SUMMARY
SIRT1 regulates the DNA damage response by deacetylating p53, thereby repressing p53 transcriptional output. Here we demonstrate that the sorting protein PACS-2 regulates SIRT1-mediated deacetylation of p53 to modulate the DNA damage response. PACS-2 knockdown cells failed to efficiently undergo p53-induced cell cycle arrest in response to DNA damage. Accordingly, p53 acetylation was reduced both in PACS-2 knockdown cells and thymocytes from Pacs-2−/− mice, thereby blunting induction of the cyclin-dependent kinase inhibitor p21 (CDKN1A). The SIRT1 inhibitor EX-527 or SIRT1 knockdown restored p53 acetylation and p21 induction as well as p21-dependent cell cycle arrest in PACS-2 knockdown cells. Trafficking studies revealed cytoplasmic PACS-2 shuttled to the nucleus where it interacted with SIRT1 and repressed SIRT1-mediated p53 deacetylation. Correspondingly, in vitro assays demonstrated PACS-2 directly inhibited SIRT1-catalyzed p53 deacetylation. Together, these findings identify PACS-2 as an in vivo mediator of the SIRT1—p53—p21 axis that modulates the DNA damage response.
INTRODUCTION
The tumor suppressor p53 is perhaps the most frequent target of genetic lesions in human cancer. Following DNA damage p53 orchestrates biological fates ranging from growth arrest to cell death and the molecular pathways leading to these various outcomes depend on several factors, including the level and type of stress as well as the cell and tissue type (Mirzayans et al., 2012; Zilfou and Lowe, 2009). p53 functions as a sequence-specific transcription factor that drives the transactivation of target genes mediating cell cycle arrest, senescence or apoptosis induced by the intrinsic pathway (Kruse and Gu, 2009). Among the p53 target genes, perhaps the best characterized is the CDK inhibitor p21, which promotes cell cycle arrest, supports DNA damage repair and impedes apoptosis (Abbas and Dutta, 2009). In addition, p53 integrates the intrinsic apoptotic pathway with the extrinsic apoptotic pathway triggered by the death ligand TRAIL. Indeed, tumor cell apoptosis can be increased when TRAIL is combined with DNA damage-inducing therapies (Ifeadi and Garnett-Benson, 2012).
The transcriptional activity of p53 is critically dependent on posttranslational modifications, including phosphorylation and acetylation, which stabilize p53 and enhance its transactivation functions, respectively (Kruse and Gu, 2009). Acetylation of p53 is catalyzed predominantly by the histone acetyltransferase p300 (Gu and Roeder, 1997). Indeed, stress-induced p53 acetylation significantly correlates with p53 activation (Kruse and Gu, 2009; Zilfou and Lowe, 2009). Accordingly, mutation of all major lysine acetylation sites blocks the ability of p53 to induce p21 and suppress cell proliferation, suggesting acetylation of p53 is indispensable for the p53-p21 pathway (Tang et al., 2008). Conversely, the class III histone deacetylase SIRT1 inhibits p53 transcriptional activation by deacetylating p53 following DNA damage (Kruse and Gu, 2009). Together, the overall balance of p300- and SIRT1-activities modulate p53 transcriptional function.
While regulation of p53 by modifying enzymes and cofactors has been extensively studied, less is known about the regulation of SIRT1. Gene expression of SIRT1 can be regulated transcriptionally and posttranscriptionally (Kwon and Ott, 2008). In addition, SIRT1 deacetylase activity can be modulated by interaction with cellular proteins (Hasegawa and Yoshikawa, 2008; Kim et al., 2007; Kim et al., 2008; Liu et al., 2011; Zhao et al., 2008). While these studies provide insight into the regulation of SIRT1, the diversity of SIRT1 substrates in pathways ranging from DNA damage and cell survival to glucose and lipid homeostasis, suggest that regulation of SIRT1 activity is complex, and likely requires additional cellular factors (Brooks and Gu, 2009).
Here we identify the multi-functional sorting protein PACS-2 as an inhibitor of SIRT1-mediated deacetylation of p53 following DNA damage. PACS-2 was initially identified by its role in mediating secretory pathway traffic and formation of contacts between the endoplasmic reticulum and mitochondria (mitochondria-associated membranes or MAMs) to regulate interorganellar communication and autophagy (Atkins et al., 2008; Dikeakos et al., 2012; Hamasaki et al., 2013; Kottgen et al., 2005; Simmen et al., 2005). In response to TRAIL, however, PACS-2 switches to a proapoptotic effector that coordinates trafficking steps leading to mitochondria membrane permeabilization and activation of executioner caspases (Aslan et al., 2009; Werneburg et al., 2012). In this study we show that, contrary to its role in TRAIL action, PACS-2 responds to DNA damage by regulating the extent of SIRT1-mediated deacetylation of p53 to induce p21-dependent cell cycle arrest. Together, these findings suggest PACS-2 is a novel regulator of the SIRT1—p53—p21 axis that modulates the DNA damage response.
RESULTS
PACS-2 mediates the p53-dependent response to DNA damage
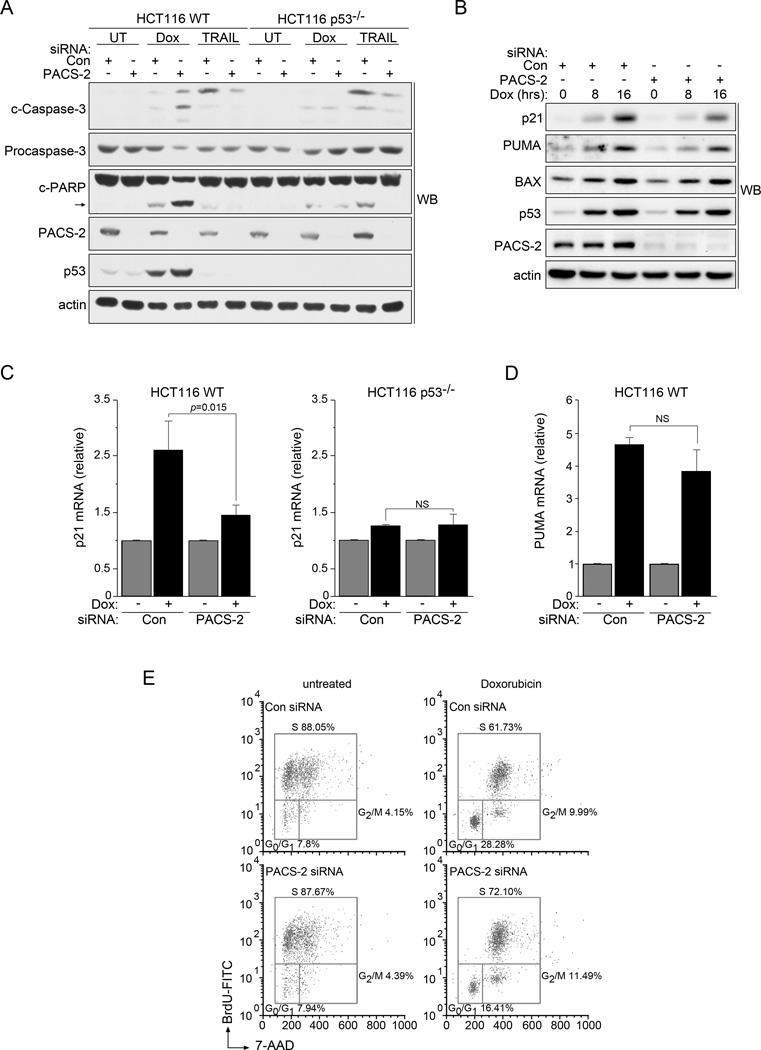
Previous studies identified an essential role for cytoplasmic PACS-2 in mediating TRAIL-induced apoptosis (Aslan et al., 2009; Werneburg et al., 2012). To determine whether this proapoptotic requirement for PACS-2 extended to DNA damage, we compared the effect of PACS-2 knockdown on TRAIL- versus Doxorubicin (Dox)-induced apoptosis in HCT116 wild-type (WT) or isogenic p53−/− cells (Fig. 1a). As expected, PACS-2 knockdown blunted TRAIL-induced apoptosis independent of p53 status. By contrast, PACS-2 knockdown sensitized HCT116 WT cells but not p53−/− cells to Dox-induced apoptosis (Figs. 1a and S1a). This effect additionally required the proapoptotic p53 target, PUMA, and was also observed using ionizing radiation (IR, Figs. S1b and S1c). These findings suggest distinct roles for PACS-2 in TRAIL- versus p53-mediated apoptosis.
Figure 1. PACS-2 deficient cells have reduced p21-dependent cell cycle arrest following DNA damage.
