Abstract
Although localized to the mineralized matrix of bone, osteocytes are able to respond to systemic factors such as the calciotropic hormones 1,25(OH)2D3 and PTH. In the present studies, we examine the transcriptomic response to PTH in an osteocyte cell model and found that this hormone regulated an extensive panel of genes. Surprisingly, PTH uniquely modulated two cohorts of genes, one that was expressed and associated with the osteoblast to osteocyte transition and the other a cohort that was expressed only in the mature osteocyte. Interestingly, PTH’s effects were largely to oppose the expression of differentiation-related genes in the former cohort, while potentiating the expression of osteocyte-specific genes in the latter cohort. A comparison of the transcriptional effects of PTH with those obtained previously with 1,25(OH)2D3 revealed a subset of genes that was strongly overlapping. While 1,25(OH)2D3 potentiated the expression of osteocyte-specific genes similar to that seen with PTH, the overlap between the two hormones was more limited. Additional experiments identified the PKA-activated phospho-CREB (pCREB) cistrome, revealing that while many of the differentiation-related PTH regulated genes were apparent targets of a PKA-mediated signaling pathway, a reduction in pCREB binding at sites associated with osteocyte-specific PTH targets appeared to involve alternative PTH activation pathways. That pCREB binding activities positioned near important hormone-regulated gene cohorts were localized to control regions of genes was reinforced by the presence of epigenetic enhancer signatures exemplified by unique modifications at histones H3 and H4. These studies suggest that both PTH and 1,25(OH)2D3 may play important and perhaps cooperative roles in limiting osteocyte differentiation from its precursors while simultaneously exerting distinct roles in regulating mature osteocyte function. Our results provide new insight into transcription factor-associated mechanisms through which PTH and 1,25(OH)2D3 regulate a plethora of genes important to the osteoblast/osteocyte lineage.
Keywords: PTH, osteocyte, transcriptional regulation, RNA-seq, ChIP-seq
1. INTRODUCTION
Osteocytes are terminally differentiated cells descended from bone-forming osteoblasts that have become embedded in mineralized matrix [1–4]. These cells are distinct from their osteoblast precursors in morphology, function and in underlying patterns of gene expression [5, 6]. Osteocytes are critical mediators of bone metabolism, transducing the stimulant of mechanical stress into the expression and secretion of local regulatory factors that control features of bone remodelling [1]. In addition to being key regulators of bone metabolism, the capacity of the osteocyte to act as an endocrine cell extends its influence beyond bone to other tissues and organs [3]. Accordingly, secretion of FGF23 from the osteocyte has distant effects on the cardiovascular system and on the kidney [7–9]. In addition, although the mechanisms are unknown, ablation of osteocytes in mice results in lymphopenia and loss of white adipose tissue, suggesting a potential endocrine role for osteocytes in lymphopoiesis and fat metabolism [4]. A level of communication also appears to exist between osteocytes and muscle [10, 11]. Finally, osteocytes have been shown to be important for hematopoietic stem/progenitor cell mobilization as mice in which osteocytes have been ablated do not mobilize hematopoietic stem/progenitor cells in response to granulocyte colony-stimulating factor [12]. Based upon these associations, the osteocyte clearly represents a dynamic skeletal component whose myriad cellular and endocrine functions are of critical importance.
Analogous to their ability to affect distant tissues, osteocytes are also recipients of systemic endocrine action as well. Two of the major calciotropic hormones that act on osteocytes are PTH and 1,25-dihydroxyvitamin D3 (1,25(OH)2D3); the activities of these hormones are mediated through the PTH receptor type 1 (PTH1R) and the VDR, respectively [13, 14]. Both of these hormones are known to upregulate the expression of receptor activator of NF-Kβ ligand (RANKL), encoded by Tnfsf11, from the osteocyte, which is now considered to be the principle mediator of bone remodeling in the skeleton [15]. RANKL acts in paracrine fashion on neighboring osteoclast precursors, both activating this class of bone resorbing cells and potentiating their differentiation [16–19]. Our recent studies show that in addition to a similar action by 1,25(OH)2D3 on this gene, the vitamin D hormone also mediates the regulation of many additional genes from the osteocyte including those directly involved in mineralization and resorption [20].
PTH, a polypeptide released by the parathyroid gland in response to low serum calcium levels, maintains serum calcium homeostasis primarily by promoting vitamin D 1α-hydroxylation in the kidney [21]. The 34 N-terminal amino-acids of the full length 84 amino-acid single polypeptide of PTH can activate the PTH1R and several downstream pathways including cAMP/protein kinase A (PKA), phospholipase C (PLC)/protein kinase C (PKC), PLC-independent PKC and Ca2+ pathways, and perhaps other as well. For example, Sost is a known primary regulatory target of PTH action in osteocytes that encodes sclerostin, a negative regulator of bone formation [22]. Indeed, overexpression of a constitutively active PTH1R in osteocytes results in a suppression of sclerostin [23], increasing bone remodeling that culminates in an elevation in bone mass, whereas deletion of PTH1R in osteocytes results in a loss of PTH regulated expression of sclerostin [24] leading to osteopenia. Interestingly, recent studies both in cells and in genetically altered mice indicate that the mechanism through which PTH mediates Sost down-regulation may involve myocyte enhancer factor 2c (Mef2c) and occurs via a Sost-linked enhancer termed ECR5 [25, 26]. Indeed, a genetic deletion which removes a large portion of the Sost downstream region that includes ECR5 results in Van Buchem disease [26]. Importantly, this regulation involves the PKA pathway but not the transcription factor CREB [27]. Regardless, identifying additional important targets of PTH in osteocytes is critical to understanding more fully the molecular basis for PTH’s effects on bone resorption and remodeling.
In recent studies, we identified genetic targets of 1,25(OH)2D3 action in osteocytes and tracked the underlying transcriptomic and epigenetic changes that occur during the osteoblast to osteocyte transition using RNA-sequencing and ChIP-sequencing methods [20]. The results of this study provided new insight into the transcriptomic changes that occur during osteocyte differentiation and revealed how genetic and epigenetic changes that occur to the genome during this process alter response to 1,25(OH)2D3. In the present study, we examined the effects of PTH on the osteocyte transcriptome and then contrasted the properties of this cohort of regulated genes with those regulated during differentiation and in response to 1,25(OH)2D3. We found that PTH and 1,25(OH)2D3 manifested similar actions to oppose differentiation-mediated changes in gene expression that occurred during the osteoblast to osteocyte transition, yet complimented positive actions on osteocyte-specific genes that were expressed exclusively in mature osteocytes. The mechanism of the former appeared to be due largely to the PKA-activated signaling component of PTH1R by virtue of the presence of pCREB at many of these genes. In contrast, a deficiency of pCREB binding at genes that were regulated by PTH in the mature osteocyte suggested the presence of alternative PTH activation pathways. These data support potentially novel actions of both PTH and 1,25(OH)2D3 on osteocyte differentiation, and are likely to provide important mechanistic insight into the molecular actions of each of these hormones on a multitude of highly regulated osteocytic genes.
2. MATERIALS AND METHODS
2.1 Reagents
PTH (1–34) (H-4835.0001) was obtained from Bachem (Bubendorf, Switzerland) and forskolin (#F3917-10mg) was obtained from Sigma-Aldrich (St. Louis, MO). An antibody to pCREB (Ser 133, 06-519) was purchased from Millipore (Darmstadt, Germany). All quantitative real-time PCR (qPCR) reagents (Fast Start SYBR Green Master Mix (with Rox)) were obtained from Roche (Indianapolis, IN) and TaqMan gene expression assays from Life Technologies (Applied Biosystems (ABI) Foster City, CA). All qPCR was conducted on the StepOnePlus from ABI. Primers for ChIP assays and recombineering were obtained from Integrated DNA Technologies, Inc (Coralville, IA) and TaqMan primers for gene expression were obtained from Life Technologies (ABI). Sequencing reagents for ChIP-seq (#11257047 RevA) were obtained from Illumina (San Diego, CA).
2.2 Cell Culture
IDG-SW3 osteoblast (d3) and IDG-SW3 osteocyte (d35) and MC3T3-E1 pre-osteoblast (d0) and MC3T3-E1 osteoblast (d15) cells were cultured as previously described [20, 28].
2.3 RNA-seq Library Preparation and Bioinformatic and Statistical Analyses
IDG-SW3 cells were differentiated for 35 days and treated 24 h prior with 100nM PTH in biological triplicate before RNA was isolated using the TRI-Reagent protocol (MRC). Subsequent preparation, bioinformatics processing, and statistical analyses have been described previously [20].
2.4 PTH Treatments and TaqMan Real Time PCR
IDG-SW3 cells were differentiated for 35 days and treated 24 hours (h) prior with 100nM PTH or vehicle before RNA was isolated using the TRI-Reagent protocol (MRC, Cincinnati, OH). RNA (1μg) was reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Life Technologies, ABI) and analyzed using TaqMan Real Time PCR as described above. Taqman Probes used are available in Supplemental (S.) Table 1, tab 1.
