Abstract
Transcriptional repressor Snail is a master regulator of epithelial–mesenchymal transition (EMT), yet the epigenetic mechanism governing Snail to induce EMT is not well understood. Here, we report that in pancreatic ductal adenocarcinoma (PDAC), elevated levels of the ubiquitin E3 ligase Ring1B and Snail, along with elevated monoubiquitination of H2A at K119 (H2AK119Ub1), are highly correlated with poor survival. Mechanistic investigations identified Ring1B as a Snail-interacting protein and showed that the carboxyl zinc fingers of Snail recruit Ring1B and its paralog Ring1A to repress its target promoters. Simultaneous depletion of Ring1A and Ring1B in pancreatic cancer cells decreased Snail binding to the target chromatin, abolished H2AK119Ub1 modification, and thereby compromised Snail-mediated transcriptional repression and cell migration. We found that Ring1B and the SNAG-associated chromatin modifier EZH2 formed distinct protein complexes with Snail and that EZH2 was required for Snail-Ring1A/B recruitment to the target promoter. Collectively, our results unravel an epigenetic mechanism underlying transcriptional repression by Snail, suggest Ring1A/B as a candidate therapeutic target, and identify H2AK119Ub1 as a potential biomarker for PDAC diagnosis and prognosis. Cancer Res; 74(16); 4353-63. ©2014 AACR
Introduction
Snail is a member of SNAG domain containing zinc finger proteins and a master regulator of epithelial–mesenchymal transition (EMT) and metastasis in various tumor types (1–3). Snail can directly bind to the E-boxes of E-cadherin gene promoter to repress its transcription and convert normal epithelial cells into mesenchymal cell phenotype (4, 5). Mechanistically, Snail recruits multiple repressive protein complexes involving histone deacetylation and methylation as well as DNA methylation to its target promoters and exerts its repressive function (6–10). However, how these protein complexes are assembled at the target chromatin regions remains elusive.
Ring1A and Ring1B belong to the RING domain containing ubiquitin E3 ligase family and are crucial components of the polycomb repressive complex 1 (PRC1) by catalyzing monoubiquitination of histone H2A at lysine 119 (H2AK119Ub1; ref. 11). H2AK119Ub1 status is associated with gene silencing (12), chromatin remodeling (13), and X chromosome inactivation (14). Genetic disruption of Ring1B in mice causes embryonic lethality due to gastrulation arrest and defective mesoderm formation (15), which are reminiscent of the Snail null mice. Snail-deficient mouse embryos also die early in gestation, displaying defects in gastrulation and mesoderm formation (16, 17). The striking genetic evidence strongly indicates that Snail very likely correlates with Ring1B in regulation of gastrulation and mesoderm formation during embryo development.
A recent study found an elevated Ring1B expression in high-grade pancreatic ductal adenocarcinoma (PDAC; ref. 18); however, no biologic and mechanistic studies on Ring1B were performed. In this present study, we report that Ring1A and Ring1B interact with Snail and are important coregulators for Snail-mediated transcriptional repression and cell migration in pancreatic cancer cells. Snail enhances the binding of Ring1A and Ring1B to the promoter region of E-cadherin to increase H2AK119Ub1 at this locus, and an elevated Snail, Ring1B, and H2AK119Ub1 modification in PDAC is highly correlated with poor prognosis.
Materials and Methods
IHC and tissues microarray
IHC staining was performed as described (18) with specific antibodies against Snail, Ring1B, or H2AK119Ub1. Tissue microarray (TMA) chips that contain 90 cases of paired tumor and peri-tumor specimens were purchased (HPan-Ade180 Sur-01; ShGnghGi Outdo Biotech Company). All specimens spotted on TMA chips were well documented including complete postoperative fellow-ups for periods from 3 to 7 years. Tumor staging was evaluated according to the TNM Classification of Malignant Tumors. After IHC staining, all specimens were strictly evaluated by a senior pathologist and only those with a tumor content of more than 50% were considered as tumor samples and the peri-tissues showing sign of chronic pancreatitis were excluded, and total of 60 cases are qualified for further analysis. Peri-tumor tissues were defined as that more than 1 cm distant from the tumor edge. TMA chips were scanned by Aperio Scanscope XT, and the whole field of each tissue spot was obtained for IHC evaluation. The immunoreactive score system (IRS; refs. 19, 20) was used to evaluate the staining of each sample as follow: negative, 0 to 1 point; mild (+), 2 to 3 points; moderate (++), 4 to 8 points; strongly positive (+++), 9 to 12 points. IHC staining showing IRS scores of more than 4 points was considered as high.
Plasmids
Ring1A and Ring1B cDNAs and their mutants were cloned into pCMV4-Flag vector between Hind III and Eco RI sites. Snail and its mutants were cloned into pCMV5-HA vector between HindIII and XbaI sites. pLKO.1-shRNAs targeting Ring1A were ATAGATCTTAGAGATCAGGGC and ATCGTTGTGGTCTGA-TCTGAC; targeting Ring1B were ATTGTGCTTGTTGAT-CCTGGC and TTCTAAAGCTAACCTCACAGC, respectively. All point mutants were made using the QuikChange Site-Directed Mutagenesis procedures (Stratagene), and were confirmed by DNA sequencing.
