Abstract
Over the past decade there has been a greater understanding of genomic complexity in eukaryotes ushered in by the immense technological advances in high-throughput sequencing of DNA and its corresponding RNA transcripts. This has resulted in the realization that beyond protein-coding genes, there are a large number of transcripts that do not encode for proteins and, therefore, may perform their function through RNA sequences and/or through secondary and tertiary structural determinants. This review is focused on the latest findings on a class of noncoding RNAs that are relatively large (>200 nucleotides), display nuclear localization, and use different strategies to regulate transcription. These are exciting times for discovering the biological scope and the mechanism of action for these RNA molecules, which have roles in dosage compensation, imprinting, enhancer function, and transcriptional regulation, with a great impact on development and disease.
Keywords: enhancers, imprinting, transcriptional silencing, chromatin, chromatin-modifying complexes, RNA polymerase II
INTRODUCTION
The RNA world hypothesis, the most widely accepted theory for the origin of life on Earth (20), envisions a world in which RNA constituted the first and only replicative molecule (30, 46). Although not without its opponents, this theory is born from the observation that RNA can replicate via base complementarity, like DNA, and can catalyze chemical reactions, like proteins. A simple hydroxyl group turns the reliable but inert deoxyribose into a less stable but more eclectic ribose, which endows RNA with the ability to fold into complex tertiary structures (176), interact specifically with proteins and other molecules, and catalyze chemical reactions (82). Therefore, it is not surprising that RNA would play central roles in all aspects of molecular biology, weaving an intricate web of regulatory mechanisms in complex multicellular organisms (21). However, for many, the RNA revolution was an unexpected twist in molecular biology. As the race to obtain the complete sequence of the human genome was in full swing, it was widely believed that the functions of RNA were limited to the central dogma. Mainly, messenger RNAs carry genetic information from the nucleus to the cytoplasm, where transfer RNAs read it and ribosomal RNAs coordinate its translation into the more sophisticated language of protein biochemistry.
The past decade has ushered in experimental evidence suggesting a much wider role for RNA in molecular biology. Widespread regulatory functions of noncoding RNAs (ncRNAs) first came to prominence in the 1990s with the discovery of RNA interference (RNAi) and its role in post-transcriptional gene silencing (55). This led to the identification of different types of small ncRNAs that guide protein complexes to mRNA targets and inhibit their translation (161). Having realized that RNA may delineate novel biological pathways, researchers began cataloging other ncRNA species using genome-wide approaches spearheaded by the technological advances in high-throughput sequencing (103). We now know that large expanses of the genome are transcribed into RNA, and only a small portion of it encodes proteins (35, 38, 40).
Given that it is more difficult to infer the existence of a ncRNA based on sequence features alone when compared to protein-coding mRNAs, most established bioinformatic approaches are not well-suited for their analysis (39), leading to greater impetus for development of new computational strategies. We need a clearer understanding of functional roles of ncRNAs before a comprehensive picture of the full scope of the ncRNA repertoire in higher eukaryotes emerges. Many excellent reviews have been written about the various aspects of ncRNA function (18, 50, 67, 89, 99, 119, 126, 136, 165, 171). Here, we focus on the large and complex category of long ncRNAs (lncRNAs) and their role in regulating gene expression by interacting with chromatin and with the protein machinery that controls its structure and function.
Classification of lncRNAs
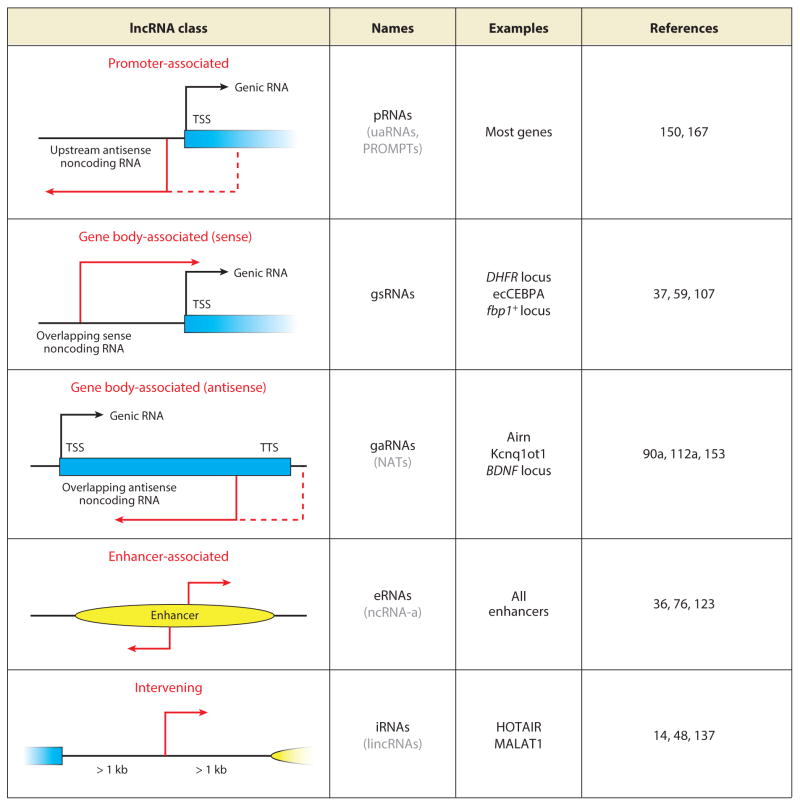
Long ncRNA have been operationally defined as transcripts that are produced by RNA polymerase II, longer than 200 nucleotides, and devoid of an open reading frame (ORF) that can be translated into a protein (14, 35, 48). It has been difficult to group lncRNAs in different classes because we have scant information regarding their precise scope of function and their overall secondary and tertiary structure. The simplest and most insightful classification of lncRNAs was recently presented by cataloging the spliced and unspliced lncRNAs in human and mouse ES cells according to their loci of origin (150). It was concluded that the large majority of lncRNAs either map to enhancer regions (~20%; termed eRNAs) or correspond to upstream antisense RNAs (uaRNAs) that originate near the transcription start site (TSS) of genic RNAs (60–70%). The remaining lncRNAs derive from antisense transcripts that overlap with annotated gene bodies (~5%) or originate from more distal, unannotated regions (~5%) (Figure 1). The latter are commonly referred to as long intervening or intergenic ncRNAs (lincRNAs) (48, 166). We note that some intervening and intragenic lncRNAs might be transcribed from as yet annotated enhancer elements (either distal to or within the gene body) and may therefore be eventually recategorized as eRNAs. The finding that a large number of lncRNAs arise from loci with proximity to protein-coding genes is consistent with previous genome-wide analyses of lncRNAs (167). A few cases of lncRNAs that originate upstream of a cognate mRNA gene and are transcribed in the sense direction have also been described (37, 107); in this case, however, the downstream sequence of the mRNA gene is often part of the final RNA, and therefore these overlapping sense RNAs are annotated as alternative isoforms and excluded from lncRNA catalogs. Because of the confusion arising from the phonetically similar lncRNA and lincRNA, for the remainder of this review we refer to the latter as intervening lncRNAs, which we propose to rename iRNAs. Similarly, to unify the nomenclature of lncRNAs, we suggest pRNAs for the promoter-associated uaRNAs, and gsRNAs or gaRNAs for gene body–associated sense and antisense transcripts, respectively (Figure 1).
Figure 1.
