Abstract
The sharing of secreted invertase by yeast cells is a well-established laboratory model for cooperation, but the only evidence that such cooperation occurs in nature is that the SUC loci, which encode invertase, vary in number and functionality. Genotypes that do not produce invertase can act as ‘cheats’ in laboratory experiments, growing on the glucose that is released when invertase producers, or ‘cooperators’, digest sucrose. However, genetic variation for invertase production might instead be explained by adaptation of different populations to different local availabilities of sucrose, the substrate for invertase. Here we find that 110 wild yeast strains isolated from natural habitats, and all contained a single SUC locus and produced invertase; none were ‘cheats’. The only genetic variants we found were three strains isolated instead from sucrose-rich nectar, which produced higher levels of invertase from three additional SUC loci at their subtelomeres. We argue that the pattern of SUC gene variation is better explained by local adaptation than by social conflict.
Keywords: cheating, cooperation, copy number variation, droplet digital PCR, Saccharomyces, SUC
Introduction
In contrast to other eukaryotes, the genome of Saccharomyces cerevisiae is compact, containing few redundant genes or pseudogenes (Goffeau et al. 1996; Lafontaine et al. 2004). The SUC genes, which encode the extracellular enzyme invertase, are exceptional. There are nine known loci for SUC genes: SUC1–SUC5 and SUC7–SUC10 (Naumov & Naumova 2010). SUC2, the ancestral locus, is located in the left arm of chromosome IX, but the other copies are all found at subtelomeric regions (Carlson & Botstein 1983; Carlson et al. 1985; Naumov & Naumova 2010). Most strains contain only a single SUC2 gene, but some contain one or more of the subtelomeric SUC loci in addition to SUC2, and others have a suc2 pseudogene and produce no invertase (Carlson & Botstein 1983; Naumov et al. 1996; Denayrolles et al. 1997).
The variation in SUC genotypes can be explained using social evolution theory (Greig & Travisano 2004). The invertase produced from SUC genes is secreted to digest extracellular sucrose into the preferred sugars glucose and fructose, which can be taken up by the cell and metabolized. Sugars diffuse readily, so cells that cannot produce invertase themselves because they lack any functional SUC genes can use the glucose and fructose produced by those that do (Gore et al. 2009). Thus, invertase production is analogous to public goods cooperation with nonproducers as cheats that can exploit and invade populations of cooperators (Greig & Travisano 2004). The phenomenon of telomeric silencing (Wyrick et al. 1999) has been used to explain subtelomeric SUC loci. If suc2 cheats retain an unexpressed but functional copy of SUC, then having invaded a colony of cooperators and depleted the public good, they could regain the ability to produce invertase from a silent subtelomeric ‘backup’ copy (Greig & Travisano 2004). Consistent with this social theory model, laboratory experiments find that the relative fitness of nonproducers can be higher or lower than that of producers, depending on factors such as density (Greig & Travisano 2004), frequency (Gore et al. 2009; Damore & Gore 2012) and sucrose concentration (Koschwanez et al. 2011). However, a recent experiment found that mixed cultures of producers and nonproducers had higher mean fitness than monocultures of producers, inconsistent with the model of nonproducers as cheats (MacLean et al. 2010).
An alternative explanation for SUC variation is that different SUC genotypes have adapted to environments with different availabilities of sucrose. For thousands of years, humans have used yeast to make alcohol, and more recently, to raise bread, to flavour foods, to study genetics and to secrete bio-engineered products such as insulin (Thim et al. 1986; Botstein & Fink 1988; Porro et al. 1995). A survey of the drinks available in a typical bar reveals some of the diverse substrates that domesticated yeast strains are grown on. Yeast produces invertase constitutively, even in the absence of sucrose, although high levels of glucose can suppress invertase production (MacLean et al. 2010). Substrates low in sucrose might favour the loss of costly invertase production and the selection of suc2 null mutants. Conversely, substrates rich in sucrose might select for additional subtelomeric copies of SUC if they were not completely silenced and could therefore contributed to increased invertase production (Denayrolles et al. 1997; Batista et al. 2004). Thus, the observed diversity in SUC genotypes may simply be due to domestication in different environments (Libkind et al. 2011). Similar increases in diversity are seen in other domesticated species, for example domesticated dogs have much greater morphological variation than wolves, their wild ancestors (Wayne 1986) indeed the range of body sizes in among breeds of this single domesticated species exceeds the range of all other wild canid species (Lindblad-Toh et al. 2005).
These two competing hypotheses can be tested by examining how individuals vary within and between habitats. The social conflict hypothesis predicts that different strains isolated from the same habitat will differ in their SUC genotypes, because some will be cheats and others will be cooperators. The sucrose adaptation hypothesis predicts that different strains from the same habitat will have the same SUC genotype, but that strains from environments with different sucrose availabilities will differ. Naumov et al. (1996) surveyed SUC gene variation in 91 strains isolated from many different environments, finding eleven invertase nonproducing strains that contained only a nonfunctional suc2 allele. Five of these came from olive processing (Vaughan & Martini 1987), and two came from human faeces (Naumov et al. 1990), environments that are low in sucrose (Marsilio et al. 2001), which is consistent with the sucrose adaptation hypothesis. But two came from wine, an environment that also provided many invertase producers, consistent with the social conflict hypothesis. The remaining nonproducer (GM51) has unknown origins (Naumov et al. 1996). Ten strains had multiple SUC genes, and all came from sucrose-rich environments (strawberry, grape, ginger wine, billi wine and palm wine, Naumov et al. 1993; Basson et al. 2010; Kim & Lee 2006) or from fermentations that are artificially supplemented with sucrose (distilling and champagne making, Naumov et al. 1996), supporting the sucrose adaptation hypothesis. These results are difficult to interpret because different lineages of S. cerevisiae are often genetically mixed (Liti et al. 2009), perhaps by the process of human domestication (Libkind et al. 2011), and because many of the strains were not systematically isolated and their origins are unclear.