(A) Control (Con) or PACS-2 siRNA-treated HCT116 WT or p53−/− cells were treated with 0.5 µM Dox for 48hr or 20 ng/mL TRAIL for 5hr prior to harvest and analyzed by western blot. The cleaved caspase-3 Ab detected 19, 17 and 15 kDa forms. Arrow denotes cleaved PARP. (B) Control (Con) or PACS-2 siRNA-treated HCT116 cells were treated with 0.5 µM Dox and analyzed by western blot. Quantification using AlphaView (Protein Simple) showed that after 16 hr Dox treatment, PACS-2 knockdown reduced p21 by 50% whereas PUMA and Bax were less affected (16% and 12% reduction, respectively). (C) Control (Con) or PACS-2 siRNA-treated HCT116 WT (left) or HCT116 p53−/− (right) cells were treated with 0.5 µM Dox (4hr) and analyzed by qRT-PCR (normalized to GAPDH). Error bars represent mean ± SD from ≥ 3 independent experiments. Statistical significance determined using Student’s t-test. (D) HCT116 WT cells from panel B were analyzed for PUMA mRNA by qRT-PCR (normalized to GAPDH). Error bars represent mean ± SD from four independent experiments. Statistical significance determined using Student’s t-test. (E) Synchronized control (Con) or PACS-2 siRNA-treated HCT116 cells were stimulated to re-enter the cell cycle in the presence of 10 µM BrdU, treated with 0.5 µM Dox (24hr) and processed for flow cytometry. Results are representative of 3 independent experiments. See also Table S1 and Figures S1 and S2. NS, not significant.
PACS-2 regulates p53-mediated cell cycle arrest following DNA damage
The increased sensitivity of PACS-2-deficient cells to p53- and PUMA-mediated apoptosis following DNA damage led us to ask whether PACS-2 knockdown altered induction of p53 target genes. Analysis by western blot showed PACS-2 knockdown reduced the p53-dependent induction of p21 but had little effect on the proapoptotic targets PUMA or Bax (Fig. 1b). These findings were consistent with qPCR analysis, which showed PACS-2 knockdown reduced the p53-dependent induction of p21 by approximately 40% (Fig 1c). MDM2 and Noxa were reduced to a lesser extent, 27% and 19%, respectively, while there was no significant reduction in PUMA, Bax, DR5, 14-3-3, or Gadd45a (Figs. 1d and S2a). Similar results were observed in U2OS cells following IR, demonstrating this effect of PACS-2 knockdown on p21 induction was not restricted to a single cell line or treatment (Fig. S2b).
The blunted induction of p21 following DNA damage in PACS-2 knockdown cells led us to test the role of PACS-2 in mediating DNA damage-induced cell cycle arrest. As expected, PACS-2 knockdown significantly reduced the percentage of HCT116 WT cells in G0/G1 compared to control cells following treatment with Dox (Fig. 1e and Table S1). As p21 has well-described roles in promoting growth arrest and repressing apoptosis following DNA damage (Mirzayans et al., 2012), we tested whether the repressed induction of p21 in PACS-2 knockdown HCT116 WT cells contributed to their increased sensitivity to DNA damage. Similar to PACS-2 knockdown, p21 knockdown sensitized HCT116 cells to Dox-induced apoptosis (Fig. S2c). Consistent with this finding, knockdown of PACS-2 in isogenic HCT116 p21−/− cells failed to further sensitize them to Dox-induced apoptosis compared to control cells (Fig. S2d). Together, these findings suggest that the disparate p53-dependent induction of p21 versus PUMA in PACS-2-depleted cells resulted in reduced G1 arrest and a corresponding increase in apoptosis. These findings further suggest that PACS-2 and p21 may participate in a common pathway to promote cell cycle arrest.
PACS-2 regulates the p53-p21 axis in vivo following DNA damage
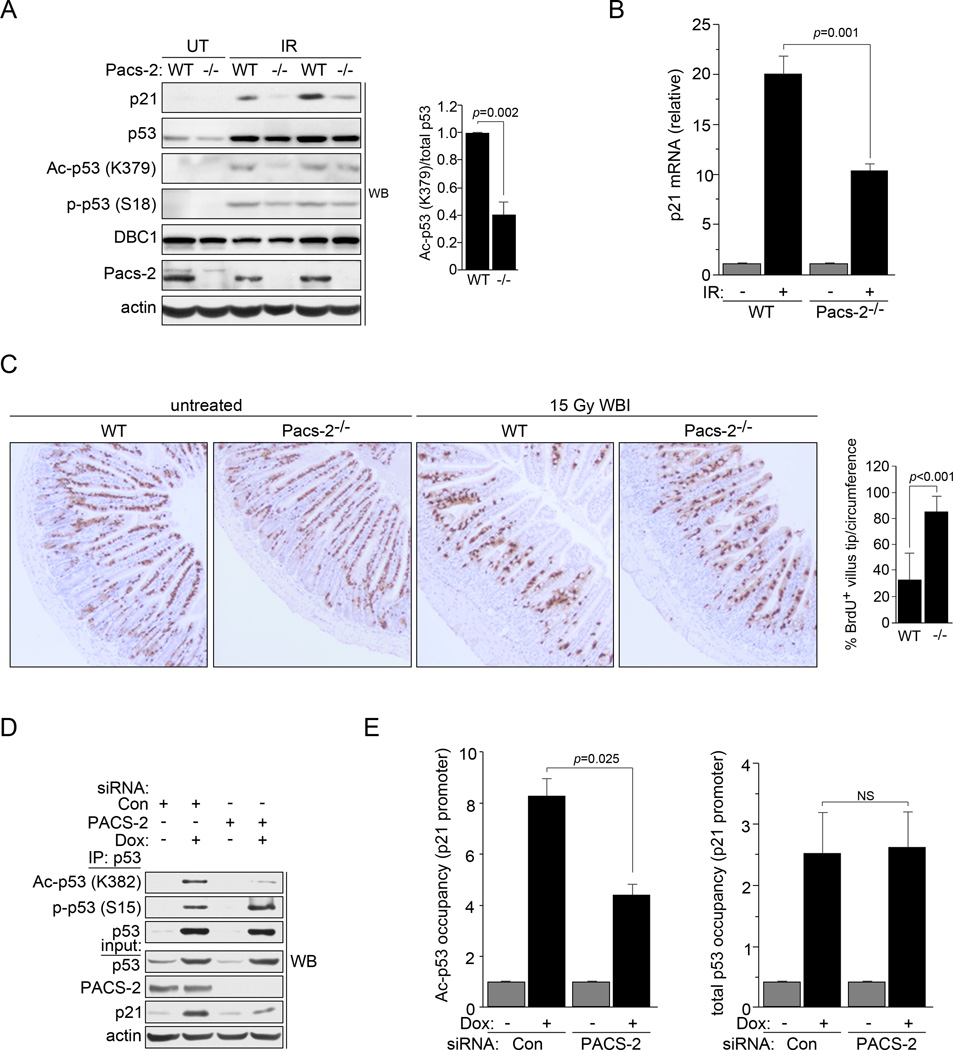
Consistent with the results from both HCT116 and U2OS cells, Pacs-2−/− thymocytes contained reduced levels of p21 protein and mRNA following exposure to IR (Figs. 2a and b and see Fig. 1). The predisposition of thymocytes to undergo apoptosis in response to IR precluded evaluation of cell cycle arrest. By contrast, p53-mediated p21 induction plays an essential role in mitigating GI toxicity following IR by inducing cell cycle arrest to restrict enterocyte migration along the crypt-villus axis (Komarova et al., 2004; Leibowitz et al., 2011; Li et al., 2005; Sullivan et al., 2012). Consistent with these studies, we found that migration of BrdU+ enterocytes was restricted in WT mice but not Pacs-2−/− mice following IR (Fig. 2c).
Figure 2. PACS-2 modulates p53 acetylation in vivo following DNA damage.
(A) Left: Thymuses from WT and Pacs-2−/− (−/−) mice exposed or not to 4.5 Gy IR (6hr) were analyzed by western blot. Right: Ac-p53 (K379) and total p53 were quantified using NIH Image J from IR-exposed WT or Pacs-2−/− mice. (B) Thymuses from WT and Pacs-2−/− mice exposed to 4.5 Gy IR (6hr) as indicated were analyzed for p21 induction by qRT-PCR (normalized to GAPDH). Error bars represent mean ± SEM from ≥ 4 mice per condition. Statistical significance determined using Student’s t-test. (C) Left: Small intestines from WT and Pacs-2−/− (−/−) mice were analyzed by IHC for BrdU 48hr following labeling and exposure to 15 Gy WBI as indicated. Right: Percentage of BrdU+ cells reaching the upper 1/4th of the villus tips per circumference was quantified. Error bars represent mean ± SD. Statistical significance determined using Student’s t-test. Quantification from untreated mice was not determined as all villus tips from these mice contained BrdU+ cells. (D) p53 was immunoprecipitated from Control (Con) or PACS-2 siRNA-treated HCT116 cells were treated with 0.5 µM Dox and analyzed by western blot. (E) Control (Con) or PACS-2 siRNA-treated HCT116 cells treated with 0.5 µM Dox (6hr) were analyzed by ChIP qPCR for the p21 promoter using antibodies specific for total p53 (DO-1) or Ac-p53 (K382). Error bars represent mean ± SEM from 3 independent experiments. Statistical significance determined using Student’s t-test. NS, not significant. See also Figure S3.
