Acute lymphoblastic leukemia (ALL) in infants less than 1 year of age is rare and the biological features are different from ALL in older children.1 Infant ALL is characterized by a high frequency of rearrangements of the MLL gene (MLL-R) and heterogeneous outcome. However overall, their event-free survival (EFS) is much worse than older children with ALL.1–5 A large collaborative trial, Interfant-99, demonstrated improved outcome, while characterizing definitively the independent prognostic variables in infant ALL.6 While cytogenetic data are reported within individual infant ALL clinical trials, the numbers are typically small and many reports are less detailed for those patients without MLL gene rearrangements (MLL-G). However, it was previously suggested that MLL-G had an important predictive influence on outcome.7,8 These observations were later confirmed in Interfant-99,6 in which MLL-G patients showed a threefold reduced risk of an event compared with MLL-R patients, although all MLL-G patients were grouped together into a single category. To better understand the association of different chromosomal abnormalities and outcome among MLL-G infants, here we have carried out detailed cytogenetic investigation of two infant ALL trials: Interfant-99 and Children’s Oncology Group (COG)-P9407.
Patients were 365 days old or less with newly diagnosed ALL without a rearrangement of the MLL gene enrolled to Interfant-99 (May 1999–December 2005; n = 110) and COG-P9407 (June 1996–October 2006; n = 52).6,9 Individual study groups obtained ethical approval, and treating physicians obtained informed consent from parents or guardians. The presence of MLL gene rearrangements was excluded using fluorescence in situ hybridization (FISH), reverse transcription (RT)-PCR and/or Southern blotting, as previously reported.6 Each national study group provided patient data, including cytogenetics, FISH and molecular results. EFS and overall survival (OS) were calculated from the date of trial enrolement to the date of the first event (induction failure, relapse, second malignancy or death) or last follow-up. Median follow-up time was 7 years.
Among 162 MLL-G patients, no cytogenetic data were available for 34 (21%), resulting in a success rate of 79%. An abnormal karyotype was detected in 90/128 (70%) patients with a successful cytogenetic result (Supplementary Table 1) with the remainder classified as normal based on the presence of at least 10 (but usually 20) normal metaphases. They were categorized according to cytogenetic risk group as previously defined for childhood ALL.10 Compared with childhood ALL (1–18 years) using data from the UKALL97/99 treatment trial,10 the frequency of good risk cytogenetic abnormalities among MLL-G infants was significantly lower (12 vs 60%, P<0.01), whereas the frequency of poor risk abnormalities (excluding MLL translocations) was similar (8 vs 10%). Although ETV6–RUNX1 fusion is present in 25% of childhood ALL, we found no ETV6–RUNX1 cases among the 75 patients tested by FISH or RT-PCR. High hyperdiploidy (HeH) was the most prevalent abnormality, although the frequency was also much lower than childhood ALL (12 vs 38%, P<0.01).10 It is possible that some cases with normal or failed cytogenetic results harbored a hidden HeH clone. Among patients for whom DNA index was available, a value of 1.13–1.2 correlated with HeH in five patients; one with no cytogenetic result had a DNA index of 1.13, likely indicating the presence of a HeH clone. The chromosome number of HeH karyotypes ranged from 53 to 64 (Supplementary Table 1). The karyotypes of patients 12 and 14 may represent doubling of low hypodiploid clones, although it was not proven. Other established translocations were observed: t(9;22)(q34;q11) (n = 2), t(1;19)(q23;p13) (n = 3) and t(7;12)(q36;p13) (n = 1). Interestingly, the incidence of t(9;22) and t(1;19) among MLL-G infants was not markedly different from childhood ALL:10 2/128 (1.6%) versus 43/1633 (2.6%) and 3/128 (2.3%) versus 50/1420 (3.5%), respectively. Abnormalities of the short arm of chromosome 9 (9p) were observed in 14 (11%) cases at a similar incidence to childhood ALL; karyotypes included dic(7;9) (n = 3) and dic(9;20)(p11 ~ 13;q11) (n = 2).