2.5 ChIP-seq Analyses
Chromatin immuno-precipitation (ChIP) was performed as described previously [29]. Briefly, samples were subjected to immuno-precipitation using either a control IgG antibody or experimental antibody (pCREB (ser 133)). The remainder of ChIP and ChIP-seq methodology including statistical information and data processing were performed as recently reported [28].
2.6 Data Access
All sequencing data is publically available in the GEO database: GSE62981
3. RESULTS
3.1 Identification of the PTH-Regulated Transcriptome
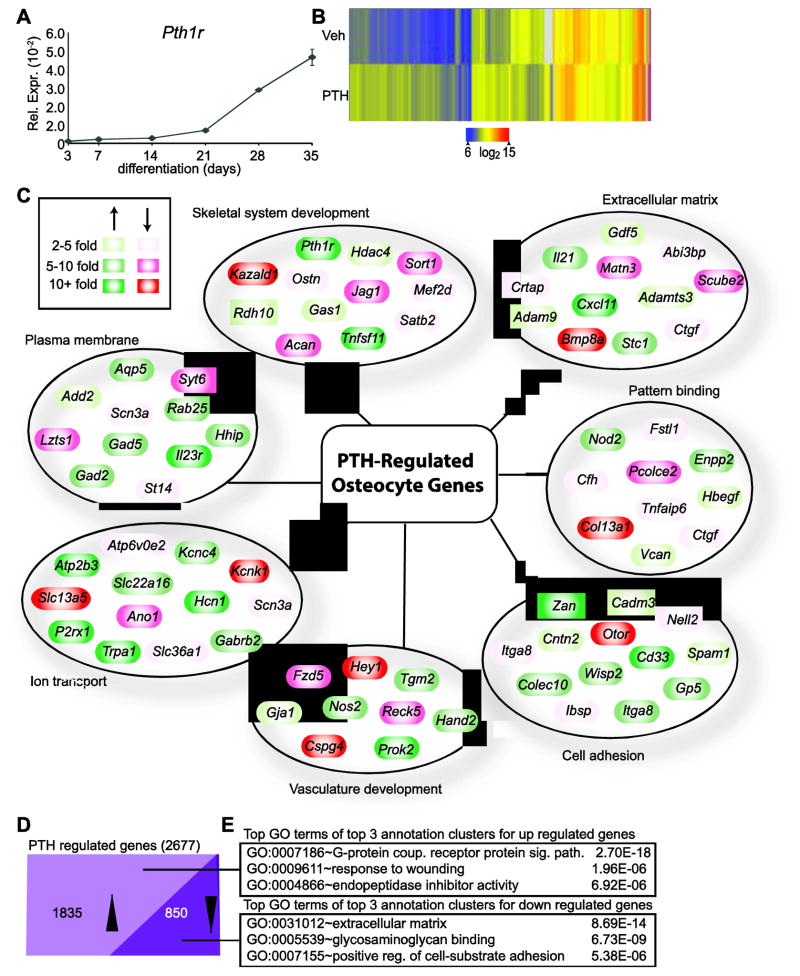
PTH plays a significant role in osteocyte function, down-regulating the expression of sclerostin to promote bone formation by sensitizing early osteoblasts to the growth-promoting effects of Wnts and other osteogenic signaling pathways and up-regulating the expression of RANKL to facilitate the coupling of bone formation to bone resorption [22]. In an earlier study of the genetic and epigenetic determinants of osteocyte differentiation, we used the IDG-SW3 cell line as a unique in vitro cell culture model to identify the many changes in gene expression that were associated with differentiation of osteocytes from their osteoblast precursors [20]. As documented in Fig. 1a, one of these up-regulated genes was Pth1r, suggesting that the osteocyte transition might be linked to a natural increase in responsiveness to PTH. We therefore explored the actions of this hormone on fully differentiated IDG-SW3 cells (osteocytes) by treating them for 24 hr with either vehicle or PTH in biological triplicate and then assessing their transcriptomic response using RNA-seq analysis. As illustrated in Fig. 1b, 2677 genes were affected >2-fold by this hormonal treatment (95% confidence, moderated T test), clustered using the Euclidean distance metric and visualized by heatmap. The top categories and a subset of representative genes from the global GO term analysis of these 2677 PTH-regulated genes are depicted in Fig. 1c. Enrichment for GO terms associated with skeletal system development, extracellular matrix, and ion transport were anticipated given previously characterized roles of PTH in osteocytes. Other cohorts of genes including those involved in vasculature development and cell adhesion were especially interesting and suggested that through these genes PTH could be involved in several novel functional aspects of osteocyte biology or in paracrine regulation of other cell types [3]. 1835 genes were up-regulated and 850 were down-regulated at this time point as documented in Fig. 1d (the sum of these two numbers is greater than 2677 due to differences in the regulation of isoforms). Importantly, gene ontology (GO) term analyses of these two cohorts of genes revealed an enrichment for a series of distinct annotation clusters. As summarized in Fig. 1e, genes up-regulated by PTH were highly enriched for G-protein coupled receptor protein signaling pathways, while down-regulated genes were most associated with extracellular matrix, highlighting the specificity of the differential actions of PTH that were likely to affect functional output. Gene lists and extended GO term results for all analyses in Fig. 1 can be found in Supplemental (S.) Table 1, tabs 2–5.
Figure 1. Osteocytes exhibit an extensive PTH-mediated transcriptome.
(A) TaqMan gene expression analysis of Pth1r during IDG-SW3 osteocyte differentiation was normalized to β-actin. Samples were analyzed in triplicate ± SEM. (B) Genes with differential gene expression at 24 hr between vehicle (Veh) and 100nM PTH (≥ 2 fold, 95% confidence interval, moderated t test) treatment were clustered using the Euclidian distance metric and visualized as a heatmap. (C) Selected, regulated genes from (B) were manually grouped by category following DAVID GO term analysis and delineated based upon their degree of PTH-regulation (green, up-regulated; red, down-regulated). (D) PTH-regulated genes from (B) were classified based upon their direction of regulation by PTH as compared to vehicle treatment. (E) Top GO term from the top 3 annotation clusters with associated P values from DAVID GO term analysis for gene groups from part (D).
PTH regulation of genes encoding sequence-specific transcription factors are also likely to contribute to the large PTH-regulated transcriptome in osteocytes, as modulation of these factors likely potentiates PTH’s secondary regulation of many additional genes. To explore this possibility, we identified PTH-regulated genes within these GO term categories that encoded sequence-specific transcription factors (GO:0003700) (Table 1). Among these was Hey1, a key transcription factor that was down-regulated by PTH ~17 fold and could influence the Notch signaling pathway [30]. Genes that encoded components of important signaling pathways in osteocytes were also of interest. PTH-regulated genes for Notch (GO:0007219), Wnt (GO:0016055), BMP (GO:0030509), and transcription cofactors (GO: 0003712) are shown in S. Table 1, tabs 6–7. Finally, as shown in S. Table 1, tab 8, PTH also regulated a series of genes whose products were associated with locomotion as well as immune function. Taken together, these results demonstrate that the regulation by PTH of transcription factors, signaling pathway components and elements whose products are capable of participating directly in cellular activity likely contribute to a highly robust and extensive hormone-modulated transcriptome that could have striking effects on the biological function of the osteocyte.
Table 1. PTH and 1,25(OH)2D3-mediated changes in expression of differentiation-related genes encoding specific transcription factors.
Fold change values for regulation by PTH (100nM PTH vs vehicle, 24 hr treatment), differentiation (IDG-SW3 osteocytes (day 35 vs osteoblasts (day 3), and 1,25D3 (100nM 1,25(OH)2D3 vs vehicle, 24 hr treatment for genes regulated by PTH by ≥ 2 fold (95% confidence, moderated t test) and present in GO category GO:0003700 for sequence specific transcription factors.