PACS-2 modulates p53 acetylation in vivo following DNA damage
The determination that PACS-2 knockdown repressed p21 induction but had no significant effect on total p53 levels (see Figs. 1b, 2a and 2d), suggested PACS-2 might modulate p53 activation by controlling p53 posttranslational modifications. Therefore, we analyzed the extent of p53 Lys382 acetylation, which correlates with p53 activation and p21 induction (Wang et al., 2008), and p53 Ser15 phosphorylation, which promotes p53 stabilization and interaction with p300 (Lee et al., 2010), in control or PACS-2 knockdown HCT116 cells following Dox treatment. Strikingly, PACS-2 knockdown reduced endogenous p53 Lys382 acetylation but not Ser15 phosphorylation (Fig. 2d). Similar results were obtained in Pacs-2−/− thymocytes where acetylation of p53 Lys379 (equivalent to human p53 Lys382) was reduced 60% while p53 pSer18 (equivalent to human p53 Ser15) was unaffected (Fig. 2a). Additionally, p53 acetylation and p21 induction were blunted in PACS-2 knockdown cells following treatment with the MDM2 antagonist, Nutlin-3a, which triggers p53-dependent induction of p21 independent of DNA damage (Fig. S3 and (Kumamoto et al., 2008)). Together, these results suggest PACS-2 mediates induction of a subset of p53 target genes by controlling p53 acetylation.
PACS-2 controls the extent of acetylated p53 bound to the p21 promoter following DNA damage
p53 acetylation is a crucial regulatory element controlling transactivation of p53 target genes. Therefore, we performed chromatin immunoprecipitation (ChIP) to analyze the amount of total and acetylated p53 bound to the p21 promoter following DNA damage (Fig. 2e). Indeed, PACS-2 knockdown reduced the extent of acetylated p53, but not total p53, bound to the p21 promoter following DNA damage. These findings suggest PACS-2 loss does not impede binding of p53 to target DNA sequences, but instead diminishes the extent to which promoter-bound p53 is acetylated and, therefore, transcriptionally active.
PACS-2 traffics to the nucleus and interacts with SIRT1
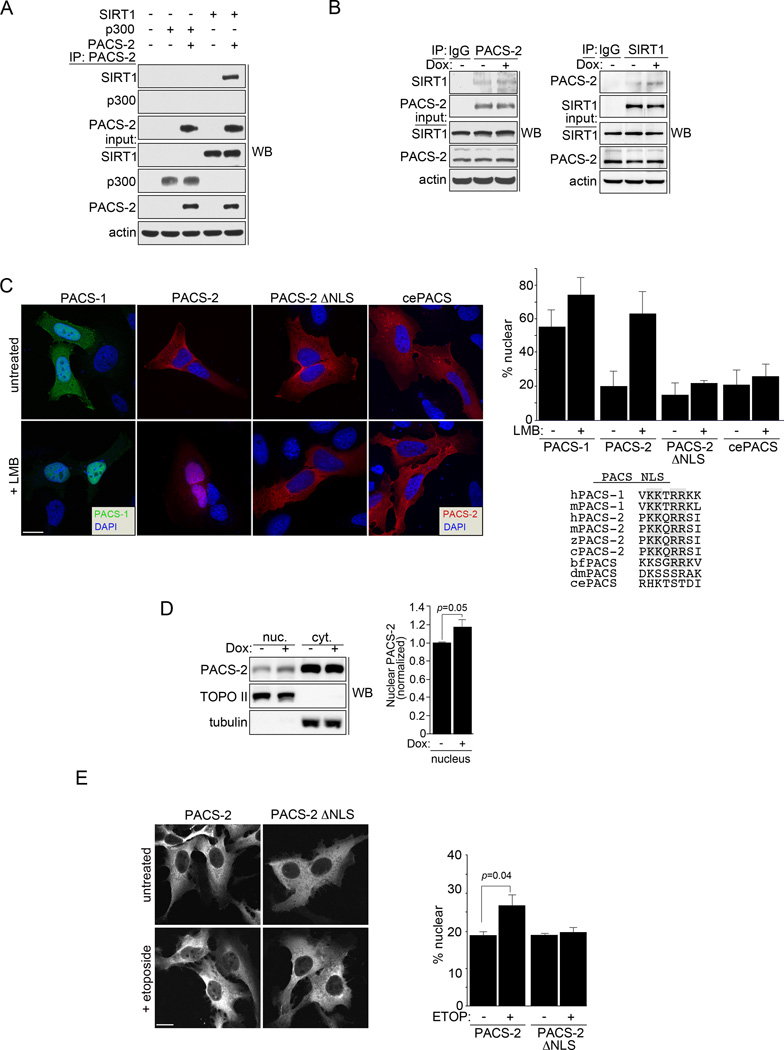
The determination that loss of PACS-2 reduced the level of acetylated p53 bound to the p21 promoter led us to ask whether PACS-2 modulates p53 acetylation by interacting with p300 or SIRT1. Co-immunoprecipitation analyses demonstrated that PACS-2 interacted with SIRT1 and that the endogenous SIRT1-PACS-2 interaction was increased following Dox treatment (Figs. 3a and b).
Figure 3. PACS-2 interacts with SIRT1 in the nucleus.
(A) PACS-2-FLAG was immunoprecipitated from HCT116 cells and co-precipitating SIRT1-V5 or p300-HA was detected by western blot. (B) HCT116 cells treated with 0.5 µM Dox (3hr) as indicated were immunoprecipitated with control IgG, anti-PACS-2 (top) or anti-SIRT1 (bottom) and co-precipitating SIRT1 (top) or PACS-2 (bottom) detected by western blot. (C) Left: U2OS cells expressing eGFP-PACS-1, mCherry-PACS-2, mCherry-PACS-2ΔNLS or mcherry C. elegans PACS (T18H9.7a) were treated with 40 nM LMB (3hr) and analyzed by deconvolution microscopy. Nuclei were stained with DAPI. Top right: Percent total cellular fluorescent protein signal in the nucleus was quantified. Error bars represent mean ± SD from >100 cells in 3 independent experiments. Bottom right: Alignment of the predicted NLS motif (https://www.predictprotein.org) fitting the consensus [PLV]K[RK]x[RK][RK][RK][PL] from human PACS-1 (sp|Q6VY07, gi|30089916) with human PACS-2 (sp|Q86VP3, gi|155029546), mouse PACS-1 (gi|54291704), mouse PACS-2 (kkqrrsiv, gi|124487181), chicken PACS-2 (gi|513196172), zebrafish PACS-2 (gi|170172595), amphioxous PACS (gi|229298623), Drosophila PACS (Krt95D, gi|24649488) and C. elegans PACS (T18H9.7a, gi|373219078). Consensus basic amino acid doublets are shaded. (D) HCT116 cells were treated or not with 0.5 µM Dox for 6 hr. PACS-2 in the nuclear and cytoplasmic fractions was detected by western blot. Error bars represent mean ± SD from 3 independent experiments. Statistical significance was determined using Student’s t-test. (E) U2OS cells expressing mCherry-PACS-2 or mCherry-PACS-2ΔNLS were treated with 100 µM Etoposide (3hr) and analyzed by confocal microscopy. Nuclei stained with DAPI. Bottom: Percent total cellular fluorescent protein signal in the nucleus was quantified. Error bars represent mean ± SD from >100 cells in 3 independent experiments. Scale bar, 10 µm.
Whereas SIRT1 deacetylates histones and transcription factors in the nucleus, PACS function has thus far been studied solely in the regulation of membrane traffic in the cytoplasm (Brooks and Gu, 2009; Youker et al., 2009). Inspection of the PACS-2 sequence, however, revealed multiple candidate leucine-rich nuclear export signals (NES, see Fig. 7a and (la Cour et al., 2004; Youker et al., 2009)). Indeed, treatment with the nuclear export inhibitor Leptomycin B (LMB) led to nuclear accumulation of PACS-2 (Fig. 3c). Whereas PACS-2 lacks a predicted nuclear localization signal (NLS), the PACS-2 paralogue, PACS-1, contains a canonical NLS (‥VKKTRRKL318‥) and readily distributes between the cytoplasm and nucleus (Fig. 3c and see Discussion). The homologous sequence in PACS-2 (‥PKKQRRSI239‥), when mutated (RR237→AA, hereafter called PACS-2ΔNLS), was unaffected by LMB, suggesting this motif is required for nuclear localization of PACS-2 (Fig. 3c). PACS sequence alignment suggests nuclear trafficking motifs appeared later in evolution, as C. elegans PACS lacks a predicted NLS and failed to accumulate in the nucleus (Fig. 3c). Treatment with Dox or Etoposide increased nuclear localization of PACS-2, suggesting a fraction of PACS-2 enters the nucleus to regulate SIRT1 following DNA damage (Figs. 3d and e). The reduced nuclear accumulation of mcherry-PACS-2 in response to DNA damage compared to LMB may reflect differences in the signaling pathways that control nuclear accumulation in response to DNA damage compared to a pharmacologic block of CRM1-dependent nuclear export.