Classes of lncRNAs (long noncoding RNAs). All known lncRNAs are divided into five classes, on the basis of their relationship with adjacent or overlapping genomic features. The initiation site for the noncoding transcript is shown in red, and the arrow indicates the direction of transcription. Less-common variants within the same class are indicated with dashed arrows. In the second column, existing names for the different classes are listed in gray within parentheses, and their corresponding names from our proposed new nomenclature are in black. Abbreviations: eRNAs, enhancer RNAs; gaRNAs, gene body–associated antisense RNAs; gsRNAs, gene body–associated sense RNAs; iRNAs, intervening long noncoding RNAs; lincRNAs, long intervening noncoding RNAs; NATs, natural antisense transcripts; ncRNAs-a, noncoding RNAs activating; pRNAs, promoter-associated RNAs; PROMPTs, promoter-associated pervasive transcripts; TSS, transcription start site; TTS, transcription termination site; uaRNAs, upstream antisense RNAs.
Although many lncRNAs are associated with previously annotated genomic regions, there remains a class of intervening lncRNAs that originate from independent transcriptional units that do not overlap with mRNA genes or enhancers. These loci have their own promoter and are marked by the same chromatin modifications found at protein-coding genes (14, 48). Well-known structural intervening lncRNAs such as MALAT1 and NEAT1 belong to this class. Most intervening lncRNAs are indistinguishable from mRNAs in that they are capped, spliced, and polyadenylated (48), and among the different classes of lncRNAs they are likely the most diverse in sequence features, evolutionary origin, and function. To distinguish intervening lncRNAs from uaRNAs, we favor a more restrictive definition for intervening lncRNAs, which requires their TSS to be located at least 1 kb away from the next closest TSS or enhancer.
It is important to gain an understanding of the properties of lncRNAs in regard to their 5′- and 3′-end processing, which might further aid in their classification. Although all studies agree that the 5′ ends of lncRNAs, like those of mRNAs, are capped by methylguanosine, their splicing status and their 3′-end processing have not been fully defined. Initial annotation of lncRNAs favored transcripts that displayed some evidence of splicing and polyadenylation and that did not overlap with protein-coding genes (14, 35, 48). However, recent findings have indicated that a large number of lncRNAs correspond to either eRNAs or uaRNAs and that they are predominantly monoexonic and nonpolyadenylated, particularly in the case of eRNAs (4, 36, 76). Unlike mRNAs, uaRNAs and eRNAs are depleted of splice motifs (2, 4). Therefore, it is likely that splice site recognition occurs with a low frequency at most lncRNA loci and that the predominant form of lncRNAs may be monoexonic and nonpolyadenylated. This contention is consistent with recent findings indicating that uaRNAs and eRNAs display shorter half-lives than do mRNAs and are subject to degradation by the nuclear exosome (4, 122).
Biological Functions of lncRNAs
Although few would doubt that small ncRNAs such as miRNAs (microRNAs) and piRNAs (PIWI-interacting RNAs) possess distinct and fundamental biological functions, understanding of lncRNA biology is at an earlier stage and still elicits controversy (80). At various points in time, it was proposed that pervasive transcription is due to low specificity of RNAPII (157) and that lncRNAs encode cryptic peptides responsible for the observed biological functions (66). Because lncRNAs exhibit low sequence conservation, even between related species (14, 48), and experience frequent gene birth and death (83), their biological significance has been questioned.
Despite early skepticism, the experimental evidence in favor of a direct biological function of lncRNAs has been steadily growing and has convincingly disposed of the criticisms above. As a set, lncRNAs exhibit the imprint of purifying selection (i.e., conservation of sequence above the genomic background), not only in the gene body but also at promoters (132). Phylogenetic comparisons confirmed that thousands of lncRNA families originated more than 90 million years ago and hundreds can be traced back 300 million years to the common ancestor of mammals and birds (120). At a biochemical level, ribosome profiling data are consistent with the notion that the vast majority of lncRNAs are not translated (51), and their localization is predominantly nuclear (35), although the latter conclusion has been challenged (165).
There is also accumulating genetic evidence in favor of the biological importance of lncRNAs. Deleting the entire locus for 5 out of 18 mouse intervening lncRNAs resulted in lethality or developmental defects (144). However, such large genetic deletions might result in the removal of regulatory sequences (e.g., enhancers) of other genes in addition to the removal of the lncRNA gene. In zebrafish, depletion of the intervening lncRNA cyrano caused developmental defects that were rescued by mammalian orthologs, despite the fact that sequence conservation was limited to short patches of nucleotides (166). Importantly, mutations introduced to disrupt the frame of presumed ORFs in the RNA sequence did not affect the rescuing ability of these lncRNAs, providing additional proof that RNA was the functional agent (166).
Taken together, phylogenetic, biochemical, and genetic evidence all support the conclusion that many lncRNAs perform biological functions independent of protein translation. However, most of lncRNAs in yeast are suppressed by a molecular pathway that enforces promoter directionality via Ndr1 and the exosome (146). Similarly, many mammalian lncRNAs that arise from divergent transcription (29, 147, 150) fail to accumulate due to a depletion of splice recognition sites in the antisense direction and removal by the nuclear exosome (2, 122), suggesting that their presence might not confer a fitness advantage (177). In an age when genome editing has become an easily implemented strategy (26), all claims to noncoding functions of RNA molecules should be subjected to rigorous genetic testing. Specifically, to prove that a select biological process is regulated by lncRNAs it should be shown that (a) loss-of-function of the lncRNA perturbs the process; (b) the defect is rescued by reinstating expression of the lncRNA (in cis or trans; see below); and (c) lncRNAs carrying mutations that would disrupt potential ORFs still rescue the defect (166).
lncRNAs AND TRANSCRIPTION
As lncRNAs can interface with the genome at the sequence level and also fold into tertiary structures capable of specific interactions with proteins, they are particularly well suited to function in regulating gene expression. Indeed, the past decade of investigation has uncovered many specific examples and general classes of lncRNAs that repress or activate transcription.
Transcriptional Interference in Yeast
As in higher eukaryotes, the genome of the simplest eukaryotic model organism, Saccharomyces cerevisiae, is extensively transcribed and this includes protein-coding and protein-noncoding regions (33). lncRNAs in yeast originate in large part from bidirectional transcription initiation and most of them are rapidly degraded [cryptic unstable transcripts (CUTs)], although some are stable [stable unannotated transcripts (SUTs)] (178). Early work from Winston and colleagues demonstrated that the regulatory region of SER3, an anabolic enzyme involved in serine biosynthesis, is transcribed into a lncRNA (SRG1) in a serine-dependent fashion (106). The presence of SRG1 correlates with repression of the SER3 ORF; however, the repressive activity was observed even when >90% of the lncRNA sequence was replaced (105), arguing in favor of a transcriptional interference model. According to this model, the act of transcription through the regulatory region is sufficient to mediate repression, and the lncRNA itself does not play a direct role (106). Similar cases have since been reported for other highly regulated loci, including GAL1–10 (61); master regulators of gametogenesis, IME4 and IME1, which are repressed in cis by transcription from the lncRNA loci RME2 and IRT1 (60, 169); and FLO11, whose expression is regulated by a pair of lncRNAs, ICR1 and PWR1, through a network of mutual transcription interference (13). Whether interference affects gene regulation at the genome-wide level in S. cerevisiae remains to be elucidated. Stabilization of Xrn1-sensitive unstable transcripts (XUTs) resulted in global changes in gene expression (168), but stabilization of a different class of lncRNAs [Ndr1-dependent unterminated transcripts (NUTs)], sensitive to the action of the termination factor Nrd1, had relatively minor effects (146).
In addition to causing cis interference, select yeast lncRNAs recruit repressive chromatin factors to silence target genes. The PHO84 locus comprises an antisense lncRNA that participates in the repression of the coding gene by recruiting histone deacetylases specifically to its promoter (17). Interestingly, this lncRNA can also function in trans (16), suggesting that transcriptional interference alone cannot explain the repressive action. Similar trans-acting lncRNAs have been observed at Ty1 retrotransposons in budding yeast (9) and in fission yeast, where a lncRNA represses the pho1 gene through RNAi-dependent formation of transient heterochromatin via H3K9me2 deposition (148).