Cell culture and transfections
HEK-293T cells and pancreatic cancer cells PanC1 and AsPC1 were obtained from the ATCC and were tested and authenticated by DNA typing at the Shanghai Jiao Tong University Analysis Core. The cells were maintained in DMEM supplemented with 10% FBS, 2 mmol/L l-glutamine, and penicillin (50 U/mL)/streptomycin (50 µg/mL) at 37°C under 5% CO2 in a humidified chamber.
Transfection of PanC1 and HEK-293T cells was performed using Lipofectamine 2000 as described (8). The viral supernatants were generated in HEK-293T cells, and were infected into PanC1 and AsPC1 cells. Puromycin was added into the media to generate stable knockdown of Ring1A and Ring1B in PanC1 and AsPC1 cells. FACS was performed to sort the cells stably expressing Flag-Snail.
Affinity purification of Snail-interacting protein complex
A Flag-tagged, full-length Snail cDNA in the pcDNA3.1-vector was stably expressed in HEK-293T cells. Single-cell clones were selected with G418 and screened by Western blot assays using anti-Flag antibody. The method used for affinity purification was previously described (8). A total of 5 × 109 cells were used for affinity purification, and the eluted proteins were resolved on 4% to 12% SDS-PAGE gels (Invitrogen) for Western blot and colloidal staining analyses. The proteins were excised from the gel and identified by standard mass spectrometry.
Coimmunoprecipitation, Western blot, immunofluorescence, and antibodies
Plasmids encoding Flag-Ring1A, Flag-Ring1B, hemagglutinin (HA)-Snail proteins were transiently expressed in HEK-293T cells, and 24 hours after transfection, cells were lysed in buffer containing 20 mmol/L Tris-HCl (pH 8.0), 150 mmol/L NaCl, 2.5 mmol/L EDTA, 0.5% NP40, 0.1 mmol/L phenylmethylsulfonylfluoride, and protease inhibitor cocktail. Method for total histones extraction was as described (12). The whole-cell extracts were precleared with protein A/G beads, and coimmunoprecipitation (co-IP) assays were performed with either Flag or HA antibodies. The methods used for Western blot and immunofluorescence were previously described (8). Antibodies for Flag (Sigma-Aldrich; F 7425), HA (COVANCE; MMS-101P), Ring1A, Ring1B, H2A, ubiquityl-Histone H2A-lys119 and E-cadherin (Cell Signaling Technology; #2820, #5694,#2578,#8240, #3195), Snail (Santa Cruz; sc-28199); and β-actin (Proteintech; 60008–1-Ig) were purchased.
Chromatin immunoprecipitation and qPCR
The chromatin immunoprecipitation (ChIP) experiments were carried out in PanC1 cells and derivatives. To prepare cells for ChIP assays, the PanC1 cells were grown in 10 cm plates to 70% to 90% confluency and were processed as described (8). The immunoprecipitated DNA fragments were detected by qPCR assays. The primer sets that amplify the DNA fragment flanking the known E-boxes in the E-cadherin promoter are as follows: forward, 5-GCAGGTGAACCCTCAGC-CAA-3; reverse, 5-CACAGGTGCTTTGCAGTTCC-3.
Total RNA was isolated from cells with TRIzol reagent (Invitrogen). qRT-PCR was performed on a 7500 Fast Realtime PCR system (Applied Biosystem) using SYBR Green agent. Primers used for qRT-PCR assay were listed in Supplementary information. All RT-PCR assays were repeated three times.
Transwell cell migration assays
PanC1 cells were harvested after serum-free starvation for 12 hours, and were resuspended in plain DMEM media. Ten thousand cells were applied to 8-µm pore transwell filters (Corning). DMEM media containing 10% FBS were added to the bottom chamber as attractants. After incubation for 24 hours, the nonmigrated cells at the top of the filter were removed and the migrated cells at the bottom of the filter were fixed with 4% paraformaldehyde and were stained with colloidal staining method. The number of migrating cells in each chamber was quantified by counting nine randomly chosen fields under ×20 magnification using a bright-field microscope. Each condition was performed in duplicate, and the average number of cells per field was represented. Experiments were repeated three times.
Statistical analysis
Data shown as mean ± SD were analyzed by the independent Student t test. The distribution of the IHC scoring results of each protein on TMA chips was analyzed by the McNemar test. The correlation between the expression of Snail and Ring1B in PDAC was analyzed by the Spearman rank correlation coefficient test. Spearman ρ are categorized as moderate to strong correlations according to Dancey and Reidy‧s categorization: 0 (zero); 0.1 to 0.3 (weak); 0.4 to 0.6 (moderate); 0.7 to 0.9 (strong); and 1 (perfect; ref. 21). The postoperative survival of patients with PDAC was analyzed by Kaplan–Meier estimator and tested by the log-rank. P value of <0.05 was considered statistically significant. Statistical analysis was performed using IBM SPSS Statistics Version 20 software.
Study approval
The study was approved by the Ethic and Research Committees of Ruijing Hospital, Shanghai Jiaotong University School of Medicine and was conducted in accordance with the Declaration of Helsinki Principles. The procedures for pancreatic tumor resection were described in detail to all patients before admission, and informed consent of patients was obtained.
