Abstract
Background & Aims
microRNAs (miRNAs) are short noncoding RNAs that regulate gene expression negatively. Although a role for aberrant miRNA expression in cancer has been postulated, the pathophysiologic role and relevance of aberrantly expressed miRNA to tumor biology has not been established.
Methods
We evaluated the expression of miRNA in human hepatocellular cancer (HCC) by expression profiling, and defined a target gene and biologically functional effect of an up-regulated miRNA.
Results
miR-21 was noted to be highly overexpressed in HCC tumors and cell lines in expression profiling studies using a miRNA microarray. Inhibition of miR-21 in cultured HCC cells increased expression of the phosphatase and tensin homolog (PTEN) tumor suppressor, and decreased tumor cell proliferation, migration, and invasion. In contrast-enhanced miR-21 expression by transfection with precursor miR-21 increased tumor cell proliferation, migration, and invasion. Moreover, an increase in cell migration was observed in normal human hepatocytes transfected with precursor miR-21. PTEN was shown to be a direct target of miR-21, and to contribute to miR-21 effects on cell invasion. Modulation of miR-21 altered focal adhesion kinase phosphorylation and expression of matrix metalloproteases 2 and 9, both downstream mediators of PTEN involved in cell migration and invasion.
Conclusions
Aberrant expression of miR-21 can contribute to HCC growth and spread by modulating PTEN expression and PTEN-dependent pathways involved in mediating phenotypic characteristics of cancer cells such as cell growth, migration, and invasion.
Hepatocellular cancer (HCC) is the most common malignancy of the liver, and is the fifth most common cause of cancer worldwide. The incidence of HCC has been increasing in the United States in recent years.1 Although the risk factors for HCC are well characterized, the molecular pathogenesis of these tumors is understood poorly.2 An improved understanding of critical pathways involved in cancer cell development and progression will facilitate the development of effective targeted therapeutic strategies for HCC.
Tissue invasion and metastases are key phenotypic characteristics of cancer cells.3 These processes require that cancer cells acquire the ability to proliferate in an unrestricted manner, as well as to migrate and invade adjacent tissues. Tumor growth and metastases can be facilitated by genetic changes in cancer cells that regulate these phenotypic changes. Many recent studies have described genetic changes occurring in experimental and clinical hepatocarcinogenesis. Moreover, gene expression patterns have been shown to be valuable in predicting survival as well as response to therapy.4 These studies have been helpful in identifying potential candidates for therapeutic intervention.
microRNAs (miRNAs) are a class of gene products that recently were implicated in several cancers.5–7 Several hundred miRNAs have been described in human beings. miRNAs can function as potent regulators of gene expression and altered miRNA levels can result in aberrant expression of gene products that may contribute to cancer biology. In many studies, the expression of miRNAs appears to be lower in malignant tissues compared with the corresponding nonmalignant tissues. However, the expression of selected miRNAs can be increased. Altered expression of several miRNAs has been described in HCC.8,9 However, the specific role of aberrantly expressed miRNAs is unknown. We postulated that aberrantly expressed miRNAs may contribute to tumor growth and spread by modulating the expression of gene products involved in phenotypic characteristics of cancer cells such as cell growth, migration, and invasion.
Materials and Methods
Cell Lines, Cultures, and HCC Tumor Tissues
Malignant hepatocyte cell lines were obtained from the American Type Culture Collection (Manassas, VA) and cultured as recommended by the supplier. Normal human hepatocytes were obtained from Sciencell (San Diego, CA). For miRNA profiling, total RNA from normal human liver tissue and from HCC was obtained from BioChain Institute, Inc. (Hayward, CA). For miR-21 expression, RNA was obtained from tumor samples with histologic evidence of HCC, and nontumoral samples from adjacent liver without histologic evidence of HCC from archival samples.
Transfections
Transfections were performed by nuclear transfection using the Nucleofector system (Amaxa Biosystems, Koln, Germany). Transfection conditions for each HCC cell type first were optimized to result in 20%–30% transfection efficiency with a cell viability of more than 80% using solution V, program H22. The transfection efficiency of the normal human hepatocytes used was 10.4% ± 1.0% within 10 passages after purchase. Cells (1–2 × 106) were suspended in 100 μL Nucleofector solution (Amaxa Biosystems) containing 33 μL of 100 nmol/L miRNA precursor, antisense miRNA inhibitor, phosphatase and tensin homolog (PTEN) small interfering RNA (siRNA), or respective controls at room temperature as previously described.10 Transfected cells then were resuspended in culture media containing 10% serum for 72 hours before study. All studies were performed in quadruplicate unless otherwise specified.
Isolation of MicroRNA
Total RNA was obtained from cell lines and tissue samples using the Totally RNA isolation kit (Ambion, Austin, TX). The miRNA fraction was obtained by flashPAGE purification using the flashPAGE Fractionator System (Ambion) as previously described.10 The size of the miRNA fractions was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Palo Alto, CA).
MiRNA Array Hybridization and Analysis
The isolated miRNAs were 3′-end labeled with Cy3 (control samples) or Cy5 (treated samples) using the mirVana miRNA Array Labeling Kit (Ambion) and the Post Labeling Reactive Dye kit (Amersham Bioscience, Pittsburgh, PA) as previously described.10 The labeled samples were washed 3 times using miRNA washing buffer, mixed in the same labeling cartridge, eluted, and stored at −70°C, or analyzed by hybridizing to miRNA arrays. miRNA arrays were generated on glass slides using the mirVana miRNA Array Probe Set (Ambion) and an OmniGrid Microarrayer (Gene Machines, San Carlos, CA). Each probe was printed in duplicate. After hybridization, the miRNA arrays were scanned using a GenePix 4000A array scanner (Axon Instruments, Union City, CA). Raw data were normalized and analyzed using GeneSpring 7.0 Software (Silicon Genetics, Redwood City, CA). Normalization was performed by expressing each miRNA replicate relative to a control miRNA (Ambion) added to each sample, thus allowing comparisons between chips. An average value of the median intensity of each replicate in 4 groups was generated. Cluster analyses were performed by using MultiExperiment Viewer software from the Institute of Genomic Research (Rockville, MD).11 Details and experimental data have been deposited in the ArrayExpress data base (www.ebi.ac.uk/arrayexpress) with the accession number E-MEXP-1125.
Real-Time Polymerase Chain Reaction Assays for Mature miRNAs
microRNA was isolated as described previously and the expression of specific mature miRNAs was confirmed using real-time polymerase chain reaction (PCR) analysis using a TaqMan Human MicroRNA Assay kit (Applied Biosystems, Foster City, CA).
Northern Blot Analysis
An aliquot (10–20 μg) of total RNA was subjected to denaturing (7 mol/L urea) polyacrylamide (15% acrylamide) gel electrophoresis, transferred to Zetaprobe GT membrane (Bio-Rad, Hercules, CA), immobilized to the membrane by ultraviolet cross-linking, and hybridized overnight at 42°C to 32P-labeled deoxyoligonucleotide antisense to mir-21 as described.8 The blots were reprobed with antisense oligo specific for 5S ribosomal RNA to show comparable RNA loading in each lane. The blots were subjected to autoradiography and the ratio of miR-21 to 5S ribosomal RNA was quantitated using Kodak Imaging software (Rochester, NY).