The translocation, t(7;12)/ETV6–HLXB9 fusion, most frequently occurs in infant acute myeloid leukemia (AML) and rarely in infant ALL.11 Although associated with a dismal outcome in AML, the outcome in ALL is unknown as numbers of cases are too low. One patient was identified in this series (36). However, as this cryptic abnormality is often linked to deletions of the long arm of chromosome 7 (7q) and gain of chromosome 19, patients 34 and 35 may also harbor this translocation. Unfortunately no material was available for molecular testing of these or the other six cases with 12p13 breakpoints, suggesting involvement of the ETV6 gene.
Chromosome 15 abnormalities with variable breakpoints were found in 12 patients. The more frequent occurrence of 15q abnormalities in infant compared with childhood ALL has been previously noted.12 The translocation, t(5;15)(p15;q11–13), was observed in two patients (21, 25), which has previously been specifically associated with infant ALL.
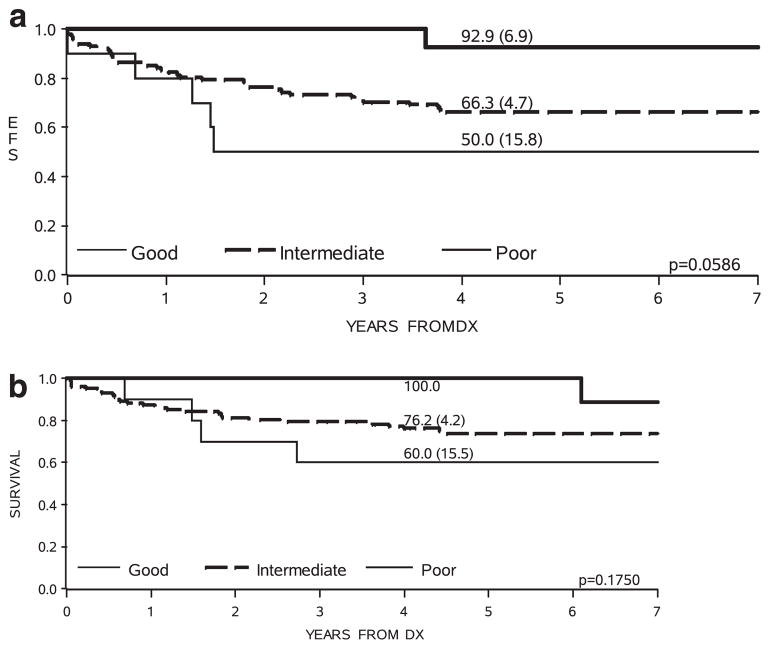
It is well established that MLL-R infants are younger than their MLL-G counterparts: in Interfant-99, 91% of infants <6 months were MLL-R compared with 66% of those 6–12 months (P<0.0001).6 Classification of MLL-G patients by cytogenetic risk group showed further correlation with age. The majority of cytogenetic good risk and all poor risk patients were >9 months old, whereas half of the cytogenetic intermediate risk patients were <9 months (Supplementary Table 2). In addition, there was evidence of outcome heterogeneity according to cytogenetic risk group (Table 1 and Figure 1). Despite the modest numbers of MLL-G cases, especially in the cytogenetic good and poor risk groups, the pattern of outcome was very similar to that observed in childhood ALL when the same classification was applied.10 However, the difference in outcome among the three cytogenetic risk groups among the MLL-G patients did not quite reach statistical significance (likely due to low numbers in the good and poor risk categories). It was of interest that two patients in the poor risk group were Philadelphia chromosome (Ph) positive. When these patients were removed from the survival analysis, there was no change to the EFS of the poor risk group, although there was an improvement in OS because one Ph-positive case was among the four patients in the cohort who died. This situation will likely change in future, as Ph-positive infants will be treated with tyrosine kinase inhibitors in the same manner as older children.
Table 1.
Incidence and outcome of 162 MLL-G infants treated on INTERFANT-99 and COG-P9407 by cytogenetic risk group
| Total |
Cytogenetic risk groupa
|
|||
|---|---|---|---|---|
| Goodb | Intermediatec | Poord | ||
| Total | 162 | 15 | 103 | 10 |
| Incidence (%) | 100 | 12 | 80 | 8 |
| Outcome variables | ||||
| Induction failure | 10 (6%) | 0 | 6 (6%) | 1 (10%) |
| Relapses | 33 (20%) | 1 (7%) | 24 (23%) | 2 (20%) |
| Death in CCR | 9 (6%) | 0 | 4 (4%) | 2 (20%) |
| Events | 52 (32%) | 1 (7%) | 34 (33%) | 5 (50%) |
| Deaths | 41 (25%) | 1 (7%) | 26 (25%) | 4 (40%) |
| EFS at 4 years (s.e.) | 67% (3.7) | 93% (6.9) | 66% (4.7) | 50% (15.8) |
| OS at 4 years (s.e.) | 77% (3.4) | 100% (—) | 76% (4.2) | 60% (15.5) |
Abbreviations: EFS, event-free survival; OS, overall survival.