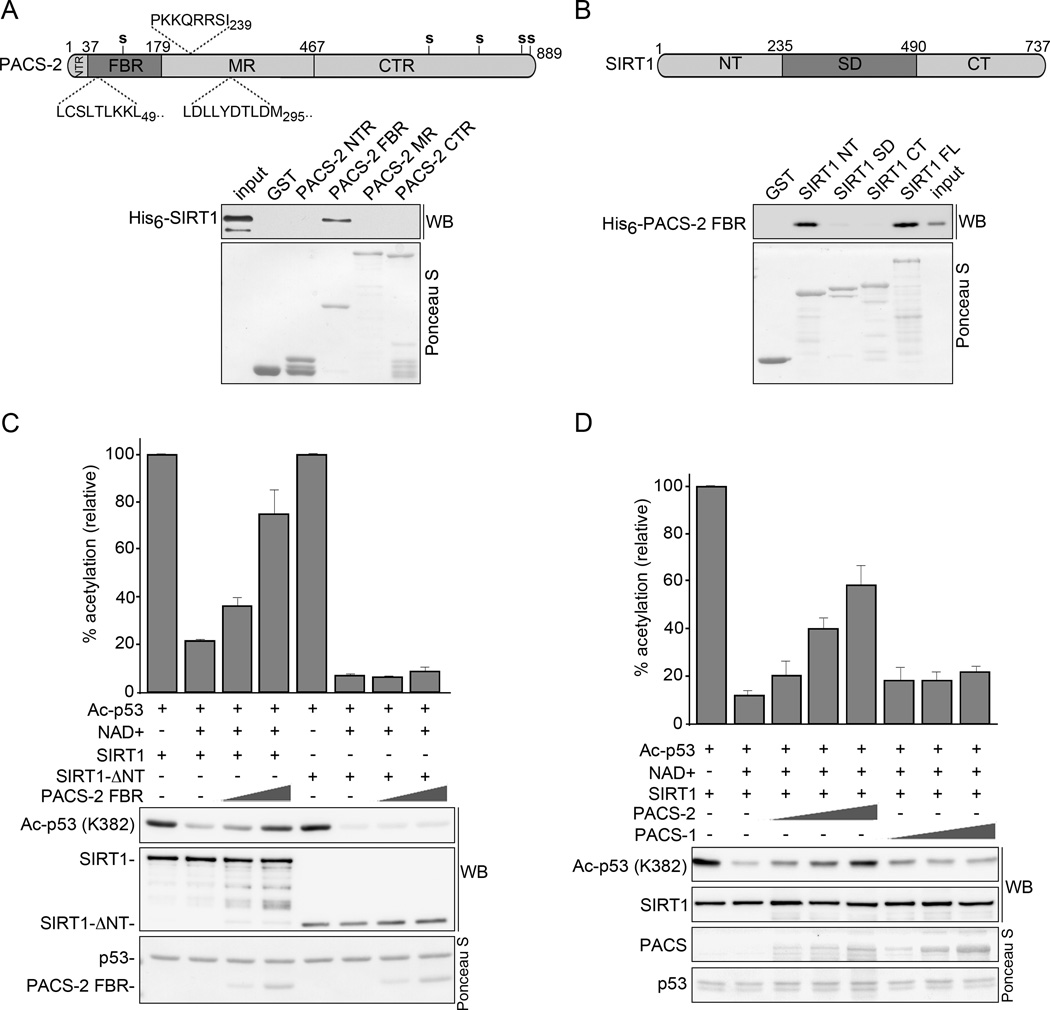
Figure 7. PACS-2 inhibits SIRT1-mediated deacetylation of p53 in vitro.
(A) Top: Schematic of hPACS-2 and the predicted NLS (above) and NES (below). Bottom: Full length His6-SIRT1Δ6–83 was incubated with GST or GST-PACS-2 constructs and interacting His6-SIRT1Δ6–83 was detected by western blot. Captured GST fusion proteins detected with Ponceau S. (B) Top: Schematic of mSIRT1. Bottom: His6-PACS-2FBR was incubated with GST or GST-SIRT1 constructs and interacting His6-PACS-2FBR was detected by western blot as in (A). (C) GST-Ac-p53 (see Methods) was incubated with 0.5 µM NAD+ and His6-tagged SIRT1 or SIRT1-ΔNT together with increasing amounts of His6-PACS-2FBR as indicated. The amount of Ac-p53 (K382) was quantified and presented as percent acetylation. Error bars represent mean ± SEM from 4 independent experiments. (D) GST- Ac-p53 was incubated with 0.5 µM NAD+ and His6-SIRT1 together with increasing amounts of His6-PACS-1 or PACS-2 as indicated. The amount of Ac-p53 (K382) was quantified as in (C). See also Figure S4.
PACS-2 regulates SIRT1-mediated deacetylation of p53 to induce p21-dependent cell cycle arrest
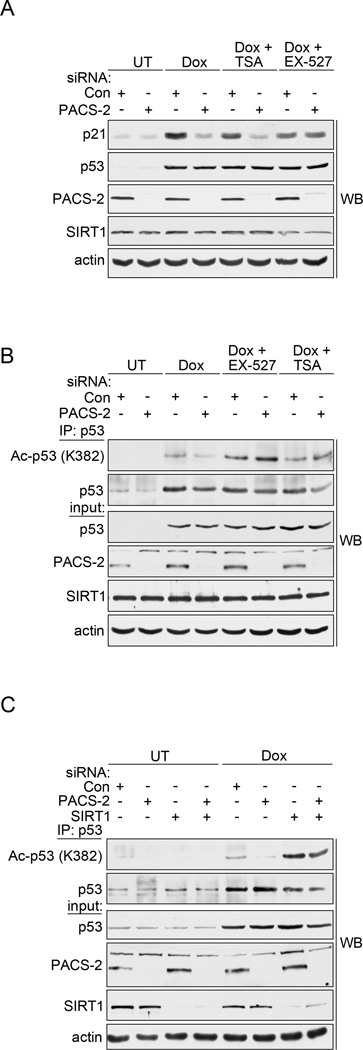
The reduced acetylation of p53 in PACS-2-deficient cells following DNA damage together with the DNA damage-induced interaction between PACS-2 and SIRT1 (Figs. 1–3), led us to ask whether PACS-2 mediates p21 induction by regulating SIRT1-dependent deacetylation of p53. Accordingly, control or PACS-2 knockdown HCT116 cells were treated with Dox in the absence or presence of the class I/II HDAC inhibitor TSA, which does not inhibit the class III HDAC SIRT1, or with the SIRT1 specific inhibitor EX-527 (Napper et al., 2005; Solomon et al., 2006). As expected (see Figs. 1 and 2), PACS-2 knockdown reduced Dox-mediated induction of p21 and this effect was abrogated by EX-527 but not TSA (Fig. 4a), suggesting the reduced p21 expression in PACS-2-depleted cells was SIRT1-dependent. Moreover, in agreement with others, the reduced p53 acetylation in PACS-2-depleted cells was reversed by EX-527 or the concomitant knockdown of SIRT1 (Figs. 4b and c and see (Cheng et al., 2003; Solomon et al., 2006)). The molecular basis underlying the ability of TSA to rescue p53 acetylation but not p21 induction is not known, but is consistent with previous studies (Cheng et al., 2003).
Figure 4. PACS-2 regulates SIRT1-mediated deacetylation of p53 in vivo.
(A) Control (Con) or PACS-2 siRNA-treated HCT116 cells were treated with 0.5 µM Dox (6hr) alone or with 0.5 µM TSA or 1 µM EX-527 and analyzed by western blot. (B) Control (Con) or PACS-2 siRNA-treated HCT116 cells were treated as in (A), total p53 immunoprecipitated with anti-p53 (DO-1) and Ac-p53 (K382) or total p53 (FL-393) detected by western blot. (C) HCT116 cells transfected with control (Con), PACS-2 or SIRT1 siRNAs were processed as in (B).