| Gene | PTH (fold) | Diff. (fold) | 1,25D3 (fold) |
|---|---|---|---|
| Crx | 18.1 | 0.4 | 11.4 |
| Pou4f2 | 14.6 | 0.9 | 5.8 |
| Nr5a2 | 12.6 | 0.1 | 6.5 |
| Myt1l | 12.1 | 6.0 | 2.0 |
| Tbx22 | 11.8 | 0.7 | 6.9 |
| Gata4 | 11.5 | 0.4 | |
| Tbr1 | 11.2 | 0.2 | 2.3 |
| Esrrg | 11.0 | 2.0 | 1.4 |
| Zic3 | 9.9 | 0.5 | 4.0 |
| Tal1 | 9.5 | 8.1 | |
| Myb | 8.4 | 0.1 | 3.1 |
| Six6 | 8.3 | 0.6 | 2.6 |
| Hnf4g | 8.1 | 0.3 | 2.4 |
| Runx1t1 | 7.5 | 0.2 | 3.4 |
| Csrnp3 | 6.5 | 1.4 | 2.5 |
| Irf4 | 6.2 | 2.2 | |
| Tbx20 | 5.8 | 0.4 | 3.8 |
| Fosl1 | 5.7 | 0.0 | 1.3 |
| Hand2 | 5.7 | 0.3 | 1.8 |
| Pou6f2 | 5.4 | 1.2 | 5.4 |
| Sox17 | 5.1 | 9.6 | |
| Lmx1a | 5.0 | 1.2 | 10.5 |
| Vsx1 | 5.0 | 0.3 | 1.0 |
| Lhx5 | 4.6 | 0.5 | 0.6 |
| Pou2f2 | 4.6 | 0.0 | 2.2 |
| Meox1 | 4.4 | 6.8 | 2.0 |
| Pax7 | 4.1 | 0.4 | 2.1 |
| Bcl3 | 4.0 | 0.4 | 0.8 |
| Pbx1 | 3.8 | 0.5 | 2.0 |
| Rarb | 3.7 | 0.8 | 1.2 |
| Ehf | 3.7 | 9.2 | 0.8 |
| Foxl1 | 3.7 | 0.5 | 2.5 |
| Noto | 3.3 | 1.1 | |
| Maff | 3.2 | 0.1 | 2.0 |
| Hdx | 2.9 | 0.4 | 2.1 |
| Tbx3 | 2.8 | 0.4 | 1.2 |
| Mafb | 2.8 | 2.1 | 1.5 |
| Foxc2 | 2.7 | 0.3 | 1.9 |
| Irf1 | 2.7 | 0.4 | 0.9 |
| Tsc22d1 | 2.7 | 1.2 | 2.3 |
| Mecp2 | 2.6 | 0.8 | 1.5 |
| Elf5 | 2.6 | 2.8 | 0.6 |
| Lef1 | 2.5 | 2.4 | 0.7 |
| Crem | 2.4 | 0.4 | 1.6 |
| Sox11 | 2.3 | 0.2 | 0.3 |
| Batf | 2.3 | 1.5 | 0.9 |
| Arid3a | 2.2 | 1.7 | 1.4 |
| Foxf1a | 2.2 | 1.5 | 3.2 |
| Hsf2 | 2.1 | 1.3 | 1.0 |
| Hopx | 2.0 | 2.9 | 0.5 |
| Nr1d2 | 2.0 | 1.2 | 1.2 |
| Cbfa2t2 | 2.0 | 0.8 | 1.3 |
| Shox2 | 0.5 | 1.7 | 1.0 |
| Zgpat | 0.4 | 0.8 | 1.2 |
| Aff3 | 0.4 | 0.6 | 0.6 |
| Mef2d | 0.4 | 0.9 | 0.5 |
| Arnt2 | 0.4 | 0.3 | 1.8 |
| Cebpa | 0.4 | 2.3 | 0.8 |
| Nfya | 0.3 | 0.9 | 0.6 |
| Hoxa13 | 0.3 | 5.6 | 0.4 |
| Atf3 | 0.3 | 0.2 | 0.7 |
| Foxd1 | 0.3 | 1.2 | 0.5 |
| Nr3c2 | 0.3 | 3.5 | 1.0 |
| Klf4 | 0.3 | 0.4 | 0.3 |
| Klf12 | 0.3 | 7.3 | 0.3 |
| Sox6 | 0.3 | 1.2 | 0.6 |
| Id1 | 0.3 | 0.2 | 0.6 |
| Satb2 | 0.3 | 3.4 | 0.6 |
| Tead3 | 0.2 | 0.3 | 0.5 |
| Creb3l1 | 0.2 | 1.4 | 0.3 |
| Smad9 | 0.2 | 1.5 | 0.5 |
| Sox8 | 0.2 | 26.5 | 2.8 |
| Dlx3 | 0.2 | 9.1 | 1.0 |
| Nfib | 0.2 | 1.0 | 0.6 |
| Nr1h4 | 0.2 | 0.8 | 0.6 |
| Egr1 | 0.2 | 0.1 | 1.0 |
| Egr2 | 0.2 | 0.1 | 1.3 |
| Dlx6 | 0.2 | 4.2 | 0.3 |
| Klf2 | 0.1 | 0.9 | 0.6 |
| Hoxc12 | 0.1 | 0.4 | 0.6 |
| Tbx2 | 0.1 | 12.8 | 0.3 |
| Tcf7 | 0.1 | 8.9 | 0.3 |
| Rcor2 | 0.1 | 56.6 | 0.2 |
| Mef2c | 0.1 | 31.2 | 0.3 |
| Tcf7l1 | 0.1 | 0.5 | 0.6 |
| Hey1 | 0.1 | 27.6 | 0.2 |
| Id3 | 0.1 | 0.6 | 0.9 |
| Irf9 | 0.0 | 2.0 | 3.2 |
3.2 Validation of Response to PTH in a Subset of Genes Expressed in Differentiated IDG-SW3 Osteocytes
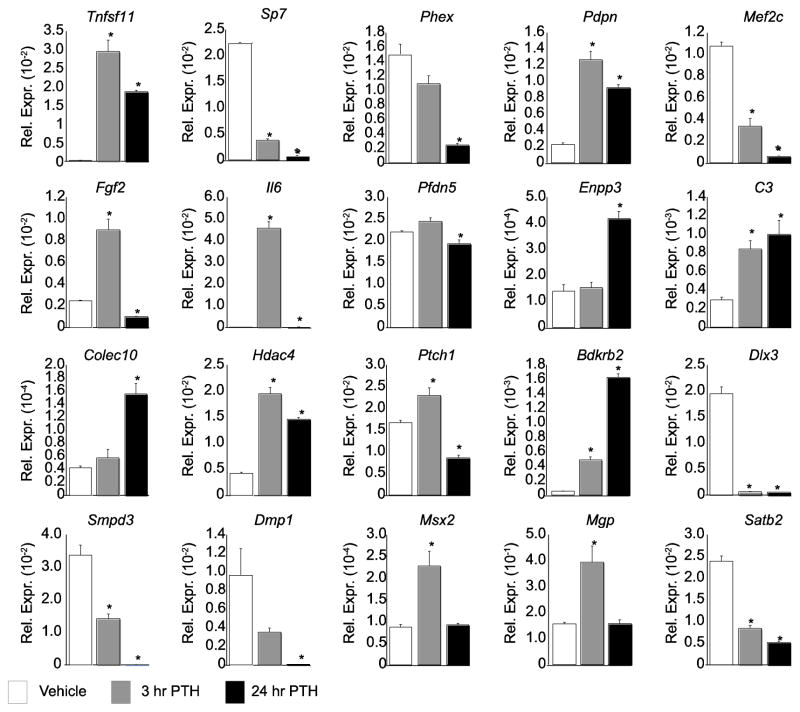
A more detailed examination of PTH-mediated transcriptional effects in IDG-SW3 osteocytes was conducted by interrogating gene expression changes following a 3 hr or 24 hr treatment with PTH, both to validate the RNA-seq data set that was acquired specifically at 24 hr and to probe the temporal nature of PTH response. As can be seen in Fig. 2, all 20 of the genes selected for examination were appropriately validated for the regulation by PTH that was obtained at 24 hr through the genome-wide analyses. Interestingly, we observed striking temporal diversity of gene expression in response to PTH. Many of the genes showed a consistent (Dlx3) or exaggerated (Sp7, Phex, Mef2c, C3, Colec10, Bdkrb2, Smpd3, Dmp1, and Satb2) response to PTH at 24 hr compared to that at 3 hr. In contrast, others were regulated in the appropriate direction, but had returned (Msx2 and Mgp) or were returning (Tnfsf11, Pdpn, Il6, and Hdac4) to baseline levels. The regulation of Enpp3 and Pfdn5 was observed only at 24 hr. While Pdfn5 was not identified as a PTH target in the RNA-seq data set, it was shown to be modestly down-regulated ~1.2 fold at 24 hr. The most dynamic temporal regulation by PTH was exemplified by Fgf2 and Ptch1, which were up-regulated at 3 hr but then down-regulated by 24 hr, suggesting the possibility of a precise, time-dependent induction by PTH followed by a potential secondary inhibitory effect at 24 hr. While the selection of the 24 hr time point was appropriate for RNA-seq, it is clear that the transient regulation of several genes by PTH may have been missed, or that the dynamic regulation of some genes such as Fgf2 and Ptch1 might not have been captured appropriately using a single time point. Nevertheless, these results generally validate the RNA-seq data analysis and suggest that PTH manifests differential patterns of regulation at specific gene targets. This diversity may reflect the ability of PTH to regulate a number of secondary pathways.
Figure 2. qPCR validates the RNA-seq data and identifies complex response to PTH.
Cells treated with vehicle (white), 3 hr PTH (grey), or 24 hr PTH (black) were evaluated for expression of the indicated gene and normalized to β-actin levels. Samples were analyzed in triplicate ± SEM (*, p<0.05 vs. vehicle).