We therefore decided to determine the frequency of invertase nonproducers in S. paradoxus, the wild relative to S. cerevisiae. S. paradoxus has several advantages over S. cerevisiae for this study. The most important is that S. paradoxus is not used in human fermentations and instead has a well-established and well-sampled natural habitat extending to several continents on oak trees (Naumov et al. 1998; Johnson et al. 2004), and on Canadian maple trees (Charron et al. 2014). Unlike S. cerevisiae, whose populations show little geographical structure and high gene flow, perhaps because humans move strains around the world and mix them (Liti et al. 2009), S. paradoxus populations have strong geographical structure, with little mixing between lineages from different places (Kuehne et al. 2007; Liti et al. 2009). These properties mean that any S. paradoxus strain is likely to have evolved in the environment from which it was isolated, and is very unlikely to be a recent immigrant adapted to a different environment or to contain genetic material from such an immigrant. To test the hypothesis that social conflict should produce SUC variation among individuals within a single type of habitat, we determined the invertase production, the SUC loci and the SUC gene copy number of a set of 80 S. paradoxus strains: 65 isolated from oak trees and 15 isolated from maple trees. We did not have similarly large sets of S. cerevisiae strains from well-defined habitats and cannot exclude the possibility that some wild-caught strains might originate from human fermentations, or be related to such feral escapees. Nevertheless, we also tested 30 S. cerevisiae strains we could find that were isolated from apparently natural sources, including 15 recent isolates from primeval forests, which form distinct lineages compared to all the other S. cerevisiae strains identified so far (Wang et al. 2012). Finally, we tested whether strains with SUC2 deleted produced invertase from their subtelomeric SUC loci or whether they were ‘silent’.
Materials and methods
All strains, their original strain numbers, references, details of their origins and inclusion in genome sequencing projects are described in Appendix S1 (Supporting information).
To determine whether SUC genotypic variation occurs within (rather than between) wild populations, it is necessary to have multiple examples of wild strains isolated from a single well-defined habitat. S. paradoxus is ideal for this because it is not domesticated and many strains have been systematically isolated from oak trees. We tested all the oak-associated strains that we could access, including 29 that we isolated ourselves in Germany, 25 from the United Kingdom, 7 from Russia, 3 from North America and 1 from Japan (see Appendix S1, Supporting information for details). More recently, Canadian maple trees have been identified as a habitat for S. paradoxus, and we included 15 strains of S. paradoxus isolated from Canadian maple trees (Charron et al. 2014). We tested all S. paradoxus strains that we could acquire, but we excluded single strains isolated from unique or poorly described habitats and those from insect vectors which might have fed on unknown substrates.
We were concerned that any S. cerevisiae strains we tested might have recently escaped from human fermentations, or might have been crossed to such feral strains. Further, the natural habitat of S. cerevisiae is less well established than that of S. paradoxus. We therefore focused primarily on S. paradoxus. However, a large set of S. cerevisiae has recently been isolated from primeval forests in China, far from human influence, and there is good evidence they represent a truly wild population (Wang et al. 2012). We were able to get hold of 15 of these strains to test (7 from rotten wood, 2 from soil, 2 from oak, 2 from beech and one each from persimmon and oriental raisin trees). The majority of other available S. cerevisiae strains have been isolated from human fermentations or associated places, such as vineyards and food processing facilities. However, we were able to find 15 additional S. cerevisiae strains from a variety of apparently natural habitats: 5 from oak, 3 from soil, 3 from Bertram palm nectar and one each from cactus, cactus fruit, fig and cocoa.
We also tested various S. cerevisiae strains as controls and for comparison purposes. Our standard control strains were C.Lab.1. and C.Lab.1.suc2::KANMX, are isogenic with strains that have been used as a ‘cooperators’ and ‘cheats’, respectively, in previous laboratory studies on cooperation (Greig & Travisano 2004; MacLean & Brandon 2008; Gore et al. 2009; MacLean et al. 2010). We included two domesticated strains, C.Ginger.wine and C.Billi.wine, as positive controls with known multiple SUC copies. Finally, we knocked out the SUC2 loci from these two strains as well as from the three wild strains that turned out to have multiple SUC copies, creating five new strains: C.Ginger.wine.suc2::NATMX, C.Billi.wine.suc2::NATMX, C.Nectar.1.suc2::NATMX, C.Nectar.2.suc2::NATMX and C.Nectar.3.suc2::NATMX.
Screening wild strains for invertase nonproducers
To determine which of our strains produced invertase, we used the Glucose (HK) Assay Kit (Sigma-Aldrich, St. Louis, MO, USA), which produces a colorimetric reaction in response to glucose. We calibrated the assay using known dilutions of purified invertase (Sigma-Aldrich). Twenty microlitre of each dilution was combined with 100 μL sodium acetate buffer (0.2 m, pH = 5.2), and 50 μL of 0.5 m sucrose added. The reaction was incubated at 37 °C for 20 min, then stopped by adding 300 μL of 0.2 m K2HPO4 and heating at 100 °C for 5 min. One-hundred and fifty microlitre of this reaction mixture was added to 1 mL glucose assay reagent provided by the kit, and the optical absorbance at 340 nm was determined after following the kit instructions. We found the assay gave a linear response between absorbances of 0.11 and 0.78 (Appendix S4, Supporting information).
We optimized the assay conditions using a laboratory strain, C.Lab.1., which produces invertase from a single SUC2 locus and has been used as a ‘cooperator’ in previous work on sociality (Greig & Travisano 2004; MacLean & Brandon 2008; Gore et al. 2009; MacLean et al. 2010). Each strain was grown in 2 mL of YEPD (1% yeast extract, 2% peptone, 2% dextrose) overnight at 30 °C. We spun down 1 mL of the culture, washed it with 1 mL of sterile water and centrifuged again. The pellet was resuspended in 1 mL of 0.9% sterile saline, and 5 μL of the cell suspension was spotted onto a YEPS plate (1% yeast extract, 2% peptone, 2% sucrose and 2.5% agar) and incubated it for 2 days at 30 °C. The resulting colony was then resuspended in 5 mL of sterile water, and a 100 μL sample was spun down and washed twice, then resuspended in 50 μL of sterile water, combined with 100 μL sodium acetate buffer and 50 μL of 0.5 m sucrose, incubated at 37 °C for 20 min and stopped with 300 μL of 0.2 m K2HPO4 heating as described above. One-hundred microlitre of the reaction mixture was added to 1 mL glucose assay reagent, and the absorbance was read. We used the same method on the isogenic ‘cheat’ strain C.Lab.1.suc2::KANMX. Pilot experiments indicated that wild strains produced so much more invertase than the laboratory ‘cooperator’ strain C.Lab.1 that they saturated the assay, so we reduced the volume of the resuspended cells from 100 to 20 μL, making the suspension up to 100 μL with 80 μL of the nonproducer C.Lab.1.suc2::KANMX prepared in the same way. Measurements were then multiplied by five to correct for this dilution. We screened the invertase production of all 110 wild strains in this way (see Appendix S1, Supporting information).
SUC alleles in whole genome sequences
Whole genome sequences were available for 29 S. paradoxus and 8 S. cerevisiae strains. For details of which strains had sequences, and where the sequences can be accessed, please see Appendix S1 (Supporting information). These genome sequences were used to determine whether a strain contained intact SUC open reading frames or suc pseudogenes. The nucleotide sequences of SUC genes that were identified in this way are listed in Appendices S5 and S6 (Supporting information).
