Abstract
Background
The inhibitor-of-apoptosis protein survivin, encoded by BIRC5, regulates apoptosis, cell division and proliferation. Several survivin splice variants have been described however, the prognostic significance of their expression has not been well defined in pediatric acute myeloid leukemia (AML).
Procedure
Quantitative expression analyses of BIRC5 mRNA (n = 306) and survivin transcript splice variants (n = 90) were performed on diagnostic bone marrow samples from children with de novo AML treated on the clinical trials CCG-2961 and AAML03P1, then correlated with disease characteristics and clinical outcome.
Results
Total BIRC5 expression did not correlate with clinical outcome. Fragment length analysis and sequencing of the entire BIRC5 transcript demonstrated three splice variants. The most prominent product, wild-type survivin, was expressed in all samples tested. Two minor transcripts were present in 90 patients treated on CCG-2961; survivin-2B and a novel variant, survivin-ΔEx2, characterized by deletion of BIRC5 exon II. A high 2B/ΔEx2 expression ratio (≥1) correlated with increased diagnostic WBC count, monocytic phenotype, +8 cytogenetics, lower complete remission (45% [n = 10] vs. 88% [n = 59], P < 0.001) and higher induction failure rates (23% [n = 5] vs. 3% [n = 2], P = 0.009). Consistent with this poor induction response, patients with a 2B/ΔEx2 ratio ≥1 had inferior 5-year survival rates (OS 36% vs. 60%, P = 0.011; EFS 23% vs. 53% at 5 years, P = 0.001) and appear to have increased relapse risk (P = 0.056). Subset analyses suggest that relative over-expression of 2B, rather than under-expression of ΔEx2 determines clinical response.
Conclusions
High survivin-2B/ΔEx2 ratios are associated with refractory disease and inferior survival in childhood AML. Survivin splice variant expression warrants prospective evaluation in clinical trials.
Keywords: Survivin, splice variant, acute myeloid leukemia, childhood AML, molecular genetics, refractory disease
Introduction
Acute myeloid leukemia (AML) is characterized by numerous molecular abnormalities impairing differentiation and promoting proliferation [1]. Although recurrent cytogenetic and molecular abnormalities are powerful predictors of relapse and survival in both adult and pediatric AML, identifying reliable prognostic markers of primary refractory disease remains a challenge. Overall, approximately 20–30% of adults (<60 years) and up to 17% of children will fail to achieve complete remission (CR) with cytarabine and anthracycline based induction chemotherapy [1,2]. The prognosis for children with primary refractory disease is extremely poor, with only a third achieving remission with a second therapeutic attempt [3]. Novel prognostic markers that predict induction failure are needed to facilitate the early introduction of intensified and/or novel therapeutic strategies.
Survivin, a small inhibitor-of-apoptosis protein (IAP) encoded by the BIRC5 gene, plays critical roles in apoptosis, cell division, and proliferation [4]. Several splice variants of survivin such as full-length survivin (-WT), survivin-2B, and survivin-ΔEx3 are well characterized [5]. Survivin-WT is encoded by four exons (I–IV), survivin 2B has an additional exon (IIB), and survivin ΔEx3 has deletion of exon III [5]. Expression of survivin and its splice variants correlates with white blood cell (WBC) count, cytogenetics, clinical response, and survival in AML [6–13]. Studies on survivin expression in childhood AML, however, have been restricted to relatively small cohorts (<50 patients) [6,9,12]. Recent preclinical data show that survivin is a transcriptional target of the FLT3-STAT pathway [14] and regulates proliferation in FLT3-mutated AML [15].
To determine whether survivin expression correlates with disease characteristics, such as FLT3 mutational status, and clinical outcome, we performed molecular and expression characterization of BIRC5 and its transcript splice variants in a large cohort of childhood AML.