Cytogenetic data were only available for 128 patients.
Good risk: all patients with high hyperdiploidy (HeH).
Intermediate—all other cases.
Poor risk: all patients with t(9;22)/BCR-ABL1, abnormal 17p and loss of 13q.
Figure 1.
Kaplan–Meier curves showing event-free (a) and overall (b) survival of 128 MLL-G infant ALL patients by cytogenetic risk group.
Recently, Kang et al.13 reported that gene expression profiles were predictive of age and outcome in 97 infants (including 17 MLL-G) with ALL treated in COG-P9407. Their statistical modeling of an outcome predictor revealed three genes (FLT3, IRX2 and TACC2) to be highly predictive of EFS beyond age and MLL status. In particular, low FLT3 expression was found to be associated with an excellent outcome, as had been previously indicated.14 Supplementary Figure 1 shows the FLT3 intensities for the entire cohort of infants tested in COG-P9407, which was validated in Interfant-99. Interestingly, the majority (10/11) of the cases with low expression were MLL-G. However, low FLT3 expression was not simply a reflection of MLL-G status, as seven of the MLL-G patients had high FLT3 expression. However, it is also thought to be linked to the very immature pro B cells, in which the MLL rearrangements often occur. These low-expressing MLL-G patients emerged from all cytogenetic groups, as shown in Supplementary Table 3. Interestingly, none of the MLL-G patients with low expression had an event, whereas all five of the events occurred in the high expressers.
We have confirmed a unique cytogenetic profile among infants with ALL. We demonstrate that the infants without MLL translocations (MLL-G) share the same cytogenetic abnormalities as older children with ALL. Generally infants with MLL-G ALL are older, have low FLT3 expression and have an improved outcome compared to their MLL-R counterparts. Despite small numbers of MLL-G infants, when classified into the same good, intermediate and poor risk cytogenetic subgroups as childhood ALL, their pattern of outcome was very similar to that observed in older children. However, their overall worse outcome likely reflects the differences in distribution of good and poor risk abnormalities: a lower incidence of the good risk abnormality, such as high hyperdiploidy and absence of ETV6–RUNX1, and a higher incidence of poor risk abnormalities. Nevertheless, these data suggested that some MLL-G infants, especially those with good risk cytogenetics, may benefit from treatment on childhood protocols, which are generally less intensive and less toxic than infant ALL regimens.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the NCI to the Children’s Oncology Group including U10 CA98543 (COG Chair’s grant), U10 CA98413 (COG Statistics & Data Center) and U24 CA114766 (COG Specimen Banking). CLW and RH were additionally supported by a Leukemia and Lymphoma Specialized Center of Research Grant in Infant Leukemia (17 372). SPH is the Ergen Family Chair in Pediatric Cancer. The study was also partially supported by Leukaemia Lymphoma Research, UK (to CJH and AVM) and Associazione Italiana Ricerca sul Cancro: Investigator Grant Code 5017 (to MGV). The Interfant99 international study operating center is partially supported by Fondazione Tettamanti and Comitato ML Verga (to MGV and PDL).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
CJH, AVM, RP and MGV designed the study. SPH as Chair of COG, ZED as Chair of the COG-P9407 trial and MD provided data from this trial. MGV and RP provided data from Interfant-99. NAH and AJC provided cytogenetic data from COG. RH and CLW provided FLT3 data. PDL, MGV and AVM carried out statistical analysis. CJH, AVM and SPH wrote the paper. All authors approved the final version of the manuscript.