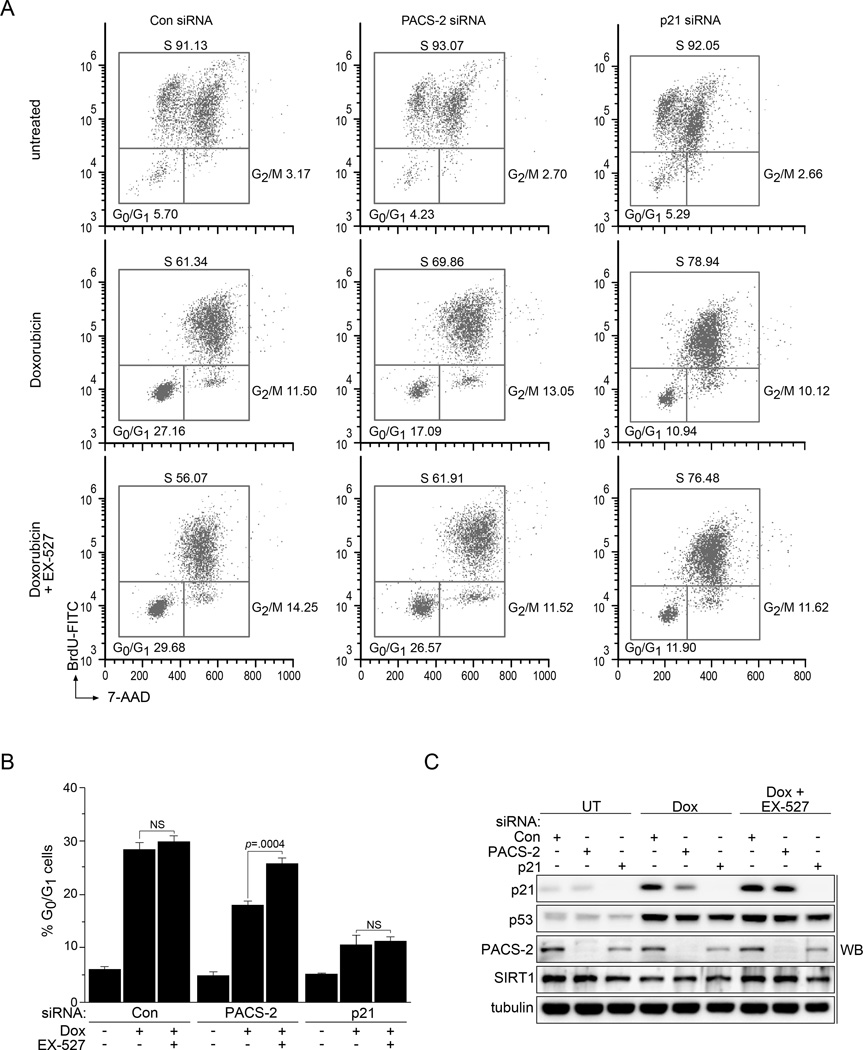
The SIRT1-dependent repression of p53-acetylation and p21 expression in PACS-2 knockdown cells suggested the subsequent reduction in p21-dependent cell cycle arrest (see Fig. 1e) might similarly be SIRT1-dependent. Indeed, treatment of PACS-2 knockdown cells with EX-527 restored p21-dependent cell cycle arrest and p21 expression (Figs. 5a–c and Table S2). Importantly, the effect of SIRT1 inhibition was dependent on PACS-2 knockdown, as both the extent of p21 induction and G1 arrest were not significantly affected by EX-527 in control cells. Moreover, EX-527 failed to increase the extent of G1 arrest in p21 knockdown cells, demonstrating the cell cycle arrest rescued by the SIRT1 inhibitor in PACS-2 knockdown cells was p21 dependent. Together, these data suggest PACS-2 regulates the ability of SIRT1 to deacetylate p53 following DNA damage, thereby promoting p21 induction and p21-dependent cell cycle arrest.
Figure 5. Inhibition of SIRT1 restores p21-dependent cell cycle arrest in PACS-2 knockdown cells.
(A) Synchronized control (Con), PACS-2 or p21 siRNA-treated HCT116 cells were stimulated to re-enter the cell cycle with 10% FBS in the presence of 10 µM BrdU and then treated or not with 0.5 µM Dox (24hr) in the presence or absence of 10 µM EX-527 and processed for flow cytometry. Results are representative of 3 independent experiments. (B) Quantitation of cells from panel A arrested in G0/G1. Error bars represent mean ± SEM from 3 independent experiments. Statistical significance determined using Student’s t-test. (C) Lysates from panel A were analyzed for the indicated proteins by western blot. NS, not significant. See also Table S2.
PACS-2 inhibits SIRT1-mediated deacetylation of p53
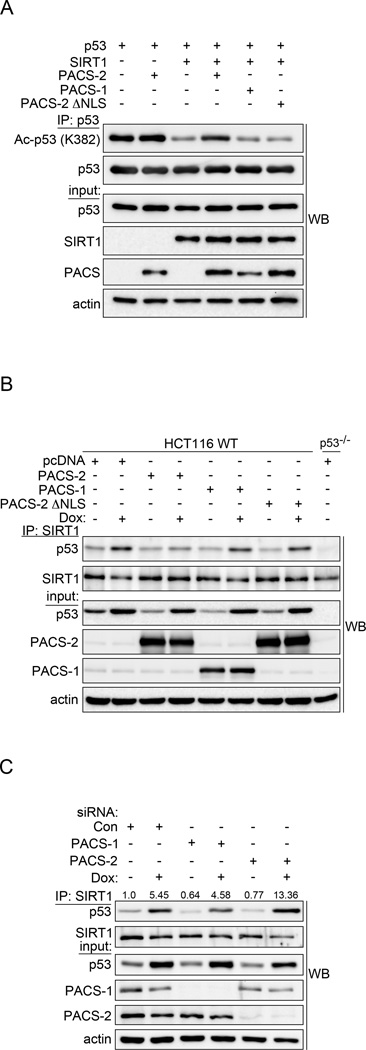
The SIRT1-dependent reduction of acetylated p53 and thus p21 expression in PACS-2 deficient cells suggested PACS-2 promotes p53 activation by regulating SIRT1 deacetylase activity. Consistent with this possibility, PACS-2, but not PACS-2ΔNLS or PACS-1, inhibited SIRT1-dependent p53 deacetylation (Fig. 6a). Interestingly, PACS-2, but not PACS-2ΔNLS or PACS-1, blocked the DNA damage-induced interaction between endogenous SIRT1 and p53 (Fig. 6b) while knockdown of PACS-2 increased the interaction (Fig. 6c). Together, these studies suggest that following DNA damage PACS-2 localizes to the nucleus where it regulates the interaction between SIRT1 and p53 to promote p53 acetylation.
Figure 6. Nuclear PACS-2 regulates the p53-SIRT1 interaction.
(A) Total p53 was immunoprecipitated from HCT116 cells and Ac-p53 (K382) analyzed by western blot. (B) Endogenous SIRT1 was immunoprecipitated from HCT116 WT or p53−/− cells treated with 0.5 µM Dox (6hr) as indicated and co-precipitating p53 analyzed by western blot. (C) HCT116 cells were treated with 0.5 µM Dox (14hr), endogenous SIRT1 immunoprecipitated and co-precipitating p53 analyzed by western blot. p53 was quantified using AlphaView (Protein Simple).
Next, we tested whether PACS-2 could bind SIRT1 and directly inhibit SIRT1-mediated deacetylation of p53 in vitro. We found that SIRT1 bound specifically to the PACS-2 cargo(furin)-binding region (PACS-2FBR, Fig. 7a). Conversely, the PACS-2FBR preferentially bound the SIRT1 N-terminal region (NT, Fig. 7b), but interacted weakly with a catalytically active SIRT1 construct lacking the NT ((SIRT1-ΔNT, Fig. S4a and see Methods). Importantly, PACS-2FBR inhibited the ability of full-length SIRT1, but not SIRT1-ΔNT, to deacetylate p53 in an NAD+- and dose-dependent manner (Fig. 7c). Moreover, full-length PACS-2 bound SIRT1, but not the p53 substrate, and inhibited the ability of SIRT1 to deacetylate p53 in vitro (Figs. 7d, S4b and S4c). By contrast, full-length PACS-1, which also bound SIRT1, had no measurable effect on SIRT1 activity (see Fig. 6). Together, these results suggest PACS-2 regulates p53-mediated p21 induction following DNA damage by controlling SIRT1-mediated deacetylation of p53.
DISCUSSION
The balance between cytostasis and apoptosis is a key determinant of cell fate following DNA damage. The selective repression of p53-mediated p21 induction shifts the DNA damage response from cytostasis to PUMA-dependent apoptosis (Haupt et al., 2003; Seoane et al., 2002). Whereas the most well-characterized role of p21 is the execution of p53-dependent cell cycle arrest, p21 also directly antagonizes PUMA- and Bax-mediated apoptosis by preventing procaspase-3 activation and by repressing procaspase-2 expression (Baptiste-Okoh et al., 2008; Hill et al., 2011; Suzuki et al., 2000; Yu et al., 2003). Thus, our determination that following DNA damage PACS-2 knockdown cells have repressed induction of p21 but not PUMA or Bax, as well as increased apoptosis, suggests PACS-2 inhibits SIRT1 to selectively promote p21-dependent cytoprotective functions of the p53-dependent DNA damage response.