It is tempting to speculate that these recruitment pathways involve specific recognition of lncRNA structure by chromatin modifiers, but an alternative explanation is that some generic feature of overlapping transcription is sufficient to mediate repression (75). For example, a molecular pathway for transcriptome surveillance might have evolved that detects the presence of converging polymerases or of unspliced, unpolyadenylated ncRNAs and guides their removal as well as imposes silencing on the region that transcribes the lncRNAs. Indeed, recent experiments in the fission yeast Schizosaccharomyces pombe have delineated a silencing pathway that is coupled to the removal of cryptic introns (91). It is important to point out that the existence of such surveillance pathways does not argue against the fact that lncRNAs might also exert a direct regulatory function. On the contrary, nonfunctional noncoding transcripts might have constituted the raw material that evolutionary forces shaped into regulatory mechanisms.
Annotation and Function of lncRNAs in Drosophila
One criticism raised against the notion that lncRNAs fulfill important biological functions argues that forward genetic screens performed in various model organisms, most notably Drosophila melanogaster, have failed to identify lncRNAs (109). An answer to this criticism stems from the observation that point mutations induced by ethyl methane sulfonate (155), the most commonly used mutagen in Drosophila, are more likely to disrupt protein function by causing missense and nonsense mutations in the protein-coding sequences, whereas similar mutations in ncRNAs may not exact similar functional consequences. Nonetheless, as early as 1919, C.B. Bridges observed a homeotic mutation in Drosophila that he named bithoraxoid (bxd) (95). This mutation maps to a cis-regulatory region of the bithorax complex that, we now know, is extensively transcribed into lncRNAs (97, 143).
Beyond the bithorax complex, lncRNAs are a general feature of the Drosophila genome. Early annotations identified 14 intervening lncRNAs, some of which were conserved with other Drosophila species and led the authors to hypothesize that up to 100 lncRNAs may be present (163). In fact, upward of 1,000 putative lncRNAs loci have since been annotated (140, 184), and many of them show evidence of purifying selection (i.e., sequence conservation) when polymorphisms across different Drosophila strains are analyzed (53). What their functions might be remains a topic of intense investigation. So far, little evidence can be summoned in favor of direct, RNA-mediated activities of Drosophila lncRNAs. Those transcribed from the bxd locus were first shown to activate transcription of the adjacent Ultrabithorax gene (142). Later, it was suggested that lncRNAs repress Ultrabithorax via transcriptional interference (128). Most recently, genetic inactivation of one such lncRNA resulted in no discernible phenotype (127). Another famous example is pgc, which was first proposed to function as lncRNA (108) but later shown to encode a 71-aa protein responsible for gene function (56). Other lncRNAs from the bxd and iab regions of the bithorax complex appear to function as miRNA precursors or via sequence-independent transcriptional interference (6, 47).
One exception to this litany of results against a direct function of lncRNAs is the process of dosage compensation. In flies, dosage compensation occurs via transcriptional upregulation of the single male X chromosome (27). It requires functional contributions from five proteins (MSL1, MSL2, MSL3, MOF, and MLE) and two lncRNAs (roX1 and roX2) (3) that are transcribed from the X chromosome and coat it in its entirety (111). The two lncRNAs can substitute for each other, but deletion of both is lethal in males (110). Despite this redundancy, very little sequence similarities exist between roX1 and roX2 (125), suggesting that functional similarities might be dictated more by structural homology than by sequence conservation. Chemical and nuclease footprinting, along with biochemical analyses, identified conserved structural motifs required for roX function, supporting a model whereby the lncRNAs participate in the assembly of the dosage compensation complex (65, 100).
Repression by lncRNAs in Plants
Plants need to respond to a changing environment for optimal resource allocation and proper timing of developmental phase transitions, and, because they are sessile, they largely utilize epigenetic mechanisms (1). Consistent with a high degree of epigenetic regulation, the genome of Arabidopsis thaliana contains tens of thousands of putative lncRNAs, including more than 30,000 antisense lncRNAs (170) and more than 6,000 intervening lncRNAs (98).
One model locus that has been valuable in dissecting the functions of lncRNA in plants is the flowering control locus FLC, which maintains the epigenetic memory of cold exposure that results in vernalization, i.e., the ability to flower in spring (154). The locus gives rise to a number of noncoding transcripts, including various splice isoforms of the downstream antisense lncRNA COOLAIR (160) as well as a nonpolyadenylated intronic lncRNA called COLDAIR (58). Although the mechanism of action of COOLAIR remains unclear, COLDAIR binds CURLY LEAF (CLF) (58), the plant homolog of EZH2 and a subunit of Polycomb repressive complex 2 (PRC2), which mediates epigenetic silencing via trimethylation of histone H3 lysine 27 (H3K27me3) (104). COLDAIR recruits PRC2 to chromatin, thus initiating the epigenetic cascade that leads to vernalization via silencing of FLC. This model is similar to those proposed for the mechanism of action of several mammalian lncRNAs, including HOTAIR (137) (see below). CLF also binds to the antisense of COLDAIR in vitro but only to the sense RNA when incubated with nuclear extracts, suggesting that additional factors contribute to specificity in vivo (58).
Plant lncRNAs also contribute to epigenetic silencing via DNA methylation. A subclass of lncRNAs is transcribed by a dedicated RNA polymerase (PolV) and is required for small ncRNAs to recognize their genomic targets in the process of RNA-dependent DNA methylation (174). A current model posits that the lncRNAs transcribed by PolV recruit chromatin remodelers [protein complexes that reposition nucleosomes along the chromatin fiber (15)] via the RNA-binding adapter IDN2. In turn, the repositioned nucleosomes facilitate DNA methylation by DRM2, thus enforcing epigenetic repression (189). Whether similar mechanisms are at work in metazoans is doubtful, as this particular pathway of DNA methylation, as well as the specialized RNA polymerases that enable it, are specific to plants (85).
lncRNA-Mediated Epigenetic Silencing in Mammals
The initial evidence for direct roles of lncRNAs in epigenetic silencing came from experiments in mammalian systems. The founding member of the lncRNA family is Xist, a ~17-kb transcript that originates from the silent X chromosome in female cells and coats it during early stages of development to establish epigenetic X inactivation (24). The highly conserved repA region of Xist is required for silencing and was reported to interact directly with the PRC2 complex (187). Consistent with this observation, recruitment of PRC2 to the X chromosome tracks that of Xist temporally and spatially, both during initial establishment and during reestablishment following transient Xist depletion (152). However, Xist and PRC2 do not colocalize on chromatin as determined by super-resolution microscopy (22), suggesting that a simple model whereby PRC2 is directly recruited by Xist might not be entirely accurate.
X inactivation is an extreme case of genomic imprinting, the process by which certain genes are expressed from only the maternal or paternal chromosomes (135). To date, parental imprinting has been observed only in plants (78) and mammals (42). There are approximately 150 parentally imprinted loci in the mouse genome (90), most of which harbor lncRNAs that are essential for proper execution of the imprinting program (90, 153). In some notable cases, it has been argued that the imprinted lncRNAs function by recruiting chromatin-modifying factors, such as EHMT2/G9A at the Air locus (118) and EHMT2, PRC2, and DNMT1 at Kcnq1 (113, 124). However, insertion of termination cassettes to truncate the Airn lncRNA and shifting of its promoter showed that the RNA itself is dispensable for transcriptional silencing, arguing against the possibility that recruitment of chromatin modifiers is mediated by specific protein-RNA interactions (88). Nonetheless, the presence of lncRNAs at nearly all imprinted loci cannot be coincidental and the fact that one of the most sophisticated types of epigenetic regulation—the allelic discrimination of identical DNA sequences—correlates with the presence of lncRNAs provides strong circumstantial evidence in support of their involvement in epigenetic processes.