3.3 PTH Regulates the Expression of Genes Associated with Osteocyte Differentiation and Function
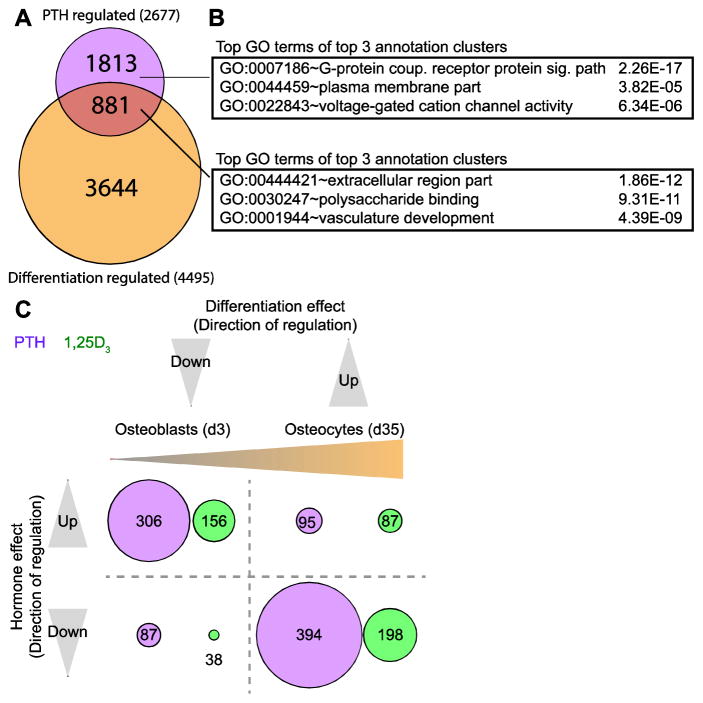
Our previous studies revealed that the differentiation of IDG-SW3 cells from their osteoblast precursors to mature osteocytes was associated with striking changes in gene expression, and that many of these changes were linked to a large cohort of differentiation-related genes that was expressed in both cell types but up- or down-regulated during differentiation relative to the osteoblast precursor [20]. A second cohort of osteocyte-specific genes was also observed, however; this group was absent in the osteoblast precursor but uniquely expressed in the osteocyte. We therefore examined whether the PTH-regulated transcriptome might in some manner overlap these two gene subsets and thus provide insight into PTH action. As can be seen in Fig. 3a, 881 of the 2677 genes whose expression was regulated by PTH were also modulated during the osteoblast to osteocyte transition. GO term analysis of these genes as documented in Fig. 3b revealed subsets that were enriched for extracellular components, polysaccharide binding, and, notably, vasculature development. In contrast, the 1813 genes that were globally regulated by PTH yet unassociated with the course of osteocyte differentiation were most closely linked to G-protein coupled receptor signaling pathways, suggesting similar PTH activities in both osteoblasts and osteocytes.
Figure 3. PTH opposes changes in gene expression that occur during the osteoblast to osteocyte transition.
(A) Venn diagram of PTH-regulated (100nM PTH vs vehicle, 24 hr treatment) and differentiation-regulated genes. Genes in both cohorts are classified as regulated if their gene expression levels are affected ≥ 2 fold, 95% confidence interval, moderated t test. (B) Top GO term from the top 3 annotation clusters with associated P values from DAVID GO term analysis for gene groups from part (A). (C) Diagram comparing direction of regulation by PTH (purple) and 1,25(OH)2D3 (green) to regulation during differentiation (osteoblast vs osteocyte) (x-axis). Genes in all cohorts are classified as regulated if their gene expression levels are affected ≥ 2 fold, 95% confidence interval, moderated t test. The number of genes within each category are labeled and represented visually by the size of the circle.
Of the 881 PTH-regulated and osteocyte differentiation-regulated genes, 401 were up-regulated and 481 were down-regulated by the hormone. Surprisingly, as seen in Fig. 3c, 306 of the 401 genes up-regulated by PTH (76%) were down-regulated during the course of differentiation while 394 of the 481 genes that were down-regulated by PTH (82%) were up-regulated during differentiation. The correlation beween PTH and this differentiation-regulated gene subset is summarized in Table 1 for transcription factors and in S. Table 1, tab 7 for key signaling components. These results suggest that PTH may exert an opposite, and perhaps negative, regulatory impact on gene expression changes that occur during the osteoblast to osteocyte transition, as identified in this in vitro model. Interestingly, only 300 of the 2677 PTH-regulated genes were found to overlap the second, osteocyte-specific cohort (S. Tab. 1, tab 9). Surprisingly, however, almost all of these (294) were up-regulated by PTH. Thus, PTH strongly reinforces rather than opposes expression of this cohort of genes which is uniquely up-regulated in the osteocyte. The expression of Saa2, for example, was largely undetectible in the IDG-SW3 osteoblast but significantly up-regulated in the IDG-SW3 osteocyte; it was then further induced ~43 fold by PTH. Interestingly, Saa2 encodes serum amyloid A2, one of the two acute phase serum proteins that are induced in the cartilage of patients with osteoarthritis [31]. Of note in Fig. 3c, however, a minor but significant number of genes associated with the osteoblast to osteocyte transition were similarly up- or down-regulated by PTH in parallel, indicating that genetic reinforcement can be seen in both gene cohorts. These data involving the expression of both functional components as well as transcription factors and signaling pathway components suggest that PTH may be linked to osteocyte differentiation from its osteoblast precursors. On the other hand, the hormone may also positively regulate genes that are both unique to the mature osteocyte and perhaps essential for its distinct morphological and functional phenotype. Gene lists and extended GO term results for all analyses in Fig. 3 can be found in Supplemental (S.) Table 1, tabs 10–13.
3.4 PTH-Regulated Genes Associated with Osteocyte Differentiation Directionally Overlap Those Regulated by 1,25(OH)2D3
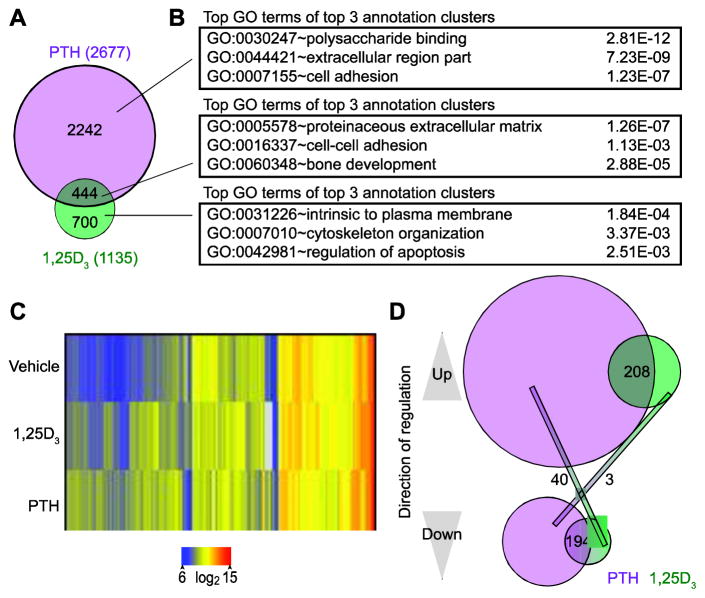
Our previous studies revealed that IDG-SW3 cell differentiation alters transcriptomic response to 1,25(OH)2D3 by restricting the 3870 genes that were regulated by the hormone in the osteoblast to 1135 genes in the osteocyte; 788 of these were unique to the osteocyte while 318 overlapped those regulated during the earlier stage of differentiation [20]. As previously stated, the sum of these two numbers is greater than 1135 due to differences in the regulation of isoforms. Interestingly, as can be seen in Fig. 4a, 444 of the 2677 genes that were regulated by PTH were also modulated by 1,25(OH)2D3 as well. Genes uniquely regulated by PTH or 1,25(OH)2D3 were associated with GO terms distinct from enriched categories for co-regulated genes, which included proteinaceous extracellular matrix, cell-cell adhesion, and bone development as seen in Fig. 4b. Genes regulated by PTH or 1,25(OH)2D3 ≥2 fold were clustered using the Euclidean distance metric (95% confidence, moderated T test) and visualized by heatmap (Fig. 4c). Interestingly, as summarized in Fig. 4d, further delineation of the co-regulated gene cohort revealed a tight coupling between PTH and 1,25(OH)2D3 regulation. Accordingly, of the 444 PTH and 1,25(OH)2D3 co-regulated genes, 208 of them were similarly up-regulated and 194 of them were similarly down-regulated by both hormones. Thus, only 43 were inversely regulated by the two hormones; 40 of these were down-regulated by PTH and up-regulated by 1,25(OH)2D3, while 3 were regulated oppositely. Tnfsf11 (RANKL), for example, was up-regulated as anticipated by both PTH and 1,25(OH)2D3 [32, 33]. To further investigate the relationship between PTH and 1,25(OH)2D3 on co-regulated genes, we analyzed IDG-SW3 osteocytes (d35) with each hormone individually or in combination for 24 hr and analyzed gene expression via qPCR for a panel of ten genes (S. Fig. 2). For Tnfsf11, Sp7, Bdkrb1, Bdkrb2, and Klf4, co-treatment of PTH and 1,25(OH)2D3 acted in an additive or synergistic manner to affect gene expression more than either hormone alone. On other genes, such as Smpd3, the dramatic effects of PTH were attenuated by the comparably modest-effects of 1,25(OH)2D3. Together, these data suggest that the majority of the genes that are regulated by the two hormones are regulated in a similar directional pattern to achieve precise levels of transcriptional regulation.