Methods
Patients and Treatment
In an initial sequencing of the entire coding sequence of BIRC5 in diagnostic specimens from 100 children with AML, no disease-associated alterations were identified. We therefore quantitatively analyzed BIRC5 mRNA expression in 306 patients with de novo AML treated on multicenter clinical trials CCG-2961 (N = 91) and AAML03P1 (N = 215). Furthermore, survivin transcript splice variant data was available for 90 patients, all of whom were treated on CCG-2961 (99%). Details and clinical outcomes of these trials have been reported previously [16,17]. CCG-2961, enrolled 901 eligible de novo patients (<21 years old) and incorporated an IdaDCTER induction regimen, followed by a randomization between IdaDCTER or fludarabine, cytarabine, and idarubicin in course 2. Patients with a matched related family donor underwent haematopoietic stem cell transplantation (HSCT) as course 3, and those without a donor received high-dose cytarabine (HDAC) and L-asparaginase followed by a randomization between one or no courses of IL-2 [16]. The CCG-2961 study failed to demonstrate any difference for any randomization, with no new agent improving outcome [16]. The AAML03P1 trial enrolled 340 eligible de novo patients (≤21 years old) and assessed the safety of adding gemtuzumab ozogamicin (GO; 3 g/m2 as a single dose during each of courses 1 and 4) to intensive chemotherapy. Patients with a matched family donor received HSCT as course 4, and those without a donor received two further courses of intensification (mitoxantrone, cytarabine, and GO followed by HDAC and l-asparaginase) [17]. Clinical characteristics were similar for patients enrolled on CCG-2961 and AAML03P1 [16,17]. Furthermore, clinical outcomes for patients treated on the AAML03P1 study were generally similar to those treated during the latter stages of CCG2961 (EFS and OS 53% and 66% vs. 46% and 57%, respectively) [16,17].
Informed consent was obtained in accordance with the Declaration of Helsinki for both participation in the clinical trials and the collection of specimens for biological studies. The institutional review boards of all participating institutions approved the clinical trials, and the COG Myeloid Disease Biology Committee approved this study. CCG-2961 and AAML03P1 are registered at ClinicalTrials.gov as NCT00002798 and NCT00070174, respectively.
BIRC5 Expression and Splice Variant Analysis
All studies were performed on Ficoll purified marrow specimens with no further selection. The median percentage of bone marrow blasts for all patients analyzed was 71% (range 2–100%). For the 91 patients enrolled on CCG-2961, the median percentage of bone marrow blasts was 77% (range 16–100%). Quantitative analysis of BIRC5 expression in AML blasts from diagnostic samples was performed by TaqMan RT-PCR, using the primer/probes set (ABI) Hs03063352_s1, which binds to BIRC5 exon IV and its 3′ UTR. Expression of BIRC5 in AML blasts was normalized to normal peripheral blood mononuclear cells (PBMCs). The control normal PBMCs were obtained from normal volunteers to create a baseline for comparison purposes as previously described [18]. The expression value in each patient was normalized against this baseline control to define variability between patient specimens. Splice variants were identified by reverse transcription, followed by PCR amplification of the entire transcript using fluorescently labeled primers. Amplified products were resolved by fragment length analysis and the length and sequence of each fragment were compared with published data [5].
Statistical Methods
The Kaplan–Meier method was used to estimate overall survival (OS), event-free survival (EFS) and disease-free survival (DFS). Overall survival was defined as time from study entry to death. Event-free survival was defined as time from study entry until death, failure to achieve CR during induction therapy, or relapse. Disease-free survival was defined as time from end of course 1 or end of course 2 for patients in complete response (CR) until relapse or death. For patients of AAML03P1, induction 1 (course 1) failures were defined as patients who went off therapy due to relapse or persistent central nervous system (CNS) disease. Patients with refractory disease and who withdrew from therapy at the end of induction 1 were also considered failures. Induction 2 (course 2) failures were defined as patients who were not in CR by the end of induction 2. Estimates of relapse risk (RR) and treatment related mortality (TRM) were obtained using methods that account for competing events. Relapse risk was defined as time from end of course 1 or course 2 for CR patients to relapse where deaths from non-progressive disease were considered competing events. Treatment related mortality was defined as time from end of course 1 or course 2 for CR patients to death from non-progressive disease where relapses were considered competing events. The significance of predictor variables was tested with the log-rank statistic for OS, EFS, and with Gray's statistic for RR and TRM. The significance of observed differences in proportions was tested using the Chi-squared test and Fisher's exact test when data were sparse. The Mann–Whitney test was used to determine the significance between differences in medians.