Southern blots for SUC loci
Whole genome sequences were not available for most of our strains, and even for the 29 S. paradoxus and 8 S. cerevisiae that had been sequenced, we could not reliably infer the SUC loci or copy numbers from the sequences because of the short reads and low sequencing coverage. Subtelomeric SUC genes are embedded in highly repetitive DNA which may not be properly assembled in genome sequencing projects. To determine the SUC loci in our wild strains, we therefore made Southern blots of whole-chromosome pulsed-field gels, and probed them with labelled SUC2 fragments. We also included controls on the pulsed-field gels: the C.Lab.1.suc2::KANMX as nonproducer containing no known SUC genes, C.Lab.1 as a producer containing a single SUC2 gene and the domesticated strains C.Ginger.wine and C.Billi.wine as positive controls previously identified as containing multiple SUC loci (Naumov et al. 1996). All strains are described in Appendix S1 (Supporting information).
We prepared chromosomal DNA plugs according to Carle & Olson (1985). S. cerevisiae CHEF DNA size standard (YNN295 strain) was used in all pulse-field gel electrophoresis runs (Bio-Rad, Hercules, CA, USA). After the pulse-field gel electrophoresis (0.5× TBE, 14 °C, 200 V for 15 h with 60-s switching time, and for 8 h with a 90-s switching time), DNA was transferred to positively charged nitrocellulose membrane (GE Healthcare, Buckinghamshire, UK).
The number and chromosomal location of each SUC locus were determined by probing the membrane with DIG-labelled probes (Eurofins, Ebersberg, Germany). S. paradoxus and S. cerevisiae probes were designed according to the most conserved 5′ regions of SUC2 gene.
Hybridization and detection reactions were carried out according to the Roche's DIG High Prime DNA Labeling and Detection Starter Kit 1 (Roche, MannHeim, Germany).
S. paradoxus SUC2 probe sequence:
CGTCTGGGGTACGCCATTGTATTGGGGCCATGCTACTTCCGATGATTTGACCCACTGGCAAGACGAACCCATTGCTATTG
S. cerevisiae SUC2 probe sequence:
ATGACAAACGAAACTAGCGATAGACCTTTGGTCCACTTCACACCCAACAAGGGCTGGATGAATGATCCAAATGG
ddPCR for SUC copy number
The Southern blots of whole-chromosome pulsed-field gels could detect SUC loci in addition to SUC2. But because each chromosome has two telomeres, and because different chromosomal bands can colocalize on the gel, it cannot be used to precisely determine SUC copy number in strains that have subtelomeric copies of SUC in addition to SUC2. We therefore used droplet digital PCR (Bio-Rad QX100 system) to determine SUC copy number in the strains that had been determined by Southern blotting to contain multiple SUC loci, as well as in the 15 Chinese S. cerevisiae strains which we received most recently (as an alternative to Southern blotting). ddPCR uses simultaneous duplex reactions for target and reference genes within a single tube that contains about 20 000 reaction microdroplets, which are individually scored as positive or negative for the presence of amplicons by TaqMan fluorescence (see Huggett et al. 2013 for an introduction to the digital PCR technology). We used prevalidated TaqMan gene probes and primers designed by Life Technologies (CA, USA) for SUC2 (VIC, Sc04134115_s1) and two reference genes RPN5 (FAM, Sc04107686_s1) and MNN1 (FAM, Sc04117288_s1).
We isolated genomic DNA (MasterPureTM Yeast DNA Purification Kit, Epicentre Biotechnologies) from C.Lab.1 as a single-copy control, C.Lab.2.suc2::KANMX as a zero copy control, the two strains identified by a previous study as having multiple SUC loci (C.Ginger.wine and C.Billi.wine; Naumov et al. 1996) as positive controls, as well as the wild strains to be tested (Please see Appendix S1, Supporting information). Genomic DNA was restricted with HindIII, as this enzyme has a conserved cut site within the SUC2, RPN5 and MNN1 open reading frames, but outside the binding regions of the TaqMan probes. In each case, 1 μg of genomic DNA, in 40-μL reactions, was digested using 5U of HindIII (BioLabs, New England) at 37 °C for 60 min, and terminated the reaction at 65 °C for 20 min. 2500 pg of restricted DNA, 10 μL of ddPCR SuperMix (Bio-Rad), 1 μL FAM reference probe/primer mixture (RPN5 or MNN1), 1 μL VIC target probe/primer mixture (SUC2) were mixed and brought up to 20 μL final volume with molecular-grade water. Twenty microlitre reaction mixture and 70 μL droplet generation oil (Bio-Rad) were loaded into droplet generation cartridges, and ∼20 000 droplets were generated in separate wells. Droplet samples (∼40 μL) were transferred into the 96-well plates, and amplifications were carried out at 95 °C for 10 min, followed by 40 cycles of 94 °C for 30 s and 56 °C (optimized earlier by a thermal gradient PCR assay) for 1 min, and deactivated at 98 °C for 10 min. The plates were then loaded onto the QX100 droplet digital reader, and copy number was estimated using the quantasoft software (Bio-Rad). All ddPCR analyses were performed using two different 1-copy-reference-gene probes (RPN5 and MNN1) on three independent DNA isolates (three biological replicates). Six data points were combined in the same graphic (see Fig. 2), as both probes gave a mixed copy number distribution, and the final results were given as mean copy number of all 6 data points.
Variation in invertase production
Our screen of wild strains (see ‘Screening wild strains for invertase nonproducers’, above) was calibrated to detect the difference between producers and nonproducers of invertase. To precisely compare the invertase produced by ten producers with different copy numbers, we modified the assay to account for possible differences in cell density between the different strains. Strains were grown up as described before, but after resuspending each colony in 5 mL of sterile water, a 100-μL sample was taken and serially diluted to determine the cell density. We further optimized the dilutions of each strain to bring its OD340 measurement within the quantitative range. Thus, between 15 and 100 μL of the cell suspension from each strain was made up to the total test volume of 100 μL with a suspension of C.Lab.1.suc2::KANMX cells, prepared in the same way. This mixture was then assayed as previously, and the resulting signal was multiplied by this additional dilution factor. We converted absorbance to mg of invertase, using the standard curve in Appendix S4 (Supporting information), and we used the cell density to generate a per-cell measure of molecular invertase production for the standard laboratory producer strain (C.Lab.1), a selection of the wild S. cerevisiae strains from different sources (C.Oak.3, C.Soil.3, C.Cactus.1, C.Fruit.1, C.Cocoa.1), the three wild strains identified as having multiple SUC copies (C.Nectar.1, C.Nectar.2 and C.Nectar.3) and two domesticated control strains previously identified (Naumov et al. 1996) as having multiple SUC copies (C.Billi.wine and C.Ginger.wine). Every strain was tested three times to allow quantitative comparisons to be made between the strains (raw data are in Appendix S3, Supporting information).