Figure 4. Regulation of gene expression by PTH and 1,25(OH)2D3 is highly correlated.
(A) Venn diagram of PTH-regulated (100nM PTH vs vehicle, 24 hr treatment) and 1,25(OH)2D3-regulated (100nM 1,25(OH)2D3 vs vehicle, 24 hr treatment) genes. Genes in both cohorts are classified as regulated if their gene expression levels are affected ≥ 2 fold, 95% confidence interval, moderated t test. (B) Top GO term from the top 3 annotation clusters with associated P values from DAVID GO term analysis for gene groups from part (A). (C) Genes from (A) were clustered using the Euclidian distance metric and visualized as a heatmap. (D) Venn diagram of PTH (purple) and 1,25(OH)2D3 (green) up- and down- regulated genes.
In a final transcriptome analysis, we examined whether genes regulated by 1,25(OH)2D3, like those controlled by PTH, were also modulated during the osteoblast to osteocyte transition. As can be seen in Fig. 3c, 156 genes that were up-regulated by 1,25(OH)2D3 were down-regulated during differentiation and 198 genes that were down-regulated by 1,25(OH)2D3 were up-regulated by the differentiation process. These results suggest that like PTH, 1,25(OH)2D3 exerts an opposite, and perhaps inverse, regulatory impact on subsets of genes that are either up- or down-regulated during the osteoblast to osteocyte transition. In the mature osteocyte, however, only 57 genes that were specific to this cell type were also regulated by 1,25(OH)2D3; like PTH, the majority of the genes in this cohort were up-regulated (51) while the remainder (6) were down-regulated. Importantly, a strong overlap was present for the genes that were regulated by both hormones during differentiation. This correlation can be seen specifically for transcription factors in Table 1 and signaling components in S. Table 1, tab 7. Of those regulated by 1,25(OH)2D3, however, only 24 overlapped those controlled by PTH. Differential regulation of osteocyte-specific genes by PTH and 1,25(OH)2D3, including those that encode transcription factors and histone modifying enzymes such as Prmt8, may contribute to a broader transcriptional response to these two hormones. On the other hand, 1,25(OH)2D3 exhibited only a modest regulatory effect on PTH-regulated genes involved in motility and immune function, as seen in S. Table 1, tab 8, highlighting a distinct difference between the two systemic modulators. Gene lists and extended GO term results for all analyses in Fig. 4 can be found in Supplemental (S.) Table 1, tabs 14–17.
3.5 The pCREB Cistrome in Differentiating and Mature Osteocytes
PTH binds to PTH1R, triggering activation of both PKA and PKC signal transduction pathways and their associated downstream transcription factors; the former favors the catabolic actions of PTH while the latter favors more anabolic outcomes. Activation of the PKA pathway is known to result in stimulation of cyclic AMP response element binding protein (CREB) and the expression of pro-resorptive regulators such as RANKL, ENPP1 and ENPP3 [34]. Based upon the central although not exclusive role of CREB in PTH action in osteoblasts, we utilized previously acquired genome-wide phospho-CREB (pCREB) cistromic data sets derived from ChIP-seq analyses in osteoblastic MC3T3-E1 cells to explore the relationship between pCREB binding and the two target gene cohorts described above in the IDG-SW3 osteocyte. The pCREB cistrome was comprised of 9398 binding sites in the absence of inducer and 17496 occupied sites following a 1 hr treatment with forskolin, as summarized in S. Fig. 1a. Thus, while pCREB binding activity at the genome is substantial, it is increased by almost 2-fold following PKA activation. Additional properties of this cistrome can be seen in S. Fig. 1b and c, where pCREB binding sites have been validated through the de novo identification of a DNA sequence motif representative of a classic CREB responsive element (CRE) and quantification of the distribution of these pCREB-occupied sites across the genome relative to nearby genes. It is worth noting here that while many transcription factors are now known to bind abundantly to sites distal to gene promoters [35, 36], pCREB binding occurs much more frequently at promoter-proximal sites (S. Fig. 1c).
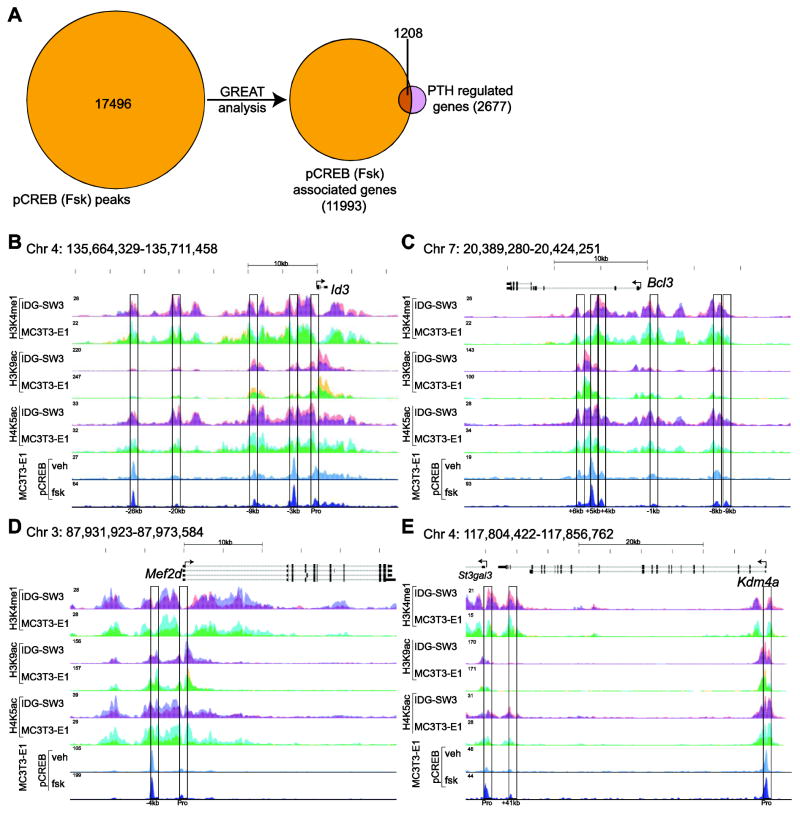
We then explored the frequency through which pCREB binding sites were associated with genes that were regulated by PTH through the application of GREAT analysis [37]. As summarized in Fig. 5a, 1208 of the 2677 (45%) PTH-regulated genes using this algorithm were associated with pCREB peaks. These results are highlighted in Fig. 5b–e which depict pCREB binding ChIP-seq tracks for four selected genes that encode transcription factors or histone modifying enzymes. At the Id3 locus, five sites of pCREB binding can be found, one near the promoter and the others at -3kb, -9kb, -20kb, and -26kb (Fig. 5b). Inhibitor of DNA binding 3 (Id3) encodes a helix-loop-helix transcription factor first identified as a component of early transcriptional response to growth factors and is also involved in the differentiation of several cell types, including regulatory T cells and myoblasts [38–40]. The Bcl3 locus has several sites of enhancer/pCREB binding, including those at -8kb, -1kb, +4kb, and +5kb (Fig. 5c). B-cell leukemia/lymphoma 3 (Bcl3) encodes BCL-3, which contributes to regulation of diverse biological processes through its interaction with NF-κB subunit p50 [41, 42]. Interestingly, myocyte enhancer factor 2d (Mef2d), closely related to Mef2c that is involved in Sost regulation, retained a small pCREB peak at the promoter and a larger peak just upstream at -4kb (Fig. 5d). Finally, the main pCREB binding peak at the Kdam4a locus was located near the promoter with additional peaks present at +41kb and at the promoter of the nearby gene St3gal3 (Fig. 5e). Lysine demethylase 4a (Kdm4a) encodes JMJD2A, which associates with class I histone deacetylases (HDACs) and retinoblastoma protein (pRb) to mediate gene suppression [43]. These specific examples and the overall genome-wide data set identifies pCREB binding sites at almost half of the genes found to be regulated by PTH suggesting a strong linkage through PKA signaling. Genes that were both PTH- and differentiation-regulated as well as genes that were PTH- and 1,25(OH)2D3-regulated also exhibited strong associations with 61% and 56% of these genes, respectively, having pCREB peaks located nearby. Interestingly, 78 genes that were regulated by both PTH and 1,25(OH)2D3 exhibited overlapping pCREB and VDR binding sites (S. Fig. 1d). In comparison, only 30 of the 300 osteocyte-specific and PTH-regulated genes were associated with pCREB occupied binding sites.
Figure 5. pCREB binding sites correlate with PTH-regulated genes and are located within epigenetically delineated enhancers.