Subgroups of patients designated as low, standard, or high risk patients were also analyzed for outcome. Low risk (LR) patients were defined as either having core-binding factor AML [with t (8;21) or inv(16) cytogenetics], NPM1 mutation, or CEBPA mutation. High risk (HR) patients were defined as patients having monosomy 7, -5/5q- cytogenetics, or with FLT3/ITD high allelic ratio (>0.4). All other patients having data for cytogenetics, FLT3/ITD, NPM1, and CEBPA who were not otherwise HR or LR were designated as standard risk (SR).
Results
BIRC5 Expression in Pediatric AML
Expression of BIRC5 was assessed by quantitative RT-PCR in diagnostic specimens from 306 patients as described under methods, and normalized to the expression in PBMCs. Expression of BIRC5 in diagnostic bone marrow samples varied widely, ranging from 0- to 61.04-fold over BIRC5 expression in normal PBMCs (Fig. 1A). The median expression was 0.24. The majority of patients (84%, n = 257) had expression lower than normal PBMCs (PBMC-normalized ratio ≤1). High BIRC5 expression (> 1.0; 16% [n = 49]) correlated with an immature phenotype (FAB M0 12% [n = 6] vs. 1% [n = 3], P = 0.001) and monosomy 7 (11% [n = 4] vs. 1% [n = 3], P = 0.008). BIRC5 expression also correlated with risk group. Twenty-six percent (n = 10) of patients with BIRC5 expression >1 were classified HR versus 12% (n = 29) of low expressors (P = 0.024); conversely, 37% (n = 86) of patients with BIRC5 expression ≤ 1 were classified LR versus 16% (n = 6) of high expressors (P = 0.011). There was no correlation between BIRC5 expression status (high or low) and recurring mutations in FLT3, NPM1, or CEBPA. Despite the correlation with poor prognostic markers such as high-risk group and monosomy 7, BIRC5 expression did not correlate with remission status, relapse rate or survival (Supplementary Tables I and II).
Fig. 1.
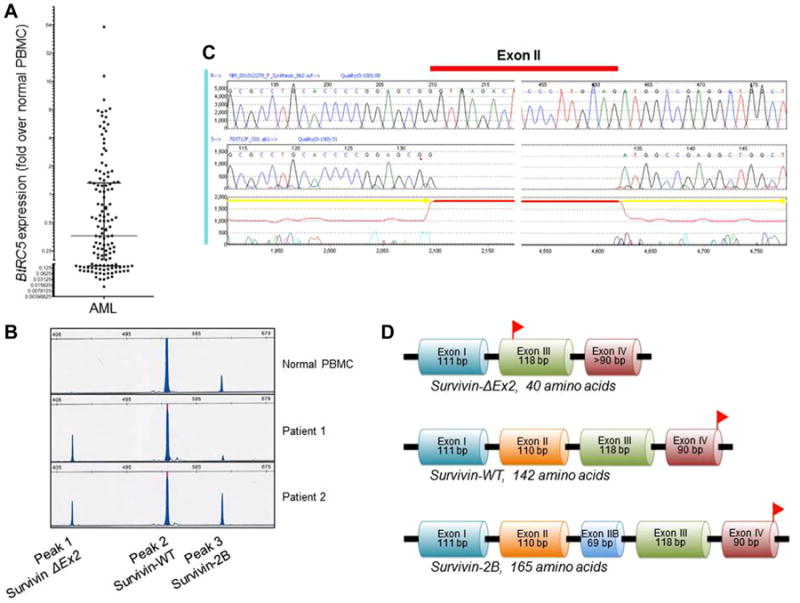
Identification of survivin-2B and the novel splice variant ΔEx2. A: Range of BIRC5 expression overall, relative to normal PBMCs. B: Capillary electropherogram from normal PBMCs and two representative patients with 2B/ΔEx2 ratios <1 (Patient 1) and >1 (Patient 2). C: Sequence analysis revealed loss of exon II from the splice variant represented by peak 1 in the electropherogram (B), and the variant was hence termed survivin-ΔEx2. D: Deletion of exon II results in a frameshift mutation, leading to a premature stop codon in exon III. Stop codons are represented by red flags. PBMCs, peripheral blood mononuclear cells; WT, wild-type; bp, base pairs.