Results
Expression of Snail and Ring1B proteins in PDAC is positively correlated
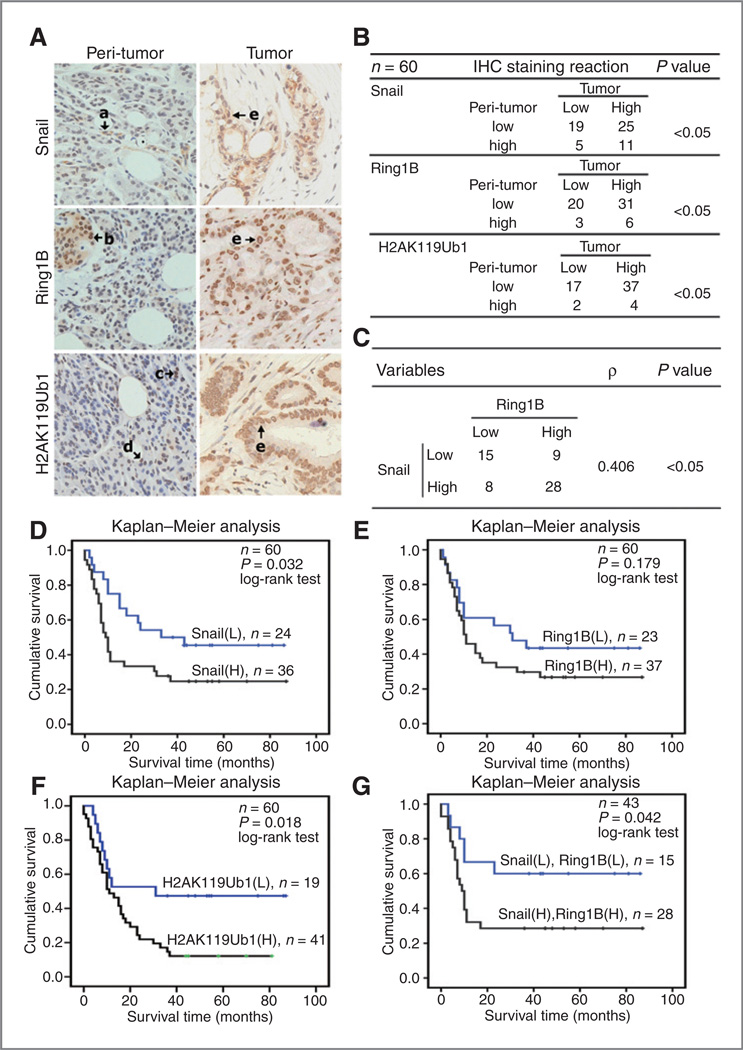
To examine the clinical relevance of Snail, Ring1B and level of H2AK119Ub1 in PDAC, IHC assays were performed on human pancreatic ductal tumor resection specimens and paired peri-tumor tissues were used as normal controls. The staining for Snail was strong in tumor cells surrounding acinus and lumen, but was barely detected in epithelial cells of normal tissues (Fig. 1A, top). Similarly, the staining for Ring1B was strong in tumor cells especially in high-grade differentiated cells, whereas in normal tissues the staining was weak or not detectable except in islet cells that showed positive staining (Fig. 1A, middle). Notably, the staining for H2AK119Ub1 was strong in tumor cells, but remained low level in islet cells and acinar cells of the normal tissues (Fig. 1A, bottom).
Figure 1.
High expression of Snai and Ring1B is positively correlated and predicts poor prognosis in PDAC. A, IHC staining to detect Snail, Ring1B, and H2AK119Ub1 in paired human PDAC specimens (tumor vs. peri-tumor). Arrows, positively stained cells. a and d, duct epithelial cells; b, islet cells; c, acinar cells; e, tumor cells. Original magnifications, ×40. B, the McNemar test of the IHC staining of 60 cases of the paired PDAC specimens on TMA chips. Each group was shown by the distribution of IHC staining scores for each case. Only IHC scores ≥ 4 point (++) was considered high. n = 60; *, statistical significance, P < 0.05. C, Spearman ρ analysis of correlation of Snail and Ring1B expression in PDAC. n = 60. ρ > 0, positive correlation; ρ < 0, negative correlation. D–G, survival analysis of patients with PDAC patients by Kaplan–Meier plots and log-rank tests. Patients were categorized by high and low expression of the Ring1B, Snail, and H2AK119Ub1 based on IHC staining scores. H, high; L, low. Average follow-up time of 60 patients was 26.2 months (range, 0–87 months). The median survival time of each group is as follows: Snail (L), 33 months; Snail (H), 9 months; Ring1B (L), 31 months; Ring1B (H), 11 months; H2AK119Ub1 (L), 31 months; H2AK119Ub1 (H), 11 months; Snai (L) + Ring1B (L), the median survival time cannot be estimated because more than 50% of cases are censored in this group; Snail (H) + Ring1B (H), 9 months.
To further elucidate the clinical correlation of Snail, Ring1B, and level of H2AK119Ub1 in PDAC, we performed IHC on TMA chips containing strictly selected 60 cases of paired tumor and peri-tumor specimens of PDAC. Of the 60 patients, 38 cases are males and 22 cases are females with an average age of 62 ± 11 years old. These patients have undergone pancreatectomy between September 2004 and December 2008, and the complete postoperative follow-up was finished on December 2011, with duration of 3 to 7 years. The overall median survival time of 60 patients after pancreatectomy was 15 months. The McNemar test was conducted to analyze the symmetrical distribution of the IHC staining for Snail, Ring1B, and H2AK119Ub1 among the 60 cases of samples according to the IRS scores (high: >4 vs. low: ≤4). We found that specimens showing high levels of Snail, Ring1B, and H2AK119Ub1 were distributed asymmetrically with much more cases found in tumor samples (Fig. 1B, P < 0.05). We next performed the Spearman rank correlation coefficient analysis to determine whether elevated expression of Snail and Ring1B in PDAC samples cooccurs. Indeed, there was a positive correlation between Snail and Ring1B (Fig. 1C, P < 0.05). Taken together, these data suggest Snail and Ring1B may functionally correlate in PDAC development and progression.
