial community patterns during disease. Our comparative analysis shows that it is possible to derive conserved bacterial footprints of diseased coral holobionts that might help in identifying key bacterial species related to the underlying etiopathology. Furthermore, our data demonstrate that similar-appearing disease phenotypes produce microbial community patterns that are consistent over coral species and oceans, irrespective of the putative underlying pathogen. Consequently, profiling coral diseases by microbial community structure over multiple coral species might allow the development of a comparative disease framework that can inform on cause and relatedness of coral diseases.
Keywords: 16S rRNA gene microarray, coral disease, microbial community, Orbicella faveolata, Orbicella franksi, Pavona duerdeni, Porites lutea, White Plague Disease (WPD), White Plague-like Disease, White Syndrome (WS)
Introduction
Corals are animals that live in a symbiotic relationship with photosynthetic dinoflagellates of the genus Symbiodinium as well as a rich bacterial community among other microorganisms that are collectively referred to as the coral holobiont (Rohwer et al. 2002). A coral's associated microbial community contributes fundamentally to the holobiont's functioning due to its role in coral nutrition (Lesser et al. 2004) and host defense (Ritchie & Smith 2004; Kelman et al. 2006; Ritchie 2006). Coral diseases are considered one of the most destructive local and geographical forces that impact corals and are responsible for major reef ecosystem declines over the past decades (Sutherland et al. 2004; Weil 2004; Willis et al. 2004; Weil et al. 2006; Harvell et al. 2007; Miller et al. 2009).
Coral disease is defined as any abnormal condition affecting the coral holobiont (Rosenberg et al. 2007), often described as a progressive loss of coral tissue due to viral, fungal, protozoan, or bacterial infections (Sutherland et al. 2004; Bourne et al. 2009) and facilitated by environmental factors (e.g. high sea surface temperatures). It usually manifests through tissue discoloration and eventually tissue loss (necrosis). While the causative agents remain unknown for most diseases (Rosenberg & Kushmaro 2011), it has been shown that compromised health in corals is accompanied by shifts in the microbial community associated with the coral holobiont (Sunagawa et al. 2009; Kimes et al. 2010; Cardenas et al. 2012; Cróquer et al. 2012; Roder et al. 2014). However, it is unclear whether infection of a single pathogen or opportunistic infections secondary to exposure to physiological stress trigger the restructuring of microbial communities in coral disease (Lesser et al. 2007). While this is mainly due to the complexity and dynamics of the host microbial assemblages (Rohwer et al. 2002; Hong et al. 2009; Littman et al. 2009), difficulty in conducting experiments underwater, an overall lack of information on the structure and composition of the ‘natural’ bacterial community of corals, and differences in applied methodologies further complicate the comparison of data. Sanger cloning-and-sequencing approaches are now being complemented by high-throughput methodologies and comparative analyses have shown that data from different platforms produce similar results (Sunagawa et al. 2009; Bayer et al. 2013). However, unequal sample read representation and the use of different 16S amplicon sites hinder a direct comparison between studies. PhyloChip™ 16S rRNA gene microarrays provide a standardized platform and have been successfully used to uncover microbial community patterns in coral disease (Sunagawa et al. 2009; Kellogg et al. 2012; Roder et al. 2014).
White Plague disease (WPD) is one of the most destructive and widespread coral diseases in Caribbean reefs (Dustan 1977; Antonius 1985; Richardson et al. 1998; Miller et al. 2009; Weil et al. 2009). It presents as a bright white band (i.e. clean skeletal structure resulting from disappearing tissue) that initiates at the base or sides of a colony and separates the living tissue from recently settled turf algae on the exposed skeleton that quickly advances across the colony surface. Depending on the type of WPD (I, II or III), progression rates vary and different coral species are affected (Sutherland et al. 2004; Weil & Rogers 2011). Aurantimonas coralicida (Denner 2003) and Thalassomonas loyana (Thompson et al. 2006) were proposed as causative agents of WPD or WPD-like in corals from the Caribbean and the Red Sea, respectively. However, subsequent studies were unable to detect either of these two putative pathogens (Pantos et al. 2003; Barash et al. 2005; Sunagawa et al. 2009; Cardenas et al. 2012; Roder et al. 2014) suggesting that different pathogens must be able to produce highly similar disease phenotypes (Weil 2004; Reshef et al. 2008; Weil & Rogers 2011). In the Great Barrier Reef and Indo-Pacific region, phenotypes of WPD-like etiopathology have been denominated White Syndrome (WS) (Willis et al. 2004), and strains of the coral-bleaching pathogen Vibrio coralliilyticus have been identified as potential infectious agents in a number of coral species (Sussman et al. 2008). Accordingly, indistinguishable disease phenotypes are produced by different pathogens (Weil & Rogers 2011), and disease nomenclature can be mistaken. For this reason, we refer to coral colonies displaying visual characteristics of White Syndrome, White Plague or White Plague-like disease as WPD, acknowledging that this neither includes nor excludes the presence of the pathogens A. coralicida, T. loyana or V. coralliilyticus. On the other hand, how this convergent phenotypic resemblance relates to similarities in shifts in the underlying microbial community structure is at present unknown.
In this study, we analysed microbial communities of healthy and WPD-affected coral tissues of Orbicella faveolata and Orbicella franksi (former genus Montastraea, Budd et al. (2012)) from the Caribbean (Puerto Rico). Subsequently, we compared these data to microbial communities of two coral species, Pavona duerdeni and Porites lutea, displaying WPD characteristics (Dustan 1977; Richardson 1998) from the Indo-Pacific (Gulf of Thailand) (Roder et al. 2014). We employed 16S rRNA gene microarrays (PhyloChips™) assaying 59 222 operational taxonomic units (OTUs) to profile microbial communities of healthy (HH) and diseased (DD) coral colonies in a standardized framework. We aimed to determine whether microbial community patterns of healthy and diseased colonies are not only consistent between species from the same site (Roder et al. 2014), but also over larger geographical distances, and how these patterns change between closely related and more distantly related coral species.