Regulation of Polycomb Complexes via lncRNAs
Among the many silencing complexes that have been reported to interact with ncRNAs, the Polycomb group complex PRC2 has been the most intensively studied. Although the biochemical output of the PRC2 complex, methylation of H3K27, is known, the input signals that determine its localization and activity at specific genes in different cells are not fully understood (104, 151). Several histone methyltransferases, including E(Z), the catalytic subunit of PRC2 in Drosophila, possess RNA binding activity in vitro (81). Canaani and colleagues ascribed the activity to a fragment of E(Z) that encompasses the SET domain; however, mapping experiments on the mammalian ortholog EZH2 identified an unstructured region comprising residues 342–368 that is required for RNA binding in vitro and whose affinity for RNA is modulated by phosphorylation (70). PRC2 has been proposed to mediate the function of many lncRNAs in mammals: HOTAIR, Kcnq1ot1, Braveheart, and Meg3/Gtl2, to name a few (69, 77, 124, 137, 186). However, the fact that most tested RNAs, including some from unrelated organisms, bind to PRC2 with high affinity and low specificity in vitro and in vivo (34) casts a veil of uncertainty over some of these results (12). Far from being an in vitro artifact, binding to RNA independently of sequence and structure might be a regulatory feature of PRC2, and competing models incorporate this observation into our understanding of PRC2 function. A junk-mail model posits that low-level transcription from escapee Polycomb target genes contributes to recruitment of PRC2 and leads to their resilencing (34), whereas a sensing model proposes that the presence of RNA causes PRC2 to curtail its repressive action on chromatin in a feedback loop that prevents inappropriate silencing of active genes (71). PRC2 also interacts with and is recruited by shorter ncRNAs through its SUZ12 subunit (72). These small ncRNAs originated upstream or near the TSS, and it is unclear to what extent they might correspond to promoter-associated RNAs (73) or to the more recently described TSS miRNAs (185). Thus, the relationship of PRC2 with long and small ncRNAs is complex (Figure 2), and more mechanistic studies are required before the full picture comes into focus.
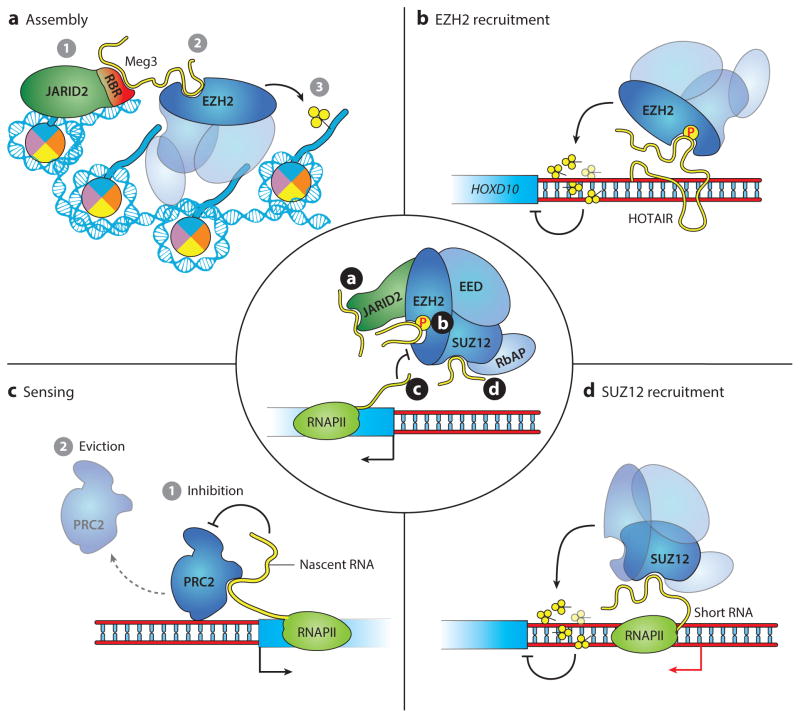
Figure 2.
Mechanisms of ncRNA (noncoding RNA)-mediated regulation of PRC2. (a) The imprinted lncRNA (long noncoding RNA) Meg3 makes contact with EZH2 and JARID2 and stimulates their interaction on chromatin, facilitating PRC2 assembly, H3K27me3 deposition, and transcriptional repression of a subset of PRC2 targets in human and mouse stem cells (69). (b) The lncRNA HOTAIR has been reported to recruit PRC2 in trans to the HOXD locus in human fibroblasts (137). Later studies revealed that HOTAIR interacts directly with EZH2 and that phosphorylation of EZH2 at T345 stimulates ncRNA binding (70). (c) In embryonic stem cells, PRC2 is found at a majority of promoters, including those of active genes. However, interactions with nascent RNAs decrease the amount of deposited H3K27me3 either by inhibiting PRC2 function or by causing its release from chromatin (71). (d) SUZ12 was reported to interact with short ncRNAs originating from the region near the transcription start site (TSS) of Polycomb target genes, leading to PRC2 recruitment, H3K27me3 deposition, and silencing (72). SUZ12 also interacts with the lncRNA Braveheart (77). The center image is a schematic of the multiple mechanisms (a–d) by which ncRNAs have been shown to interact with PRC2.
PRC2 is not the only Polycomb complex that has functional links with noncoding RNAs. CBX7 is a mammalian homolog of the Polycomb protein (PC) in Drosophila, which is a core component of PRC1 and binds to H3K27me3 via its chromodomain (43). CBX7 is expressed in mouse embryonic stem cells, where it regulates pluripotency, whereas other PC homologs such as CBX2 and CBX4 are expressed during lineage commitment and are required for differentiation (115). The chromodomains of PC, CBX4, and CBX7 bind RNA in vitro, and RNA interactions are required for chromatin localization, at least in the case of CBX7 (8). Importantly, the chromodomain of CBX2 does not bind to RNA, although it retains specificity for H3K27me3, suggesting that the two functions can be uncoupled (8).
In vivo, CBX7 interacts with ANRIL, an antisense lncRNA from the INK4b-ARF-INK4a tumor suppressor locus (183). On the basis of the observations that knockdown of ANRIL causes derepression and that RNase A treatment releases PRC1 from this locus, it is possible that ANRIL participates in PRC1 recruitment and Polycomb-mediated repression (183). CBX4 (also known as PC2) binds to at least two lncRNAs in vivo, TUG1 and MALAT1 (180). Selectivity between the two is determined by the methylation status of a lysine residue within the SUMO binding motif of CBX4 and results in differential localization of the protein and its associated chromatin targets within the three-dimensional space of the nucleus (180).
Similar to the case of PRC2, which interacts with RNA not only via core complex subunits [EZH2 (70) and SUZ12 (72)] but also through accessory regulatory components, such as JARID2 (69), PRC1 also appears to make multiple contacts with RNA. We recently discovered an RNA-binding region within SCML2 (11), a homolog of the Sex comb on midleg (SCM) protein, which is a substoichiometric subunit of the PRC1 complex in flies (149) and humans (45, 94). SCML2 mutants lacking the RNA-binding region cannot bind to chromatin and result in lower levels of PRC1 recruitment, potentially due to dominant negative effects (11).