High level of H2AK119Ub1 predicts poor survival of patients with PDAC
To determine whether there is a correlation between high levels of Snail, Ring1B, H2AK119Ub1 and survival rate of postoperative patients with PDAC, Kaplan–Meier analysis was conducted. We found that patients with PDAC expressing high level of Snail and high level of H2AK119Ub1 showed tendency of higher mortality and was positively correlated with poor survival (Fig. 1D, median survival time: 9 months; Fig. 1F, median survival time: 11 months); expression level of Ring1B displayed similar tendency, but was not statistically significant (Fig. 1E). Remarkably, patients with PDAC showing both high level of Snail and Ring1B strongly predicted poor survival (Fig. 1G, median survival time: 9 months). Taken together, these data indicate that high level of H2AK119Ub1 in PDAC is a potential epigenetic biomarker for prediction of PDAC prognosis.
Ring1A and Ring1B are Snail-interacting proteins
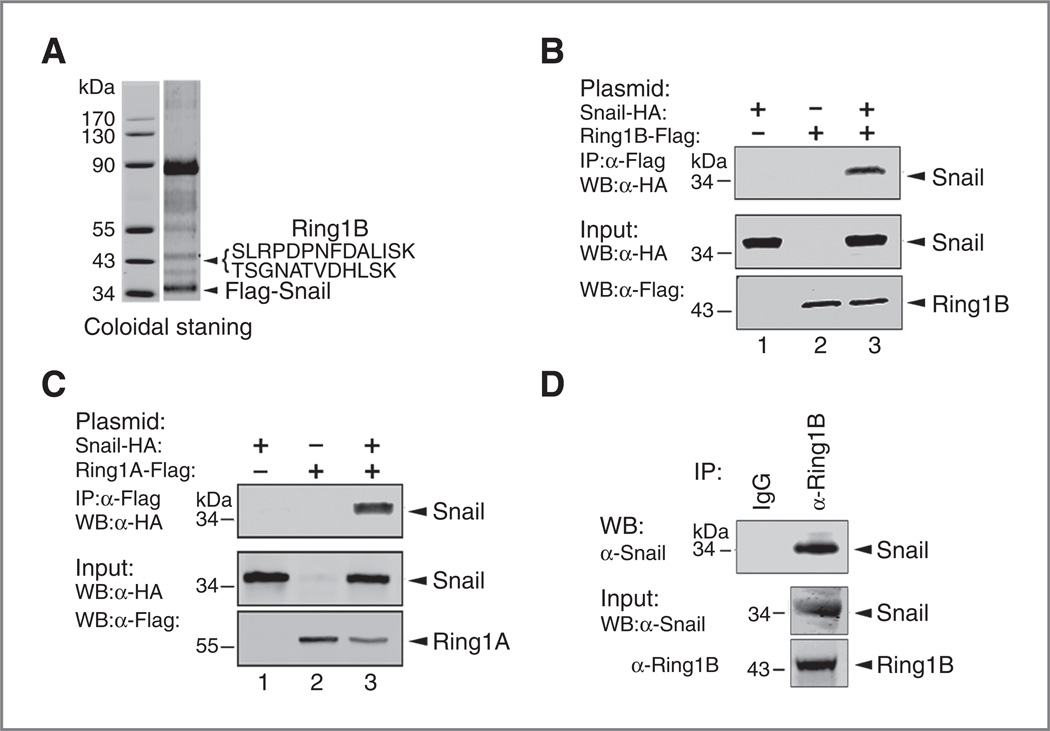
To examine if there is a physical interaction between Snail and Ring1B, we stably expressed N-terminal epitope-tagged Snail protein (Flag-Snail) in HEK-293T cells, and performed affinity purification of the Snail-interacting proteins with Flag antibody. The coeluted proteins along with Snail were identified by mass spectrometry analysis. Notably, Ring1B was found in the protein band migrating at 43 kDa (Fig. 2A). Next, we coexpressed HA-Snail and Flag-Ring1B in HEK-293T cells and performed co-IP assays to verify their interaction. Indeed, Snail and Ring1B interacted with each other (Fig. 2B). Ring1A also interacted with Snail (Fig. 2C). To further confirm the interaction of Snail with Ring1B, we attempted to detect the endogenous interaction. The endogenous Ring1B and Snail were readily detected by Western blot assays using specific antibodies in PanC1 cells. Co-IP assays indicated Ring1B interacted with Snail at endogenous level (Fig. 2D). Collectively, these data clearly demonstrate that Ring1A and Ring1B are novel Snail-interacting proteins.
Figure 2.
Ring1A and Ring1B are Snail-interacting proteins. A, mass spectrometry analysis identifies Ring1B as a Snail-interacting protein. Affinity purification of lysate from HEK-293T cell stably expressing Flag-Snail was performed with Flag antibody. SDS-PAGE and colloidal staining show proteins coeluted with Snail. Mass spectrometry analysis discovered the band migrating at 43 kDa contains Ring1B and the major band migrating at 34 kDa is Flag-Snail protein. B, Ring1B interacts with Snail. Lysates from HEK-293T cells transfected with plasmids encoding HA-Snail and/or Flag-Ring1B are incubated with Flag M2 beads, and the coeluted proteins were blotted with HA antibody. C, Ring1A interacts with Snail. D, Snail interacts with Ring1B at endogenous level. Lysates from PanC1 cells were incubated with anti-Ring1B, and the coeluted proteins were blotted with Snail antibody. Ten percent of the input was loaded. α, anti; WB, Western blot.