Material and methods
Study site and sample collection
Sampling took place on 5 and 6 September 2011 at Weimberg reef (between N 17°53′17.40/W 66°59′52.90 and N 17°53′25.40/W 66°59′19.00) off the southwest coast of Puerto Rico. Two coral species (Orbicella faveolata and O. franksi) were sampled via SCUBA between 16 and 22 m depth. From both species, tissue samples from three healthy colonies (displaying no visible signs of stress) and three colonies with WPD phenotype were collected. All corals were of similar size. All healthy samples were collected from the upper, nonshaded surface of the coral colony using hammer and chisel and were immediately transferred to sterile Whirl-pak bags. Samples of corals displaying signs of WPD were taken directly from the interface of healthy and diseased tissue. All samples were kept on ice during transportation.
Sample processing and data generation
Upon return to the laboratory, samples were rinsed with filtered seawater (0.22 μm) to remove loosely associated microbes. Rinsed samples were subsequently flash-frozen in liquid nitrogen and ground to powder using mortar and pestle. Samples were then processed as described in Roder et al. (2014). Briefly, DNA was extracted from the coral powder using the DNeasy Plant Kit (Qiagen, Hilden, Germany). After quantification of DNA using a NanoDrop 2000C spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and a Qubit fluorometer (Quant-IT dsDNA Broad Range Assay Kit; Invitrogen, Carlsbad, CA, USA), DNA was shipped on dry ice to Second Genome Inc. (San Bruno, CA, USA) for hybridization to the PhyloChip™ G3 platform as described in Hazen et al. (2010). Up to 500 ng of PCR product was applied to each PhyloChip™ G3 following previously described procedures (Hazen et al. 2010). Hybridized arrays were washed, stained, and scanned as previously described (Hazen et al. 2010). Array fluorescence intensities (HybScores) (Table S1, Supporting information) were Loess-normalized using the normalize.loess function in the affy package (Gautier et al. 2004) in the R statistical environment (R Development Core Team 2008) to obtain abundance data for OTUs present in O. faveolata and O. franksi (n = 11 256 OTUs). A microbial taxon was regarded present if it was identified in two of three replicates of any species/condition combination (O. faveolata HH, O. faveolata DD, O. franksi HH, O. franksi DD) (Table S2, Supporting information). Comparisons of these data to PhyloChip™ results from healthy and WPD-affected samples of Porites lutea and Pavona duerdeni collected in the Indo-Pacific (Roder et al. 2014) were made on Loess-normalized HybScores of shared OTUs (n = 7200 OTUs). An OTU was considered present if it was detected in two of three replicates for any species/condition combination (i.e. O. faveolata HH, O. faveolata DD, O. franksi HH, O. franksi DD, P. lutea HH, P. lutea DD, P. duerdeni HH, P. duerdeni DD).
Data analysis
We calculated OTU richness for the total data set as well as for any species/condition combination. To derive total richness of both data sets, that is, from the Caribbean (this study) and the Indo-Pacific (Roder et al. 2014), the amount of all OTUs present was determined, and OTUs shared between both, unique to either data set, or shared between conditions were compared.
Statistical evaluation of the Caribbean data set included all bacterial OTUs present (n = 11 256 OTUs). First, differentially abundant OTUs between species (O. faveolata vs. O. franksi), condition (HH vs. DD), and their interactions were determined based on normalized HybScores applying a two-way factorial analysis of variance (anova) (Table S3, Supporting information) using the TM4 software (Saeed et al. 2003). P-values were adjusted applying a 10% False Discovery Rate (FDR) using the QVALUE software package in R (Storey 2002). Log2 fold-changes in OTU abundance between different conditions were derived by averaging the abundance estimates (i.e. HybScores) for the 3 replicates and subsequent subtraction of HH from DD and division by 1000. Note that HybScores are log2 transformed fluorescence intensity values ranging from 1 to 65 536 (20–216) that are multiplied by 1000 yielding a range of 0–16 000.
Relationships between samples from the two combined data sets (Caribbean and Indo-Pacific) were illustrated using multidimensional scaling (MDS) based on Bray–Curtis distances between samples using MASS and vegan library in R (R Development Core Team 2008), in which the stress value depicts the accuracy of the ordination. Effects of the three factors ‘site’ (Caribbean vs. Indo-Pacific), ‘species’ (P. duerdeni vs. P. lutea vs. O. faveolata vs. O. franksi) and ‘condition’ (HH vs. DD) were calculated using the permutational multivariate analysis of variance (PERMANOVA) add-on in PRIMER-E (Anderson 2005). In the PERMANOVA design, we nested the factor ‘species’ within the factor ‘site’, as different species pairs were sampled at the two study sites. 999 permutations (only 998 unique for ‘condition’, only 997 unique for interaction term ‘site’בcondition’ and only 997 unique for interaction term ‘condition’בspecies(site)’) of residuals under a reduced model were conducted (Anderson & Legendre 1999; Anderson & ter Braak 2003). The resulting Pseudo-F and associated P-values can be interpreted as equal to results of univariate anovas, however, are based on the multivariate Bray–Curtis distance measures (Anderson 2005).
Differentially abundant OTUs for the combined data from the Caribbean and Indo-Pacific were determined via two-way anova, as described for the Caribbean samples above. We classified those OTUs that were more than 2-fold differentially abundant in the same direction (HH or DD) in all four species (P. duerdeni, P. lutea, O. faveolata, O. franksi) as footprint bacterial species in WPD. These respective microbial key players were additionally sorted according to their respective families. Finally, we compared phylogenetic relationships of the coral species to differences in bacterial assemblages (n = 7200 OTUs) in HH samples. Phylogenetic relationships between the four coral species (O. faveolata, O. franksi, P. lutea and P. duerdeni) (Romano & Palumbi 1996; Fukami et al. 2008; Budd et al. 2012) were compared to dendrograms based on similarities in bacterial assemblages. Dendrograms were constructed by averaging normalized OTU HybScores over samples for all HH species and subsequent application of Euclidean distance clustering (average linkage) with 1000 bootstraps using the TM4 software (Saeed et al. 2003). Trees were visualized using TreeView (Page 1996).