Many Repressive Chromatin Components Interact with lncRNAs
Although PRCs have received a large amount of attention, many other chromatin-associated factors and complexes tested have displayed an affinity for RNA in vitro and in vivo (49, 74, 81). One of the methyltransferases that deposit repressive methylation marks on H3K9 (EHMT2/G9A) as well as the reader that recognizes them (HP1) requires an RNA component for proper recruitment to certain target sites (101, 118, 124). Specifically, RNA is required for the accumulation of HP1α at DAPI-rich pericentromeric heterochromatin foci (101), presumably due to the RNA-binding activity of the hinge region located between the chromodomain and the chromo-shadow domain (117). This is consistent with the observation that the HP1 chromodomain alone does not bind RNA in vitro (8). The histone demethylase LSD1 binds to the lncRNA HOTAIR (162) and, judging by RNA immunoprecipitation experiments performed with its complex partner, CoREST, to many more lncRNAs (74), which remain to be thoroughly analyzed. Finally, lncRNAs from the CCND1 5′ regulatory sequence stimulate the inhibitory function of the RNA-binding protein TLS toward the coactivators CBP and p300, thus mediating transcriptional silencing via allosteric regulation of a repressor (173).
lncRNAs and Transcriptional Activation
Although the long list of examples above seems to imply that a majority of lncRNAs regulate transcription via repression, this is most likely the result of a historical tendency toward studying mechanisms involved in epigenetic repression in X inactivation and imprinting. In fact, recent results have indicated an activating function for a large number of lncRNAs (86) (Figure 3). Two independent lines of investigation converged to highlight a critical role for lncRNAs in activation of transcription. Functional studies following depletion of a number of lncRNAs revealed a predominant role in activating their neighboring protein-coding genes (123, 172). Many of these lncRNAs were derived from genomic regions that were previously denoted as distal enhancer elements (123, 172). Concomitantly, analysis of stimulus-dependent activation of enhancers in neuronal cells and macrophages revealed the presence of enhancer-derived lncRNAs (eRNAs; see above) (36, 76). Subsequent experiments revealed that active enhancers are transcribed and that eRNAs mediate transcriptional activation of the target promoters (54, 87, 96). In fact, genome-wide analyses of a large set of primary human cells and tissues revealed that the transcription of enhancers is the best diagnostic measure of their activity in any cell type (4). Several lncRNAs have been recovered in association with chromatin-modifying complexes that activate transcription. For example, lncRNAs HOTTIP and Mistral both bind to the MLL complex and activate transcription of neighboring HOX genes in cis via interactions with WDR5 and MLL, respectively (10, 172, 181). Activating lncRNAs were also shown to associate with the transcriptional coactivator complexes Mediator and Cohesin to promote enhancer-promoter communication through DNA looping (86, 96) (Figure 3a).
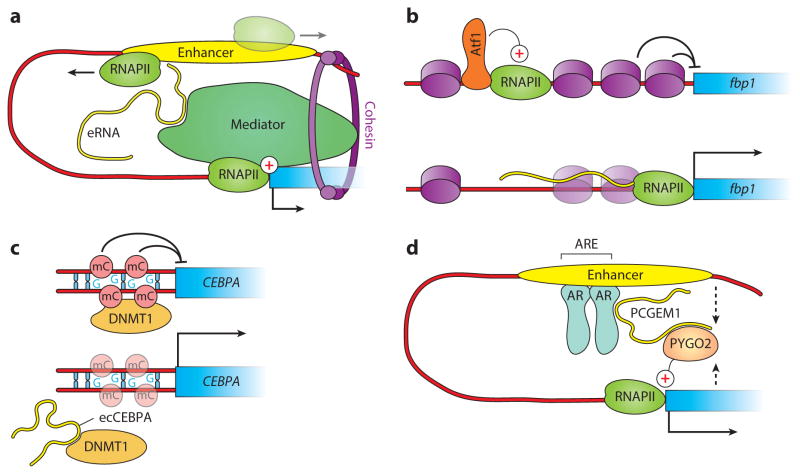
Figure 3.
Transcriptional activation by lncRNAs (long noncoding RNAs). (a) Several lncRNAs transcribed from a distal enhancer directly interact with subunits of Mediator and facilitate enhancer-promoter communication by promoting loop formation (86), which requires the function of cohesin (96). (b) In Schizosaccharomyces pombe, activation of fbp1 requires transcription of lncRNAs originating from an upstream transcription start site (TSS). This is followed by promoter remodeling via nucleosome depletion, which requires the transcribing polymerase (59). (c) Transcription of CEBPA in human cells is activated by an upstream sense lncRNA (ecCEBPA), which binds DNMT1 and sequesters it from chromatin, causing hypomethylation of the promoter. (d) Activation by an androgen receptor (AR) requires a complex cascade of post-translational modifications and lncRNA-protein interactions that culminates with the recruitment of PYGO2 via the lncRNA PCGEM1. In turn, PYGO2 stimulates transcription by strengthening enhancer-promoter looping (dashed arrows) (179).
Noncoding transcripts that traverse promoter chromatin can also participate in the activation of the adjacent coding gene, as seen at the fbp1+ locus of fission yeast (59) and the PHO5 gene of S. cerevisiae (164) (Figure 3b). Similar activating read-through transcripts have been observed at the human CEBPA locus, although in this case the molecular mechanism involves sequestration of DNMT1, which results in decreased levels of inhibitory DNA methylation and subsequent activation of CEBPA transcription (37) (Figure 3c). Finally, lncRNAs can contact transcription factors directly and stimulate their function by recruiting additional coactivators or facilitating enhancer-promoter looping, as seen in the case of the androgen receptor (179) (Figure 3d).
MECHANISTIC CONSIDERATIONS
The lack of details on the biogenesis and mechanism of action of most lncRNAs is the major hurdle to progress in the field. We are still at the early stages of understanding the RNA code involved in the activating and repressing functions of lncRNAs. A key to deciphering how RNA nucleotide sequences are utilized to convey functional information is to uncover the precise structural and biochemical requirements for the action of lncRNAs in transcriptional regulation.
Structure and Function
Important steps toward a mechanistic dissection of the function of lncRNAs have been taken by studies of the dosage compensation complex in Drosophila. Two reports detailed the network of interactions between roX RNAs and two protein components, MLE and MSL2, of the complex (65, 100). The authors analyzed in detail the secondary structure of these RNAs using traditional footprinting experiments and more modern technologies such as selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), which determines the RNA structure based on chemical reactivity (112), and individual nucleotide resolution UV cross-linking and immunoprecipitation (iCLIP), which utilizes UV cross-linking to detect protein-RNA contacts (79). Although the two studies reached somewhat different conclusions, they agree that the RNA helicase activity of MLE plays a critical role in the assembly of the dosage compensation ribonucleoprotein complex.
Much work remains to be done to reach a structural understanding of lncRNA function. Several techniques are available to probe RNA structure in vitro and in vivo, each with its own disadvantages. A thorough comparison of SHAPE, dimethyl sulfate (DMS) (28), in-line probing (134), and RNAse footprinting for the analysis of the secondary structure of the lncRNA SRA, revealed that the most accurate information can currently be obtained by combining these techniques (121). Among them, DMS probing might be the most promising, as this chemical easily enters cells and can probe RNA structure in vivo (138), although its reactivity is limited to As and Cs.
Biochemical approaches that investigate the protein side of the interaction are also useful. We and others have argued that the best way to address the function of protein-RNA interactions in the chromatin space is that of defining the protein regions or domains responsible for RNA binding and deleting them (11). Such analysis will be more powerful following the identification of the specific residues required for the RNA contacts and interrogation of point mutations that disrupt such interactions (181, 183). Often these RNA-binding surfaces do not resemble the well-characterized RNA recognition motif (RRM) (25); rather, they are unstructured stretches of charged residues rich in lysines and arginines (11, 69, 70). The lack of predicted secondary structure should not be interpreted as lack of function, as induced fit is a common feature of protein-RNA interactions (175). For example, the fragile X syndrome protein FMR1/FMRP comprises an arginine-rich stretch that is disordered in solution but becomes structured upon binding to RNA (129). An arginine-rich motif is also responsible for binding of the HIV protein REV to its response element (RRE) within the RNA genome of the virus and becomes ordered only upon RNA binding (19). Similar to the case of some RNA-binding regions in chromatin-associated proteins, such as SCML2 (11), the arginine-rich motif of REV binds to a variety of RNA sequences and structures in vitro but is strictly specific for RRE in vivo (31). Structural studies have suggested that this specificity is acquired via its multimerization and precise geometrical orientation in the functional hexamer (32).