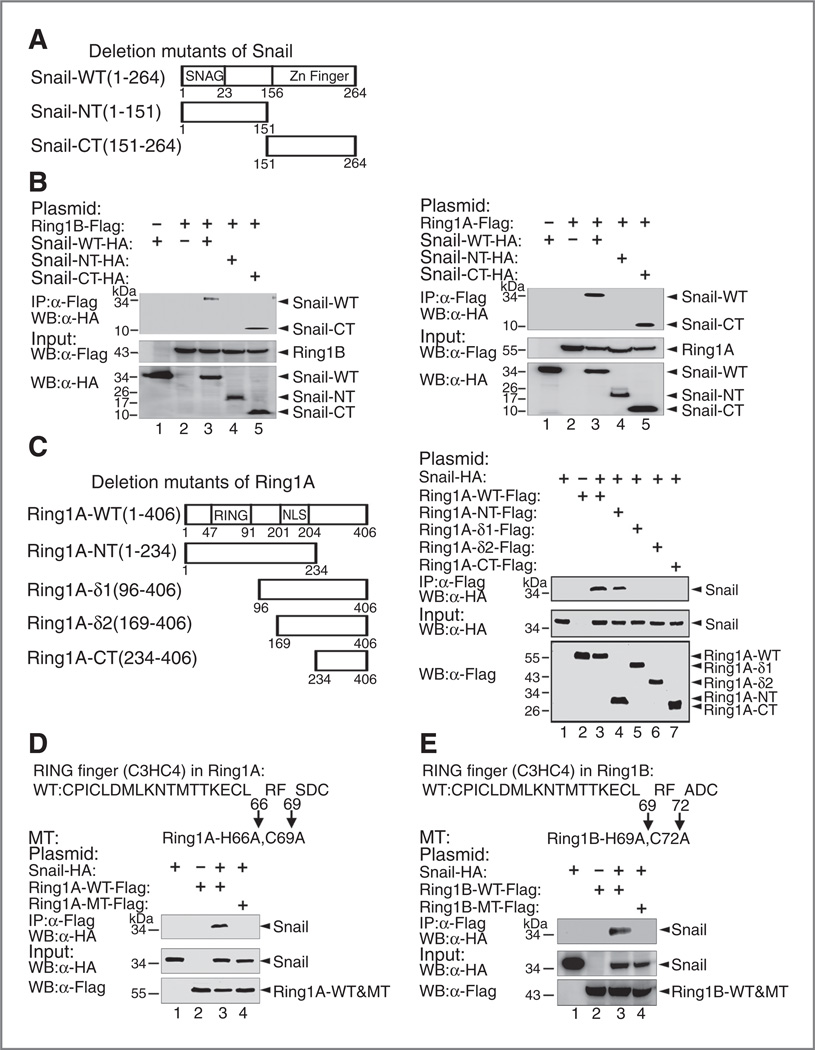
Ring1A and Ring1B bind to the carboxyl tandem zinc fingers of Snail
To identify the regions in the Snail protein that interact with Ring1A and Ring1B, plasmids encoding the N-terminal SNAG domain (Snail-NT) or C-terminal zinc fingers (Snail-CT) were constructed respectively and were expressed in HEK-293T cells alone with Ring1A or Ring1B (Fig. 3A). Co-IP assays demonstrated that the zinc finger region retained the binding activity with Ring1A and Ring1B (Fig. 3B), whereas the SNAG domain and the linker region of Snail showed no binding activity.
Figure 3.
A, the RING domains of Ring1A and Ring1B and the zinc fingers of Snail are required for their interaction. Schematic representation of deletion mutants of Snail. B, the C-terminal zinc fingers of Snail interact with Ring1A and Ring1B. Lysates prepared from HEK-293T cells transfected with plasmids encoding HA-Snail and its truncation mutants, together with Flag-Ring1A or Flag-Ring1B were incubated with anti-Flag M2 beads, and coeluted proteins were analyzed by anti-HA. C, N-terminal (1–96 aa) of Ring1A interacts with Snail schematic representation of deletion mutants of Ring1A. N-terminus of Ring1A contains a conserved RING finger domain. D, mutation of the 66th histidine and 69th cysteine in the RING finger of Ring1A abolishes the binding between Snail and Ring1A. E, mutation of the 69th histidine and No. 72 cystenie to alanines in the RING finger of Ring1B abolishes the interaction of Snail and Ring1B. WT, wild-type; MT, mutant-type; NT, Amino terminus; CT, carboxyl terminus.
The RING domains of Ring1A and Ring1B are required for the interaction with Snail
To identify the regions of Ring1A and Ring1B responsible for the interaction with Snail, we generated a series of deletion mutants of Ring1A (Fig. 3C, left). Next, we coexpressed the deletion mutants for Ring1A and Snail in HEK-293T cells and performed co-IP assays with Flag antibody. We observed that the N-terminal Ring1A containing the conserved RING domain retained the binding ability to Snail (Fig. 3C, right), but the C-terminal region of Ring1A was unable to bind Snail.