Determining the contribution of subtelomeric SUC loci to invertase production
We deleted the open reading frame of SUC2 in the three wild strains identified as having multiple SUC copies (C.Nectar.1, C.Nectar.2, C.Nectar.3), and two domesticated control strains previously identified (Naumov et al. 1996) as having multiple SUC copies (C.Billi.wine, C.Ginger.wine). We used PCR-mediated gene replacement (Wach 1996) with the selectable drug resistance marker NATMX and the following PCR primers:
Forward primer for all strains:
CAAGCAAAACAAAAAGCTTTTCTTTTCACTAACGTATATGCGTACGCTGCAGGTCGAC
Reverse primer for C.Nectar.1, C.Nectar.2 and C.Nectar3:
CTTTTGAAAAAAATAAAAAAGACAATAAGTTTTATGACCTATCGATGAATTCGAGCTCG
Reverse primer for C.Ginger.wine and C.Billi.wine:
GCTTTTGAAAAAAATAAAAAGACAATAAGTTTTATAACCTATCGATGAATTCGAGCTCG
To test candidate transformants, we performed PCRs with two sets of diagnostic primers. Set 1 amplifies the region between upstream gene YIL163C and SUC2 5′ region. Set 2 amplifies the region between the 3′ end of SUC2 and downstream gene YIL161W.
Primers used for diagnostic PCR are as follows:
- Set 1 – upstream region:
- Suc1F CGATCCATTATGAGGGCTTC
- Suc1R GCCAAAAGGAAAAGGAAAGC
- Set 2 – downstream region:
- Suc2F GAACATGACCACTGGTGTCG
- Suc2R GAGTTCCTTCGTTTCCCAAA
We also confirmed that SUC2 was deleted from these five strains using the CHEF Southern blot (see Fig. S1, Supporting information). We then performed quantitative invertase production assays using the conditions described above (under ‘Variation in invertase production’) on the five wild-type strains (C.Nectar.1, C.Nectar.2 and C.Nectar.3; C.Ginger.wine and C.Billi.wine) and the five SUC2 knockouts derived from them (C.Nectar.1.suc2::NATMX, C.Nectar.2.suc2::NATMX, C.Nectar.3.suc2::NATMX, C.Ginger.wine.suc2::NATMX, C.Billi.wine.suc2::NATMX, respectively). As mentioned before, three independent replicates were made of each assay, allowing quantitative comparisons to be made (raw data are on Appendix S3, Supporting information).
Results
No wild strains were invertase nonproducers
Figure 1 shows that all the 110 wild strains that we tested produce more invertase than the standard invertase nonproducer or ‘cheater’ used in several previous experiments about cooperation (Greig & Travisano 2004; MacLean & Brandon 2008; Gore et al. 2009; MacLean et al. 2010). None of the 110 wild strains produced invertase at a level low enough to fall within the 95% confidence interval around the residual invertase activity of the standard nonproducer laboratory strain, C.Lab.1.suc2::KANMX. In fact, all the wild strains also had higher invertase activity than the 95% confidence interval around the invertase activity of the standard laboratory producer strain, C.Lab.1. We applied a Tukey post-hoc test to a one-way anova on these three groups and found that the 110 wild strains, as a group, produced significantly more invertase than both nonproducer and producer laboratory strains (F2,113 = 125.5, P < 0.0001).
Figure 1.
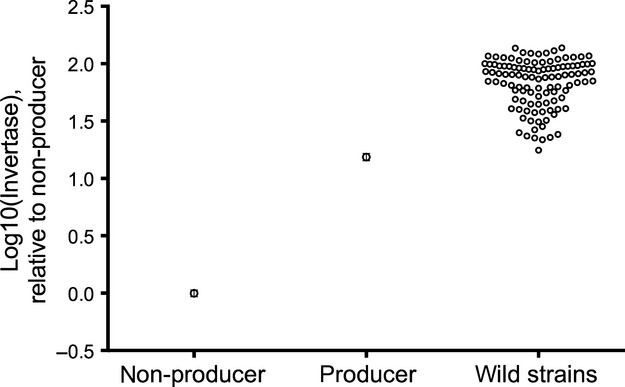
Screening of wild strains for invertase nonproducers. The invertase production of the 110 wild strains screened, as well as the standard laboratory invertase-producer strain C.Lab.1, is shown, relative to the production of the standard laboratory invertase nonproducer strain C.Lab.1.suc2::KANMX. All strains are described in Appendix S1 (Supporting information), and all data are listed in Appendix S2 (Supporting information).
No suc2 pseudogenes were detected in wild strains
Whole genome sequences existed for 29 S. paradoxus strains (Liti et al. 2009; Bergström et al. 2014). Consistent with their ability to produce invertase, we found intact open reading frames (ORFs) homologous to the reference S. cerevisiae strain (s288c/C.Lab.1) in all these strains. The length of the ORF was identical among all 29 S. paradoxus strains. Also for 8 S. cerevisiae strains, we found intact ORFs homologous to the reference strain (SGRP1: Liti et al. 2009; SGRP2: Bergström et al. 2014). There were no frameshift or nonsense mutations in any of the wild strains for which sequence was available (see Appendices S5 and S6, Supporting information for the SUC2 nucleotide sequences identified in the wild strains used in this study).
Three S. cerevisiae strains contained additional SUC genes
Our Southern blots showed that all the wild S. paradoxus strains isolated from oak and maple trees contained just a single SUC locus, SUC2, located on chromosome IX. All 27 S. cerevisiae strains isolated from nature also contained SUC2 on chromosome IX, but three S. cerevisiae strains (C.Nectar.1, C.Nectar.2 and C.Nectar.3) contained additional SUC loci on chromosome II (SUC3), on chromosome X (SUC8) and on chromosome XIV (SUC9) (Figs S1 and S2, Supporting information). ddPCR (Fig.2) shows that the SUC copy number of the three wild strains with multiple loci is closest to four, corresponding to one SUC open reading frame for each chromosome with a SUC locus (SUC2, plus the extra loci SUC3, SUC8 and SUC9). All three of these wild strains were isolated from the same environment: Bertam palm (Eugeissona tristis) nectars in West Malaysia (Liti et al. 2009).
Figure 2.