Real-Time PCR Assays for PTEN and Matrix Metalloproteases
RNA was isolated using an RNA isolation kit (Bio-Rad), and cDNA was generated by reverse transcription using 1 μg of total RNA and the reverse transcription kit (Invitrogen, Carlsbad, CA). Quantitative realtime PCR was performed on a MX 3000P PCR Instrument (Stratagene, San Diego, CA) and using SYBR Green as the detection fluorophore as described previously.12 Forward (F) and reverse (R) primers used were as follows: matrix metalloprotease (MMP)-1-F 5′-AGCTAGCTCAGGATGACATTGATG-3′, MMP-1-R 5′-GCCGATGGGCTGGACAG-3′; MMP-2-F 5′-TGGCGATGGATACCCCTTT-3′, MMP-2-R 5′-TTCTCCCAAGGTCCATAGCTCAT-3′; MMP-3-F 5′-TGG CATTCAGTCCCTCTATGG-3′, MMP-3-R 5′-AGGACAAAGCAGGATCACAGTT-3′; MMP-9-F 5′-CCTGGGCAGATTCCAAACCT-3′, MMP-9-R 5′-GCAAGTCTTCCGAGTAGTTTTGGA T-3′; MMP-11-F 5′-TGACTTCTTTGGCTGTGCC-3′, MMP-11-R 5′-GTTGTCATGGTGGTTGTACCC-3′; β-actin-F 5′-CCAAGGCCAACCGCG AGAAGATGAC-3′, and β-actin-R: 5′-AGGGTACATGGTGGTGCCGCCAGAC-3′. PCR parameters were as follows: 10 minutes at 95°C, and then 40 cycles of 30 seconds at 95°C, 1 minute at 60°C Each sample was tested in triplicate. Threshold values were determined for each sample/primer pair and average and standard error values were calculated. The PCR products were verified by melting curve analysis as well as by 1.8% agarose gel electrophoresis of the PCR product. The mRNA level of β-actin was used as an internal control, and gene-specific mRNA expression was normalized against β-actin expression. For PTEN, realtime PCR was performed using the 5′- CGGCAGCATCAAATGTTTCAG-3′ and 5′-AACTGGCAGGTAGAAGGCAACTC-3′ and an annealing temperature of 55°C.
Cell Proliferation Assay
Cell proliferation was assessed using the CellTiter 96 AQueous assay kit (Promega, Madison, WI). After transfection, cells (10,000/well) were plated in 96-well plates (BD Biosciences, Rockville, MD) and incubated at 37°C, and cell proliferation was assessed after 72 hours as previously described.13
Cell Motility Assay
Normal human hepatocytes, SK-HEP-1 cells, and SNU-182 cells (5 × 104 cells) were placed into the top chamber of a BD Falcon HTS FluoroBlok insert with a membrane containing 8-μm pores (BD Biosciences) in 300 μL of serum-free Dulbecco’s modified Eagle medium in triplicate. The inserts were placed into the bottom chamber wells of a 96-well plate containing Dulbecco’s modified Eagle medium media and fetal bovine serum (5%) as chemoattractant. Cells that migrated through the pores of the membrane to the bottom chamber were labeled with 8 μg/mL calcein-AM (Molecular Probes, Eugene, OR) in phosphate-buffered saline (PBS) for 30 minutes at 37°C. The fluorescence of migrated cells was quantified using a fluorometer at excitation wavelengths of 485 nm and emission wavelengths of 530 nm and expressed as arbitrary fluorescence units. Data are expressed as mean ± standard error of 4 separate determinations.
Invasion Assays
Cell invasion was assessed across a solid gel of basement membrane proteins prepared from the Engelbreth Holm-Swarm mouse tumor using the QCM 96-well cell Invasion assay kit (Chemicon International, Temecula, CA).
Western Blotting
Cells grown in 100-mm dishes were washed twice with ice-cold PBS before lysis by incubation for 20 minutes in 1 mL of ice-cold cell lysis buffer (1% Nonidet P-40, 50 mmol/L HEPES, pH 7.4, 150 mmol/L NaCl, 2 mmol/L ethylenediaminetetraacetic acid, 2 mmol/L phenylmethylsulfonyl fluoride, 1 mmol/L sodium vanadate, 1 mmol/L sodium fluoride, and 1× protease inhibitor mixture). Modified cell lysis buffers (without the phosphatase inhibitors sodium vanadate and sodium fluoride) were used for focal adhesion kinase (FAK) [P-Tyr516/517] immunoblots only. The protein concentrations of the lysates were measured using a Bradford protein assay kit (Bio-Rad). Equivalent amounts of protein were mixed with 6× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, electrophoresed in a 4%–20% linear gradient Tris-HCl-ready gel (Bio-Rad), and then transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline, pH 7.4, containing .05% Tween 20, and were incubated with primary antibodies and IRDye700- and IRDye800-labeled secondary antibodies (Rockland, Gilbertsville, PA) according to the manufacturer’s instructions. The protein of interest was visualized and quantitated using the LI-COR Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE).
Immunocytochemistry
Cells were cultured to confluency on 1.5% gelatin-coated culture chambers (Chemicon International). Monolayers were washed twice and fixed in 100% ice-cold acetone for 10 minutes, then blocked in PBS containing 2% rabbit serum and 5% mouse serum for 1 hour at room temperature before incubation with primary antibodies, mouse anti-PTEN (1:100), and rabbit anti–P-FAK (1:100) overnight at 4°C followed by secondary fluorescein isothiocyanate–conjugated anti-rabbit (1:200) and Texas Red–conjugated anti-mouse IgG (1:200) for 1 hour at room temperature. After 3 washes, monolayers were mounted on glass slides with ProLong antifade mounting medium with DAPI (Molecular Probes). Images were viewed and captured using an Axiovert 200 Motorized Fluorescent Microscope Imaging System from Carl Zeiss MicroImaging Inc. (Thornwood, NY).
Luciferase Reporter Assay
The pGL3-PTEN-3′-UTR construct, which contains the putative binding site for mir-21 downstream of the stop codon in the pGL3 Firefly luciferase reporter, was constructed as reported.10 SK-HEP-1 and SNU-182 cells were plated (2 × 106 cells/well) in 6-well plates. One microgram of pGL3-PTEN-3′-UTR construct was cotransfected with 1 μg of a Renilla luciferase expression construct pRL-TK (Promega), using Trans-It (Mirus, Madison, WI). Luciferase assays were performed 48 hours after transfection using the dual-luciferase reporter assay system (Promega). Firefly luciferase activity was normalized to renilla luciferase expression for each sample.
Reagents
Pre-miR miRNA precursors and anti-miR miRNA-specific inhibitors of miR-21, miR-132, along with control precursor and inhibitor miRNA were purchased from Ambion. Peroxidase-conjugated anti-rabbit and anti-mouse secondary antibodies were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Antibodies against PTEN and phospho-FAK [Tyr516/517] were from Santa Cruz Biotechnology Inc.; phospho-Akt [Ser473] from Cell Signaling Inc. (Beverly, MA); and α-tubulin from Sigma (St. Louis, MO). PTEN and control siRNAs were obtained from Cell Signaling.
Statistical Analysis
Data are expressed as the mean ± standard error from at least 3 separate experiments performed in triplicate, unless otherwise noted. The differences between groups were analyzed using a double-sided Student t test when only 2 groups were present and the null hypothesis was rejected at the .05 level unless otherwise specified.