To further determine if the RING domain contributes to the interaction with Snail, simultaneous mutations of the 66th histidine (H66A) and 69th cysteine (C69A) to alanines in Ring1A were made to disrupt the RING structure (Fig. 3D). Co-IP assays showed that the double mutation of the key residues H66 and C69 of Ring1A resulted in loss of the binding activity to Snail (Fig. 3D), suggesting that the RING domain is critical for Snail binding. Similarly, mutation of H69 and C72 to alanines in the RING domain of Ring1B abolished its binding to Snail (Fig. 3E). Further, we showed that mutations of these core amino acid residues in the RING domains did not change the subcellular localization of Ring1A and Ring1B proteins (Supplementary Fig. S1). Taken together, these results demonstrate that the RING domains of Ring1A and Ring1B are required for their interaction with Snail.
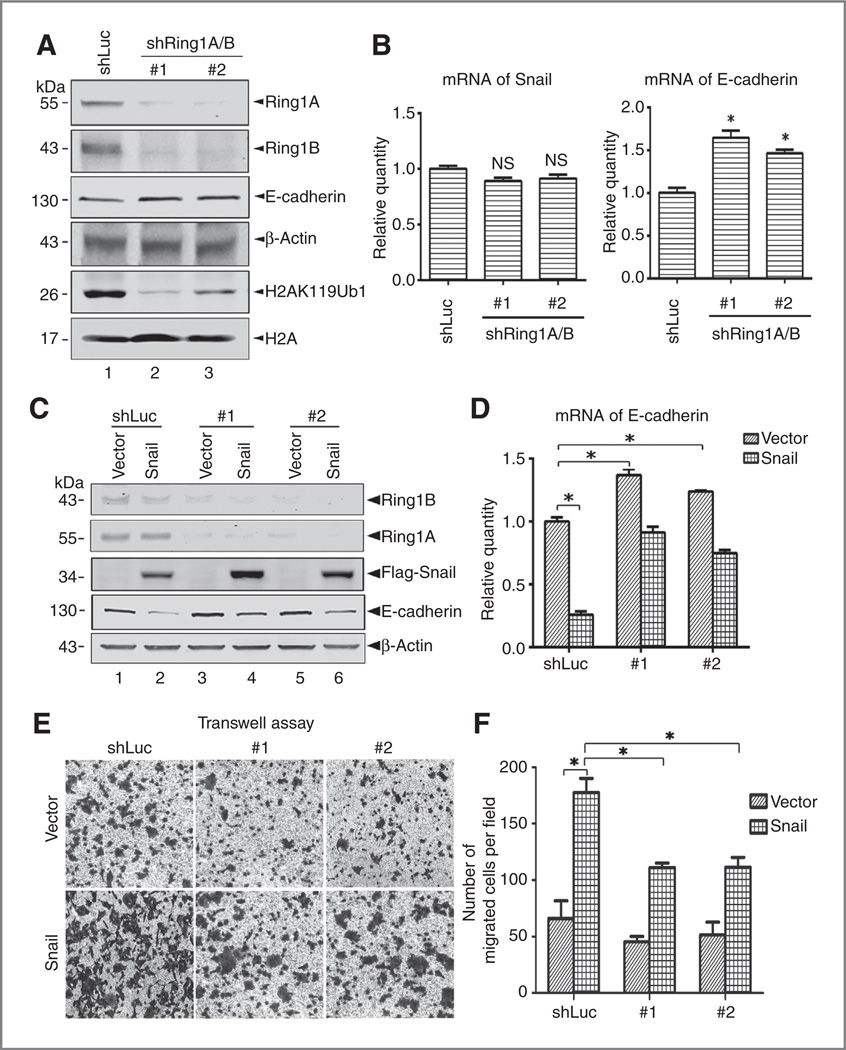
Ring1A and Ring1B are required for Snail-mediated E-cadherin gene repression and cell migration
To examine the role of Ring1A and Ring1B in Snail-mediated transcriptional repression, we first depleted their expression in PanC1 cells, a widely used pancreatic cancer cells, using specific shRNAs targeting Ring1A and Ring1B simultaneously to overcome the redundancy effect (Fig. 4A). Western blot assays indicated that depletion of Ring1A and Ring1B resulted in global decrease of H2AK119Ub1, but no apparent change on H3K27Me3 (Fig. 4A). qRT-PCR assays showed that expression of the Snail target genes E-cadherin and Cyclin D2 and the known Ring1A/B target genes HOXC5 and HOXB4 was increased in PanC1-shRing1A/B cells (Fig. 4B and Supplementary Fig. S2). Next, we stably expressed Flag-Snail in PanC1-shLuc or PanC1-shRing1A/B cells using lentiviral system (Fig. 4C). The expression of Snail, Ring1A, and Ring1B was validated by Western blot assays. The protein and mRNA levels of E-cadherin were examined by Western blot and qRT-PCR approaches. As expected, expression of Snail markedly reduced both the protein and mRNA levels of E-cadherin in PanC1-shLuc cells, but still slightly decreased the E-cadherin expression in PanC1-shRing1A/B cells (Fig. 4C and D). In addition, we observed similar effect of depletion of Ring1A and Ring1B on Snail-mediated transcriptional repression in another pancreatic cancer AsPC1 cells (Supplementary Fig. S3).
Figure 4.