Identification of the Novel Survivin Splice Variant ΔEx2
The entire coding region of BIRC5 was amplified and analyzed for transcript variants. Three distinct splice variants were identified in diagnostic specimens from 90 patients treated on the CCG-2961 study and characterized by fragment length analysis and sequencing (Fig. 1B–D and Supplementary Fig. 1). Wild type (WT) survivin transcript, with 429 base pairs (bp) and 142 amino acids (aa), was variably expressed in all samples. In addition to the WT transcript, two additional variants, a smaller and a larger product were identified (Fig. 1B). Sequencing of the two products showed that the larger isoform was that of previously described survivin-2B, characterized by an additional exon II, comprising 498 bp and 165 aa in total.
The smaller isoform revealed a novel BIRC5 variant with loss of BIRC5 exon II; hence we designated this splice variant as survivin-ΔEx2 (Fig. 1C). Exon II has 110 bp, with the last two bases (adenosine [A] and thymidine [T]) normally combining with the first base of exon III (A) in survivin-WT or the first base of exon IIB (T) in survivin-2B. In both survivin-WT and survivin-2B, the resulting codon translates to isoleucine. Since exon I has 111 bp, deletion of exon II results in a frameshift mutation in exon III, with the first codon becoming AGA (adenosine–guanosine–adenosine; translating to arginine) and a premature stop codon (TAA) arising after just 9 bp in exon III. As a result, the translated peptide would comprise only 40 aa (Fig. 1D and Supplementary Fig. 1).
Survivin-2B/ΔEx2 Ratio and Disease Characteristics
Although survivin-WT was the predominant isoform, expression levels of the two survivin isoforms varied significantly. Since survivin-WT expression did not predict clinical outcome, we sought to determine whether expression of the two variants might have biologic and clinical implications. The ratio of the survivin-2B and survivin-ΔEx2 splice variants (2B/ΔEx2) was therefore used as a measure of expression of the two variants. Ratio of 2B to ΔEx2 ranged from 0 (primarily ΔEx2 product) to >10 (primarily 2B product). Clinical and laboratory characteristics and outcomes were compared for patients with isoform ratio ≥1 and those with lower ratios. All 90 patients with available survivin-2B and -ΔEx2 isoform data were enrolled on the CCG-2961 study. The 22 (24%) patients with a 2B/ΔEx2 ratio ≥1 had a higher WBC count at diagnosis (median WBC 69.4 vs. 19.4 × 103/μl, P = 0.025) and an increased incidence of monocytic phenotype (FAB M5; 36% [n = 8] vs. 12% [n = 8]; P = 0.02) and +8 cytogenetics (46% [n = 6] vs. 0% [n = 0], P < 0.001), compared with those with a 2B/ΔEx2 ratio <1. A low 2B/ΔEx2 ratio (<1) was more frequent in patients with wild-type FLT3 (78% [n = 39] vs. 55% [n=11], P = 0.054), compared with FLT3-ITD or FLT3-TKD positive patients combined. There was no correlation between Survivin-2B/ΔEx2 ratio and FLT3-ITD or FLT3-TKD mutational status when analyzed separately.
The Ratio of Survivin-2B/ΔEx2 Correlates With Induction Failure, Refractory Disease, and Poor Outcome
The correlation between 2B/ΔEx2 and clinical outcome was of most interest (Tables I and II). Patients with a 2B/ΔEx2 ratio ≥ 1 had significantly lower CR rates and higher induction failure rates than those with a ratio < 1 after course 1 (45% [n = 10] vs. 88% [n = 59], P < 0.001 and 23% [n = 5] vs. 3% [n = 2], P = 0.009, respectively). Importantly, the correlation between a high 2B/ΔEx2 ratio (≥ 1) and refractory disease remained significant after course 2 of induction chemotherapy, with only 57% (n=12) achieving CR, compared with 80% (n = 47) of those with a low ratio (<1; P = 0.044). Criteria for induction failure after course 2 were met by 24% (n = 5) of high ratio patients, compared with only 5% (n = 3) of low ratio patients (P = 0.026).