Results
Bacterial richness in healthy and diseased corals
Of the 59 222 microbial OTUs assayed on the PhyloChip™ G3 microarray, 11 256 OTUs were present in the coral samples collected from the Caribbean. OTU numbers were similar for both species (Orbicella faveolata and O. franksi) with more than a 50% increase in OTU richness in diseased corals compared with healthy specimens (Table 1). The numbers of detected OTUs were similar to results from a previous study in the Indo-Pacific, where we assayed healthy and WPD-affected colonies in Pavona duerdeni and Porites lutea (Roder et al. 2014). In both surveys, we observed increased richness in DD samples compared with HH. Combining OTU richness from both studies, we found a total of 18 269 distinct OTUs over the four different coral species. Of these, around 40% (7200 OTUs) were shared between coral species from both regions, while the remaining OTUs were distributed unevenly between corals from the Caribbean (4056 nonshared OTUs) and the Indo-Pacific (7013 nonshared OTUs). Of all OTUs present in either study, more than three times as many OTUs (7122) were found in diseased samples in comparison with OTUs from healthy samples (2335). Aurantimonas coralicida or Thalassomonas loyana, the two proposed causative agents of WPD, were not detected in any sample from both data sets analysed with the PhyloChip™ platform. Further, strains of Vibrio coralliilyticus, identified as proposed WS pathogens by Sussman et al. (2008), were not represented on the PhyloChip™ platform, and accordingly, not assayed.
Table 1.
OTU richness from PhyloChip™ hybridizations investigating WPD in four species from the Caribbean (Puerto Rico) and the Indo-Pacific (Gulf of Thailand)
| PhyloChip | # Bacterial OTUs |
|---|---|
| This study – Puerto Rico, Caribbean | |
| Total | 11 256 |
| In Orbicella faveolata HH | 4336 |
| In Orbicella faveolata DD | 6791 |
| In Orbicella franksi HH | 4538 |
| In Orbicella franksi DD | 7448 |
| Roder et al. (2014) – Gulf of Thailand, Indo-Pacific | |
| Total | 14 213 |
| In Pavona duerdeni HH | 2756 |
| In Pavona duerenii DD | 4434 |
| In Porites lutea HH | 7580 |
| In Porites lutea DD | 10 848 |
| Both studies | |
| Total | 18 269 |
| Unique Puerto Rico (Caribbean) | 4056 |
| Unique Gulf of Thailand (Indo-Pacific) | 7013 |
| Shared (Puerto Rico and Gulf of Thailand) | 7200 |
| Associated with HH (Caribbean and Indo-Pacific) | 2335 |
| Associated with DD (Caribbean and Indo-Pacific) | 7122 |
OTU richness is shown for all four species/condition combinations from the Caribbean (O. faveolata HH, O. faveolata DD, O. franksi HH, O. franksi DD) and from the Indo-Pacific (P. duerdeni HH, P. duerdeni DD, P. lutea HH, P. lutea DD). Bacterial taxa were counted present when detected in 2 of 3 replicates of any species/condition combination.
Differentially abundant OTUs in healthy and diseased corals from the Caribbean
A two-way anova was conducted to test for significant differences in OTU abundances between conditions (HH vs. DD), species (O. faveolata vs. O. franksi) and combinations thereof (Table 2). While no bacterial taxon differed significantly in abundance between the two coral species, 2411 OTUs were significantly different between HH and DD samples. Log2 fold-changes (FC) in bacterial abundance ranged from −5.03 (OTU 72172, genus Fusobacterium: ∼32-fold higher in HH) to +3.10 (OTU 61563, family Rhodobacteraceae: ∼8-fold higher in DD) in O. faveolata (Table S3, Supporting information). In O. franksi, log2 bacterial abundance changes ranged from −6.29 (OTU 76854, genus Spirochaeta: >70-fold higher in HH) to +5.08 (OTU 51567, phylum Acidobacteria, class PAUC37f: ∼30-fold higher in DD) (Table S3, Supporting information). Average log2 fold-changes were higher in O. franksi (log2FC = 2.42 ± 0.41) in comparison with O. faveolata (log2FC = 1.21 ± 0.70).
Table 2.
Summary statistics of two-way anova separating species and condition effects between Orbicella faveolata and Orbicella franksi
| Two-way anova (11 256 OTUs, FDR <0.1) | # Bacterial OTUs |
|---|---|
| Species significant (O. franksi vs. O. faveolata) | 0 |
| Condition significant (HH vs. DD) | 2411 |
| Interaction significant: species × condition | 1 |
Data based on normalized HybScores of 11 256 OTUs present over all coral samples.
Approximately half of the OTUs significantly different between HH and DD samples were more abundant in HH compared with DD for both Orbicella species, about 40% were more abundant in DD, and <10% of the bacterial OTUs either increased or decreased when assessing condition-specific OTUs per coral species (Table S3, Supporting information). Only a single bacterial taxon, Jannaschia sp., was found to differ significantly in abundance in a species-and-condition-type interaction confirming that bacterial abundance patterns in healthy and diseased corals seem to be conserved and distinct from coral species-specific bacteria for the majority of OTUs analysed, as found by Roder et al. (2014).