Ring1A and Ring1B are required for Snail-mediated E-cadherin gene repression and cell migration. A, simultaneous depletion of Ring1A and Ring1B in PanC1 cells results in global decrease of H2AK119Ub1. Double depletion of Ring1A and Ring1B in PanC1 cells was made using two sets of specific shRNAs targeting Ring1A and Ring1B, respectively, and created two pools of PanC1-shRing1A/B cells. Western blots were performed to examine the expression of Ring1A, Ring1B, H2AK119Ub1, and H3K27Me3. B, E-cadherin is upregulated in PanC1-shRing1A/B cells. Total RNAs were isolated from the resulting PanC1 cells, and expression of Snail and E-cadherin was examined by qRT-PCR. #1 and #2, different cell pools generated from Ring1A and Ring1B double knocking down by shRNAs; GAPDH was used as loading reference. Data, mean ± SD from three independent experiments. *, statistical significance, P < 0.05. NS, statistical insignificance, P > 0.05. C, Snail was stably expressed in PanC1-shLuc or PanC1-shRing1A/B cells. Plu-Snail-Flag was introduced into the resulting PanC1 cells vial lentiviral infection, and the cells were sorted by FACS. The expression of Snail-Flag, Ring1A, Ring1B, and E-cadherin was examined by Western blot assays. D, depletion of Ring1A and Ring1B compromises Snail’s repression on E-cadherin. qRT-PCR assays were conducted to examine the mRNA of E-cadherin. GAPDH was used as loading reference. E, Snail induces cell migration in transwell assays. F, statistical analysis for the migrating cells from transwell assays was shown in the bar graphs. Nine randomly chosen fields were counted for statistic analysis. Data, mean ± SD from three independent experiments. #1 and #2, different cell pools generated from Ring1A and Ring1B double knocking down by shRNAs; *P < 0.05.
To examine the role of Ring1A and Ring1B in Snail-induced cell migration, we performed standard transwell assays to measure the migration capability of the PanC1 cells and its derivatives. Expression of Snail robustly enhanced migration capability of the PanC1-shLuc cells. However, expression of Snail in PanC1-shRing1A/B cells increased cell migration, but at lesser extent compared with that of in PanC1-shLuc-Snail cells (Fig. 4E and F). Collectively, these data suggest that Ring1A and Ring1B are essential for Snail to maximally repress E-cadherin and to induce cell migration.
Snail recruits Ring1A and Ring1B to the promoter locus of E-cadherin
To examine if Snail recruits Ring1A and Ring1B to the E-cadherin promoter, we performed ChIP assays in PanC1-derivated cells with antibodies specific to Snail, Ring1A, Ring1B, or H2AK119Ub1. The coeluted DNA fragments were amplified using primer pairs flanking the E-boxes located in the proximal promoter of E-cadherin (Fig. 5A). In PanC1-shLuc cells, expression of Snail resulted in an increased binding of Snail, Ring1A, and Ring1B to the E-cadherin promoter, and a concomitant increase in H2AK119Ub1 modification at this locus (Fig. 5B–E). Depletion of Ring1A and Ring1B simultaneously resulted in loss of their binding to the E-cadherin promoter, diminished H2AK119Ub1 modification, and decreased binding of Snail to the E-cadherin promoter, suggesting Ring1A and Ring1B may stabilize Snail/DNA complexes at the target promoter. However, depletion of Ring1A and Ring1B did not affect EZH2 binding and H3K27 methylation at the E-cadherin promoter locus (Fig. 5F and G). Taken together, these data demonstrate that Snail recruits Ring1A and Ring1B to modify H2A at K119 at the target chromatin region.
Figure 5.
A, Snail recruits Ring1A and Ring1B to modify H2A at K119 at the target chromatin region. Schema shows PCR primers used for amplifying the E-cadherin promoter flanking the functional E-boxes. B–G, ChIP DNA fragments from PanC1cells were analyzed by qPCR assays. For comparison between cells, PCR amplifications were normalized to their inputs respectively. The units on the y axis were arbitrary; data, mean ± SD from three independent experiments. *P < 0.05. NS, statistical insignificance, P > 0.05. Enrichment of Snail (B), H2AK119Ub1 (C), Ring1A (D), and Ring1B (E) at the E-cadherin promoter is increased when Snail is expressed, whereas depletion of Ring1A and Ring1B results in loss of Snail occupancy and diminished H2AK119Ub1 modification at the E-cadherin promoter. Snai increases EZH2 binding and H3K27Me3atE-cadheirn promoter locus, but depletion of Ring1A and Ring1B does not affect EZH2 binding (F) and H3K27Me3 modification (G).