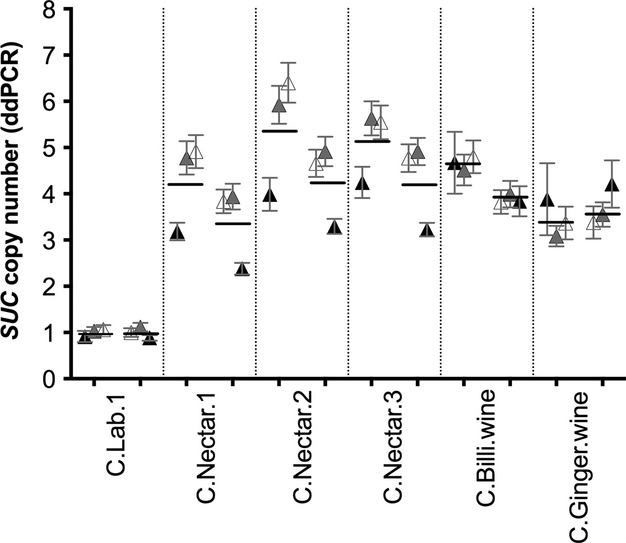
SUC gene copy number detection using droplet digital PCR (ddPCR) in five multilocus strains, normalized to a known single-copy control C. Lab.1 (first column). Three different symbol tones (dark, grey and empty) represent three different biological replicates. Copy number estimates calculated against RPN5 reference probe are on the left-hand side of each column, and copy number estimates calculated against MNN1 are on the right-hand side of each column. Black bars show the means of each set of three biological replicates.
Producers vary in their invertase production
We found that 11 different S. cerevisiae strains isolated from nine different domestic and wild environments varied significantly in their invertase production (Fig.3; F10,22 = 39.92, P < 0.0001). Post-hoc Tukey tests (letter above the bars in Fig.3) found that some, but not all, strains with four SUC copies produced significantly more invertase than strains with a single copy; some strains also produced significantly more invertase than other strains that had the same number of SUC copies. When grouped by number of SUC copies, the five strains with multiple SUC copies produced significantly more invertase than the six strains containing only SUC2 (Student's t-test, P = 0.0023, t = 3.32, DF = 31), but this difference was driven by two strains (laboratory strain C.Lab.1 and domesticated strain C.Ginger.wine): when the analysis was repeated on the wild strains alone, no significant difference was detected between the strains with 1 SUC copy and the strains with four copies (Student's t-test, P = 0.0713, t = 1.8951, DF = 22). Thus, it was unclear whether or not additional subtelomeric copies of SUC contributed to the variation in invertase production, or whether it was caused simply by variation in SUC2 expression. We therefore decided to test directly, by knocking out SUC2, whether the additional subtelomeric copies of SUC are expressed or whether they function only as silent backup copies for ‘cheats’.
Figure 3.
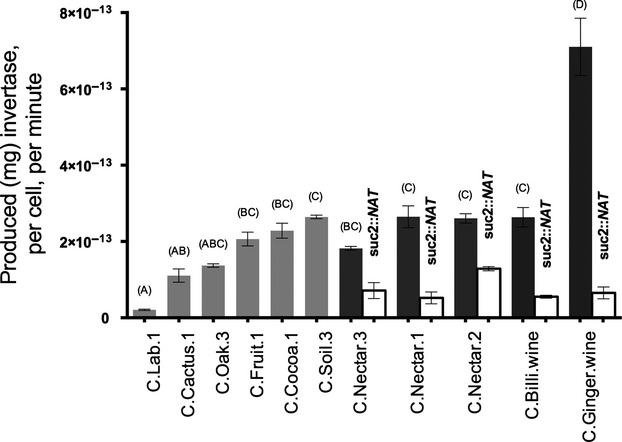
Light grey bars show the mean invertase production per cell for the single-copy standard producer and 5 other single-copy S. cerevisiae strains from different wild habitats. Dark grey bars show the production for the three strains with subtelomeric SUC loci isolated from Bertram palm nectar, and two strains from domesticated origins with subtelomeric loci as controls. Letters above the filled bars indicate which strains differ from each other with respect to their wild-type invertase production: strains with a letter in common are not significantly different. Open bars show the residual production of invertase from subtelomeric loci after SUC2 was knocked out. Three replicate assays were made for each strain; error bars show the standard error of the mean.
Subtelomeric SUC copies are not silent
Figure 3 shows that SUC2 contributes much more to total invertase production than subtelomeric copies of SUC. Knocking out SUC2 in the five strains with multiple copies reduces invertase production in every case, a statistically significant effect (P = 0.0312, paired sign test). The average reduction in invertase when SUC2 was deleted was 64%, suggesting that each of three subtelomeric SUC genes contributes only about 12% to total invertase production. But subtelomeric copies are far from silent: the SUC2 knockouts all produce more invertase than the standard laboratory producer strain C.Lab.1 (Fig.3).
Discussion
Our results do not support the hypothesis that natural variation in SUC genes is caused by social conflict (Greig & Travisano 2004). It is more likely that different SUC genotypes are selected by habitats with different availabilities of sucrose (Naumov et al. 1996), but our survey does not contain enough variation for us to be certain.
Invertase nonproducers
Our main aim was to determine whether invertase nonproducers existed in the same natural habitats as producers, which would be required in order for nonproducers to cheat. We found no nonproducers among the 65 oak-associated S. paradoxus strains that we tested nor among the 15 strains from maple trees. Unfortunately, the other habitats included in the survey had only a few strains available from each, so we might not find both producers and nonproducers cooccurring in the same type of habitat even if they were there. Nevertheless, we found no nonproducers at all among a total of 110 different wild strains (Fig.1). There is therefore no evidence to support the idea that nonproducing cheats occur among wild strains.
Our results stand in contrast to Naumov et al.'s (1996) finding of 11 nonproducers among a sample of 91 S. cerevisiae strains. One explanation is that Naumov et al. (1996) surveyed strains from a wider range of environments, which might select for or against the production of invertase according to sucrose adaptation hypothesis. Another is that most of Naumov's strains were associated with humans, whereas ours came only from natural sources. Artificial selection on domesticated species can increase diversity (Vila et al. 1999), as well as allowing loss of functions that would be important for survival in the wild (e.g. loss of pigmentation in domestic pigs and horses, Andersson & Georges 2004). It is therefore possible that invertase nonproducing mutants that would be eliminated by natural selection in the wild can persist by drift or even be selected in anthropogenic environments that are abundant in sugars other than sucrose or which lack producers as competitors. Thus, the variation observed in human-associated strains may be due to changes in environment, demography and population structure resulting from domestication. It is also possible that some domesticated environments produce conditions that allow cheating, for example by increasing population densities or environmental stability, compared to those conditions that would exist naturally, and thus, variation in domesticated strains could be due to the social conflict hypothesis. Because the evolutionary history of human-associated strains is obscure, it would be difficult to disentangle these explanations for the variation among domesticated yeast, but as there is no evidence for nonproducers and producers occupying the same habitat and abundant evidence for variability in sucrose availability, we, like Naumov et al. (1996), favour the sucrose adaptation hypothesis for domesticated strains as well as for the wild strains we describe here.