Results
Expression of miRNAs in HCC
We first isolated and compared the miRNA expression profile from normal liver and HCC tumor tissue (Figure 1). Eight microarray hybridization studies were performed on 3 different pairs of tumor and normal human liver- derived RNA. The miRNAs that are overexpressed differentially in HCC included miR-21, miR-34a, miR-221, miR-213, miR-376a, miR-222, miR-373, miR-210, and miR-294 (Table 1). Of the miRNAs that had decreased expression, the greatest changes were noted for miR-92-1 and miR-199a-1. A bootstrapping hierarchal cluster analysis confirmed a highly significant increase in expression for miR-21 and miR-34a, with unchanged or minimally increased expression in 151 miRNA, and decreased expression of 44 miRNA in tumor tissue compared with normal liver. The group of miRNA that are altered significantly in expression may contribute to the pathogenesis or phenotypic behavior of HCC. Moreover, these miRNAs also are potential markers that may be useful for the diagnosis of HCC.
Figure 1.
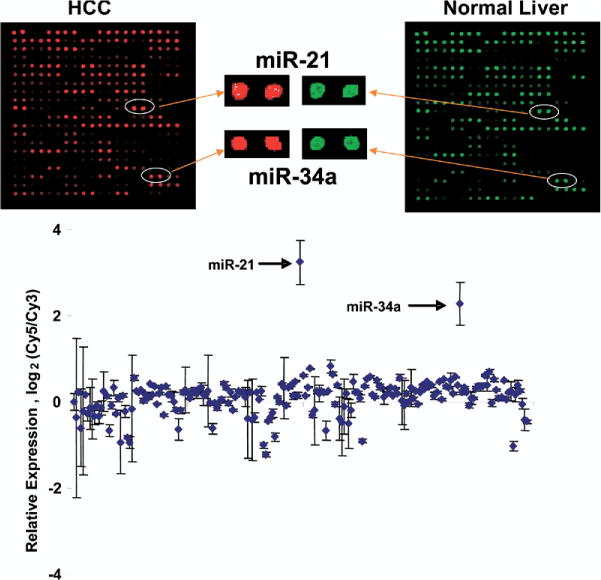
miRNA expression profiles in human malignant and nonmalignant liver tissues and cells. miRNA was isolated and profiling was performed as described in the Materials and Methods section by hybridization to miRNA-specific probes on epoxy-coated slides. Samples from normal liver tissues were labeled with Cy3, whereas HCC tumor samples were labeled with Cy5 (n = 8). Representative chip images and spots for selected miRNA that have increased expression are illustrated. The data in the bottom panel represent the average ± standard errors of the log2 of the ratios of Cy5/Cy3 fluorescence intensity for each specific miRNA.
Table 1.
miRNAs Expressed Differentially in HCC Compared With Normal Liver Tissue
| miRNA | Mean ± SE | P value | Chromosome location | Potential targets |
|---|---|---|---|---|
| Increased expression >1.5-fold | ||||
| miR-21 | 9.43 ± .15 | <.001 | 17q23.2 | PTEN, RECK |
| miR-34a | 4.66 ± .39 | <.01 | 1p36.23 | Cadherin, TP53BP2 |
| miR-221 | 1.77 ± .05 | <.05 | Xp11.3 | FAT2 |
| miR-213 | 1.70 ± .31 | <.01 | 1q31.3 | SATB2 |
| miR-376a | 1.61 ± .06 | <.05 | 14q32.31 | VHLL |
| miR-222 | 1.57 ± .15 | <.05 | Xp11.3 | FAT2 |
| miR-373 | 1.54 ± .10 | <.05 | 19q13.41 | TP53INP1, CLOCK |
| miR-210 | 1.53 ± .12 | <.05 | 11p15.5 | PCDH17 |
| miR-294 | 1.50 ± .16 | <.05 | random | MECP |
| Decreased expression <.6-fold | ||||
| miR-92-1 | .49 ± .02 | <.01 | 13q31.3 | KRAS-2, HIPK3 |
| miR-199a-1 | .50 ± .11 | <.01 | 19p32.2 | KRAS |
| miR-122a | .52 ± .01 | <.01 | 18q21.31 | EDN1, VAV3 |
| miR-125b-1 | .52 ± .02 | <.01 | 11q24.1 | VEGF, Akt3 |
| miR-292-3p | .54 ± .05 | <.05 | random | VAMP2 |
| miR-125a | .57 ± .04 | <.05 | 19q13.33 | VEGF, EDN1 |
miRNAs Are Expressed Differentially in Human Malignant Hepatocytes
Studies on the analysis of miRNA expression from whole tumors may be confounded by the presence of cell types other than malignant hepatocytes. Indeed, miRNA expression in biliary epithelia is distinct from that in hepatocytes.10 To study the potential contribution of aberrant miRNA in pathophysiologic processes involved in HCC, we began by profiling the expression of miRNAs in normal and tumoral tissues and in several HCC cell lines (Supplementary Figure 1; supplementary material online at www.gastrojournal.org). In each cell type, there was a wide range in miRNA expression varying from undetectable to greater than 24-fold relative to that of an internal standard control. Notably, the expression patterns varied with different cell types. After normalization, an increased expression of several miRNAs was seen in vitro as well as in vivo compared with the expression of an internal control miRNA. These include miR-21, let7a-1, let7c, let7d, let7f, miR-16-1, miR-17-5p, miR-23, miR-24, and miR-320. A greater than 5-fold increase in expression was noted for miR-21 and selected members of let-7 family, whereas decreased expression was noted for let-7b, miR-127, miR-149, miR-154, miR-221, and miR-329 for all cell lines and for tumor tissue. Because the pattern of miRNA expression can occur in a tissue-specific manner, these specific miRNAs may reflect markers of a hepatic lineage. miR-122 is abundant and specific for hepatic tissue, and was noted to be decreased in SK-HEP-1, SNU-182, and SNU-449 cells but not in HepG2 and PLC/PRF-5. Indeed, the pattern of expression for SK-HEP-1, SNU-182, and SNU-449 cells was similar to each other and distinct from Hep-G2 and PLC/PRF-5 cells. These differences may reflect variations in the developmental origins of these cell lines.
Next we compared miRNA expression between malignant and nonmalignant hepatocytes and confirmed them with the patterns discovered in vivo. The pattern of miRNA expression in malignant cells was markedly different from that of normal human hepatocytes. In the malignant cells, the expression of most miRNAs was decreased when compared with the nonmalignant cells. We determined the percentage of all miRNAs from the total set of 197 miRNAs that were increased more than 2-fold compared with expression of a control standard miRNA. An increase in expression was noted for 25.6% of miRNAs in normal hepatocytes, greater than that noted in Hep-G2 (18.8%), SK-HEP-1 (12.2%), SNU-182 (9.8%), SNU-449 (10.3%), or PLC/PRF-5 (10.9%) cells. These data are similar to our in vivo findings that show an increased expression of 23.8% of miRNAs in normal liver tissues but only 12.4% in HCC tissues. The expression of a group of miRNA including human miR-21, miR-34a, and miR-20 was increased by greater than 2-fold in at least 4 HCC cell lines compared with normal human hepatocytes (Table 2). These miRNAs may play an important role in malignant transformation or tumor cell behavior.
Table 2.