SIRT1 is frequently reported to delay aging by promoting cell survival (Brooks and Gu, 2009; Cheng et al., 2003; Cohen et al., 2004). Thus, our determination that PACS-2 knockdown leads to a SIRT1-dependent reduction in p53-acetylation and p21 expression as well as sensitization of cells to DNA damage-induced apoptosis appears inconsistent with a cytoprotective role for SIRT1. The well-established anti-aging roles for SIRT1, however, may not necessarily correlate with a cytoprotective role following DNA damage. Indeed, deletion of p21 improves stem cell maintenance and increases lifespan whereas induction of p21 protects adult stem cells from acute genotoxic stress but impairs stem cell survival in the context of age-associated chronic DNA damage (Ju et al., 2007; Mantel and Broxmeyer, 2008; Sperka et al., 2012). To what extent these findings in stem cells extend to cancer cells warrants investigation.
The role of SIRT1 in tumor cell survival and cancer progression is controversial as SIRT1 has both tumor promoting and tumor suppressive functions (Herranz and Serrano, 2010; Roth and Chen, 2013). These opposing roles for SIRT1 are likely manifest through extensive SIRT1 substrates together with the ability of SIRT1 to differentially affect the cellular response to chemotherapeutics. For example, in cells exposed to high concentrations of chemotherapeutic, overexpressed SIRT1 represses induction of p53 target genes and reduces apoptosis (Luo et al., 2001; Vaziri et al., 2001). By contrast, and similar to our findings reported here, viability of HCT116 cells and other cell lines exposed to therapeutic doses of Doxorubicin is reduced by SIRT1 overexpression or treatment with SIRT1 activators but is increased by addition of EX-527 (Shin et al., 2014). Together, these findings suggest the roles of SIRT1 in the cellular response to DNA damage are complex and manifest in a dose- and context-dependent manner.
In contrast to other sirtuin family members, SIRT1 additionally possesses N- and C-terminal extensions containing autoregulatory domains and binding sites for SIRT1 modulators (Hasegawa and Yoshikawa, 2008; Kang et al., 2011; Kim et al., 2007; Kim et al., 2008; Liu et al., 2011; Zhao et al., 2008). The SIRT1 C-terminal extension contributes to the Km for NAD+ and contains an autoregulatory segment that binds the core domain, which is required for catalytic activity (Kang et al., 2011; Pan et al., 2012). The SIRT1 N-terminal extension contributes to Kcat and regulates allosteric binding of sirtuin-activating compounds ((STACs), (Hubbard et al., 2013)). Like PACS-2, the SIRT1 activator AROS binds the SIRT1 N-terminal region but has an opposing role on p53 regulation. Whereas PACS-2 inhibits SIRT1 to increase p53 acetylation, p21 induction and p21-dependent cell cycle arrest, AROS activates SIRT1 to decrease p53 deacetylation and consequently blunt p21 induction and cell cycle arrest (Kim et al., 2007). By contrast, DBC1 binds the SIRT1 catalytic (sirtuin) domain and, unlike PACS-2, siRNA knockdown of this inhibitor blunts the induction of several proapoptotic p53 targets, including PUMA, Bax, and Bim, and paradoxically increases in the cell cycle inhibitor GADD45a (Kim et al., 2008; Yuan et al., 2012; Zhao et al., 2008). Correspondingly, DBC1 knockdown reduces apoptosis, albeit with high concentrations of chemotherapeutic. However, the extent to which the cytoprotective effects of DBC1 knockdown following DNA damage result from increased SIRT1 versus the selective reduction of proapoptotic genes warrants further investigation. Moreover, the blunted acetylation of p53 in Pacs-2−/− thymocytes following DNA damage despite similar expression of DBC1 (Fig. 2a), suggests PACS-2 and DBC1 regulate SIRT1 independently and have dynamic and concerted roles in resolving the DNA damage response.
Acetylation-dependent transactivation of p53 involves a C-terminal cluster of six lysine residues and three additional sites within the core domain (Brooks and Gu, 2011). p300 acetylates the p53 C-terminal cluster, including Lys382, preventing MDM2-mediated ubiquitiylation and promoting p53 transactivation (Lee et al., 2010; Luo et al., 2004; Tang et al., 2008; Wang et al., 2008). However, the PACS-2 mediated acetylation of p53 Lys382 alone cannot explain the requirement for PACS-2 in the expression of a subset of p53 target genes. Interestingly, the DNA damage-mediated induction of p21 but not Bax additionally requires recruitment of BRCA1 and CARM1, which methylates p300 to stabilize the transcriptional complex (Lee et al., 2011). In contrast to p21, the p53-dependent induction of proapoptotic PUMA requires Tip60 or MOF-mediated acetylation of Lys120 (Sykes et al., 2006; Tang et al., 2006). As SIRT1 also regulates these two acetyltransferases, it will be important to determine whether PACS-2 regulates the action of SIRT1 against one or both enzymes or whether PACS-2 mediates recruitment of BRCA1/CARM1 to induce p21 (Peng et al., 2012).
Cytoplasmic PACS-2 mediates MAM formation, autophagy and protein traffic in the secretory and endocytic pathways (Atkins et al., 2008; Betz et al., 2013; Dikeakos et al., 2012; Hamasaki et al., 2013; Kottgen et al., 2005; Simmen et al., 2005). In response to TRAIL, PACS-2 switches to a proapoptotic effector independent of p53 status that coordinates trafficking steps leading to Bim- and Bid-dependent lysosomal and mitochondria membrane permeabilization, respectively, to trigger activation of executioner caspases (Aslan et al., 2009; Werneburg et al., 2012). It was therefore surprising that PACS-2 has seemingly opposing roles in TRAIL- versus DNA damage-induced apoptosis where in response to DNA damage, nuclear PACS-2 promotes p53 transactivation to induce p21-dependent cell cycle arrest (Figs. 4–6). Recent studies, however, suggest TRAIL selectively kills arrested cells (Ehrhardt et al., 2013). Thus, PACS-2-mediated cell cycle arrest may enhance PACS-2-dependent TRAIL-induced apoptosis. Therefore, it will be important to determine the stoichiometry of nuclear PACS-2 and SIRT1 that promotes p21 induction as well as the extent to which SIRT1 interaction involves pre-existing nuclear PACS-2 versus PACS-2 translocated from the cytoplasm in response to DNA damage.
Our data identify PACS-2 as a new member of an emerging collection of proteins with established roles in regulating both secretory pathway traffic and nuclear gene expression (Copley, 2012). Phylogenetic studies indicate the PACS genes appeared first in metazoans (Youker et al., 2009). While lower metazoans express a single PACS gene, separate PACS-1 and PACS-2 genes are predicted to have arisen in higher metazoans by gene duplication. Sequence alignment suggests PACS-1 and PACS-2 underwent evolutionary adaptation with the acquisition of nuclear trafficking motifs (see Fig. 3c). Consistent with this model, C. elegans PACS failed to accumulate in the nucleus in LMB-treated cells (Fig. 3c). This evolutionary acquisition of nuclear trafficking functions parallels the role of p53 in directing cell cycle arrest. p53 was found to promote apoptosis in worms and f lies, but in higher metazoans was found to additionally induce p21-dependent cell cycle arrest (Lu et al., 2009). These findings raise the possibility that the ability of PACS-2 to traffic to the nucleus resulted from evolutionary adaptation required to support the more complex demands of p53 to resolve the DNA damage response.
In summary, our data demonstrate that the multi-functional protein PACS-2 shuttles to the nucleus and directly inhibits SIRT1-mediated deacetylation of p53, thereby controlling the SIRT1—p53—p21 axis to modulate the DNA damage response. Our findings, together with the broad and important role of SIRT1 in controlling pathways associated with cancer, aging and metabolism, suggest a more complete understanding of the regulation of SIRT1 is warranted to better understand the complex role of this deacetylase in controlling homeostasis and disease.
EXPERIMENTAL PROCEDURES
Experimental animals
The OHSU Department of Comparative Medicine approved all animal studies. Pacs-2−/− mice (Aslan et al., 2009) were maintained on a C57BL/6 background with >10 backcrosses. Mice were irradiated using a J.L. Shepherd Mark I Model 30 Cesium irradiator (1.475 Gy/min) in a plexiglass pieplate rotating at 7 RPM. Where indicated, WT and Pacs-2−/− mice were injected with 50 mg/kg BrdU and immediately exposed to 15 Gy WBI. At 48hr following labeling, a 3-cm segment of ileum just proximal to the cecum was processed for IHC by the OHSU Histopathology Core.