A) 17496 forskolin-induced pCREB binding sites were associated with 11993 genes as assessed using the GREAT algorithm. ≥ 1208 of the 2677 genes associated with the PTH-regulated transcriptome in IDG-SW3 osteocytes (24 hr at 100nM PTH vs vehicle, 2 fold, 95% confidence interval, moderated t test). contained pCREB. B) ChIP-seq tag density tracks (normalized to 107 tags) for pCREB binding in MC3T3-E1 preosteoblast and for selected histone modifications in IDG-SW3 osteoblasts (red), IDG-SW3 osteocytes (blue with overlap in purple), MC3T3-E1 preosteoblasts (yellow) and MC3T3-E1 preosteoblasts (blue with overlap in green) at the gene loci Id3 (B), Bcl3 (C), Mef2d (D) and Kdm4a (E) loci.
In a final analysis, we compared the pCREB cistrome to the published VDR cistrome in IDG-SW3 d35 osteocyte cells [20]. Strikingly, we found 1080 overlapping sites for the MC3T3-E1 pCREB (fsk) and IDG-SW3d35 (1,25(OH)2D3) cistromes, which were associated with 1485 genes through GREAT analysis (Fig. 6a; S. Tab. 1, tabs 19–20). We next compared these genes associated with overlapping pCREB and VDR binding sites to the 444 genes regulated by both PTH and 1,25(OH)2D3 in osteocytes (d35) and found that over 10% (46 genes) of the PTH and 1,25(OH)2D3 co-regulated genes (Fig. 4a) had overlapping pCREB and VDR binding sites (Fig. 6a; S. Tab. 1, tab 20). As an example, ChIP-seq tracks for pCREB and VDR are shown for the gene encoding ectodysplasin A receptor, Edar in Fig. 6b. These results demonstrate the potential for overlap between PTH- and 1,25(OH)2D3- mediated transcriptional regulation through pCREB and VDR, respectively.
Figure 6. pCREB and VDR exhibit overlapping cistromes.
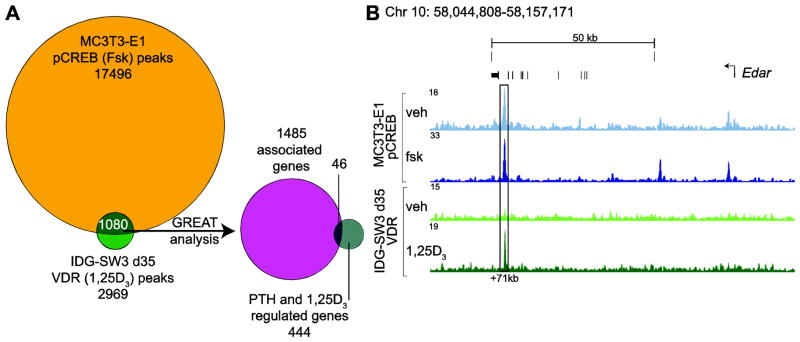
A) 17496 forskolin-induced pCREB binding sites in MC3T3-E1 cells were compared to 2969 1,25(OH)2D3-induced VDR binding sites in IDG-SW3 d35 cells. 1080 sites overlapped and these were associated with 1485 genes through the GREAT algorithm. 46 of the 444 genes associated with the PTH- and 1,25(OH)2D3-regulated transcriptome in IDG-SW3 osteocytes (24 hr at 100nM PTH or 1,25(OH)2D3 vs vehicle, ≥ 2 fold, 95% confidence interval, moderated t test) were among the associated genes. B) ChIP-seq tag density tracks (normalized to 107 tags) for pCREB binding in MC3T3-E1 preosteoblasts and VDR binding in IDG-SW3 d35 osteocytes at the gene locus for Edar.
3.6 Epigenetic Enhancer Signature Marks Highlight pCREB Binding Sites Genome-wide
Our previous epigenetic studies have shown that regulatory enhancers are established as components of the genome early in the osteoblast lineage [20, 44]. Thus, while the enrichment levels of these marks are dynamic at specific genes as a function of either cellular differentiation or through regulation by secondary factors such as 1,25(OH)2D3, only a few of these marks appear or are removed de novo during osteocyte formation. This conclusion is supported by the results documented in Fig. 5b–e, which depicts the data tracks of histone 3 lysine 4 mono-methylation (H3K4me1), histone 3 lysine 9 acetylation (H3K9ac), and/or histone 4 lysine 5 acetylation (H4K5ac) enrichment across loci for Id3, Bcl3, Mef2d, and Kdm4a in IDG-SW3 osteoblasts and osteocytes as well as MC3T3-E1 preosteoblasts and osteoblasts. These genes also highlight the tight epigenetic associations that are seen on a genome-wide scale across the several osteoblast genomes. Thus, as observed in S. Fig. 1e, histone enrichment for H3K4me1, H4K5ac, and H3K9ac is present in each of these four osteoblast lineage cells 76%, 79%, and 94% of the time as compared to that seen in MC3T3-E1 preosteoblasts, respectively. The results in Fig. 5b–e also document the close relationship that exists between these histone marks and binding sites for pCREB at the four representative target gene loci in the IDG-SW3 cell line as well. Importantly, this observation is similarly documented on a genome-wide scale, with enrichment for H3K4me1, H4K5ac, and H3K9ac in each of these four osteoblast lineage cells occurring in close association with pCREB binding 93%, 89% and 62% of the time as well (S. Fig. 1f). It should be noted, however, that pCREB binding sites may align directly over a histone-marked enhancer, such as is seen at Id3 -26kb (Fig. 5b) and Kdm4a +41kb (Fig. 5e). or alternatively, can be flanked by an activating histone mark, such as can be seen at Id3 -20kb and -9kb (Fig. 5b) and Bcl3 -1kb and -8kb (Fig. 5c). It has been suggested that the latter profile is due to specific histone displacement. We conclude that these data clearly establish the locations of pCREB binding sites at genes that are regulated by PTH. As pCREB binding was found to be deficient at genes that were uniquely up-regulated by PTH in mature IDG-SW3 osteocytes, the histone marks that are found at these genes may reflect the presence of enhancers controlled by any one of the myriad of other transcription factors including the VDR that are likely active in this cell type at this time.
4. DISCUSSION
The studies described herein document the actions of PTH and 1,25(OH)2D3 on osteoblast differentiation and function as assessed on a genome-wide scale in the IDG-SW3 cell model. We show that PTH regulates an extensive transcriptome in differentiated IDG-SW3 osteocytes and that the genes sensitive to this hormone belong to two separate cohorts: those such as Slc1a1 and Phex that are up- or down-regulated during the differentiation process and those such as Prmt8 that are uniquely expressed only in the osteocyte. Further examination of the effects of PTH reveal that while PTH both stimulates and suppresses various members of the differentiation-associated transcriptome, these actions largely, although not exclusively, oppose changes in gene expression that highlight the transition. These genes include Satb1 and Hey1. In contrast, the majority of the genes that are up-regulated and uniquely expressed in the osteocyte, such as Saa2, are not suppressed by PTH, but rather further up-regulated. These findings support an hypothesis in which PTH functions to retard not only osteoblast formation from mesenchymal precursors, but their further differentiation into osteocytes. PTH’s positive actions on genes that are expressed in fully differentiated osteocytes, on the other hand, could imply that in contrast, PTH may represent a direct positive modulator of mature osteocyte structure and function. Specific studies in vivo will be necessary to further substantiate or refute these hypotheses.
Interestingly, we found that 1,25(OH)2D3 exerted actions on the expression of genes regulated both during osteocyte differentiation as well as in an osteocyte-specific manner that were similar. Accordingly, 1,25(OH)2D3 opposed the expression of genes differentially regulated during the osteoblast to osteocyte transition similar to that of PTH, yet largely reinforced the expression of genes unique to the mature osteocyte. A direct comparison of the 1,25(OH)2D3- and PTH-regulated genes in the two cohorts indicated the presence of a significant overlap. This suggests that the two hormones may have similar functions to limit osteocyte differentiation from its osteoblast precursors. Like PTH, 1,25(OH)2D3 may play a negative role in osteoblast differentiation from its mesenchymal precursors as well. Surprisingly, while 1,25(OH)2D3 induced further a subset of the cohort of genes that were expressed in mature osteocytes, the majority of these genes were unrelated to those up-regulated by PTH. Thus, we speculate that at least a subset of the actions of the two hormones to regulate mature osteocyte function may be different. Despite these potential differences, the two hormones are known to regulate in parallel the expression of sclerostin [20, 45], RANKL [46, 47] and Fgf23 [48]. Additional studies will be necessary to bolster the proposed diverse actions of these two hormones on osteocyte development and function in vivo. It is worth noting, of course, that despite the large number of genes whose expression levels are regulated by PTH and 1,25(OH)2D3 during the transition, the linkage betwee their expression and the process of osteocyte differentiation itself is unknown.