Comparison of bacterial patterns in WPD from the Caribbean and the Indo-Pacific
We combined data from this study with data from a previous study in the Indo-Pacific (Roder et al. 2014) to compare bacterial abundance patterns in healthy and diseased corals across oceans (i.e. the Caribbean and the Indo-Pacific) and coral species (i.e. O. franksi, O. faveolata, P. duerdeni, P. lutea). For this analysis, we only considered OTUs present in both data sets (n = 7200). Relationships based on OTU diversity of the samples were displayed in an MDS ordination (Fig. 1), and the relative contributions of ‘site’, ‘species’, and ‘condition’ were tested via PERMANOVA (Table 3). While all factors contributed highly significant to the structure of the data (P < 0.001), the denominator ‘site’ was the most pronounced factor as indicated by the highest Pseudo-F value (Pseudo-F = 10.40). The contribution of the factor ‘site’ was more than twice as high as that of ‘condition’ (Pseudo-F = 4.90) or ‘species’ (Pseudo-F = 2.82). As the combined data sets did not comprise the same coral species in both regions, the factor ‘species’ was nested within the factor ‘site’ and therefore yielded a lower Pseudo-F, even though the sum of squares was higher than for the factor ‘condition’ (20.72 vs. 18.02). Furthermore, interaction terms of ‘site’ and ‘condition’ were significant, while there was no significant interaction between the factors ‘condition’ and ‘species’. This pattern recaptures what we saw when analysing diseased corals from the Indo-Pacific: a strong separation by condition and a slightly stronger separation by species, but no interaction between species and condition (Roder et al. 2014). PERMANOVA results were corroborated by the MDS plot where the largest separation was between the two sites (i.e. HH samples of either ocean were distributed towards the edges of the plane), while all DD samples were arranged amid the HH samples. This indicates that bacterial communities associated with different corals, irrespective of species or site, are more similar when affected by WPD than when corals are healthy (Fig. 1).
Figure 1.
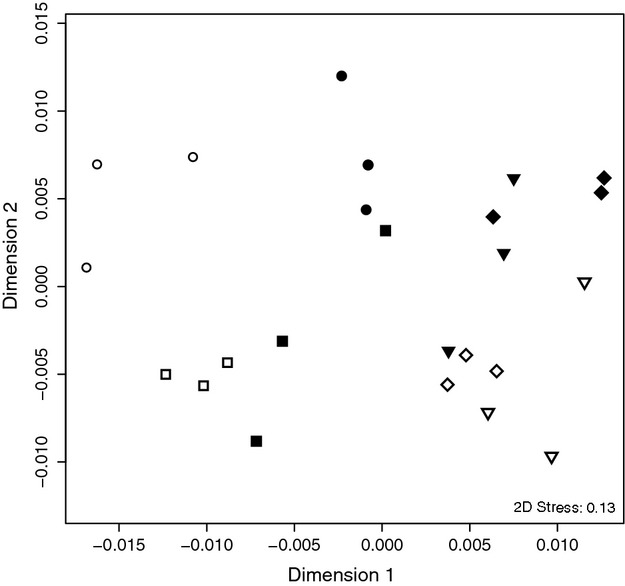
Similarities between coral species, health state and ocean basin based on microbial communities. Multidimensional scaling (MDS) plot derived from Bray–Curtis distances of normalized PhyloChip™ HybScores (n = 7200 OTUs). Healthy (open) and WPD-affected (filled) specimens of the corals P. duerderni (square), Porites lutea (circle), Orbicella faveolata (triangle) and Orbicella franksi (diamond) are shown. Stress represents the goodness of fit of the data onto the MD ordination.
Table 3.
Factors influencing bacterial communities of healthy and WPD-affected corals
| df | SS | MS | Pseudo-F | P perm | Permutations | |
|---|---|---|---|---|---|---|
| Site | 1 | 38.28 | 38.28 | 10.40 | 0.001 | 999 |
| Condition | 1 | 18.02 | 18.02 | 4.90 | 0.001 | 998 |
| Species (site) | 2 | 20.72 | 10.36 | 2.82 | 0.001 | 999 |
| Site × condition | 1 | 11.33 | 11.33 | 3.08 | 0.002 | 997 |
| Condition × species (site) | 2 | 10.28 | 5.14 | 1.40 | n.s. | 997 |
| Residuals | 16 | 58.87 | 3.68 | |||
| Total | 23 | 157.50 |
PERMANOVA summary statistics for factors affecting bacterial communities in healthy and WPD-affected corals from the Caribbean (i.e. Puerto Rico) and the Indo-Pacific (i.e. Gulf of Thailand).
Df, degrees of freedom; SS, sum of squares; MS, mean squares; n.s., not significant.
To identify key bacterial species and community shifts (i.e. bacterial footprints) within the microbiomes of healthy and diseased corals, we conducted a two-way anova and determined all those OTUs that were more than 2-fold different between HH and DD in all four species and assorted them to bacterial families (Fig. 2). For those taxa >2-fold more abundant in HH corals, only 2 bacterial families (Lachnospiraceae, Prevotellaceae) were represented by more than 1 OTU, whereas 5 bacterial families (Desulfomicrobiaceae, Lactobacillaceae, Rikenellaceae, Streptococcaceae, Xanthomonadaceae) were each represented by 1 OTU. Except for Desulfomicrobium orale (OTU 57563), none of the OTUs could be assigned to a described bacterial species. Those bacterial taxa with more >2-fold higher abundances in DD corals were comprised of 4 OTUs from 4 families (Alteromonadaceae, Flavobacteriaceae, Phyllobacteriaceae, Rhodospirillaceae) and 12 OTUs belonging to the family Rhodobacteraceae. Accordingly, the family Rhodobacteraceae contributed 75% of the key OTUs in DD samples and also represented the highest fold-changes. The only OTU assigned to the species level from this bacterial family was Rhodobacter sphaeroides (OTU 61632), while all other bacterial taxa were taxonomically unclassified at the species level.
Figure 2.