Antibodies, chemicals, plasmids, siRNAs
Cell harvest and tissue processing
HCT116 cell lines (provided with permission from B. Vogelstein), U2OS and 293T cells were cultured in DMEM + 10% FBS. For most experiments, cells were harvested in mRIPA (50 mM Tris pH 8.0 + 1% NP-40, 1% deoxycholate, 150 mM NaCl, protease inhibitors (0.5 mM PMSF, 0.1 µM each of aprotinin, E-64, and leupeptin) and phosphatase inhibitors (1 mM Na3VO4, 20 mM NaF). For monitoring Ac-p53, cells were harvested in Ac Lysis Buffer (20 mM Tris pH 7.6 + 0.5% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 5 µM TSA, protease inhibitors and phosphatase inhibitors). Mouse tissues were homogenized in RIPA [50 mM Tris pH 8.0 + 150 mM NaCl, 1% NP-40, 1% deoxycholate, 0.1% SDS, and protease/phosphatase inhibitors] using a motorized teflon-glass homogenizer. Protein concentration was determined according to manufacturer’s instructions (#500 – 0006; Bio-Rad).
Flow cytometry analyses
Flow cytometry was performed on FACSCalibur or Accuri C6 (BD) flow cytometers. Data were analyzed using FCS Express (v 3.0). Cell cycle analysis: HCT116 cells were nucleofected (Amaxa) on days 0 and 2 with the indicated siRNAs. On day 4 the nucleofected cells were synchronized by serum deprivation (0.1% FBS) for 24 hr and stimulated to re-enter the cell cycle by addition of 10% FBS in the presence of 10 µM BrdU and treated or not with 0.5 µM Dox or 10 µM EX-527 for an additional 24 hr as indicated. EX-527 was used in these experiments because it could selectively inhibit SIRT1 following serum deprivation, thereby avoiding nutrient-affected changes in SIRT1 prior to DNA damage (Nemoto et al., 2004). Cells were processed according to manufacturer’s instructions (#559619BD; Pharmingen) and flow cytometry performed as above.
qRT-PCR analysis
Cell culture studies performed using sub-confluent HCT116 cells, synchronized with 1% serum transfected with siRNAs and treated with serum in the absence or presence of Dox as indicated. RNA was isolated according to manufacturer’s instructions using Trizol (#15596-026; Sigma) for mouse tissues and RNeasy (#74104; Qiagen) for cell culture. RNA was reverse transcribed using Superscript III first strand cDNA synthesis kit (#18080-051; Invitrogen). Reactions were performed in an ABI StepOne Real-Time PCR System with the Power SYBR GREEN PCR Master Mix (#4368706; Applied Biosystems) using the primer pairs listed in Supplemental Procedures. All reactions were performed in triplicate.
ChIP analysis
ChIP experiments were performed according to (Mahata et al., 2012) with modifications as detailed in Supplementary Procedures.
Baculovirus
Full-length hPACS-1 and hPACS-2 coding sequences containing a C-terminal His-tag were subcloned into pVL1392 (BD Biosciences) to create high-titer recombinant baculoviruses (BV:His-PACS-2) in Sf9 insect cells according to the manufacturer’s protocol (BD Biosciences). Sf9 cells were cultured for 72hr with BV:His-PACS-2 or BV:His-PACS-1 and dounce homogenized in harvest buffer (50mM NaHPO4 pH 7.4, 300 mM NaCl). Lysates were clarified by centrifugation (20,000 RPM, 20 min). His-tagged proteins were purified using Ni2+-NTA agarose (Qiagen) and dialyzed against PBS.
In vitro binding
GST proteins were pre-incubated with glutathione-Sepharose 4B (GE Healthcare) in binding buffer (20 mM Tris pH 7.9, 150 mM NaCl, 0.1% NP-40, 0.1 mM EDTA, 0.3 mM DTT), mixed with His6-constructs (30 min, RT), washed 3× (20 mM Tris pH 7.9, 300 mM NaCl, 0.1% NP-40, 0.1 mM EDTA, 3 mM DTT), and analyzed by western blot using anti-His6. For SIRT1 and full-length PACS interactions, His6-PACS-1 or PACS-2 expressed in baculovirus-infected Sf9 cells was mixed with GST or GSTSIRT11–737 pre-incubated with glutathione sepharose 4B and processed as above.
In vitro deacetylase assays
Substrate preparation and in vitro deacetylation assays were performed as described (Aslan et al., 2009; Kang et al., 2009; Simmen et al., 2005). Briefly, purified GST-hp53373–385 was acetylated by His6-PCAF352–832 in HAT buffer [50mM Tris pH 8.0, 10% glycerol, 0.1 mM EDTA, 1.0 M DTT] with 5 mM acetyl CoA (Millipore). For assays with full-length PACS proteins, purified His6-mSIRT1 was pre-incubated with GST-Ac-p53 and full-length His6-PACS-1 or PACS-2 in deacetylase buffer [50 mM Tris pH 8.0 + 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, and protease inhibitors] for 30 min. Assays using PACS-2FBR were conducted in binding buffer (above). Reactions were initiated with 0.5 mM NAD+ (Millipore) and run for 1 hr to overnight at 37° C.
Microscopy
U2OS cells were transfected (Fugene 6) with the indicated plasmids and images were acquired on a high-resolution, wide-field Core Deltavision system (Applied Precision) as described (Dikeakos et al., 2012). Nuclear and cytoplasmic fluorescent signal was determined using softWoRx Explorer 2.0 (Applied Precision) also as described (Dikeakos et al., 2012). Briefly, for each cell counted, the mcherry or DAPI fluorescent signal was quantified from a precise region of interest over the nucleus, cytoplasm and extracellular space. The fluorescent signal in each channel was automatically captured and the signal from the extracellular space was subtracted from the nuclear and cytoplasmic signals. The nuclear signal was then divided by the total (nuclear + cytoplasmic) signal. Statistical significance was determined using a two-sided unpaired Student’s t test with unequal variance.
Nuclear fractionation
Cells were harvested in Buffer 1 [50 mM Tris pH 7.9 + 10 mM KCl, 1 mM EDTA, 0.2% NP-40, 10% glycerol, and protease inhibitors] and centrifuged (6K RPM, 3 min, 4°C). The nuclear pellet lysed with Buffer 2 [20 mM Hepes pH 7.9 + 400 mM NaCl, 10 mM KCl, 1% NP-40, 20% glycerol, 1 mM EDTA, and protease inhibitors] for 20 min at 4°C, then centrifuged (13.2K RPM, 10 min). Soluble nuclear and cytoplasmic fractions were immunoprecipitated with anti-FLAG-agarose or anti-SIRT1 (H-300) antibodies, immune complexes captured with Protein G Sepharose (Invitrogen) for 2hr at 4°C, washed 3× in Wash Buffer [20 mM Tris pH 7.6 + 0.5% NP-40, 150 mM NaCl, 1 mM EDTA] and eluted in SDS sample buffer prior to western blot.
Supplementary Material
HIGHLIGHTS.
PACS-2 is required for p53- and p21-dependent cell cycle arrest
PACS-2 modulates DNA damage-induced p53 acetylation to control p21 induction
PACS-2 traffics to the nucleus where it regulates SIRT1-mediated p53 deacetylation
PACS-2 binds SIRT1 in vitro and directly inhibits SIRT1-catalyzed p53 deacetylation
ACKNOWLEDGEMENTS
The authors thank B. Vogelstein, L. Zhang, J. Yu, D. Sieburth, G.V. Thomas, P. Stork, and J. Denu for reagents and J. Yu, M. Dai, V. Procaccia, T. Smithgall, J. A. Tanyi and J. Moroco for advice and assistance with experiments. This work was supported by NIH R01 CA151564 and awards from the Knight Cancer Institute and the University of Pittsburgh (to GT). KMA is a Rubinstein Radiation Research Scholar and was supported by RSNA RMS1112 and the N.L. Tartar Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
KMA, LLT, JB, LT, SA, JY, and JDD conceived and designed the experiments, KMA, LLT, JB, LT, SA and JY performed the experiments, KMA, LLT, JB, LT, SA, JY, HK, JHC and JDD analyzed the data, GT wrote the paper, KMA, LLT and JB edited the paper, LT assembled the artwork and all the authors approved the final version of the manuscript.