Copy number variation
A secondary aim of the project was to determine whether variation in SUC copy number was consistent with social evolution.
According to the social conflict hypothesis as originally formulated (Greig & Travisano 2004), subtelomeric SUC loci could act as transcriptionally silenced backups which can be stochastically de-repressed (Gottschling et al. 1990; Louis 1995) or which could restore function to a suc2 pseudogene by gene conversion (analogous to mating-type switching using silent telomeric copies of the hidden mating-type, HM, loci) (Naumov & Tolstorukov 1973; Hicks & Herskowitz 1977). Silent copies of SUC could allow cheats to switch back to invertase production when there are no cooperators to exploit, a form of ‘facultative cheating’ (Gore et al. 2009). This part of the social conflict hypothesis is now much less plausible because subsequent research has shown that subtelomeric silencing is predominantly a haploid phenomenon (Mercier et al. 2005). Indeed, the three wild strains we found with multiple SUC copies produced invertase at a high level, and they continued to do so even when SUC2 was knocked out, showing that the remaining subtelomeric SUC loci are transcriptionally active and are not silenced backup copies (Fig.3). Further, all the strains with subtelomeric copies came from the same environment, Bertram palm nectar, and all strains from this environment contained three subtelomeric SUC alleles in addition to SUC2: there was no genotypic variation within the environment as predicted by the social conflict hypothesis. This could simply be because our tiny sample contained only three strains, but it is also most consistent with the sucrose adaptation hypothesis. Sucrose is the major carbon source in most plant nectars (Corbet 2003; Pacini et al. 2003; Dupont et al. 2004; Wiens et al. 2006; Peay et al. 2012), and Bertam palm nectars contain high and stable concentrations of sucrose (∼10%; Wiens et al. 2006). However, these strains are very closely related: C. Nectar.1 differs from C.Nectar.2 by just 0.0059% of nucleotides across the whole genome, and from C. Nectar.3 by 0.019%; C.Nectar.2 and C.Nectar.3 differ by 0.012% (Liti et al. 2009). Given the small sample size, the high genetic relatedness and the likelihood that all three strains inherited their subtelomeric SUC genes by common descent, we cannot exclude the possibility that the expansion of the SUC gene family in this environment is due to neither social evolution nor environmental selection, but simply genetic drift.
The expression of invertase from strains with subtelomeric SUC loci shows that they are not ‘cheats’. However, a social model could still be used to explain their evolution if the originally proposed roles of cooperator and cheat were reversed. If strains with more SUC copies produce more invertase, they could be considered cooperators instead of cheats, and they could feed other, cheating, strains that have only SUC2 and produce less. As under the original social explanation for SUC genetic variation, we would predict that cheats and cooperators should occur in the same environment. Whilst we might not expect to detect such copy number variation among only three strains from Bertram palm nectar, we would expect to find variation within the well-sampled oak-tree and maple-tree habitats, but we did not. Instead, we find copy number variation between (but not within) environments that differ in sucrose availability. Whilst we must be cautious not to overgeneralize from just three closely related strains, the little copy number variation we do find in our survey is clearly better explained by the sucrose adaptation hypothesis than by the social conflict hypothesis.
Is invertase production a cooperative trait?
We previously proposed the social conflict hypothesis to explain variation in SUC genotypes among S. cerevisiae strains (Greig & Travisano 2004). But because S. cerevisiae is domesticated, and isolates came from many different sources, it was difficult to know whether different genotypes evolved in a common environment that would permit social cheating. In this survey of wild strains, we find very little variation of SUC genotypes, and the limited variation we do find occurs between, and not within, environments. The genetic variation is therefore better explained by adaptation to different environmental levels of sucrose than by social conflict. However, given the lack of variation in our samples, we have very limited power to differentiate between the two hypotheses. The ideal survey would test the invertase production and the SUC genotype of multiple strains isolated from at last two different natural habitats that differed in their sucrose availability. Such a design would have the best chance of being able to definitively distinguish the difference between the two hypotheses explaining variation for SUC. If different SUC genotypes are selected by the local availability of sucrose, then the two environments will be fixed for different genotypes. If social conflict produces variation, then we would expect more variation within the high-sucrose environment than within the low-sucrose environment. Unfortunately, such well-sampled natural habitats differing in sucrose availability do not exist, but we hope that as research in yeast natural history progresses, such a survey may be possible in the future.
Authors have previously cited the variation in SUC genotypes as evidence that cheating occurs in nature (Greig & Travisano 2004; MacLean & Brandon 2008; Gore et al. 2009), but here we show that the evidence has been misinterpreted. This has significant consequences for the use of invertase production as an experimental model of cooperation. Cooperative traits are properly defined not merely as those traits that benefit others, which would be nonsensically overinclusive, but those traits that evolved because of the benefits they convey to others (West et al. 2007). Thus, it is important to show that cooperation occurs in the environment in which a putative cooperative trait evolved, and the existence of natural genetic variation was presented as evidence that invertase production evolved in nature as a cooperative trait. It is worth noting as an aside, though, that the existence of natural cheats is not sufficient to prove a trait as cooperative: we would not consider scatter-hoarding of nuts by squirrels to be a cooperative trait, even though hoarded nuts are often eaten by scroungers and not by the squirrel that buried them (Stapanian & Smith 1978). To prove that invertase production evolved as a cooperative trait, one would need to show that not only that social conflict over invertase sharing occurs in nature, but also that invertase sharing was actually selected. Surveys like ours cannot therefore determine whether or not invertase production is a cooperative trait. Even if the natural variation for SUC copy number is not caused by social conflict, social conflict may nonetheless underlie other forms of genetic variation for invertase production (for example, Fig.3 shows there is considerable and significant variation in invertase production even among strains containing only SUC2). And even if social conflict does not cause any natural genetic variation in invertase production, it is still possible that invertase production evolved as a cooperative trait in nature. And even if it did not evolve as cooperative trait in nature, invertase sharing in an experimental setting could still be a useful model for cooperation. We are mindful, though, of the words of G.C. Williams: ‘Adaptation should be attributed to no higher a level of organization than is demanded by the evidence’ (Williams 1996). In our opinion, a trait should not be called cooperative until more parsimonious explanations for its evolution have been rejected.