Table I. Response to Two Courses of Induction Chemotherapy, According to the Survivin-2B/ΔEx2 Ratio.
| Survivin-2B/ΔEx2 <1 (n = 68) | Survivin-2B/ΔEx2 ≥ 1(n = 22) | ||||
|---|---|---|---|---|---|
|
|
|
||||
| N | % | N | % | P-value | |
| Phase 1 (course 1) response | |||||
| CR | 59 | 88 | 10 | 45 | <0.001 |
| PR | 3 | 4 | 3 | 14 | 0.158 |
| Fail | 2 | 3 | 5 | 23 | 0.009 |
| Death | 3 | 4 | 4 | 18 | 0.060 |
| Not evaluable | 1 | 0 | |||
| Phase 2 (course 2) cumulative response | |||||
| CR | 47 | 80 | 12 | 57 | 0.044 |
| PR | 3 | 5 | 0 | 0 | 0.563 |
| Fail | 3 | 5 | 5 | 24 | 0.026 |
| Death | 6 | 10 | 4 | 19 | 0.442 |
| Not evaluable | 9 | 1 | |||
CR, complete remission; PR, partial remission.
Table II. Outcome for All Patients According to the Survivin-2B/ΔEx2 Ratio.
| Survivin-2B/ΔEx2 <1 (n = 68) | Survivin-2B/ΔEx2 ≥ 1 (n = 22) | ||||
|---|---|---|---|---|---|
|
|
|
||||
| N | % ± 2SE% | N | % ± 2SE% | P-value | |
| 5-year OS from study entry | 68 | 60 ± 12 | 22 | 36 ± 21 | 0.011 |
| 5-year EFS from study entry | 68 | 53 ± 12 | 22 | 23 ± 18 | 0.001 |
| 5-year OS from end of course 1 | 59 | 65 ± 13 | 10 | 60 ± 31 | 0.665 |
| 5-year DFS from end of course 1 | 59 | 57 ± 13 | 10 | 40 ± 31 | 0.323 |
| 5-year RR from end of course 1 | 59 | 31 ± 12 | 10 | 60 ± 31 | 0.061 |
| 5-year TRM from end of course 1 | 59 | 12 ± 9 | 10 | 0 ± 0 | 0.263 |
| 5-year OS from end of course 2 | 47 | 70 ± 14 | 12 | 50 ± 29 | 0.134 |
| 5-year DFS from end of course 2 | 47 | 61 ± 14 | 12 | 33 ± 27 | 0.066 |
| 5-year RR from end of course 2 | 47 | 32 ± 14 | 12 | 58 ± 28 | 0.056 |
| 5-year TRM from end of course 2 | 47 | 6 ±7 | 12 | 8 ± 16 | 0.834 |
OS, overall survival; EFS, event-free survival; DFS, disease-free survival; RR, relapse rate; TRM, treatment-related mortality; SE, standard error.
Consistent with the poor response to induction, patients with a 2B/ΔEx2 ratio ≥ 1 also had significantly lower overall survival (OS; 36% vs. 60% at 5 years, P = 0.011) and event-free survival (EFS; 23% vs. 53% at 5 years, P = 0.001) than those with a ratio < 1 (Table II and Fig. 2). By risk group, this correlation remained significant for SR (n = 34) and HR (n = 13) patients, but not LR patients.
Fig. 2.