Highly Expressed miRNA in Human HCC Cell Lines
| SK-HEP-1
|
SNU-182
|
SNU-449
|
PLC/PRF-5
|
Hep-G2
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | Mean ± SE | miRNA | Mean ± SE | miRNA | Mean ± SE | miRNA | Mean ± SE | miRNA | Mean ± SE |
| miR-21 | 10.0 ± .2 | let-7i | 8.1 ± 1.1 | miR-21 | 8.6 ± .6 | miR-20 | 12.56 ± 1.2 | miR-192 | 8.5 ± 1.6 |
| miR-221 | 6.7 ± .6 | miR-221 | 8.1 ± 1.1 | let-7f-2 | 6.6 ± .8 | miR-122a | 10.45 ± 2.1 | miR-34a | 6.8 ± .3 |
| let-7i | 5.8 ± .6 | miR-222 | 6.1 ± .6 | let-7a-1 | 5.7 ± .8 | miR-192 | 6.8 ± 1.3 | miR-20 | 6.3 ± .5 |
| miR-222 | 4.6 ± .3 | miR-29b | 5.0 ± .3 | let-7f-1 | 5.3 ± .5 | miR-21 | 6.1 ± .3 | miR-106a | 5.8 ± .8 |
| let-7a-1 | 4.5 ± 1.1 | miR-21 | 4.7 ± .1 | let-7c | 4.6 ± .6 | miR-106a | 5.8 ± 1.1 | miR-92-1 | 5.0 ± .3 |
| let-7c | 3.9 ± 1.0 | miR-181b1 | 4.7 ± .6 | miR-23a | 4.6 ± .2 | miR-221 | 5.5 ± .8 | miR-17-5p | 4.7 ± .6 |
| miR-125b-1 | 3.8 ± .3 | miR-20 | 4.6 ± .5 | let-7d | 4.1 ± .5 | miR-194-1 | 5.3 ± 1.0 | miR-194-1 | 4.0 ± .8 |
| let-7f-2 | 3.4 ± .7 | miR-92-1 | 4.3 ± .3 | miR-221 | 3.9 ± .4 | miR-17-5p | 4.9 ± .8 | miR-148a | 3.9 ± .7 |
| miR-34a | 2.6 ± .1 | miR-106a | 4.0 ± .6 | miR-16-1 | 3.8 ± .2 | miR-23a | 4.6 ± .3 | miR-30b | 3.8 ± .3 |
| let-7f-1 | 2.6 ± .5 | miR-34a | 3.6 ± .2 | miR-23b | 3.8 ± .3 | miR-16-1 | 3.8 ± .5 | miR-26a-1 | 3.3 ± .2 |
| miR-29b | 2.5 ± .1 | miR-16-1 | 3.4 ± .2 | miR-29b | 3.7 ± .2 | miR-15a | 3.7 ± .7 | miR-93 | 3.3 ± .3 |
| miR-23a | 2.4 ± .1 | let-7a-1 | 3.4 ± .2 | miR-181b1 | 3.5 ± .4 | miR-222 | 3.6 ± .3 | miR-25 | 2.7 ± .1 |
| miR-92-1 | 2.4 ± .3 | let-7c | 3.1 ± .4 | miR-222 | 3.2 ± .3 | miR-26a-1 | 3.6 ± .3 | miR-29b | 2.6 ± .3 |
| let-7d | 2.4 ± .7 | miR-23a | 3.1 ± .2 | miR-30a-5p | 3.0 ± .3 | miR-92-1 | 3.3 ± .1 | miR-146 | 2.6 ± .4 |
| miR-20 | 2.3 ± .2 | miR-138-1 | 2.8 ± .4 | miR-27a | 2.9 ± .2 | miR-23b | 3.2 ± .3 | miR-186 | 2.5 ± .1 |
| miR-16-1 | 2.3 ± .2 | miR-17-5p | 2.8 ± .4 | miR-24-1 | 2.8 ± .1 | miR-27a | 3.1 ± .1 | miR-126* | 2.3 ± .2 |
| miR-106a | 2.0 ± .1 | miR-100 | 2.8 ± .4 | miR-93 | 2.8 ± .3 | miR-93 | 3.1 ± .4 | miR-200b | 2.2 ± .3 |
| miR-27a | 1.7 ± .1 | miR-27a | 2.8 ± .1 | miR-25 | 2.6 ± .2 | miR-29b | 2.8 ± .2 | miR-340 | 2.1 ± .1 |
| miR-138-1 | 1.7 ± .1 | let-7d | 2.7 ± .4 | let-7i | 2.6 ± .3 | miR-22 | 2.7 ± .2 | miR-103-1 | 2.1 ± .3 |
| miR-26a-1 | 1.6 ± .1 | miR-196a-1 | 2.8 ± .3 | miR-98 | 2.5 ± .2 | miR-197 | 2.6 ± .3 | miR-128a | 2.0 ± .2 |
NOTE. Expression data are normalized to that in normal human hepatocytes.
miR-21 Is Up-Regulated in Primary Human HCCs
miR-21 was selected for further study because it was the most overexpressed miRNA in our profiling studies in HCC cell lines, moreover, its expression is increased in liver tumors induced in rats fed folate/methyl-deficient diets.8 Furthermore, miR-21 is overexpressed in several other cancers.10,14,15 To determine whether miR-21 is up-regulated in primary human HCCs, Northern blot analysis was performed using total RNA and 32P-labeled deoxyoligonucleotide anti-sense to mir-21. The results showed that among the 20 HCC samples analyzed, miR-21 was up-regulated significantly 2-fold or more (2-to 65-fold) in 14 samples (samples 1, 5, 9, and 10–20) compared with the matching nontumoral liver tissues (Figure 2). These results show that miR-21 expression is indeed increased frequently in human primary HCCs.
Figure 2.
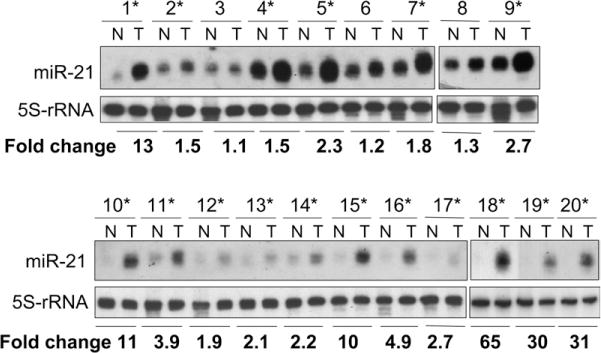
miR-21 is up-regulated in human primary HCC. Total RNA was isolated from HCC (T) and matching control (N), and Northern blot analysis was performed as described in the Materials and Methods section. The signal in each lane was quantified by Kodak Imaging software and the ratio of miR-21 to 5S ribosomal RNA was determined. Asterisks denote human primary HCC in which mir-21 is up-regulated. In the first 9 samples the signal is higher because more RNA (20 μg) was loaded. For samples 10–20, 10 μg of RNA was loaded.
miR-21 Modulates Cell Proliferation
To investigate the pathophysiologic role of miR-21 in HCC, we first verified the expression of miR-21 in HCC cell lines by real-time PCR analysis for expression of mature miR-21. The findings were similar to the pattern of expression observed using the miRNA microarray (Figure 3). To characterize the effect of miR-21 on cell proliferation we performed both overexpression studies using miR-21 precursor and inhibition studies using miR-21–specific antisense oligonucleotide inhibitor (anti–miR-21). In HepG2, PLC/PRF-5, SK-HEP-1, and SNU-182 cell lines, anti–miR-21 decreased cell proliferation (Figure 4A). In contrast, cell proliferation was not altered significantly by anti–miR-132 (data not shown), whose expression was not altered in malignant hepatocytes. Moreover, an increase in proliferation occurred in cells transfected with miR-21 precursors (Figure 4B). These studies indicate that tumor cell growth can be modulated by expression of miR-21.
Figure 3.