Mode of Action: Trans versus Cis
Another important, and in most cases unanswered, question is whether lncRNAs act in cis or trans fashion. In the former case, RNA molecules would affect only the structure and function of neighboring chromatin and/or chromatin-associated proteins. This may result from the tethering of the lncRNA to either the transcribing polymerase (cotranscriptional action) or a chromatin component after transcription is complete. Many imprinted lncRNAs, dosage compensation lnc-RNAs, and most lncRNAs with activating functions, such as eRNAs, seem to act in cis (96, 123, 172). It has been argued that cis function is a specialized task that could be accommodated better by ncRNAs, as they can fold into functional structures during transcription, without requiring translation in the cytoplasm (89). The cis mechanism of action requires lncRNAs to function in an allele-specific manner, but to date this critical aspect of lncRNA biology has only been addressed in dosage compensation.
HOTAIR, an intervening lncRNA involved in silencing of HOX genes, is thought to act in trans (23, 137). Trans-mediated function was also reported for linc-p21 (63), linc-Firre (52), and, at least in part, Meg3 (69). The observation that some lncRNAs might be trans-acting begs the question of how they reach their intended targets in the genome. It is easy to envision RNA-RNA interactions or protein-RNA interactions, but less so how RNAs could bind to specific sequences within double-stranded genomic DNA. Although formation of a triplex with DNA has been reported (107, 145), strand invasion is also possible (93) and can be guided by ncRNAs, as observed in the CRISPR-Cas9 system (156).
Finally, we note that cis and trans are not the only two possibilities. lncRNAs might be locally confined by being tethered to their locus of origin and still function at sites that are distant in a two-dimensional representation of DNA sequences but proximal in three-dimensional space, as discussed in the next section.
lncRNAs and Nuclear Organization
A set of recent observations suggests that the function of lncRNAs might be linked to the three-dimensional organization of the mammalian nucleus. The idea that chromatin is not freely diffusing in the nucleoplasmic space and that some structural organization might be required for genome function is not new (7) but has been exceedingly difficult to prove (133). Whether or not a static structural scaffold exists, the three-dimensional organization of chromatin within the nucleus plays a key role in genome function. At the kilobase scale it supports the formation of loops that bring distant regulatory regions, enhancers, and their specific targets into contact (68, 86, 96). At the larger megabase scale, it affects the degree of higher-order compaction of chromosomal domains, varying their accessibility to nuclear factors (114, 131). A proposal has also been put forward that subnuclear positioning of chromatin affects the activation status of embedded genes (188). To date, little is known about how spatial organization of chromatin is achieved, and it is likely that lncRNAs might play a key role in genomic architecture.
Several subnuclear structures contain and/or can be nucleated by well-known lncRNAs, such as NEAT1 (102) and MALAT1 (64). In further support of a connection between lncRNAs and nuclear organization, Xist interacts with HNRNPU (also known as SAFA), a component of the nuclear matrix (57). Moreover, spreading of Xist from its site of transcription during the initial phases of X inactivation is dictated by the three-dimensional organization of the X chromosome rather than by sequence motifs or other molecular features of target sites (41). Similarly, the lncRNA Firre, implicated in mouse adipogenesis (158), participates in nuclear organization by interacting with the nuclear matrix protein HNRNPU and by gathering otherwise distant genes whose products are involved in energy metabolism and adipogenesis, potentially exposing them to a shared local environment that might facilitate coregulation (52).
The Chromatin Organizer Model for lncRNA Function
We propose a “chromatin organizer” model for the function of at least a subset of lncRNAs (Figure 4). According to this model, lncRNAs utilize 3D conformation as well as sequence information to spatially organize chromatin and chromatin-associated factors within the nucleus. This function resembles that of cable organizers made of Velcro, which bundle and separate electronic cables by establishing homotypic and heterotypic interactions with themselves and other neighboring organizers.
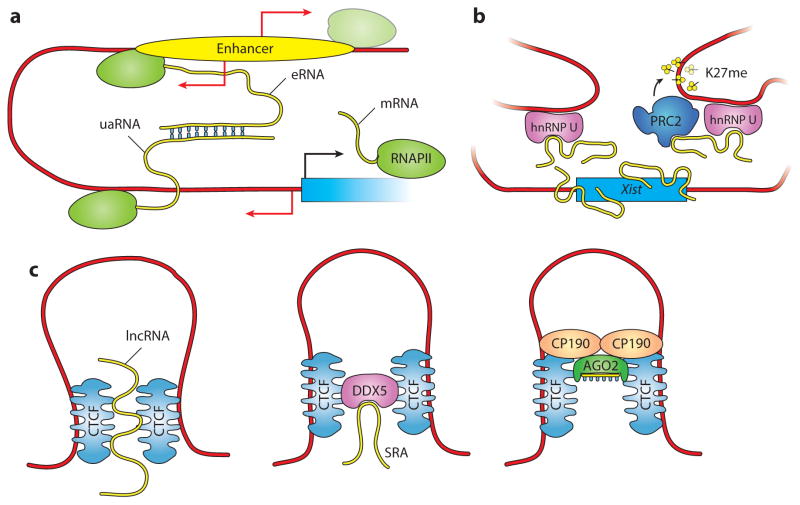
Figure 4.
Potential roles of lncRNAs (long noncoding RNAs) in nuclear organization. (a) In this speculative model, eRNAs (enhancer RNAs) mediate enhancer-promoter communication directly via base-pair interactions with uaRNAs (upstream antisense RNAs) and possibly facilitate chromatin looping. (b) The lncRNA Xist utilizes the three-dimensional organization of chromatin to spread its silencing activity over the whole X chromosome. Interactions with chromatin might be mediated via the matrix protein hnRNP U (41). At the target sites, Xist facilitates H3K27me3 deposition via PRC2 recruitment (187). (c) Roles of ncRNAs (noncoding RNAs) in CTCF function. (Left) Human CTCF binds RNA directly and RNA interactions mediate multimerization, which might guide chromatin looping and organization (141). (Middle) Human CTCF also binds RNA indirectly through its interactions with the RNA helicase DDX5, here bound to the lncRNA SRA (182). (Right) Drosophila CTCF and CP190 insulator proteins interact physically and genetically with AGO2, a small ncRNA-binding protein and key component of the RNAi machinery (92, 116).
If true, our model would explain the role of lncRNAs in a variety of contexts. Activating eRNAs might function by looping chromatin locally to bring enhancers in close proximity with promoters and the transcription machinery (Figure 4a). This could be accomplished by interactions with protein complexes such as Mediator (86), but also by direct, possibly sequence-specific, interactions with uaRNAs that are produced by divergent transcription at genic TSSs. The prediction that eRNAs might interact with cognate uaRNAs is consistent with the observation that enhancers and the promoters under their control often share sequence motifs (4) and, therefore, patches of complementary sequence.
The chromatin organizer model is also consistent with the observations that Xist and linc-Firre organize chromatin at a large scale to enforce X inactivation or the regulation of functionally related genes, respectively (41, 52). In these cases, the lncRNAs bring together more distant genomic regions with the purpose of subjecting them to a common regulatory environment (Figure 4b). A similar scenario might also explain the role of the lncRNA Meg3 in PRC2 regulation. Meg3 binds at least two subunits of PRC2 and helps its assembly on chromatin, but it does so only at a small number of selected sites disseminated in trans throughout the genome (69). These Meg3-dependent PRC2 targets, although distant in the primary sequence, might be brought together in the three-dimensional space of the nucleus, possibly to coordinate their regulation. Finally, human CTCF, the insulator protein that functions as master weaver of the genome (130), interacts with RNA directly (141, 159) and indirectly (182) via the DEAD-box helicase p68 (DDX5) (Figure 4c). Along the same lines, insulator proteins in Drosophila interact physically and genetically with the RNAi machinery (92, 116) (Figure 4c), suggesting a conserved role for ncRNAs in chromatin organization and nuclear structure.