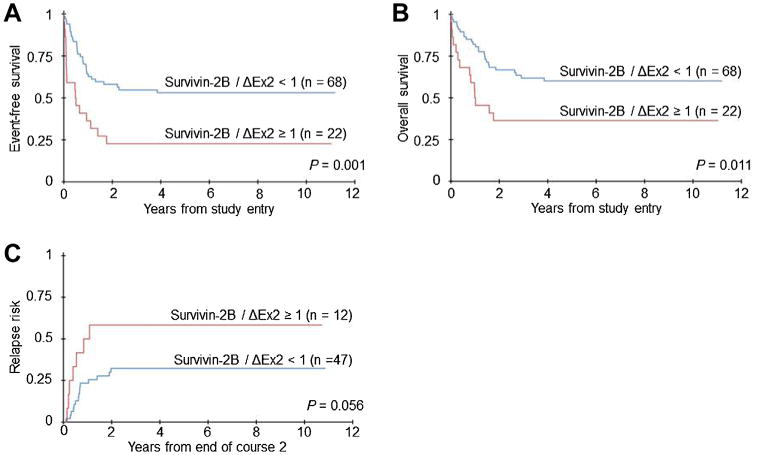
Survivin-2B/ΔEx2 expression and clinical outcome. Event-free survival (A) and overall survival (B) for patients according to the 2B/ΔEx2 ratio. C: Relapse risk, according to the 2B/ΔEx2 ratio, calculated from the end of course 2 induction therapy.
Children with a high 2B/ΔEx2 ratio also appear to be at an increased risk of relapse. A clear trend toward increased risk of relapse was evident for high 2B/ΔEx2 ratio patients compared with low ratio patients when analyzed from the end of course 1 (60% vs. 31%, P = 0.061) and course 2 (58% vs. 32% P = 0.056; Table II and Fig. 2). The impact of HSCT on relapse risk and survival for children with a 2B/ΔEx2 ratio ≥1 was not assessed.
To assess which minor splice variant was the primary determinant of outcome, clinical outcomes were compared to the individual expression of survivin-2B and survivin-ΔEx2. Consistent with the observation that a relatively high 2B/ΔEx2 ratio was associated with refractory disease, expression of survivin-2B above the median was significantly associated with inferior CR rates after course 1 (but not course 2) induction therapy (66% [n = 29] vs. 89% [n = 40], P = 0.009). Despite an association with lower CR, high expression of survivin-2B did not correlate with relapse rate or survival. Conversely, there was no association between expression of survivin-ΔEx2 above or below the median and response to induction chemotherapy (CR 70% [n = 31] vs. 84% [n = 38], P = 0.114 and 64% [n = 25] vs. 83% [n = 34], P = 0.056 after courses 1 and 2, respectively). Curiously, survivin-ΔEx2 expression above the median was associated with inferior survival (5-year OS 42% vs. 66%, P = 0.027; 5-year EFS 33% vs. 57%, P = 0.017).
Discussion
The report demonstrates a survivin splice variant with deletion of exon II, which we termed survivin-ΔEx2. Because a stop codon is introduced in exon III, the resultant peptide is expected to have only 40 aa. The structure and function of such a small peptide is unknown although like survivin-2B, the predicted Baculoviral IAP Repeat (BIR) domain would be disrupted, since it normally consists of aa 16–87 in survivin-WT, encoded by exons I, II, and III. The BIR domain is highly conserved amongst IAPs, yet its anti-apoptotic function is unclear [5]. Survivin has only one BIR domain which is disrupted in all known splice variants except survivin-WT and survivin-3B (characterized by an additional part of exon III and no exon IV). A second motif of the survivin protein is the predicted coiled-coiled domain, comprising aa 97–141 of survivin-WT and aa 120–164 of survivin-2B. Unlike survivin-WT and survivin-2B, survivin-ΔEx2 would not have this coiled-coiled domain. The biological activity of survivin-ΔEx2 remains to be assessed. It is possible that survivin-ΔEx2 somehow modulates the antiapoptotic activity of survivin-2B, since a lower 2B/ΔEx2 ratio is associated with better outcome. However, individual analyses showing that 2B, but not ΔEx2, expression is associated with response to induction therapy suggest that 2B may be exerting a dominant antiapoptotic function, rather than ΔEx2 having an important protective or modulatory role.