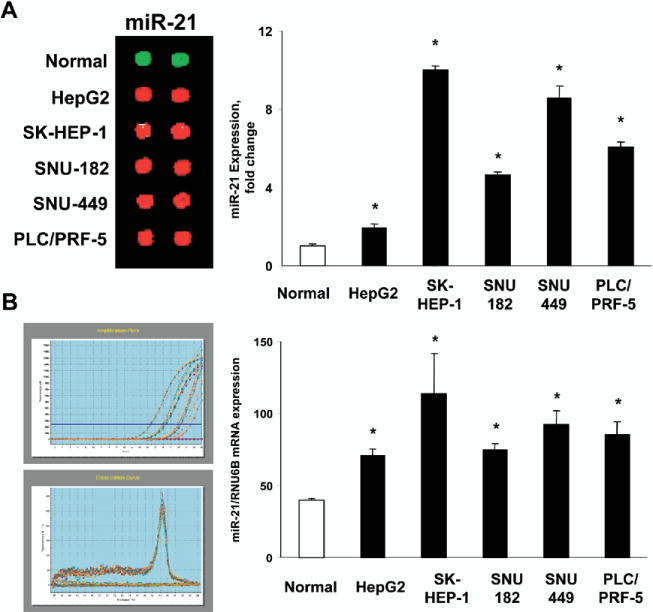
Increased expression of miR-21 in HCC cell lines. miRNA were isolated from normal human hepatocytes and from 5 HCC cell lines. (A) miRNA was labeled and analyzed by miRNA microarray. Representative hybridization spots for miR-21 are illustrated on the left whereas quantitative expression data from 4 separate studies, each in duplicate, is shown in the right panel. (B) Quantitative real-time PCR for miR-21 was performed using a TaqMan microRNA Assay kit. Representative amplification plots and disassociation curves are shown on the left, whereas quantitative data representing the mean and SD from 3 experiments performed in triplicate are presented in the bar graph on the right. The expression of miR-21 was normalized to that of the U6B small nuclear RNA gene (RNU6B) control. *P < .05 compared with expression in normal hepatocytes.
Figure 4.
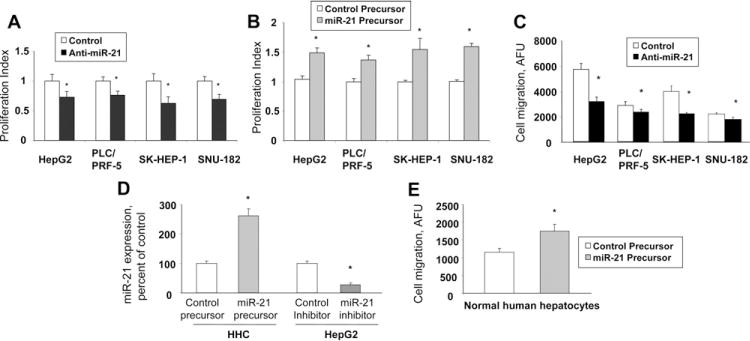
Modulation of cell proliferation and migration by miR-21. (A) HCC cells were transfected with either 30 nmol/L miR-21–specific inhibitor (■) or control miRNA inhibitors (□), and the proliferation index was assessed after 72 hours. (B) HCC cells were transfected with either 30 nmol/L miR-21 precursors (grey bars), or control miRNA precursors (□), and the proliferation index was assessed after 72 hours. (C) HCC cells were transfected with anti–miR-21 (■) or control inhibitor (□). Cell migration across a membrane with 8-μm pores was assessed as described in the Materials and Methods section and is expressed as arbitrary fluorescence units (AFU). (D) miR-21 expression was assessed by real-time PCR in normal human hepatocytes (HHC) transfected with either control or mir-21 precursors, or in HepG2 cells transfected with control or anti–miR-21 inhibitors. (E) Normal human hepatocytes were transfected with mir-21 precursor (grey bars) or control miRNA precursor (□), and cell migration was assessed. The mean and standard error from 4 separate experiments are illustrated. *P < .05 when compared with controls.
miR-21 Regulates Cell Migration and Invasion
We assessed the role of miR-21 on cell migration, a key determinant of malignant progression and metastases. HCC cell lines or normal hepatocytes were transfected with either control precursor or inhibitor, and vertical migration was assessed. A significant difference in cell migration was found between anti–miR-21 and control inhibitor-transfected HCC cells (Figure 4C). Moreover, cell migration was increased in normal human hepatocytes transfected with precursor miR-21 compared with cells transfected with controls (P = .011) (Figure 4E). However, cell migration was not altered significantly by the transfection of normal human hepatocytes with precursor miR-132 (P = .48), or of SNU-182 cells with anti–miR-132 (P = .37). Next, we determined the effect of miR-21 on cell invasion across an extracellular matrix. The HCC cell lines varied in their invasive potential, with SK-HEP-1, SNU-182, and Hep-G2 having the greatest invasive activity (Figure 5A). Four HCC cell lines were transfected with control or miR-21–specific inhibitors and their invasive potential was assessed after 72 hours. We found that anti–miR-21 decreased the invasion index in all cell lines by 35%–41% (Figure 5B), whereas anti–miR-132 failed to show a significant effect (data not shown). These results support a functional role for miR-21 in mediating cell proliferation, migration, and invasion in malignant hepatocytes, and suggest a mechanism by which overexpression of miR-21 may contribute to tumor metastasis in human HCC.
Figure 5.
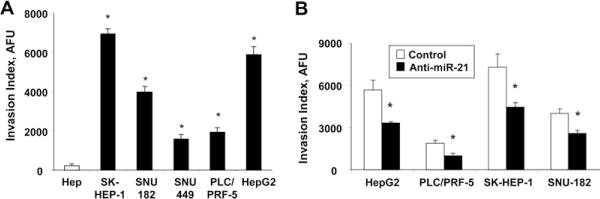
Regulation of cell invasion by miR-21. (A) Normal human hepatocytes (HEP) or HCC cells (5 × 104) were seeded in 96-well plates precoated with extracellular matrix, and cell invasion was assessed as described in the Materials and Methods section. The invasion index is expressed as arbitrary fluorescence units (AFU). The cell lines varied in their ability to invade extracellular matrix. (B) HCC cells were transfected with 30 nmol/L anti–miR-21 (■) or control inhibitor (□), and cell invasion was assessed after 72 hours. Anti–miR-21 decreased cell invasion in all 4 cell lines. The results shown represent the mean ± standard error from 4 independent experiments. *P < .05 when compared with controls.
PTEN Is a Potential Target of miR-21
We previously identified the protein tyrosine phosphatase PTEN as a potential target for miR-21 using a bioinformatics approach.10 To assess whether miR-21 directly can alter the expression of PTEN in HCC cells, a fragment of the 3′-UTR of PTEN mRNA, containing the putative miR-21–binding sequence, was cloned into a firefly luciferase reporter construct, and cotransfected with a control Renilla luciferase reporter construct into SK-HEP-1, SNU-182, HepG2, or PLC/PRF-5 cells along with control or miR-21–specific inhibitor. An increase in relative luciferase activity was noted with anti–miR-21 in all HCC cells, indicating that miR-21 can modulate gene expression directly at the PTEN 3′-UTR (Figure 6).
Figure 6.