Interestingly, while Sost is a well known target of PTH [49], this gene was not identified through our RNA-seq analyses in the group of 2677 PTH-regulated genes (≥2-fold PTH regulation, 95% confidence, moderated T test) because the dramatic effects of PTH suppressed transcript abundanace to undetectable levels in two of three triplicate samples, thus preventing a statistical evaluation of the regulation. Thus, the small group of genes with either a) not statistically valid basal values upon sequencing, thus preventing a calculation of up-regulation by PTH or b) basal activities which are entirely suppressed by PTH, thus preventing a similar statistical assessment of PTH mediated down-regulation, such as Sost, needs to be explored further using qPCR.
Despite the fact that the PKA-activated pCREB cistromes were obtained from MC3T3-E1 derived osteoblasts and not from IDG-SW3 cells (due largely to the challenges of conducting ChIP-seq analysis with pCREB antibody in the latter cell line), a subset of these pCREB binding sites correlated directly with genes that were modulated by PTH during the osteoblast to osteocyte transition. That these binding sites are both functional and capable of regulating nearby genes is reinforced by the fact that 93% of these sites were enriched for the key histone mark H3K4me1, an epigenetic signature of a gene-regulating enhancer, in all four cell types (IDG-SW3 osteoblasts and osteocytes, and MC3T3-E1 preosteoblasts and osteoblasts). Perhaps as important, these data provide considerable insight into not only diverse gene targets, but the locations of the regulatory regions on a gene by gene basis that mediate PTH action. Thus, they provide the opportunity for future investigation of the mechanism(s) through which several hundreds genes could be explored for regulation by PTH. We interpret our findings to suggest further that the predominant actions of PTH on this cohort may be due largely, although not exlusively, to activation of the PKA signaling pathway initiated by this known arm of the receptor for PTH. Interestingly, a much smaller number of genes up-regulated uniquely by PTH in osteocytes contained pCREB binding sites. We speculate that this might reflect the fact that these genes are more frequently regulated through activation of alternative signaling pathways include ones initiated through PKC. Interestingly, one of several genes discussed earlier whose expression is down-regulated by PTH and 1,25(OH)2D3 is Sost. While this suppression by PTH in particular is well established, the mechanism remains to be clarified, although it is known to involve activity of the transcription factor Mef2c [45]. The absence of pCREB at this site is consistent with the view that PTH activation involves PKA-mediated control of HDAC5 translocation to the nucleus, where its interacts directly with Mef2c to downregulate Sost expression [27]. It is worth noting, however, that functional correlations such as those made here between active genes and the presence of DNA-bound regulatory factors are difficult, primarily because functional linkage between these two entities must be unequivocally established via additional experimental efforts [46, 50, 51].
We have concluded from our earlier studies as well as the present work that the evolution of enhancer elements begins early in the osteoblast lineage [20, 28, 44]. Thus, while the level of histone modifications at enhancers is potentially dynamic over a wide range of cellular conditions, the appearance of new enhancers associated with progressive stages of differentiation appear to be infrequent. Thus, for example, only a few novel H3K4me1 marks appeared in the osteocyte that were not apparent in its osteoblast precursor [20]. These observations have led us to conclude that the osteoblast to osteocyte transition is driven largely by temporal and sequential activation of transcription factors and their ability to recruit chromatin regulators that participate in the modification of chromatin architecture necessary for altered gene output rather than the appearance of new regulatory components acting in cis. If this interpretation is correct, the impact of PTH as well as 1,25(OH)2D3 and perhaps other ligands on their cognate transcription factors would seem likely to play key roles not only in orchestrating osteocyte differentiation but in regulating the functional activity of the mature cells as well. An important question still remains, however, which relates to identification of the signaling pathway(s) and definition of its activating components that drive osteocyte differentiation. Some progress is being made in that regard as factors that are associated with mineralization such as phosphate, DMP-1, and other phosphoproteins as well as the mineralization process itself are being considered. Elucidation of these regulatory components will be important in understanding osteocyte differentiation.
Although studies in the IDG-SW3 cell line may not reflect perfectly the process of osteocyte differentiation that occurs in vivo, our studies have identified several regulatory concepts that are important, but will require examination in vivo. This assessment is not likely to be trivial, however, due to the associated complexity of the stage-specific differentiation of osteocytes in bone from their osteoblast precursors, and their heterogeneity as well as location in highly mineralized matrix in the skeleton. The use of genetically modified mouse models wherein key genes are conditionally deleted will likely be required, as has been accomplished in studies of other signaling pathways and their activities of osteoblast differentiation. We anticipate, however, that these studies will be not only of considerable interest but also of value in understanding the complex regulatory nature of systemic hormones such as PTH and 1,25(OH)2D3 on osteoblast lineage cells.
In conclusion, we report the identification of a PTH-regulated transcriptome in IDG-SW3 derived osteocytes. The major components of this transcriptome largely oppose the expression of genes that undergo change during osteocyte differentiation, but reinforce those that are uniquely up-regulated in the mature osteocyte. This suggests that PTH may play dual roles controlling both the differentiation of the osteocyte as well as its functional activity. We also find that the activity of 1,25(OH)2D3 parallels the actions of PTH during the differentiation process, but appears to positively regulate a unique set of genes in the mature osteocyte. These and the additional findings in this study provide clues for further research on the development and function of this interesting cell type.
Supplementary Material
Highlights.
PTH regulated genes in IDG-SW3 osteocytes were identified by RNA-sequencing analysis.
The transcriptional effects of PTH largely opposed gene expression changes during differentiation.
PTH and 1,25(OH)2D3 transcriptional regulation was highly correlated for co-regulated genes.
A pCREB cistrome was characterized using ChIP-sequencing.
pCREB sites were associated with PTH regulated genes and marked by a stable epigenetic landscape.
Acknowledgments
We thank members of the Pike Lab for their helpful discussions and contributions to this manuscript. We also acknowledge members of the University of Wisconsin DNA Sequencing Facility in the UW Biotechnology Center. This work was supported by NIAMS grant AR-064424 and NIDDK grant DK-072281 to JWP
Footnotes
Supplemental information is included with the manuscript
Discloure: The authors declare no conflict of interest
6. DATA ACCESS
All sequencing data is publically available in the GEO database: GSE62981
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bonewald LF. The amazing osteocyte. J Bone Miner Res. 2011;26:229–38. doi: 10.1002/jbmr.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling. Bone. 2008;42:606–15. doi: 10.1016/j.bone.2007.12.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dallas SL, Prideaux M, Bonewald LF. The osteocyte: an endocrine cell … and more. Endocr Rev. 2013;34:658–90. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sato M, Asada N, Kawano Y, Wakahashi K, Minagawa K, Kawano H, Sada A, Ikeda K, Matsui T, Katayama Y. Osteocytes regulate primary lymphoid organs and fat metabolism. Cell Metab. 2013;18:749–58. doi: 10.1016/j.cmet.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Paic F, Igwe JC, Nori R, Kronenberg MS, Franceschetti T, Harrington P, Kuo L, Shin DG, Rowe DW, Harris SE, Kalajzic I. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45:682–92. doi: 10.1016/j.bone.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo SM, Rosser J, Dusevich V, Kalajzic I, Bonewald LF. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J Bone Miner Res. 2011;26:2634–46. doi: 10.1002/jbmr.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faul C, Amaral AP, Oskouei B, Hu MC, Sloan A, Isakova T, Gutiérrez OM, Aguillon-Prada R, Lincoln J, Hare JM, Mundel P, Morales A, Scialla J, Fischer M, Soliman EZ, Chen J, Go AS, Rosas SE, Nessel L, Townsend RR, Feldman HI, St John Sutton M, Ojo A, Gadegbeku C, Di Marco GS, Reuter S, Kentrup D, Tiemann K, Brand M, Hill JA, Moe OW, Kuro-O M, Kusek JW, Keane MG, Wolf M. FGF23 induces left ventricular hypertrophy. J Clin Invest. 2011;121:4393–408. doi: 10.1172/JCI46122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gutiérrez OM, Wolf M, Taylor EN. Fibroblast growth factor 23, cardiovascular disease risk factors, and phosphorus intake in the health professionals follow-up study. Clin J Am Soc Nephrol. 2011;6:2871–8. doi: 10.2215/CJN.02740311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quarles LD. Role of FGF23 in vitamin D and phosphate metabolism: implications in chronic kidney disease. Exp Cell Res. 2012;318:1040–8. doi: 10.1016/j.yexcr.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brotto M, Johnson ML. Endocrine crosstalk between muscle and bone. Curr Osteoporos Rep. 2014;12:135–41. doi: 10.1007/s11914-014-0209-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mo C, Romero-Suarez S, Bonewald L, Johnson M, Brotto M. Prostaglandin E2: from clinical applications to its potential role in bone- muscle crosstalk and myogenic differentiation. Recent Pat Biotechnol. 2012;6:223–9. doi: 10.2174/1872208311206030223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asada N, Katayama Y, Sato M, Minagawa K, Wakahashi K, Kawano H, Kawano Y, Sada A, Ikeda K, Matsui T, Tanimoto M. Matrix-embedded osteocytes regulate mobilization of hematopoietic stem/progenitor cells. Cell Stem Cell. 2013;12:737–47. doi: 10.1016/j.stem.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Boivin G, Mesguich P, Pike J, Bouillon R, Meunier P, Haussler M, Dubois P, Morel G. Ultrastructural immunocytochemical localization of endogenous 1,25-dihydroxyvitamin D3 and its receptors in osteoblasts and osteocytes from neonatal mouse and rat calvaria. Bone Miner. 1987;3:125–36. [PubMed] [Google Scholar]