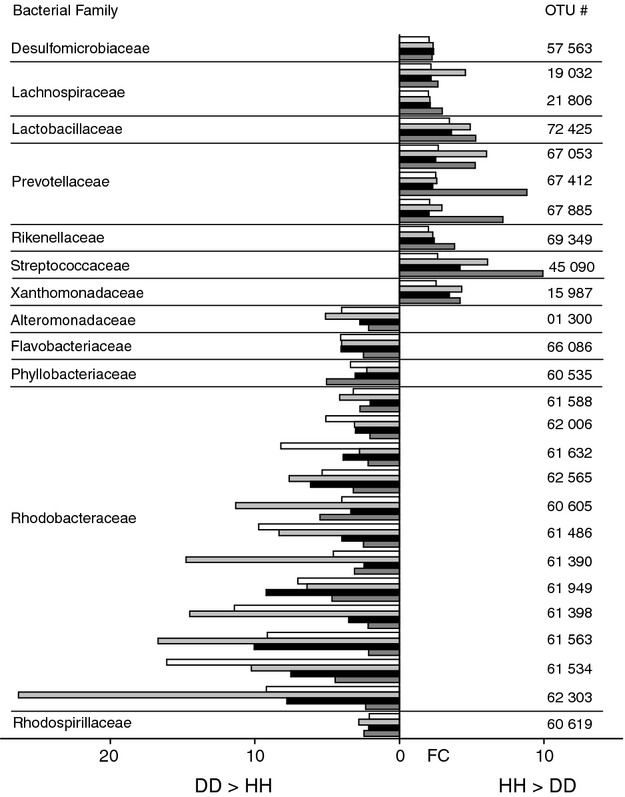
Bacterial footprints of WPD. Displayed are bacterial families and OTUs that showed a >2-fold abundance difference between HH and DD over samples from Pavona duerdeni (white), Porites lutea (light grey), Orbicella faveolata (black) and Orbicella franksi (dark grey). FC, fold change.
Phylogenetic position of the coral host and bacterial community structure
While the main factor structuring all eight species/condition combinations appeared to be region, bacterial community profiles of DD samples were closely related to each other irrespective of species or site (Fig. 1). To further understand the contribution of phylogenetic positioning of a coral species to its microbial abundance pattern, HH samples were clustered based on microbial diversity and compared to the phylogeny between the four coral species (Fig. 3). Samples from healthy colonies recaptured phylogenetic positioning of the coral species. Furthermore, similarity between corals decreased with increasing phylogenetic distance, that is, healthy specimens of the two closely related Orbicella species harboured a more similar microbial abundance pattern than the more distantly related P. duerdeni and P. lutea. The greatest difference between HH bacterial abundance patterns of all four species was between complex and robust corals that are evolutionarily separated by >200 million years (Romano & Palumbi 1996). However, species pairs from a given site in our study (i.e. O. franksi and O. faveolata from the Caribbean, P. duerdeni and P. lutea from the Indo-Pacific) coincided with affiliation to the robust (O. franksi and O. faveolata) and complex (P. duerdeni and P. lutea) clades, so we were not able to disentangle whether the differences arose from phylogenetic or ocean basin (site) affiliation.
Figure 3.
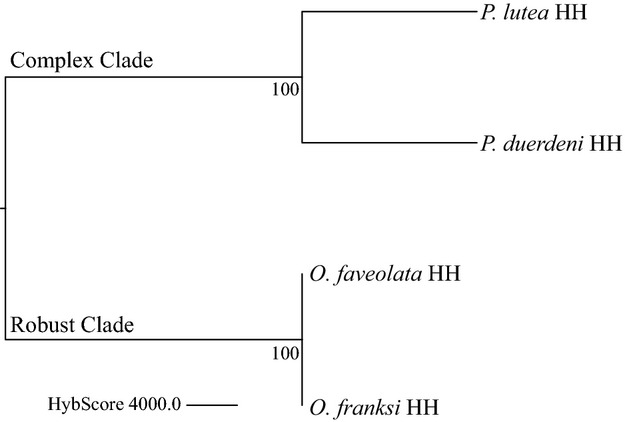
Grouping of corals based on underlying bacterial community structures. Relationship between healthy coral species in regard to differences in their bacterial community based on Euclidean distance of bacterial abundance (i.e. HybScores) from 7200 shared OTUs (1000 bootstraps). The dendrogram recaptures the phylogenetic relationship of the corals Orbicella franksi, Orbicella faveolata, Pavona duerdeni and Porites lutea.
Discussion
Bacterial richness in healthy and diseased corals
The numbers of OTUs detected in samples from the Caribbean were comparable to those detected in other studies applying PhyloChips™ in corals (Sunagawa et al. 2009; Kellogg et al. 2012; Roder et al. 2014), but higher than those in sequencing-based surveys (e.g. Cardenas et al. 2012). Richness and diversity of bacterial taxa associated with two Orbicella species were similar within the same condition (HH and DD) as has been shown for microbial communities of closely related sponge species (Schoettner et al. 2013). We also found higher numbers of bacterial taxa associated with diseased samples, which concurs with previous studies on coral disease (Pantos et al. 2003; Sunagawa et al. 2009; Cróquer et al. 2012; Roder et al. 2014). This is suggested to be a consequence of bacterial colonization from the surrounding environment on compromised and vulnerable coral tissues (Lesser et al. 2007; Sunagawa et al. 2009; Roder et al. 2014) and supported in this work by a larger number of shared OTUs, and an increase in richness of unclassified bacteria in DD samples. It is worthwhile noting that bacterial diversity has been reported to be similar and overlapping across different oceans (Martiny et al. 2006). Accordingly, colonization of bacteria from the surrounding water column would result in similar microbial profiles of diseased tissue even between geographically distinct regions.