Our findings that survivin-WT is the major splice variant, and that relative over-expression of survivin-2B correlates with inferior outcomes are consistent with a smaller study reported by Wagner et al. A semi-quantitative analysis of 31 children and 51 adults (treated on AML-BFM93 and AMLCG92 protocols, respectively) demonstrated that survivin-WT was the predominant splice variant, with low-level expression of survivin-2B and survivin-ΔEx3 (ref. 9). Adults with higher survivin-2B expression (relative to GAPDH) had poorer OS and EFS than those with lower survivin-2B expression (mean 9 vs. 19 months, P = 0.05 and mean 10 vs. 27 months, respectively, P ≤ 0.01) [9]. There was a similar trend in children (mean OS 26 vs. 42 months), but it was not statistically significant. In vitro transfection studies of survivin-2B in 293 (human embryonic kidney) cells demonstrated that exogenous overexpression of survivin-2B blocks apoptosis induced by Bax overexpression [9].
Our analysis of 306 children with de novo AML is the largest on survivin expression in pediatric AML to date. Although expression of survivin correlated with high-risk group status and the presence of monosomy 7, we did not identify a correlation between total survivin expression and clinical outcome. This is in contrast to previous studies of survivin expression in children and adults. Children positive for survivin expression at diagnosis on the AML-BFM93 protocol had a lower OS than those negative for survivin expression (mean OS 27 [n = 10] vs. 41 months [n = 34], P < 0.05) [12]. In a study of 511 adults with newly diagnosed AML, reverse-phase protein array showed that survivin expression levels were significantly higher in bone marrow than peripheral blood [8]. Using a multivariate Cox proportional hazards model, higher survivin expression predicted significantly poorer OS and EFS (hazard ratio 1.17, 95% CI 1.03–1.33, P = 0.016 and 1.15, 95% CI 1.02–1.30, P = 0.023, respectively) [8]. Survivin expression was also significantly higher in CD34+38− AML stem/progenitor cells than bulk blasts or total CD34+ AML cells, suggesting survivin plays a role in AML leukemogenesis. Unlike our study, there was no correlation between survivin expression and FLT3-mutational status, clinical response, or relapse risk [8].
In summary, our data suggest that a high survivin-2B/ΔEx2 ratio is strongly associated with refractory disease and inferior outcome in children with de novo AML. The impact of this ratio appears primarily determined by relative over-expression of survivin-2B, rather than under-expression of survivin-ΔEx2, although functional studies are required to delineate this. The expression of alternative survivin splice variants, particularly survivin-2B, may modulate the antiapoptotic functions of survivin, reducing the ability of cytotoxic chemotherapy to induce cell death in AML. Prospective clinical studies are also needed to confirm the prognostic impact of survivin splice variant expression. If the poor prognostic impact is confirmed, patients with relative over-expression of survivin-2B may benefit from intensified induction therapy or novel therapeutic strategies. Whether the apparent increased relapse risk and inferior survival can be mitigated by HSCT in first CR is unknown and warrants further evaluation.
Supplementary Material
Acknowledgments
The study was sponsored by Chair's Grant (U10-CA98543), Statistics and Data Center Grant (U10-CA98413), Children's Oncology Group (U24-CA114766), and NCI (R01-CA114563) to S.M. We thank the AML Reference Laboratories of the COG for providing diagnostic specimens and Dr. V. Sanker for scientific editing.
Grant sponsor: Chair's Grant; Grant number: U10-CA98543; Grant sponsor: Statistics and Data Center; Grant number: U10-CA98413; Grant sponsor: Children's Oncology Group; Grant number: U24-CA114766; Grant sponsor: NCI; Grant number: R01-CA114563
Footnotes
Conflict of interest: Nothing to declare.