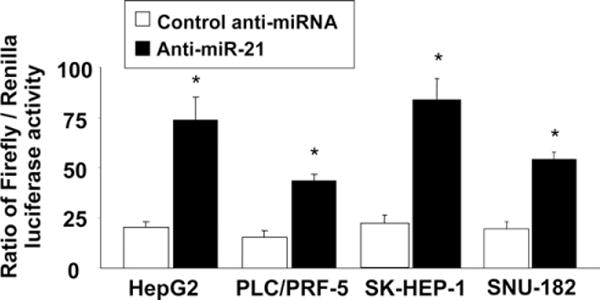
PTEN is a target for miR-21. HCC cells were plated in 6-well plates. Cells were transfected with 1 μg of a Renilla luciferase expression construct pRL-TK and 1 μg of the pGL3-PTEN-3′-UTR firefly luciferase expression construct, along with either anti–miR-21 or control inhibitor. An increase in relative firefly luciferase activity in the presence of anti-miR-21 indicates that the 3′-UTR of PTEN contains a target that is modulated by miR-21. Data represent mean ± standard error from 8 separate determinations. *P < .05 when compared with controls.
miR-21 Modulates Both PTEN Expression and FAK Phosphorylation
PTEN is a tumor-suppressor gene and its role in tumor biology is well characterized.16 We assessed cellular expression of the PTEN in human HCC tumors, normal human hepatocytes, and in several HCC cell lines. First, we assessed PTEN protein expression by immunohistochemistry in 4 human HCC tumor samples, but could not detect any expression using 2 different antibodies. Next we measured PTEN mRNA by real-time PCR in 11 HCC tumoral and nontumoral samples (Figure 7A). An inverse correlation with miR-21 expression and PTEN mRNA expression was seen in some but not all HCC samples. By immunoblot analysis, PTEN expression was decreased in all malignant cell lines compared with normal hepatocytes (Figure 7B). Moreover, there was an increase in constitutive tyrosine phosphorylation of FAK, an established downstream target of PTEN17,18 FAK is a protein tyrosine kinase involved in the regulation of cell-cycle progression, cell survival, and cell migration. Expression of PTEN leads to dephosphorylation of FAK and inhibition of cell migration. We next assessed the role of miR-21 on the expression of PTEN and tyrosine phosphorylation of FAK. Transfection of normal human hepatocytes with miR-21 precursor decreased PTEN protein expression and induced tyrosine phosphorylation of FAK at Tyr516/517 (Figure 7C). Moreover, inhibition of miR-21 in HCC cell lines significantly reduced the phosphorylation of FAK and Akt, downstream targets of PTEN that are key mediators of tumor cell survival and invasion (Figure 7D).
Figure 7.
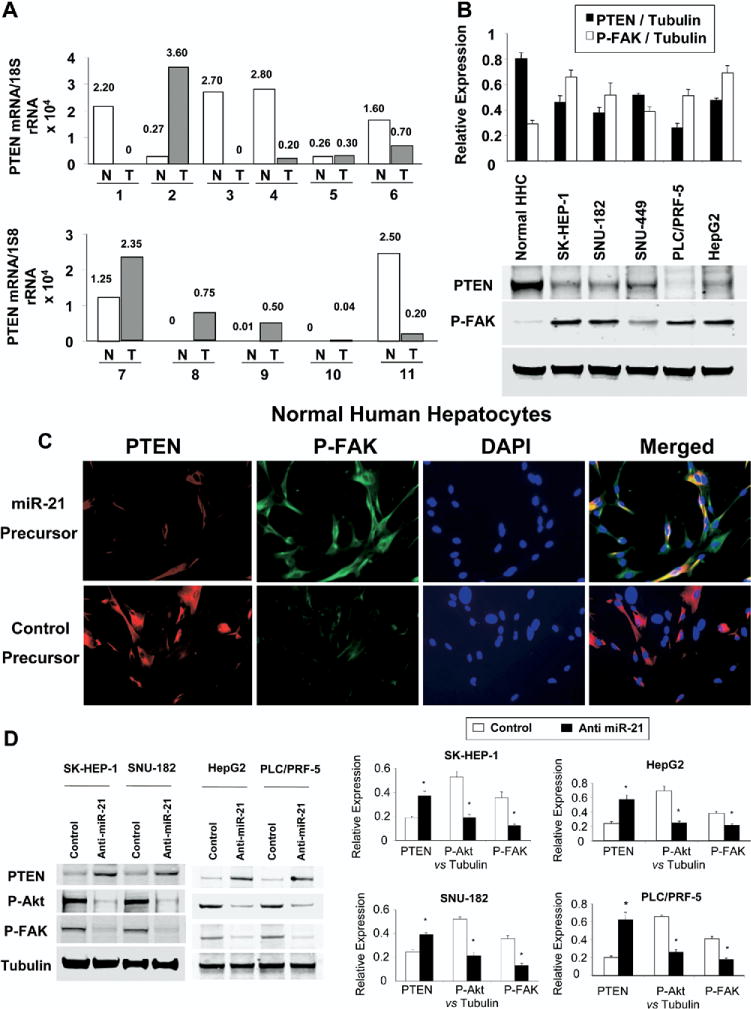
miR-21 regulates expression of PTEN and downstream kinases. (A) Real-time PCR for PTEN expression was assessed in RNA obtained from 11 HCC tumors (gray bars) and matching nontumoral sections (□). The normalized ratio of PTEN expression is indicated above each bar. (B) Cell lysates were obtained from normal human hepatocytes and HCC cell lines cultured in 100-mm culture dishes. Immunoblot analysis was performed for PTEN and for FAK activation using phosphorylation-state dependent antibodies. The blots were stripped and reprobed for α-tubulin as a loading control and for quantitation. Representative immunoblots are shown along with quantitative data showing the mean ± standard error from 4 separate blots. *P < .05 relative to expression in normal hepatocytes. (C) Normal human hepatocytes were transfected with 30 nmol/L miR-21 precursor or control. Immunocytochemistry for PTEN and phosphorylated FAK was performed after 72 hours. A decrease in PTEN expression along with an increase in FAK phosphorylation is observed in cells transfected with miR-21 precursor compared with controls. (D) HCC cells were transfected with 30 nmol/L anti–miR-21 or control inhibitor. Cell lysates were obtained after 72 hours for immunoblot analysis of PTEN protein expression and phosphorylation of its downstream target kinases Akt and FAK. Representative immunoblots and quantitative data (mean ± standard error) from 4 separate blots are shown. *P < .05 relative to expression in controls.
PTEN suppresses the expression of several MMPs through FAK dephosphorylation.18,19 To confirm the functional relevance of miR-21–dependent modulation of PTEN, we assessed the expression of MMPs involved in cell invasion. Transfection of normal human hepatocytes with miR-21 precursor increased MMP-9 mRNA expression (Figure 8A). Compared with expression in normal liver tissue, the expression of both MMP-2 and MMP-9 was increased in HCC tumors by 21.7- ± 8.7-fold and 17.5- ± 8.6-fold, respectively. Furthermore, the expression of both MMP-2 and MMP-9 were decreased after transfection with anti–miR-21 in SK-HEP-1, SNU-182, and Hep-G2 HCC cell lines (Figure 8B). Our findings provide evidence of a link between miR-21 and the expression of mediators of cell invasion in HCC cell lines. These data suggest that altered expression of miR-21 in malignant hepatocytes can contribute to metastasis by functional deregulation of activity of key signaling intermediates.
Figure 8.
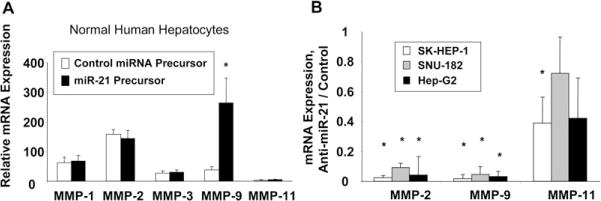
miR-21 modulates mRNA expression of MMPs. (A) Normal human hepatocytes were transfected with miR-21 precursors or controls. Quantitative real-time PCR was performed for MMP-1, MMP-2, MMP-3, MMP-9, MMP-11, and β-actin mRNA expression. Enhanced expression of miR-21 increases MMP-9 mRNA expression in normal human hepatocytes. *P< .05 relative to control. (B) SK-HEP-1, SNU-182, and Hep-G2 cells were transfected with anti–miR-21. MMP-2, MMP-9, and MMP-11 mRNA expression was assessed by real-time PCR and normalized to expression of β-actin mRNA. The data are summarized from 3 experiments performed in quadruplicate. Inhibition of miR-21 reduced MMP-2 and MMP-9 mRNA expression in HCC cell lines compared with controls. *P < .05 relative to control.