Towards elucidation of bacterial disease footprints and microbiomes
A great advantage of the PhyloChip™ microarray is that it represents a standardized platform in which data over different studies can be easily integrated, comprehensively analysed and compared in a common framework. While our recent analysis of two coral species from the same reef in the Indo-Pacific indicated that microbial community patterns of health and disease are conserved over coral species boundaries (Roder et al. 2014), we were not able to test whether these patterns hold true over geographical distances (i.e. regionally and globally) in coral species. The experimental design of this study matches and complements that of Roder et al. (2014), enabling us to integrate and compare healthy and diseased states of four coral species (Pavona duerdeni, Porites lutea, Orbicella faveolata, Orbicella franksi) comprising three genera (Pavona, Porites, Orbicella) from two distinct regions (Caribbean and Indo-Pacific). Most interestingly, almost half of all the bacterial taxa found in either data set were shared (n = 7200 OTUs), even though we analysed different coral species and different regions at different depths. We found that region is the strongest separating factor between the two data sets. However, this measure took inadvertently the closer phylogenetic relationship between O. faveolata and O. franksi into account, so that we were not able to differentiate between similarity due to common reef of origin and similarity due to phylogenetic relatedness. Profiling bacterial diversity in healthy and diseased tissues from the same coral species over geographical distances will unequivocally resolve the relative contribution of phylogenetic similarity vs. health condition (e.g. data from this study and Kellogg et al. (2013)).
Between closely related coral species (i.e. O. faveolata and O. franksi), microbial abundance patterns were indistinguishable (as indicated by no significant different OTUs between species in the two-way anova) and support the notion that there is a close relationship between a coral host and its bacterial assemblage (Sunagawa et al. 2010). Indeed, grouping of corals based on bacterial community structure of HH specimens (Fig. 3) indicates that microbial assemblage resembles phylogenetic position of the coral host. Besides, diseased corals were more similar to each other than to healthy ones as indicated by the MDS plot (Fig. 1). Last, we detected no bacteria in the data set from the Indo-Pacific (Roder et al. 2014) and only one OTU in the data set from the Caribbean (this study) that were significantly different between species and at the same time significantly different between conditions. These data substantiate the previous notion that shared disease-specific microbial community patterns exist that are distinct form nonshared species-specific bacterial assemblages (Roder et al. 2014).
Bacterial taxa that were consistently found in WPD-affected coral species analysed here comprised only few families and support the hypothesis of a bacterial abundance pattern in WPD that is structured by opportunists. For instance, Pelagibacteraceae are abundant in ocean surface bacterioplanktion communities (Morris et al. 2002), and Rhodospirillaceae have been reported to become abundant in heterotrophic environments (Landa et al. 2013). Most notable, OTUs from the family Rhodobacteraceae made up 75% of all OTUs more than 2-fold enriched in DD, and fold-changes were more drastic than for any other bacterial family. As none of the members of the Rhodobacteraceae family are known to be potentially pathogenic, we hypothesize that the reason for their colonization success must either lie in the specific characteristics of this family (e.g. competence for rapid proliferation) or be due to prevailing environmental conditions (e.g. high abundance in environment). Many members of the Rhodobacteraceae are photoheterotrophs, and their ability to accumulate in response to availability of organic substrate (Landa et al. 2013) might present optimal conditions for effective growth in or on compromised coral tissues. Accumulation of opportunistic bacteria has been suggested to result in phenotypically similar mortality patterns (diseases) that might not necessarily have the same cause (Lesser et al. 2007). WS, WPD or WPD-like infections have been reported for a variety of coral species and regions (Richardson 1998; Sutherland et al. 2004; Willis et al. 2004; Bourne et al. 2009; Kellogg et al. 2013), but only three bacterial species have been proposed as causative agents (Denner 2003; Thompson et al. 2006; Sussman et al. 2008). Also, rates of the disease's progression have been shown to be different (Dustan 1977; Richardson et al. 1998, 2001). Conversely, while it is likely that distinct pathogens might cause WPD in different coral species, similarity in phenotypes and etiopathologies might be driven by opportunistic bacteria that are conserved over species boundaries and that structure microbial abundance patterns.
Taken together, our data indicate that (i) disease-specific microbial abundance patterns exist, which are (ii) conserved across coral species and oceans, and which are (iii) largely different from species-specific abundance patterns. While the increase in bacterial diversity from WPD specimens over ocean boundaries renders the hypothesis of colonization of opportunistic bacteria a likely scenario, the trigger of opportunistic colonization remains unknown and might well be of varying origin and importance. Conserved microbial community patterns provide the opportunity to derive bacterial families and species whose relative abundance can serve as indicators for health, stress and disease. The elucidation of commonalities in coral diseases beyond species and sites would allow the establishment of ‘bacterial footprints’, that is, shared community profiles for a given disease that can inform on the state of the coral holobiont irrespective of the coral species and/or region under study. Furthermore, such bacterial footprints would allow placing different coral diseases into a comparative framework and specifically test evolutionary hypotheses in regard to cause, origin and relatedness of different coral diseases.
Acknowledgments
We would like to thank Todd DeSantis and SecondGenome, Inc. for discussion and support regarding PhyloChip™ data structure and transformation. This project was partially funded by NSF IOS # 1017510 and OCE -1105143 to EW. The Department of Marine Sciences, University of Puerto Rico Mayaguez provided partial funding for boat and diving, logistical support, and laboratory space. Remaining financial support was provided by KAUST baseline funds to CRV. We also thank the comments from the editor and two anonymous reviewers that helped validating data and improving the manuscript.
Biography
C.R.V. conceived and designed the experiments. C.R., C.A., C.D., E.W. generated data. C.R.V. and C.R. analysed and interpreted data. C.R.V. and C.R. wrote the article. All authors read the article and approved the final version.
Conflict of interests
The authors declare that no competing commercial or other interests in relation to this work exist.
Data accessibility
PhyloChip abundance data, OTU presence/absence data, anova data: uploaded as online supporting information.