Acknowledgments
We thank Leonid Kruglyak for four natural S. cerevisiae strains, Feng-Yan Bai for 15 wild S. cerevisiae strains isolated from China, Christian R. Landry for 15 S. paradoxus strains isolated from maple trees in Québec (Canada) and Eric L. Miller for providing the Plön (Germany) S. paradoxus strains and for his comments on the manuscript. We thank David W. Rogers for his recommendations to improve our invertase assays, Mathias Wegner for helping interpret our Southern blot analysis, and Zeljka Pezer for helping us with the ddPCR. Jai Denton and Laurence Hurst read an early draft of the manuscript, and the latter recommended Molecular Ecology. The study was greatly improved thanks to positive criticism from three anonymous reviewers. This work was supported by the Max Planck Society.
Biography
D.G. designed the study. G.O.B. performed the research. D.G. and G.O.B. analysed data and wrote the paper.
Data accessibility
The available SUC2 nucleotide sequences, in FASTA format, for 29 S. paradoxus and 8 S. cerevisiae strains are accessible on Appendices S5 and S6 (Supporting information), respectively. Raw data for all invertase assay experiments, including absorbance reads and calculations, are accessible on Appendices S2, S3 and S4 (Supporting information).
Supporting Information
Additional supporting information may be found in the online version of this article.
List of all strains used in this study with strain names and details.
Invertase producer screening results for each wild strain, producer strain, and nonproducer strain. Wild strains were tested once; control strains were tested as 3 replicates.
Invertase assay result for 11 strains and 5 suc2 knockout strains. Three measurements were performed for each strain.
Invertase assay results performed using commercial invertase enzyme to determine the linear response range and to calculate the invertase production using the equation of the linear regression (R2 = 0.948, six data points, three replicates each). None of our measurements gave more glucose reading over the top point of this linear regression line.
S. paradoxus SUC2 nucleotide sequences available for 29 strains.
S. cerevisiae SUC2 nucleotide sequences available for eight strains. Sequence files include ∼130 base pairs upstream and 200 base pairs downstream of the start and the stop codons, respectively.
Southern blot confirmation of SUC2 knockouts in the multiple-copied strains, and common laboratory strain C.Lab.1 (S288c).
CHEF gel and corresponding Southern blot assay showing four different chromosomal locations of SUC genes in C.Nectar.1, C.Nectar.2. and C.Nectar.3 strains. This CHEF gel was ran longer (0.5× TBE, 14 °C, 200 V for 30 h with 60-s switching time, and for 12.5 h with a 90-s switching time) to further separate chromosome II, XIV, X bands.
References
- Andersson L, Georges M. Domestic-animal genomics: deciphering the genetics of complex traits. Nature Reviews Genetics. 2004;5:202–212. doi: 10.1038/nrg1294. [DOI] [PubMed] [Google Scholar]
- Basson CE, Groenewald J-H, Kossmann J, Cronjé C, Bauer R. Sugar and acid-related quality attributes and enzyme activities in strawberry fruits: invertase is the main sucrose hydrolysing enzyme. Food Chemistry. 2010;121:1156–1162. [Google Scholar]
- Batista AS, Miletti LC, Stambuk BU. Sucrose fermentation by Saccharomyces cerevisiae lacking hexose transport. Journal of Molecular Microbiology and Biotechnology. 2004;8:26–33. doi: 10.1159/000082078. [DOI] [PubMed] [Google Scholar]
- Bergström A, Simpson JT, Salinas F, et al. A High-definition view of functional genetic variation from natural yeast genomes. Molecular Biology and Evolution. 2014;31:872–888. doi: 10.1093/molbev/msu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Fink G. Yeast: an experimental organism for modern biology. Science. 1988;240:1439–1443. doi: 10.1126/science.3287619. [DOI] [PubMed] [Google Scholar]
- Carle GF, Olson MV. An electrophoretic karyotype for yeast. Proceedings of the National Academy of Sciences, USA. 1985;82:3756. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Organization of the SUC Gene Family in Saccharomyces. Molecular and Cellular Biology. 1983;3:351–359. doi: 10.1128/mcb.3.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Celenza JL, Eng FJ. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Molecular and Cellular Biology. 1985;5:2894–2902. doi: 10.1128/mcb.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G, Leducq J-P, Bertin C, Dube AK, Landry CR. Exploring the northern limit of the distribution of Saccharomyces cerevisiae and Sacchromyces paradoxus in North America. FEMS Yeast Research. 2014;14:281–288. doi: 10.1111/1567-1364.12100. [DOI] [PubMed] [Google Scholar]
- Corbet SA. Nectar sugar content: estimating standing crop and secretion rate in the field. Apidologie. 2003;34:1–10. [Google Scholar]
- Damore JA, Gore J. Understanding microbial cooperation. Journal of Theoretical Biology. 2012;299:31–41. doi: 10.1016/j.jtbi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayrolles M, de Villechenon E, Funel LA, Aigle M. Incidence of SUC-RTM telomeric repeated genes in brewing and wild wine strains of Saccharomyces. Current Genetics. 1997;31:457–461. doi: 10.1007/s002940050230. [DOI] [PubMed] [Google Scholar]
- Dupont Y, Hansen D, Rasmussen J, Olesen J. Evolutionary changes in nectar sugar composition associated with switches between bird and insect pollination: the Canarian bird-flower element revisited. Functional Ecology. 2004;18:670–676. [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, et al. Life with 6000 genes. Science. 1996;274:546–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Gore J, Youk H, Van Oudenaarden A. Snowdrift game dynamics and facultative cheating in yeast. Nature. 2009;459:253–256. doi: 10.1038/nature07921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Greig D, Travisano M. The Prisoner's Dilemma and polymorphism in yeast SUC genes. Proceedings. Biological sciences/The Royal Society. 2004;271:S25–S26. doi: 10.1098/rsbl.2003.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JB, Herskowitz I. Interconversion of yeast mating types II. Restoration of mating ability to sterile mutants in homothallic and heterothallic strains. Genetics. 1977;85:373–393. doi: 10.1093/genetics/85.3.373b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggett JF, Foy CA, Benes V, et al. Minimum information for publication of quantitative digital PCR experiments. Clinical Chemistry. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- Johnson LJ, Koufopanou V, Goddard MR, Hetherington R, Schafer SM, Burt A. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43. doi: 10.1534/genetics.166.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Lee Y-C. Changes in some quality factors of frozen ginger as affected by the freezing storage conditions. Journal of the Science of Food and Agriculture. 2006;86:1439–1445. [Google Scholar]