PTEN Is Involved in miR-21–Dependent Effects in HCC Invasion and Migration
Loss of hepatocyte-specific PTEN has been associated with the formation of HCC.20 Although loss of PTEN can occur as a result of mutations, these are rare in HCC. Changes in PTEN expression potentially may reflect aberrant expression of miR-21. PTEN silencing by RNA interference in nonmalignant human hepatocytes results in an increase in AKT and FAK phosphorylation, similar to those increases observed with enforced expression of miR-21 by transfection with miR-21 precursor (Figure 9A). Moreover, cell migration is increased in cells transfected with PTEN siRNA compared with control siRNA (Figure 9B).
Figure 9.
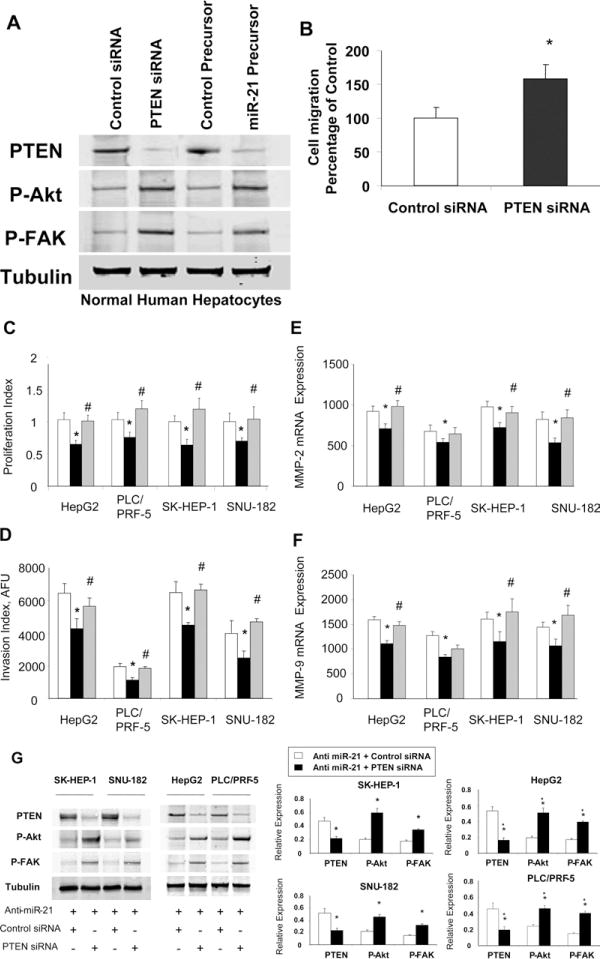
Down-regulation of PTEN attenuates the effects of anti–miR-21 on HCC cell growth and invasion. (A) Nonmalignant human hepatocytes were transfected with control or PTEN siRNA, or control or miR-21 precursor and immunoblot analysis for PTEN, phospho-Akt Ser(P)473, phospho-FAKTyr(P)516/517, and tubulin were performed. (B) Cell migration was assessed in human hepatocytes incubated with control or PTEN miRNA. (C–F) HCC cells (5 × 104/well) in 96-well plates were cotransfected with either siRNA to PTEN or control siRNA, along with 30 nmol/L anti-miR-21. (□, control anti-miRNA + control siRNA; ■, anti-miR-21 + control siRNA; grey bars, anti–miR-21 + PTEN siRNA). (C) Cell proliferation or (D) invasion was assessed after 72 hours as described in the Materials and Methods section. The mean and standard error from 4 separate experiments are shown. mRNA expression of (E) MMP-2 and (F) MMP-9 were quantitated by real-time PCR, and expressed relative to that of β-actin mRNA concurrently assessed in the same samples. (G) Cells were cotransfected with anti–mir-21 and either control or PTEN siRNA. Immunoblot analysis of PTEN, phospho-Akt Ser(P)473, and phospho-FAK Tyr(P)516/517, and tubulin was performed. Representative immunoblots are shown along with quantitative data that show the mean ± standard error from 4 separate blots. *P < .05 when compared with control siRNA-transfected cells.
To further evaluate the contribution of PTEN to biological effects of miR-21, we assessed the impact of PTEN silencing by RNA interference, and hence PTEN expression on miR-21–dependent cell proliferation and invasion in HepG2, PLC/PRF-5, SK-HEP-1, and SNU-182 cells. The effect of anti–miR-21 in decreasing both cell proliferation as well as cell invasion was prevented by the presence of siRNA to PTEN in all 4 HCC cell lines (Figure 9C and D), indicating that these effects were dependent on PTEN. We further investigated the ability of PTEN to modulate mir-21–dependent repression of MMP-2 and MMP-9 mRNA expression. Consistent with our observations in miR-21–mediated cell invasion, siRNA to PTEN enhanced MMP-2 and MMP-9 mRNA expression in all cell lines transfected with anti–miR-21 (Figure 9E and F). These data indicate that miR-21–dependent up-regulation of MMP-2 and MMP-9 involves a PTEN-dependent signaling pathway. In addition, siRNA to PTEN increased FAK tyrosine phosphorylation, which is consistent with our previous findings that FAK is dephosphorylated in vitro by anti–miR-21 inhibition. Similarly, siRNA to PTEN abrogated the reduction in Akt phosphorylation by anti–miR-21 (Figure 9G). In combination, these studies define an important role for PTEN as a mediator of the biological effects of miR-21 on cell proliferation and invasion in human HCC.
Discussion
Although a role for miRNAs in cancer has been proposed, the molecular mechanisms by which miRNA can modulate tumor growth or metastases are unknown. We show that miR-21 expression is increased in malignant hepatocytes, and in human HCC compared with matching nontumoral tissue. Moreover, we show that miR-21 promotes cell invasion, migration, and growth via repression of PTEN expression and downstream effects involving the phosphorylation of FAK and Akt, and the expression of MMP-2 and MMP-9. The identification of miR-21 as an important regulator of tumor cell proliferation, migration, and invasion in vitro emphasizes an essential role of this miRNA in mediating hepatic oncogenesis and tumor behavior, and provides insight into the contribution of altered miRNA expression in contributing to the tumor phenotype.
miR-21 has been shown to be overexpressed in many different solid tumors, including breast, colon, lung, pancreas, prostate, stomach, as well as in cholangiocarcinoma cell lines. Thus, it is highly likely that miR-21 plays a fundamental role in tumor cell behavior and malignant transformation, and our findings potentially may be relevant to other tumors in which miR-21 is overexpressed. Inhibition of miR-21 also has been shown to increase cell growth in other cancer cell types.21 Moreover, miR-21 has been reported to have anti-apoptotic properties in glioblastoma and cholangiocarcinoma.10,14 Thus, altered expression of miR-21 can have several diverse effects in tumor cells.