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- 1.Pui CH, Carroll WL, Meshinchi S, et al. Biology, risk stratification, and therapy of pediatric acute leukemias: An update. J Clin Oncol. 2011;29:551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29:487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 3.Gorman MF, Ji L, Ko RH, et al. Outcome for children treated for relapsed or refractory acute myelogenous leukemia (rAML): A Therapeutic Advances in Childhood Leukemia (TACL) Consortium study. Pediatr Blood Cancer. 2010;55:421–429. doi: 10.1002/pbc.22612. [DOI] [PubMed] [Google Scholar]
- 4.Altieri DC. Survivin cancer networks and pathway-directed drug discovery. Nat Rev Cancer. 2008;8:61–70. doi: 10.1038/nrc2293. [DOI] [PubMed] [Google Scholar]
- 5.Sampath J, Pelus LM. Alternative splice variants of survivin as potential targets in cancer. Curr Drug Discov Technol. 2007;4:174–191. doi: 10.2174/157016307782109652. [DOI] [PubMed] [Google Scholar]
- 6.Adida C, Recher C, Raffoux E, et al. Expression and prognostic significance of survivin in de novo acute myeloid leukaemia. Br J Haematol. 2000;111:196–203. doi: 10.1046/j.1365-2141.2000.02328.x. [DOI] [PubMed] [Google Scholar]
- 7.Paydas S, Tanriverdi K, Yavuz S, et al. Survivin and aven: Two distinct antiapoptotic signals in acute leukemias. Ann Oncol. 2003;14:1045–1050. doi: 10.1093/annonc/mdg277. [DOI] [PubMed] [Google Scholar]
- 8.Carter BZ, Qiu Y, Huang X, et al. Survivin is highly expressed in CD34(+)38(—) leukemic stem/ progenitor cells and predicts poor clinical outcomes in AML. Blood. 2012;120:173–180. doi: 10.1182/blood-2012-02-409888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner M, Schmelz K, Wuchter C, et al. In vivo expression of survivin and its splice variant survivin-2B: Impact on clinical outcome in acute myeloid leukemia. Int J Cancer. 2006;119:1291–1297. doi: 10.1002/ijc.21995. [DOI] [PubMed] [Google Scholar]
- 10.Oto OA, Paydas S, Tanriverdi K, et al. Survivin and EPR-1 expression in acute leukemias: Prognostic significance and review of the literature. Leuk Res. 2007;31:1495–1501. doi: 10.1016/j.leukres.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Invernizzi R, Travaglino E, Lunghi M, et al. Survivin expression in acute leukemias and myelodysplastic syndromes. Leuk Lymphoma. 2004;45:2229–2237. doi: 10.1080/10428190412331283251. [DOI] [PubMed] [Google Scholar]
- 12.Tamm I, Richter S, Oltersdorf D, et al. High expression levels of x-linked inhibitor of apoptosis protein and survivin correlate with poor overall survival in childhood de novo acute myeloid leukemia. Clin Cancer Res. 2004;10:3737–3744. doi: 10.1158/1078-0432.CCR-03-0642. [DOI] [PubMed] [Google Scholar]
- 13.Ibrahim AM, Mansour IM, Wilson MM, et al. Study of survivin and X-linked inhibitor of apoptosis protein (XIAP) genes in acute myeloid leukemia (AML) Lab Hematol. 2012;18:1–10. doi: 10.1532/LH96.11005. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Bi C, Janakakumara JV, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113:4052–4062. doi: 10.1182/blood-2008-05-156422. [DOI] [PubMed] [Google Scholar]
- 15.Fukuda S, Singh P, Moh A, et al. Survivin mediates aberrant hematopoietic progenitor cell proliferation and acute leukemia in mice induced by internal tandem duplication of Flt3. Blood. 2009;114:394–403. doi: 10.1182/blood-2008-11-188714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lange BJ, Smith FO, Feusner J, et al. Outcomes in CCG-2961, a children's oncology group phase 3 trial for untreated pediatric acute myeloid leukemia: A report from the children's oncology group. Blood. 2008;111:1044–1053. doi: 10.1182/blood-2007-04-084293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper TM, Franklin J, Gerbing RB, et al. AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: A report from the Children's Oncology Group. Cancer. 2012;118:761–769. doi: 10.1002/cncr.26190. [DOI] [PubMed] [Google Scholar]
- 18.Ho PA, Alonzo TA, Gerbing RB, et al. The prognostic effect of high diagnostic WT1 gene expression in pediatric AML depends on WT1 SNP rs16754 status: Report from the Children's Oncology Group. Pediatr Blood Cancer. 2013 doi: 10.1002/pbc.24700. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