FUTURE DIRECTIONS
The road ahead before we arrive at a full understanding of the molecular mechanisms responsible for lncRNA-mediated regulation of transcription is still very long; the models discussed above will need to be tested and new technologies developed. Current annotations of lncRNAs need to be integrated across different projects (e.g., the human lincRNA catalog of the Broad Institute and the GENCODE annotation) and phylogenetic information should be taken into account whenever possible and should focus on related species and even polymorphism within the same species, in addition to the analysis of conservation across long evolutionary distances (53, 120). Such new annotation should consider the wealth of recent findings, which indicates that a large number of lncRNAs might not be polyadenylated and function predominantly in enhancer-promoter communication. The discovery of this new class of RNAs, mainly eRNAs and uaRNAs, suggests the presence of as yet undiscovered machinery involved in their processing and mode of function. Technologies to detect protein-RNA interactions should also be improved, as they continue to suffer from suboptimal signal-to-noise ratio (12, 44). Possibly the most challenging task ahead is to understand lncRNA function in the context of nuclear organization. New technologies may also help toward this goal. Notable advances have been made in single-molecule RNA-FISH (fluorescence in situ hybridization) (5, 84), proximity labeling (139), and super-resolution microscopy (62). The synergy between these new experimental approaches and tried-and-true biochemical work will ultimately pave the way to a complete understanding of the functional roles of lncRNAs in the nucleus.
Acknowledgments
The authors wish to thank Kristin Ingvarsdottir and Danny Reinberg for their comments on the manuscript. R.S. is supported by R01 GM 078455 and R01 GM 105754.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Roberto Bonasio, Email: rbon@mail.med.upenn.edu.
Ramin Shiekhattar, Email: rshiekhattar@med.miami.edu.
LITERATURE CITED
- 1.Allis CD, Jenuwein T, Reinberg D. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harb. Lab. Press; 2007. p. 502. [Google Scholar]
- 2.Almada AE, Wu X, Kriz AJ, Burge CB, Sharp PA. Promoter directionality is controlled by U1 snRNP and polyadenylation signals. Nature. 2013;499:360–63. doi: 10.1038/nature12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–69. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- 4.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batish M, Raj A, Tyagi S. Single molecule imaging of RNA in situ. Methods Mol Biol. 2011;714:3–13. doi: 10.1007/978-1-61779-005-8_1. [DOI] [PubMed] [Google Scholar]
- 6.Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berezney R, Coffey DS. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–17. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–69. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–26. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertani S, Sauer S, Bolotin E, Sauer F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol Cell. 2011;43:1040–46. doi: 10.1016/j.molcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, Reinberg D. Interactions with RNA direct SCML2 to chromatin where it represses PRC1 target genes. eLife. 2014;3:e02637. doi: 10.7554/eLife.02637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockdorff N. Noncoding RNA and polycomb recruitment. RNA. 2013;19:429–42. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci USA. 2009;106:18321–26. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–98. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 16.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–45. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–17. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100–12. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Casu F, Duggan BM, Hennig M. The arginine-rich RNA-binding motif of HIV-1 Rev is intrinsically disordered and folds upon RRE binding. Biophys J. 2013;105:1004–17. doi: 10.1016/j.bpj.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cech TR. The RNA worlds in context. Cold Spring Harb Perspect Biol. 2012;4:a006742. doi: 10.1101/cshperspect.a006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cech TR, Steitz JA. The noncoding RNA revolution: trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, et al. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci USA. 2014;111:2235–40. doi: 10.1073/pnas.1312951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–75. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cléry A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–98. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–23. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2011;13:123–34. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 28.Cordero P, Kladwang W, VanLang CC, Das R. Quantitative dimethyl sulfate mapping for automated RNA secondary structure inference. Biochemistry. 2012;51:7037–39. doi: 10.1021/bi3008802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–48. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crick FH. The origin of the genetic code. J Mol Biol. 1968;38:367–79. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 31.Daugherty MD, D’Orso I, Frankel AD. A solution to limited genomic capacity: using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol Cell. 2008;31:824–34. doi: 10.1016/j.molcel.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daugherty MD, Liu B, Frankel AD. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol. 2010;17:1337–42. doi: 10.1038/nsmb.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–25. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20:1250–57. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLOS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–76. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eddy SR. Computational genomics of noncoding RNA genes. Cell. 2002;109:137–40. doi: 10.1016/s0092-8674(02)00727-4. [DOI] [PubMed] [Google Scholar]
- 40.ENCODE Proj. Consort. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet. 2011;12:565–75. doi: 10.1038/nrg3032. [DOI] [PubMed] [Google Scholar]
- 43.Fischle W, Wang Y, Jacobs S, Kim Y, Allis C, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedersdorf MB, Keene JD. Advancing the functional utility of PAR-CLIP by quantifying background binding to mRNAs and lncRNAs. Genome Biol. 2014;15:R2. doi: 10.1186/gb-2014-15-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, et al. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell. 2012;45:344–56. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert W. Origin of life: the RNA world. Nature. 1986;319:618. [Google Scholar]
- 47.Gummalla M, Maeda RK, Castro Alvarez JJ, Gyurkovics H, Singari S, et al. abd-A regulation by the iab-8 noncoding RNA. PLOS Genet. 2012;8:e1002720. doi: 10.1371/journal.pgen.1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guttman M, Amit I, Garber M, French C, Lin MF, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–27. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haerty W, Ponting CP. Mutations within lncRNAs are effectively selected against in fruitfly but not in human. Genome Biol. 2013;14:R49. doi: 10.1186/gb-2013-14-5-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hah N, Murakami S, Nagari A, Danko CG, Kraus WL. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–23. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hannon GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 56.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–33. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–76. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 58.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 59.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–34. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 60.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–45. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 61.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–95. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ilik IA, Quinn JJ, Georgiev P, Tavares-Cadete F, Maticzka D, et al. Tandem stem-loops in roX RNAs act together to mediate X chromosome dosage compensation in Drosophila. Mol Cell. 2013;51:156–73. doi: 10.1016/j.molcel.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat Rev Genet. 2009;10:833–44. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 68.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–35. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaneko S, Bonasio R, Saldana-Meyer R, Yoshida T, Son J, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaneko S, Li G, Son J, Xu CF, Margueron R, et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle–regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–20. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20:1258–64. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–88. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–88. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 74.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim T, Xu Z, Clauder-Munster S, Steinmetz LM, Buratowski S. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell. 2012;150:1158–69. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–87. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kohler C, Wolff P, Spillane C. Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol. 2012;63:331–52. doi: 10.1146/annurev-arplant-042811-105514. [DOI] [PubMed] [Google Scholar]
- 79.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature. 2012;482:310–11. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- 81.Krajewski WA, Nakamura T, Mazo A, Canaani E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25:1891–99. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–57. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 83.Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLOS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kwon S. Single-molecule fluorescence in situ hybridization: quantitative imaging of single RNA molecules. BMB Rep. 2013;46:65–72. doi: 10.5483/BMBRep.2013.46.2.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lahmy S, Bies-Etheve N, Lagrange T. Plant-specific multisubunit RNA polymerase in gene silencing. Epigenetics. 2010;5:4–8. doi: 10.4161/epi.5.1.10435. [DOI] [PubMed] [Google Scholar]
- 86.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, et al. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature. 2013;498:511–15. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338:1469–72. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 89.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–39. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 90.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–23. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 90a.Lee MP, DeBaun MR, Mitsuya K, Galonek HL, Brandenburg S, et al. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc Natl Acad Sci USA. 1999;96:5203–8. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee NN, Chalamcharla VR, Reyes-Turcu F, Mehta S, Zofall M, et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013;155:1061–74. doi: 10.1016/j.cell.2013.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–41. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- 93.Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506:249–53. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–78. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lewis EB. C.B. Bridges’ repeat hypothesis and the nature of the gene. Genetics. 2003;164:427–31. doi: 10.1093/genetics/164.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li W, Notani D, Ma Q, Tanasa B, Nunez E, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–20. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lipshitz HD, Peattie DA, Hogness DS. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987;1:307–22. doi: 10.1101/gad.1.3.307. [DOI] [PubMed] [Google Scholar]