- 14.Fermor B, Skerry TM. PTH/PTHrP receptor expression on osteoblasts and osteocytes but not resorbing bone surfaces in growing rats. J Bone Miner Res. 1995;10:1935–43. doi: 10.1002/jbmr.5650101213. [DOI] [PubMed] [Google Scholar]
- 15.O’Brien CA. Control of RANKL gene expression. Bone. 2010;46:911–9. doi: 10.1016/j.bone.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nerenz RD, Martowicz ML, Pike JW. An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-kappaB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Mol Endocrinol. 2008;22:1044–56. doi: 10.1210/me.2007-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–76. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 18.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med. 2011;17:1235–41. doi: 10.1038/nm.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Q, Jilka RL, Manolagas SC, O’Brien CA. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J Biol Chem. 2002;277:48868–75. doi: 10.1074/jbc.M208494200. [DOI] [PubMed] [Google Scholar]
- 20.St John HC, Bishop KA, Meyer MB, Benkusky NA, Leng N, Kendziorski C, Bonewald LF, Pike JW. The osteoblast to osteocyte transition: epigenetic changes and response to the vitamin d3 hormone. Mol Endocrinol. 2014;28:1150–65. doi: 10.1210/me.2014-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prosser D, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29:664–73. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Canalis E. Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat Rev Endocrinol. 2013;9:575–83. doi: 10.1038/nrendo.2013.154. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien CA, Plotkin LI, Galli C, Goellner JJ, Gortazar AR, Allen MR, Robling AG, Bouxsein M, Schipani E, Turner CH, Jilka RL, Weinstein RS, Manolagas SC, Bellido T. Control of bone mass and remodeling by PTH receptor signaling in osteocytes. PLoS One. 2008;3:e2942. doi: 10.1371/journal.pone.0002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powell WF, Barry KJ, Tulum I, Kobayashi T, Harris SE, Bringhurst FR, Pajevic PD. Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. J Endocrinol. 2011;209:21–32. doi: 10.1530/JOE-10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collette NM, Genetos DC, Economides AN, Xie L, Shahnazari M, Yao W, Lane NE, Harland RM, Loots GG. Targeted deletion of Sost distal enhancer increases bone formation and bone mass. Proc Natl Acad Sci U S A. 2012;109:14092–7. doi: 10.1073/pnas.1207188109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loots GG, Kneissel M, Keller H, Baptist M, Chang J, Collette NM, Ovcharenko D, Plajzer-Frick I, Rubin EM. Genomic deletion of a long-range bone enhancer misregulates sclerostin in Van Buchem disease. Genome Res. 2005;15:928–35. doi: 10.1101/gr.3437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wein MN, Spatz J, Nishimori S, Doench J, Root D, Babij P, Nagano K, Baron R, Brooks D, Bouxsein M, Pajevic PD, Kronenberg HM. HDAC5 controls MEF2C-driven sclerostin expression in osteocytes. J Bone Miner Res. 2014 doi: 10.1002/jbmr.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer MB, Benkusky NA, Pike JW. The RUNX2 cistrome in osteoblasts: characterization, downregulation following differentiation and relationship to gene expression. J Biol Chem. 2014 doi: 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer MB, Goetsch PD, Pike JW. VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol. 2012;26:37–51. doi: 10.1210/me.2011-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zanotti S, Canalis E. Notch signaling in skeletal health and disease. Eur J Endocrinol. 2013;168:R95–103. doi: 10.1530/EJE-13-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallon R, Freuler F, Desta-Tsedu N, Robeva A, Dawson J, Wenner P, Engelhardt P, Boes L, Schnyder J, Tschopp C, Urfer R, Baumann G. Serum amyloid A (apoSAA) expression is up-regulated in rheumatoid arthritis and induces transcription of matrix metalloproteinases. J Immunol. 2001;166:2801–7. doi: 10.4049/jimmunol.166.4.2801. [DOI] [PubMed] [Google Scholar]
- 32.Kondo H, Guo J, Bringhurst FR. Cyclic adenosine monophosphate/protein kinase A mediates parathyroid hormone/parathyroid hormone-related protein receptor regulation of osteoclastogenesis and expression of RANKL and osteoprotegerin mRNAs by marrow stromal cells. J Bone Miner Res. 2002;17:1667–79. doi: 10.1359/jbmr.2002.17.9.1667. [DOI] [PubMed] [Google Scholar]
- 33.Suda T, Ueno Y, Fujii K, Shinki T. Vitamin D and bone. J Cell Biochem. 2003;88:259–66. doi: 10.1002/jcb.10331. [DOI] [PubMed] [Google Scholar]
- 34.Bellido T, Saini V, Pajevic PD. Effects of PTH on osteocyte function. Bone. 2013;54:250–7. doi: 10.1016/j.bone.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pike JW, Meyer MB. Fundamentals of vitamin D hormone-regulated gene expression. J Steroid Biochem Mol Biol. 2014;144PA:5–11. doi: 10.1016/j.jsbmb.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pike JW, Lee SM, Meyer MB. Regulation of gene expression by 1,25-dihydroxyvitamin D3 in bone cells: exploiting new approaches and defining new mechanisms. Bonekey Rep. 2014;3:482. doi: 10.1038/bonekey.2013.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christy BA, Sanders LK, Lau LF, Copeland NG, Jenkins NA, Nathans D. An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc Natl Acad Sci U S A. 1991;88:1815–9. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maruyama T, Li J, Vaque JP, Konkel JE, Wang W, Zhang B, Zhang P, Zamarron BF, Yu D, Wu Y, Zhuang Y, Gutkind JS, Chen W. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat Immunol. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohamed JS, Lopez MA, Cox GA, Boriek AM. Ankyrin repeat domain protein 2 and inhibitor of DNA binding 3 cooperatively inhibit myoblast differentiation by physical interaction. J Biol Chem. 2013;288:24560–8. doi: 10.1074/jbc.M112.434423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatada EN, Nieters A, Wulczyn FG, Naumann M, Meyer R, Nucifora G, McKeithan TW, Scheidereit C. The ankyrin repeat domains of the NF-kappa B precursor p105 and the protooncogene bcl-3 act as specific inhibitors of NF-kappa B DNA binding. Proc Natl Acad Sci U S A. 1992;89:2489–93. doi: 10.1073/pnas.89.6.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wulczyn FG, Naumann M, Scheidereit C. Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-kappa B. Nature. 1992;358:597–9. doi: 10.1038/358597a0. [DOI] [PubMed] [Google Scholar]
- 43.Gray SG, Iglesias AH, Lizcano F, Villanueva R, Camelo S, Jingu H, Teh BT, Koibuchi N, Chin WW, Kokkotou E, Dangond F. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J Biol Chem. 2005;280:28507–18. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- 44.Meyer MB, Benkusky NA, Lee CH, Pike JW. Genomic Determinants of Gene Regulation by 1,25-Dihydroxyvitamin D3 During Osteoblast-Lineage Cell Differentiation. J Biol Chem. 2014 doi: 10.1074/jbc.M114.578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leupin O, Kramer I, Collette N, Loots G, Natt F, Kneissel M, Keller H. Control of the SOST bone enhancer by PTH using MEF2 transcription factors. J Bone Miner Res. 2007;22:1957–67. doi: 10.1359/jbmr.070804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol. 2006;26:6453–68. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim S, Yamazaki M, Zella LA, Shevde NK, Pike JW. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26:6469–86. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haussler MR, Whitfield GK, Kaneko I, Haussler CA, Hsieh D, Hsieh JC, Jurutka PW. Molecular mechanisms of vitamin D action. Calcif Tissue Int. 2013;92:77–98. doi: 10.1007/s00223-012-9619-0. [DOI] [PubMed] [Google Scholar]
- 49.Bellido T, Ali AA, Gubrij I, Plotkin LI, Fu Q, O’Brien CA, Manolagas SC, Jilka RL. Chronic elevation of parathyroid hormone in mice reduces expression of sclerostin by osteocytes: a novel mechanism for hormonal control of osteoblastogenesis. Endocrinology. 2005;146:4577–83. doi: 10.1210/en.2005-0239. [DOI] [PubMed] [Google Scholar]
- 50.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1alpha,25-dihydroxyvitamin D3. J Biol Chem. 2010;285:15599–610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galli C, Zella LA, Fretz JA, Fu Q, Pike JW, Weinstein RS, Manolagas SC, O’Brien CA. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149:146–53. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.