- Koschwanez JH, Foster KR, Murray AW. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biology. 2011;9:e1001122. doi: 10.1371/journal.pbio.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehne H, Murphy H, Francis C, Sniegowski P. Allopatric divergence, secondary contact, and genetic isolation in wild yeast populations. Current Biology. 2007;17:407–411. doi: 10.1016/j.cub.2006.12.047. [DOI] [PubMed] [Google Scholar]
- Lafontaine I, Fischer G, Talla E, Dujon B. Gene relics in the genome of the yeast Saccharomyces cerevisiae. Gene. 2004;335:1–17. doi: 10.1016/j.gene.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Libkind D, Hittinger CT, Valério E, et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. PNAS. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- MacLean CR, Brandon C. Stable public goods cooperation and dynamic social interactions in yeast. Journal of Evolutionary Biology. 2008;21:1836–1843. doi: 10.1111/j.1420-9101.2008.01579.x. [DOI] [PubMed] [Google Scholar]
- MacLean RC, Fuentes-Hernandez A, Greig D, Hurst LD, Gudelj I. A mixture of “Cheats” and ‘Co-Operators’ can enable maximal group benefit. PLoS Biology. 2010;8:e1000486. doi: 10.1371/journal.pbio.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsilio V, Campestre C, Lanza B, De Angelis M. Sugar and polyol compositions of some European olive fruit varieties (Olea europaea L.) suitable for table olive purposes. Food Chemistry. 2001;72:485–490. [Google Scholar]
- Mercier G, Berthault N, Touleimat N, et al. A haploid-specific transcriptional response to irradiation in Saccharomyces cerevisiae. Nucleic Acids Research. 2005;33:6635–6643. doi: 10.1093/nar/gki959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov G, Naumova E. Polygenic control for fermentation of β-fructosides in the yeast Saccharomyces cerevisiae: new genes SUC9 and SUC10. Microbiology. 2010;79:160–166. [Google Scholar]
- Naumov GI, Tolstorukov II. Comparative genetics of yeast. X. Reidentification of mutators of mating types in Saccharomyces. Genetika. 1973;9:81–91. [PubMed] [Google Scholar]
- Naumov G, Turakainen H, Naumova E, Aho S, Korhola M. A new family of polymorphic genes in Saccharomyces cerevisiae: alpha-galactosidase genes MEL1-MEL7. Molecular & General Genetics: MGG. 1990;224:119–128. doi: 10.1007/BF00259458. [DOI] [PubMed] [Google Scholar]
- Naumov G, Naumova E, Sancho E, Korhola M. Taxogenetics of the Saccharomyces sensu stricto yeasts from western and South Africa. Cryptogamie. Mycologie. 1993;4:263–270. [Google Scholar]
- Naumov GI, Naumova ES, Sancho ED, Korhbla MP. Polymeric SUC genes in natural populations of Saccharomyces cerevisiae. FEMS Microbiology Letters. 1996;135:31–35. doi: 10.1111/j.1574-6968.1996.tb07962.x. [DOI] [PubMed] [Google Scholar]
- Naumov GI, Naumova ES, Sniegowski PD. Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of North American oaks. Canadian Journal of Microbiology. 1998;44:1045–1050. [PubMed] [Google Scholar]
- Pacini E, Nepi M, Vesprini J. Nectar biodiversity: a short review. Plant Systematics and Evolution. 2003;238:7–21. [Google Scholar]
- Peay KG, Belisle M, Fukami T. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proceedings. Biological sciences/The Royal Society. 2012;279:749–758. doi: 10.1098/rspb.2011.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porro D, Brambilla L, Ranzi B, Martegani E, Alberghina L. Development of metabolically engineered Saccharomyces cerevisiae cells for the production of Lactic-Acid. Biotechnology Progress. 1995;11:294–298. doi: 10.1021/bp00033a009. [DOI] [PubMed] [Google Scholar]
- Stapanian MA, Smith CC. A model for seed scatterhoarding: coevolution of fox squirrels and black walnuts. Ecology. 1978;59:884–896. [Google Scholar]
- Thim L, Hansen MT, Norris K, et al. Secretion and processing of insulin precursors in yeast. Proceedings of the National Academy of Sciences, USA. 1986;83:6766–6770. doi: 10.1073/pnas.83.18.6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan MA, Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie van Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- Vila C, Maldonado JE, Wayne RK. Phylogenetic relationships, evolution, and genetic diversity of the domestic dog. Journal of Heredity. 1999;90:71–77. doi: 10.1093/jhered/90.1.71. [DOI] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wang Q-M, Liu W-Q, Liti G, Wang S-A, Bai F-Y. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Molecular Ecology. 2012;21:5404–5417. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- Wayne RK. Limb morphology of domestic and wild canids: the influence of development on morphologic change. Journal of morphology. 1986;187:301–319. doi: 10.1002/jmor.1051870304. [DOI] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A. Social semantics: altruism, cooperation, mutualism, strong reciprocity and group selection. Journal of Evolutionary Biology. 2007;20:415–432. doi: 10.1111/j.1420-9101.2006.01258.x. [DOI] [PubMed] [Google Scholar]
- Wiens F, Zitzmann A, Hussein NA. Fast food for slow lorises: is low metabolism related to secondary compounds in high-energy plant diet? Journal of Mammalogy. 2006;87:790–798. [Google Scholar]
- Williams GC. Adaptation and Natural Selection: A Critique of Some Current Evolutionary Thought. Princeton, New Jersey: Princeton University Press; 1996. [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of all strains used in this study with strain names and details.
Invertase producer screening results for each wild strain, producer strain, and nonproducer strain. Wild strains were tested once; control strains were tested as 3 replicates.
Invertase assay result for 11 strains and 5 suc2 knockout strains. Three measurements were performed for each strain.
Invertase assay results performed using commercial invertase enzyme to determine the linear response range and to calculate the invertase production using the equation of the linear regression (R2 = 0.948, six data points, three replicates each). None of our measurements gave more glucose reading over the top point of this linear regression line.
S. paradoxus SUC2 nucleotide sequences available for 29 strains.
S. cerevisiae SUC2 nucleotide sequences available for eight strains. Sequence files include ∼130 base pairs upstream and 200 base pairs downstream of the start and the stop codons, respectively.
Southern blot confirmation of SUC2 knockouts in the multiple-copied strains, and common laboratory strain C.Lab.1 (S288c).
CHEF gel and corresponding Southern blot assay showing four different chromosomal locations of SUC genes in C.Nectar.1, C.Nectar.2. and C.Nectar.3 strains. This CHEF gel was ran longer (0.5× TBE, 14 °C, 200 V for 30 h with 60-s switching time, and for 12.5 h with a 90-s switching time) to further separate chromosome II, XIV, X bands.
Data Availability Statement
The available SUC2 nucleotide sequences, in FASTA format, for 29 S. paradoxus and 8 S. cerevisiae strains are accessible on Appendices S5 and S6 (Supporting information), respectively. Raw data for all invertase assay experiments, including absorbance reads and calculations, are accessible on Appendices S2, S3 and S4 (Supporting information).