PTEN is a ubiquitous tumor-suppressor gene and the functional inactivation of PTEN by regulation of its expression is relevant to many solid tumors. PTEN has been implicated as a key contributor to HCC pathogenesis and growth. Targeted deletion of PTEN in murine hepatocytes leads to the development of HCC.20 Although mutation of PTEN is an infrequent event, PTEN protein expression frequently is decreased or absent in human HCC.22 Moreover, decreased PTEN correlates with tumor progression and poor prognosis in HCC.20,23 Loss of functional PTEN leads to increased activity of AKT and the mammalian target of rapamycin (mTOR) kinase pathways, which can promote both cell survival and proliferation through phosphorylation and inactivation of several downstream mediators.24 Activation of FAK has been observed in human HCC, and an increase in FAK activity correlates with more aggressive tumor behavior.25,26 Meanwhile, MMP-2 and MMP-9 activity have been shown to play an important role in invasion in HCC.27,28 Thus, modulation of expression of PTEN in HCC can impact on the activity of critical downstream mediators of tumor progression and metastases.
Our observations indicating that PTEN can reverse many of the biological effects of miR-21 implicate it as a predominant target of miR-21 in these processes. However, an individual miRNA has the potential ability to modulate the expression of many mRNA. In view of our experimental findings, we speculate that other mRNA that may be targeted by miR-21 may either serve unrelated functions, or may serve to modulate the effect of PTEN. Based on target prediction algorithms, PTEN potentially could be targeted by miRNA other than miR-21, but PTEN needs to be verified experimentally as a bona fide target for these miRNA, and a biologically relevant role shown.
Recurrence and metastasis are associated commonly with a poor prognosis, and therefore targeting these mechanisms may lead to more effective treatment for HCC patients. There is considerable interest in therapy specifically targeting key signaling pathways involved in tumorigenesis and tumor growth such as the phosphoinositide-3 kinase pathway, which is regulated by PTEN. A practical application of our observations could be in the use of miR-21 expression as a potential predictor of tumors that may be more likely to respond to pathway-targeting therapies.
The expression of downstream mediators of tumor growth and metastasis could be modulated by targeting miR-21. Therapeutic strategies to decrease miR-21 therefore potentially may be useful to limit HCC growth and metastasis. Further work is warranted to evaluate the role of miR-21 and the identified downstream targets and to develop therapeutic strategies targeting miR-21 in vivo. The ability to therapeutically manipulate miRNA expression is feasible, and recent proof-of-concept studies have shown that miRNA antagonists targeted to the liver can modulate expression of downstream genes.29,30 Moreover, aberrantly expressed miRNA such as miR-21 may be useful to establish a diagnosis and for assessing prognosis in HCC. Recent advances in techniques for the identification of miRNA should enable the use of clinical material for this purpose. Knowledge of specific processes such as tumor cell proliferation, invasion, and migration that are regulated by miRNA, and the identification of critical targets for individual miRNAs such as PTEN and FAK, provide novel insights into the mechanisms of tumorigenesis and metastasis in HCC.
Supplementary Material
Acknowledgments
Supported by the Scott and White Hospital Foundation, and by grants DK069370 and CA122694 from the National Institutes of Health.
Abbreviations used in this paper
- FAK
focal adhesion kinase
- miRNA
microRNA
- MMP
matrix metalloprotease
- PCR
polymerase chain reaction
- PTEN
phosphatase and tensin homolog
- siRNA
small interfering RNA
Appendix: Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1053/j.gastro.2007.05.022.
References
- 1.El Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 2.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Thorgeirsson SS, Lee JS, Grisham JW. Functional genomics of hepatocellular carcinoma. Hepatology. 2006;43:S145–S150. doi: 10.1002/hep.21063. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 7.McManus MT. MicroRNAs and cancer. Semin Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 8.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;15:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 10.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 11.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 12.Meng F, Yamagiwa Y, Ueno Y, Patel T. Over-expression of interleukin-6 enhances cell survival and transformed cell growth in human malignant cholangiocytes. J Hepatol. 2006;44:1055–1065. doi: 10.1016/j.jhep.2005.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, Tadlock L, Gores GJ, Patel T. Inhibition of interleukin 6-mediated mitogen-activated protein kinase activation attenuates growth of a cholangiocarcinoma cell line. Hepatology. 1999;30:1128–1133. doi: 10.1002/hep.510300522. [DOI] [PubMed] [Google Scholar]
- 14.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 15.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim RH, Mak TW. Tumours and tremors: how PTEN regulation underlies both. Br J Cancer. 2006;94:620–624. doi: 10.1038/sj.bjc.6602994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gautam A, Li ZR, Bepler G. RRM1-induced metastasis suppression through PTEN-regulated pathways. Oncogene. 2003;22:2135–2142. doi: 10.1038/sj.onc.1206232. [DOI] [PubMed] [Google Scholar]
- 18.Park MJ, Kim MS, Park IC, Kang HS, Yoo H, Park SH, Rhee CH, Hong SI, Lee SH. PTEN suppresses hyaluronic acid-induced matrix metalloproteinase-9 expression in U87MG glioblastoma cells through focal adhesion kinase dephosphorylation. Cancer Res. 2002;62:6318–6322. [PubMed] [Google Scholar]
- 19.Akahane T, Akahane M, Shah A, Connor CM, Thorgeirsson UP. TIMP-1 inhibits microvascular endothelial cell migration by MMP-dependent and MMP-independent mechanisms. Exp Cell Res. 2004;301:158–167. doi: 10.1016/j.yexcr.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, Mizuno K, Hasegawa G, Kishimoto H, Iizuka M, Naito M, Enomoto K, Watanabe S, Mak TW, Nakano T. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113:1774–1783. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan XW, Jiang M, Cao HF, He YQ, Liu SQ, Qiu XH, Wu MC, Wang HY. The alteration of PTEN tumor suppressor expression and its association with the histopathological features of human primary hepatocellular carcinoma. J Cancer Res Clin Oncol. 2003;129:100–106. doi: 10.1007/s00432-002-0410-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu TH, Huang CC, Lin PR, Chang HW, Ger LP, Lin YW, Changchien CS, Lee CM, Tai MH. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer. 2003;97:1929–1940. doi: 10.1002/cncr.11266. [DOI] [PubMed] [Google Scholar]
- 24.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 25.Itoh S, Maeda T, Shimada M, Aishima S, Shirabe K, Tanaka S, Maehara Y. Role of expression of focal adhesion kinase in progression of hepatocellular carcinoma. Clin Cancer Res. 2004;10:2812–2817. doi: 10.1158/1078-0432.ccr-1046-03. [DOI] [PubMed] [Google Scholar]
- 26.Fujii T, Koshikawa K, Nomoto S, Okochi O, Kaneko T, Inoue S, Yatabe Y, Takeda S, Nakao A. Focal adhesion kinase is overexpressed in hepatocellular carcinoma and can be served as an independent prognostic factor. J Hepatol. 2004;41:104–111. doi: 10.1016/j.jhep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto H, Itoh F, Adachi Y, Sakamoto H, Adachi M, Hinoda Y, Imai K. Relation of enhanced secretion of active matrix metalloproteinases with tumor spread in human hepatocellular carcinoma. Gastroenterology. 1997;112:1290–1296. doi: 10.1016/s0016-5085(97)70143-4. [DOI] [PubMed] [Google Scholar]
- 28.Arii S, Mise M, Harada T, Furutani M, Ishigami S, Niwano M, Mizumoto M, Fukumoto M, Imamura M. Overexpression of matrix metalloproteinase 9 gene in hepatocellular carcinoma with invasive potential. Hepatology. 1996;24:316–322. doi: 10.1053/jhep.1996.v24.pm0008690399. [DOI] [PubMed] [Google Scholar]
- 29.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