Supporting Information
Additional supporting information may be found in the online version of this article.
HybScores for all present OTUs (n = 11 256) of PhyloChip™ microarrays from Orbicella faveolata HH, DD and Orbicella franksi HH, DD.
Presence/Absence calls of 11 256 OTUs over all samples on PhyloChip™ microarrays (0 = absent, 1 = present).
Two-way anova on 11 256 OTUs that were detected present in healthy and diseased specimens of the corals Orbicella faveolata and Orbicella franksi.
References
- Anderson MJ. PERMANOVA: A FORTRAN Computer Program for Permutational Multivariate Analysis of Variance. New Zealand: Department of Statistics, University of Auckland; 2005. [Google Scholar]
- Anderson MJ, Legendre P. An empirical comparison of permutation methods for tests of partial regression coefficients in a linear model. Journal of Statistical Computation and Simulation. 1999;62:271–303. [Google Scholar]
- Anderson MJ, ter Braak CJF. Permutation tests for multi-factorial analysis of variance. Journal of Statistical Computation and Simulation. 2003;73:85–113. [Google Scholar]
- Antonius A. Coral diseases in the Indo-Pacific: a first record. Marine Ecology. 1985;6:197–218. [Google Scholar]
- Barash Y, Sulam R, Loya Y, Rosenberg E. Bacterial Strain BA-3 and a filterable factor cause a white plague-like disease in corals from the Eilat coral reef. Aquatic Microbial Ecology. 2005;40:183–189. [Google Scholar]
- Bayer T, Neave MJ, Alsheikh-Hussain A, et al. The microbiome of the Red Sea Coral Stylophora pistillata is dominated by tissue-associated Endozoicomonas bacteria. Applied and Environmental Microbiology. 2013;79:4759–4762. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne DG, Garren M, Work TM, et al. Microbial disease and the coral holobiont. Trends in Microbiology. 2009;17:554–562. doi: 10.1016/j.tim.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Budd AF, Fukami H, Smith ND, Knowlton N. Taxonomic classification of the reef coral family Mussidae (Cnidaria: Anthozoa: Scleractinia) Zoological Journal of the Linnean Society. 2012;166:465–529. [Google Scholar]
- Cardenas A, Rodriguez RL, Pizarro V, Cadavid LF, Arevalo-Ferro C. Shifts in bacterial communities of two Caribbean reef-building coral species affected by white plague disease. ISME Journal. 2012;6:502–512. doi: 10.1038/ismej.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cróquer A, Bastidas C, Elliott A, Sweet M. Bacterial assemblages shifts from healthy to yellow band disease states in the dominant reef coral Montastraea faveolata. Environmental Microbiology Reports. 2012;5:90–96. doi: 10.1111/j.1758-2229.2012.00397.x. [DOI] [PubMed] [Google Scholar]
- Denner EBM. Aurantimonas coralicida gen. nov., sp. nov., the causative agent of white plague type II on Caribbean scleractinian corals. International Journal of Systematic and Evolutionary Microbiology. 2003;53:1115–1122. doi: 10.1099/ijs.0.02359-0. [DOI] [PubMed] [Google Scholar]
- Dustan P. Vitality of reef coral populations off Key Largo, Florida - recruitment and mortality. Environmental Geology. 1977;2:51–58. [Google Scholar]
- Fukami H, Chen CA, Budd AF, et al. Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (Order Scleractinia, Class Anthozoa, Phylum Cnidaria) PLoS ONE. 2008;3:e3222. doi: 10.1371/journal.pone.0003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Harvell D, Jordán-Dahlgren E, Merkel S, et al. Coral disease, environmental drivers, and the balance between coral and microbial associates. Oceanography. 2007;20:172–195. [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- Hong MJ, Yu YT, Chen CA, Chiang PW, Tang SL. Influence of species specificity and other factors on bacteria associated with the coral Stylophora pistillata in Taiwan. Applied and Environmental Microbiology. 2009;75:7797–7806. doi: 10.1128/AEM.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg CA, Piceno YM, Tom LM, et al. PhyloChip microarray comparison of sampling methods used for coral microbial ecology. Journal of Microbiological Methods. 2012;88:103–109. doi: 10.1016/j.mimet.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Kellogg CA, Piceno YM, Tom LM, et al. Comparing bacterial community composition between healthy and White Plague-like disease states in Orbicella annularis using PhyloChipTM G3 microarrays. PLoS ONE. 2013;8:e79801. doi: 10.1371/journal.pone.0079801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman D, Kashman Y, Rosenberg E, Kushmaro A, Loya Y. Antimicrobial activity of Red Sea corals. Marine Biology. 2006;149:357–363. [Google Scholar]
- Kimes NE, Van Nostrand JD, Weil E, Zhou J, Morris PJ. Microbial functional structure of Montastraea faveolata, an important Caribbean reef-building coral, differs between healthy and yellow-band diseased colonies. Environmental Microbiology. 2010;12:541–556. doi: 10.1111/j.1462-2920.2009.02113.x. [DOI] [PubMed] [Google Scholar]
- Landa M, Cottrell MT, Kirchman DL, Blain S, Obernosterer I. Changes in bacterial diversity in response to dissolved organic matter supply in a continuous culture experiment. Aquatic Microbial Ecology. 2013;69:157–168. [Google Scholar]
- Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Bythell JC, Gates RD, Johnstone RW, Hoegh-Guldberg O. Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. Journal of Experimental Marine Biology and Ecology. 2007;346:36–44. [Google Scholar]
- Littman RA, Willis BL, Pfeffer C, Bourne DG. Diversities of coral-associated bacteria differ with location, but not species, for three acroporid corals on the Great Barrier Reef. FEMS Microbiology Ecology. 2009;68:152–163. doi: 10.1111/j.1574-6941.2009.00666.x. [DOI] [PubMed] [Google Scholar]