- 98.Liu J, Jung C, Xu J, Wang H, Deng S, et al. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–45. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–34. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 100.Maenner S, Muller M, Frohlich J, Langer D, Becker PB. ATP-dependent roX RNA remodeling by the helicase maleless enables specific association of MSL proteins. Mol Cell. 2013;51:174–84. doi: 10.1016/j.molcel.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 101.Maison C, Bailly D, Peters AHFM, Quivy J-P, Roche D, et al. Higher-order structure in peri-centric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–34. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 102.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mardis ER. Next-generation sequencing platforms. Annu Rev Anal Chem. 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 104.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–49. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–77. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 106.Martens JA, Wu P-YJ, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 108.Martinho RG, Kunwar PS, Casanova J, Lehmann R. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 2004;14:159–65. doi: 10.1016/j.cub.2003.12.036. [DOI] [PubMed] [Google Scholar]
- 109.Mattick JS. The genetic signatures of noncoding RNAs. PLOS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Meller VH, Rattner BP. The roX genes encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J. 2002;21:1084–91. doi: 10.1093/emboj/21.5.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–57. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- 112.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–31. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 112a.Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012;30:453–59. doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development. 2010;137:2493–99. doi: 10.1242/dev.048181. [DOI] [PubMed] [Google Scholar]
- 114.Morey C, Da Silva NR, Perry P, Bickmore WA. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development. 2007;134:909–19. doi: 10.1242/dev.02779. [DOI] [PubMed] [Google Scholar]
- 115.Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, et al. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell. 2012;10:47–62. doi: 10.1016/j.stem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 116.Moshkovich N, Nisha P, Boyle PJ, Thompson BA, Dale RK, Lei EP. RNAi-independent role for Argonaute2 in CTCF/CP190 chromatin insulator function. Genes Dev. 2011;25:1686–701. doi: 10.1101/gad.16651211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Muchardt C, Guilleme M, Seeler J-S, Trouche D, Dejean A, Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep. 2002;3:975–81. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 119.Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 120.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–40. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 121.Novikova IV, Hennelly SP, Sanbonmatsu KY. Structural architecture of the human long non-coding RNA, steroid receptor RNA activator. Nucleic Acids Res. 2012;40:5034–51. doi: 10.1093/nar/gks071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ntini E, Jarvelin AI, Bornholdt J, Chen Y, Boyd M, et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat Struct Mol Biol. 2013;20:923–28. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 123.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–46. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 125.Park SW, Kang Y, Sypula JG, Choi J, Oh H, Park Y. An evolutionarily conserved domain of roX2 RNA is sufficient for induction of H4-Lys16 acetylation on the Drosophila X chromosome. Genetics. 2007;177:1429–37. doi: 10.1534/genetics.107.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12:136–49. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pease B, Borges AC, Bender W. Noncoding RNAs of the Ultrabithorax domain of the Drosophila bithorax complex. Genetics. 2013;195:1253–64. doi: 10.1534/genetics.113.155036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, et al. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–21. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, et al. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–95. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–65. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Razin SV, Iarovaia OV, Vassetzky YS. A requiem to the nuclear matrix: from a controversial concept to 3D organization of the nucleus. Chromosoma. 2014;123:217–24. doi: 10.1007/s00412-014-0459-8. [DOI] [PubMed] [Google Scholar]
- 134.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 135.Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet. 2005;6:403–10. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 136.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–10. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–97. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Saldana-Meyer R, Gonzalez-Buendia E, Guerrero G, Narendra V, Bonasio R, et al. CTCF regulates the human p53 gene through direct interaction with its natural antisense transcript, Wrap53. Genes Dev. 2014;28:723–34. doi: 10.1101/gad.236869.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–23. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 143.Sánchez-Herrero E, Akam M. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development. 1989;107:321–29. doi: 10.1242/dev.107.2.321. [DOI] [PubMed] [Google Scholar]
- 144.Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–69. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schulz D, Schwalb B, Kiesel A, Baejen C, Torkler P, et al. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell. 2013;155:1075–87. doi: 10.1016/j.cell.2013.10.024. [DOI] [PubMed] [Google Scholar]
- 147.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, et al. Divergent transcription from active promoters. Science. 2008;322:1849–51. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Shah S, Wittmann S, Kilchert C, Vasiljeva L. lncRNA recruits RNAi and the exosome to dynamically regulate pho1 expression in response to phosphate levels in fission yeast. Genes Dev. 2014;28:231–44. doi: 10.1101/gad.230177.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, et al. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- 150.Sigova AA, Mullen AC, Molinie B, Gupta S, Orlando DA, et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc Natl Acad Sci USA. 2013;110:2876–81. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 152.Simon MD, Pinter SF, Fang R, Sarma K, Rutenberg-Schoenberg M, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–69. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–13. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 154.Song J, Angel A, Howard M, Dean C. Vernalization: a cold-induced epigenetic switch. J Cell Sci. 2012;125:3723–31. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- 155.St Johnston D. The art and design of genetic screens: Drosophila melanogaster. Nat Rev Genet. 2002;3:176–88. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 156.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–5. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 158.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, et al. Long noncoding RNAs regulate adipo-genesis. Proc Natl Acad Sci USA. 2013;110:3387–92. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–51. doi: 10.1016/j.cell.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 161.Tijsterman M, Ketting RF, Plasterk RH. The genetics of RNA silencing. Annu Rev Genet. 2002;36:489–519. doi: 10.1146/annurev.genet.36.043002.091619. [DOI] [PubMed] [Google Scholar]
- 162.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, et al. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci USA. 2005;102:5495–500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci USA. 2007;104:8011–16. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLOS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–17. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 169.van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170–81. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wang H, Chung PJ, Liu J, Jang IC, Kean MJ, et al. Genome-wide identification of long noncoding natural antisense transcripts and their responses to light in Arabidopsis. Genome Res. 2014;24:444–53. doi: 10.1101/gr.165555.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–24. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang X, Arai S, Song X, Reichart D, Du K, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–30. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–34. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–37. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 176.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–39. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 177.Wu X, Sharp PA. Divergent transcription: a driving force for new gene origination? Cell. 2013;155:990–96. doi: 10.1016/j.cell.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–37. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang L, Lin C, Jin C, Yang JC, Tanasa B, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yang L, Lin C, Liu W, Zhang J, Ohgi KA, et al. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–88. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang YW, Flynn RA, Chen Y, Qu K, Wan B, et al. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Yao H, Brick K, Evrard Y, Xiao T, Camerini-Otero RD, Felsenfeld G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev. 2010;24:2543–55. doi: 10.1101/gad.1967810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, et al. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol. 2012;4:427–42. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zamudio JR, Kelly TJ, Sharp PA. Argonaute-bound small RNAs from promoter-proximal RNA polymerase II. Cell. 2014;156:920–34. doi: 10.1016/j.cell.2014.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–53. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–56. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–79. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhu Y, Rowley MJ, Bohmdorfer G, Wierzbicki AT. A SWI/SNF chromatin-remodeling complex acts in noncoding RNA–mediated transcriptional silencing. Mol Cell. 2013;49:298–309. doi: 10.1016/j.molcel.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]