References
- 1.Biondi A, Cimino G, Pieters R, Pui CH. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96:24–33. [PubMed] [Google Scholar]
- 2.Reaman GH, Sposto R, Sensel MG, Lange BJ, Feusner JH, Heerema NA, et al. Treatment outcome and prognostic factors for infants with acute lymphoblastic leukemia treated on two consecutive trials of the Children’s Cancer Group. J Clin Oncol. 1999;17:445–455. doi: 10.1200/JCO.1999.17.2.445. [DOI] [PubMed] [Google Scholar]
- 3.Hilden JM, Dinndorf PA, Meerbaum SO, Sather H, Villaluna D, Heerema NA, et al. Analysis of prognostic factors of acute lymphoblastic leukemia in infants: report on CCG 1953 from the Children’s Oncology Group. Blood. 2006;108:441–451. doi: 10.1182/blood-2005-07-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chessells JM, Harrison CJ, Watson SL, Vora AJ, Richards SM. Treatment of infants with lymphoblastic leukaemia: results of the UK Infant Protocols 1987–1999. Br J Haematol. 2002;117:306–314. doi: 10.1046/j.1365-2141.2002.03442.x. [DOI] [PubMed] [Google Scholar]
- 5.Frankel LS, Ochs J, Shuster JJ, Dubowy R, Bowman WP, Hockenberry-Eaton M, et al. Therapeutic trial for infant acute lymphoblastic leukemia: the Pediatric Oncology Group experience (POG 8493) J PediatrHematol Oncol. 1997;19:35–42. doi: 10.1097/00043426-199701000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Pieters R, Schrappe M, De Lorenzo P, Hann I, De RG, Felice M, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370:240–250. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 7.Chessells JM, Harrison CJ, Kempski H, Webb DK, Wheatley K, Hann IM, et al. Clinical features, cytogenetics and outcome in acute lymphoblastic and myeloid leukaemia of infancy: report from the MRC Childhood Leukaemia working party. Leukemia. 2002;16:776–784. doi: 10.1038/sj.leu.2402468. [DOI] [PubMed] [Google Scholar]
- 8.Heerema NA, Sather HN, Ge J, Arthur DC, Hilden JM, Trigg ME, et al. Cytogenetic studies of infant acute lymphoblastic leukemia: poor prognosis of infants with t(4;11)—a report of the Children’s Cancer Group. Leukemia. 1999;13:679–686. doi: 10.1038/sj.leu.2401413. [DOI] [PubMed] [Google Scholar]
- 9.Salzer WL, Jones TL, Devidas M, Hilden JM, Winick N, Hunger S, et al. Modifications to induction therapy decrease risk of early death in infants with acute lymphoblastic leukemia treated on Children’s Oncology Group P9407. Pediatric Blood Cancer. 2012;59:834–839. doi: 10.1002/pbc.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moorman AV, Ensor HM, Richards SM, Chilton L, Schwab C, Kinsey SE, et al. Prognostic effect of chromosomal abnormalities in childhood B-cell precursor acute lymphoblastic leukaemia: results from the UK Medical Research Council ALL97/99 randomised trial. Lancet Oncol. 2010;11:429–438. doi: 10.1016/S1470-2045(10)70066-8. [DOI] [PubMed] [Google Scholar]
- 11.von Bergh AR, van DE, van Wering ER, van Zutven LJ, Hainmann I, Lonnerholm G, et al. High incidence of t(7;12)(q36;p13) in infant AML but not in infant ALL, with a dismal outcome and ectopic expression of HLXB9. Genes ChromosomesCancer. 2006;45:731–739. doi: 10.1002/gcc.20335. [DOI] [PubMed] [Google Scholar]
- 12.Heerema NA, Sather HN, Sensel MG, La MK, Hutchinson RJ, Nachman JB, et al. Abnormalities of chromosome bands 15q13-15 in childhood acute lymphoblastic leukemia. Cancer. 2002;94:1102–1110. [PubMed] [Google Scholar]
- 13.Kang H, Wilson CS, Harvey RC, Chen IM, Murphy MH, Atlas SR, et al. Gene expression profiles predictive of outcome and age in infant acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119:1872–1881. doi: 10.1182/blood-2011-10-382861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stam RW, Schneider P, de Lorenzo P, Valsecchi MG, den Boer ML, Pieters R. Prognostic significance of high-level FLT3 expression in MLL-rearranged infant acute lymphoblastic leukemia. Blood. 2007;110:2774–2775. doi: 10.1182/blood-2007-05-091934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.