- Martiny JB, Bohannan BJ, Brown JH, et al. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Miller J, Muller E, Rogers C, et al. Coral disease following massive bleaching in 2005 causes 60% decline in coral cover on reefs in the US Virgin Islands. Coral Reefs. 2009;28:925–937. [Google Scholar]
- Morris RM, Rappe MS, Connon SA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Page RD. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Pantos O, Cooney RP, Le Tissier MDA, et al. The bacterial ecology of a plague-like disease affecting the Caribbean coral Montastrea annularis. Environmental Microbiology. 2003;5:370–382. doi: 10.1046/j.1462-2920.2003.00427.x. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Reshef L, Ron E, Rosenberg E. Genome analysis of the coral bleaching pathogen Vibrio shiloi. Archives of Microbiology. 2008;190:185–194. doi: 10.1007/s00203-008-0388-0. [DOI] [PubMed] [Google Scholar]
- Richardson LL. Coral diseases: what is really known? Trends in Ecology & Evolution. 1998;13:438–443. doi: 10.1016/s0169-5347(98)01460-8. [DOI] [PubMed] [Google Scholar]
- Richardson L, Goldberg WM, Carlton G, Halas JC. Coral disease outbreak in the Florida Keys: Plague Type II. Revista de Biología Tropical. 1998;46:187–198. [Google Scholar]
- Richardson L, Smith G, Ritchie K, Carlton R. Integrating microbiological, microsensor, molecular, and physiologic techniques in the study of coral disease pathogenesis. Hydrobiologia. 2001;460:71–89. [Google Scholar]
- Ritchie KB. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Marine Ecology Progress Series. 2006;322:1–14. [Google Scholar]
- Ritchie KB, Smith GW. Microbial Communities of Coral Surface Mucopolysaccharide Layers. In: Rosenberg E, Loya Y, editors. Coral Health and Disease. Berlin: Springer-Verlag; 2004. pp. 259–263. [Google Scholar]
- Roder C, Arif C, Bayer T, et al. Bacterial profiling of White Plague Disease in a comparative coral species framework. ISME Journal. 2014;8:31–39. doi: 10.1038/ismej.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Marine Ecology Progress Series. 2002;243:1–10. [Google Scholar]
- wRomano S, Palumbi S. Evolution of scleractinian corals inferred from molecular systematics. Science. 1996;271:640–642. [Google Scholar]
- Rosenberg E, Kushmaro A. Microbial diseases of corals: pathology and ecology. In: Dubinsky Z, Stambler N, editors. Coral Reefs: An Ecosystem in Transition. New York: Springer; 2011. pp. 451–464. [Google Scholar]
- Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. The role of microorganisms in coral health, disease and evolution. Nature Reviews Microbiology. 2007;5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Schoettner S, Hoffmann F, Cardenas P, et al. Relationships between host phylogeny, host type and bacterial community diversity in cold-water coral reef sponges. PLoS ONE. 2013;8:e55505. doi: 10.1371/journal.pone.0055505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B. 2002;64:479–498. [Google Scholar]
- Sunagawa S, DeSantis TZ, Piceno YM, et al. Bacterial diversity and White Plague Disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME Journal. 2009;3:512–521. doi: 10.1038/ismej.2008.131. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PLoS ONE. 2010;5:e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman M, Willis B, Victor S, Bourne D. Coral pathogens identified for White Syndrome (WS) epizootics in the Indo-Pacific. PLoS ONE. 2008;3:e2393. doi: 10.1371/journal.pone.0002393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KP, Porter JW, Torres C. Disease and immunity in Caribbean and Indo-Pacific zooxanthellate corals. Marine Ecology Progress Series. 2004;266:273–302. [Google Scholar]
- Thompson FL, Barash Y, Sawabe T, et al. Thalassomonas loyana sp. nov., a causative agent of the white plague-like disease of corals on the Eilat coral reef. International Journal of Systematic and Evolutionary Microbiology. 2006;56:365–368. doi: 10.1099/ijs.0.63800-0. [DOI] [PubMed] [Google Scholar]
- Weil E. Coral Reef Diseases in the Wider Caribbean. In: Rosenberg E, Loya Y, editors. Coral Health and Disease. Berlin, Heidelberg: Springer; 2004. pp. 35–64. [Google Scholar]
- Weil E, Rogers CS. Coral Reef Diseases in the Atlantic-Caribbean. In: Dubinsky Z, Stambler N, editors. Coral Reefs: An Ecosystem in Transition. New York: Springer; 2011. pp. 465–491. [Google Scholar]
- Weil E, Smith G, Gil-Agudelo DL. Status and progress in coral reef disease research. Diseases of Aquatic Organisms. 2006;69:1–7. doi: 10.3354/dao069001. [DOI] [PubMed] [Google Scholar]
- Weil E, Croquer A, Urreiztieta I. Temporal variability and impact of coral diseases and bleaching in La Parguera, Puerto Rico from 2003-2007/Ernesto Weil, Aldo Croquer, Isabel Urreiztieta. Caribbean Journal of Science. 2009;45:221–246. [Google Scholar]
- Willis BL, Page CA, Dinsdale EA. Coral disease on the Great Barrier Reef. In: Rosenberg E, Loya Y, editors. Coral Health and Disease. Berlin: Springer-Verlag; 2004. pp. 69–104. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HybScores for all present OTUs (n = 11 256) of PhyloChip™ microarrays from Orbicella faveolata HH, DD and Orbicella franksi HH, DD.
Presence/Absence calls of 11 256 OTUs over all samples on PhyloChip™ microarrays (0 = absent, 1 = present).
Two-way anova on 11 256 OTUs that were detected present in healthy and diseased specimens of the corals Orbicella faveolata and Orbicella franksi.
Data Availability Statement
PhyloChip abundance data, OTU presence/absence data, anova data: uploaded as online supporting information.