REFERENCES
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nature Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan JE, You H, Williamson DM, Endig J, Youker RT, Thomas L, Shu H, Du Y, Milewski RL, Brush MH, et al. Akt and 14-3-3 control a PACS-2 homeostatic switch that integrates membrane traffic with TRAIL-induced apoptosis. Mol Cell. 2009;34:497–509. doi: 10.1016/j.molcel.2009.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins KM, Thomas L, Youker RT, Harriff MJ, Pissani F, You H, Thomas G. HIV-1 Nef binds PACS-2 to assemble a multikinase cascade that triggers major histocompatibility complex class I (MHC-I) down-regulation: analysis using short interfering RNA and knock-out mice. J Biol Chem. 2008;283:11772–11784. doi: 10.1074/jbc.M707572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptiste-Okoh N, Barsotti AM, Prives C. Caspase 2 is both required for p53-mediated apoptosis and downregulated by p53 in a p21-dependent manner. Cell Cycle. 2008;7:1133–1138. doi: 10.4161/cc.7.9.5805. [DOI] [PubMed] [Google Scholar]
- Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. Feature Article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci USA. 2013;110:12526–12534. doi: 10.1073/pnas.1302455110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nature Rev Cancer. 2009;9:123–128. doi: 10.1038/nrc2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein & cell. 2011;2:456–462. doi: 10.1007/s13238-011-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- Copley SD. Moonlighting is mainstream: paradigm adjustment required. BioEssays. 2012;34:578–588. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]
- Dikeakos JD, Thomas L, Kwon G, Elferich J, Shinde U, Thomas G. An interdomain binding site on HIV-1 Nef interacts with PACS-1 and PACS-2 on endosomes to downregulate MHC-I. Mol Biol Cell. 2012;23:2184–2197. doi: 10.1091/mbc.E11-11-0928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt H, Wachter F, Grunert M, Jeremias I. Cell cycle-arrested tumor cells exhibit increased sensitivity towards TRAIL-induced apoptosis. Cell Death & Dis. 2013;4:e661. doi: 10.1038/cddis.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. J Neurosci. 2008;28:8772–8784. doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S, Louria-Hayon I, Haupt Y. P53 licensed to kill? Operating the assassin. J Cell Biochem. 2003;88:76–82. doi: 10.1002/jcb.10311. [DOI] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nature Rev Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill R, Madureira PA, Waisman DM, Lee PW. DNA-PKCS binding to p53 on the p21WAF1/CIP1 promoter blocks transcription resulting in cell death. Oncotarget. 2011;2:1094–1108. doi: 10.18632/oncotarget.378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hubbard BP, Gomes AP, Dai H, Li J, Case AW, Considine T, Riera TV, Lee JE, E SY, Lamming DW, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifeadi V, Garnett-Benson C. Sub-lethal irradiation of human colorectal tumor cells imparts enhanced and sustained susceptibility to multiple death receptor signaling pathways. PloS one. 2012;7:e31762. doi: 10.1371/journal.pone.0031762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Z, Choudhury AR, Rudolph KL. A dual role of p21 in stem cell aging. Annal New York Acad Sci. 2007;1100:333–344. doi: 10.1196/annals.1395.036. [DOI] [PubMed] [Google Scholar]
- Kang H, Jung JW, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PloS one. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Suh JY, Jung YS, Jung JW, Kim MK, Chung JH. Peptide switch is essential for Sirt1 deacetylase activity. Mol Cell. 2011;44:203–213. doi: 10.1016/j.molcel.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28:277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Komarova EA, Kondratov RV, Wang K, Christov K, Golovkina TV, Goldblum JR, Gudkov AV. Dual effect of p53 on radiation sensitivity in vivo: p53 promotes hematopoietic injury, but protects from gastro-intestinal syndrome in mice. Oncogene. 2004;23:3265–3271. doi: 10.1038/sj.onc.1207494. [DOI] [PubMed] [Google Scholar]
- Kottgen M, Benzing T, Simmen T, Tauber R, Buchholz B, Feliciangeli S, Huber TB, Schermer B, Kramer-Zucker A, Hopker K, et al. Trafficking of TRPP2 by PACS proteins represents a novel mechanism of ion channel regulation. EMBO J. 2005;24:705–716. doi: 10.1038/sj.emboj.7600566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto K, Spillare EA, Fujita K, Horikawa I, Yamashita T, Appella E, Nagashima M, Takenoshita S, Yokota J, Harris CC. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Ott M. The ups and downs of SIRT1. Trends Biochem Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S. Analysis and prediction of leucine-rich nuclear export signals. PEDS. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci USA. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Bedford MT, Stallcup MR. Regulated recruitment of tumor suppressor BRCA1 to the p21 gene by coactivator methylation. Genes & Dev. 2011;25:176–188. doi: 10.1101/gad.1975811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz BJ, Qiu W, Liu H, Cheng T, Zhang L, Yu J. Uncoupling p53 functions in radiation-induced intestinal damage via PUMA and p21. Molecular cancer research : MCR. 2011;9:616–625. doi: 10.1158/1541-7786.MCR-11-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hassan GS, Williams TM, Minetti C, Pestell RG, Tanowitz HB, Frank PG, Sotgia F, Lisanti MP. Loss of caveolin-1 causes the hyper-proliferation of intestinal crypt stem cells, with increased sensitivity to whole body gamma-radiation. Cell Cycle. 2005;4:1817–1825. doi: 10.4161/cc.4.12.2199. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang D, Zhao Y, Tu B, Zheng Z, Wang L, Wang H, Gu W, Roeder RG, Zhu WG. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proc Natl Acad Sci USA. 2011;108:1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WJ, Amatruda JF, Abrams JM. p53 ancestry: gazing through an evolutionary lens. Nature Rev Cancer. 2009;9:758–762. doi: 10.1038/nrc2732. [DOI] [PubMed] [Google Scholar]
- Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Mahata B, Sundqvist A, Xirodimas DP. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2012;31:3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- Mantel C, Broxmeyer HE. Sirtuin 1, stem cells, aging, and stem cell aging. Curr Opin Hematol. 2008;15:326–331. doi: 10.1097/MOH.0b013e3283043819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: implications for cancer therapy. J Biomed Biotech. 2012;2012:170325. doi: 10.1155/2012/170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Pan M, Yuan H, Brent M, Ding EC, Marmorstein R. SIRT1 contains N- and C-terminal regions that potentiate deacetylase activity. J Biol Chem. 2012;287:2468–2476. doi: 10.1074/jbc.M111.285031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Ling H, Yuan Z, Fang B, Bloom G, Fukasawa K, Koomen J, Chen J, Lane WS, Seto E. SIRT1 negatively regulates the activities, functions, and protein levels of hMOF and TIP60. Mol Cell Biol. 2012;32:2823–2836. doi: 10.1128/MCB.00496-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene. 2013 doi: 10.1038/onc.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J, Le HV, Massague J. Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature. 2002;419:729–734. doi: 10.1038/nature01119. [DOI] [PubMed] [Google Scholar]
- Shin DH, Choi YJ, Park JW. SIRT1 and AMPK mediate hypoxia-induced resistance of non-small cell lung cancers to cisplatin and doxorubicin. Cancer Res. 2014;74:298–308. doi: 10.1158/0008-5472.CAN-13-2620. [DOI] [PubMed] [Google Scholar]
- Simmen T, Aslan JE, Blagoveshchenskaya AD, Thomas L, Wan L, Xiang Y, Feliciangeli SF, Hung CH, Crump CM, Thomas G. PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 2005;24:717–729. doi: 10.1038/sj.emboj.7600559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JM, Pasupuleti R, Xu L, McDonagh T, Curtis R, DiStefano PS, Huber LJ. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka T, Wang J, Rudolph KL. DNA damage checkpoints in stem cells, ageing and cancer. Nature Rev Mol Cell Biol. 2012;13:579–590. doi: 10.1038/nrm3420. [DOI] [PubMed] [Google Scholar]
- Sullivan JM, Jeffords LB, Lee CL, Rodrigues R, Ma Y, Kirsch DG. p21 protects "Super p53" mice from the radiation-induced gastrointestinal syndrome. Rad Res. 2012;177:307–310. doi: 10.1667/rr2545.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Kawano H, Hayashida M, Hayasaki Y, Tsutomi Y, Akahane K. Procaspase 3/p21 complex formation to resist fas-mediated cell death is initiated as a result of the phosphorylation of p21 by protein kinase A. Cell Death Diff. 2000;7:721–728. doi: 10.1038/sj.cdd.4400706. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhao Y, Li L, McNutt MA, Wu L, Lu S, Yu Y, Zhou W, Feng J, Chai G, et al. An ATM- and Rad3-related (ATR) signaling pathway and a phosphorylation-acetylation cascade are involved in activation of p53/p21Waf1/Cip1 in response to 5-aza-2'-deoxycytidine treatment. J Biol Chem. 2008;283:2564–2574. doi: 10.1074/jbc.M702454200. [DOI] [PubMed] [Google Scholar]
- Werneburg NW, Bronk SF, Guicciardi ME, Thomas L, Dikeakos JD, Thomas G, Gores GJ. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) protein-induced lysosomal translocation of proapoptotic effectors is mediated by phosphofurin acidic cluster sorting protein-2 (PACS-2) J Biol Chem. 2012;287:24427–24437. doi: 10.1074/jbc.M112.342238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youker RT, Shinde U, Day R, Thomas G. At the crossroads of homoeostasis and disease: roles of the PACS proteins in membrane traffic and apoptosis. Biochem J. 2009;421:1–15. doi: 10.1042/BJ20081016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proc Natl Acad Sci USA. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Liu T, Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes & Dev. 2012;26:791–796. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilfou JT, Lowe SW. Tumor suppressive functions of p53. CSH Persp Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.