Depletion of EZH2 reduces Snail/Ring1A/B multiprotein complex binding at the E-cadherin promoter locus
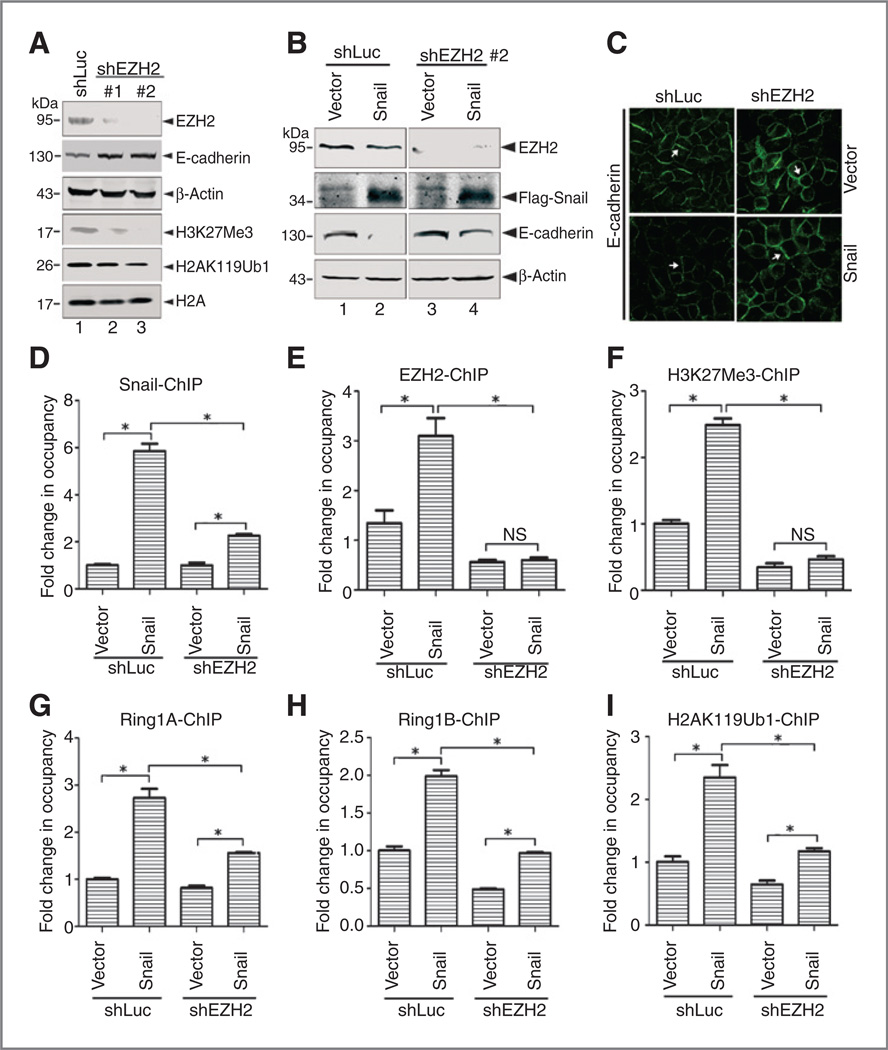
Previous studies demonstrated that EZH2 binds to the SNAG domain of Snail to repress E-cadherin expression by trimethylation of H3 at K27 (22, 23). To examine if EZH2 affects Ring1A/B-mediated H2AK119Ub1 modification, we generated PanC1-shEZH2 and PanC1-shLuc cells. Depletion of EZH2 resulted in markedly global decrease of H3K27Me3 and slight decrease of H2AK119Ub1 (Fig. 6A). Similar to that observed in PanC1-shRing1A/B cells, depletion of EZH2 increased E-cadherin expression as evidenced by Western blot and indirect immunofluorescence assays (Fig. 6B and C). Consistently, expression of Snail in PanC1-shLuc cells effectively repressed E-cadherin, but only showed weak repression on E-cadherin expression in PanC1-shEZH2 cells (Fig. 6B and C), indicating EZH2 is important for Snail-mediated E-cadherin repression. Next, ChIP assay was performed in these resulting cells. Depletion of EZH2 in PanC1 cells decreased the binding of Snail, Ring1A, and Ring1B to the E-cadherin promoter, diminished H3K27Me3, and greatly reduced H2AK119Ub1 modifications induced by Snail at this locus (Fig. 6D–I). These observations indicate that EZH2 functions upstream of Ring1A and Ring1B and promotes Snail to recruit Ring1A and Ring1B more efficiently at the E-cadherin promoter locus.
Figure 6.
Depletion of EZH2 reduces Snail/Ring1A/B multiprotein complex binding at the E-cadherin promoter locus. A, depletion of EZH2 in PanC1 cells by specific shRNAs results in global decrease of H3K27Me3 and elevated E-cadherin. Western blot showed EZH2 is effectively depleted in PanC1 cells, global H3K27Me3 is reduced, and H2AK119Ub1 is partially decreased in these cells. #1 and #2, different cell pools generated from EZH2 knocking down by two specific shRNAs. B, Snail-Flag is stably expressed in PanC1-shLuc or PanC1-shEZH2 (pool #2 in A) cells. Expression of Snail effectively represses E-cadherin in PanC1-shLuc cells, but only weakly represses E-cadherin in PanC1-shEZH2 cells. Whole-cell lysate (100 µg) was loaded and β-actin was used as loading control. C, immunofluorescence staining (green) of E-cadherin. E-cadherin is increased in PanC1-shEZH2 cells. Expression of Snail markedly decreases E-cadherin in PanC1-shLuc cells, but only slightly reduces E-cadherin in PanC1-shEZH2 cells. Arrows, E-cadherin localization on cell boundary. Original magnification, x63. D–I, ChIP DNA fragments from PanC1-shLucorPanC1-shEZH2 cells overexpressing Snail were analyzed by qPCR assays. PCR amplifications were normalized to their inputs respectively. Enrichment of Snail enhances the binding of EZH2 (E), Ring1A (G), and Ring1B (H) to E-cadherin promoter, increases H3K27Me3 (F) and H2AK119Ub1 (I) levels, whereas depletion of EZH2 decreases binding of Snail, Ring1A, and Ring1B to E-cadherin promoter, abolishes H3K27Me3, and decreases H2K119Ub1. The units on the y axis are arbitrary. Data, mean ± SD from three independent experiments. *P < 0.05. NS, without statistical significance, P > 0.05.
EZH2 and Ring1B form distinct complexes with Snail
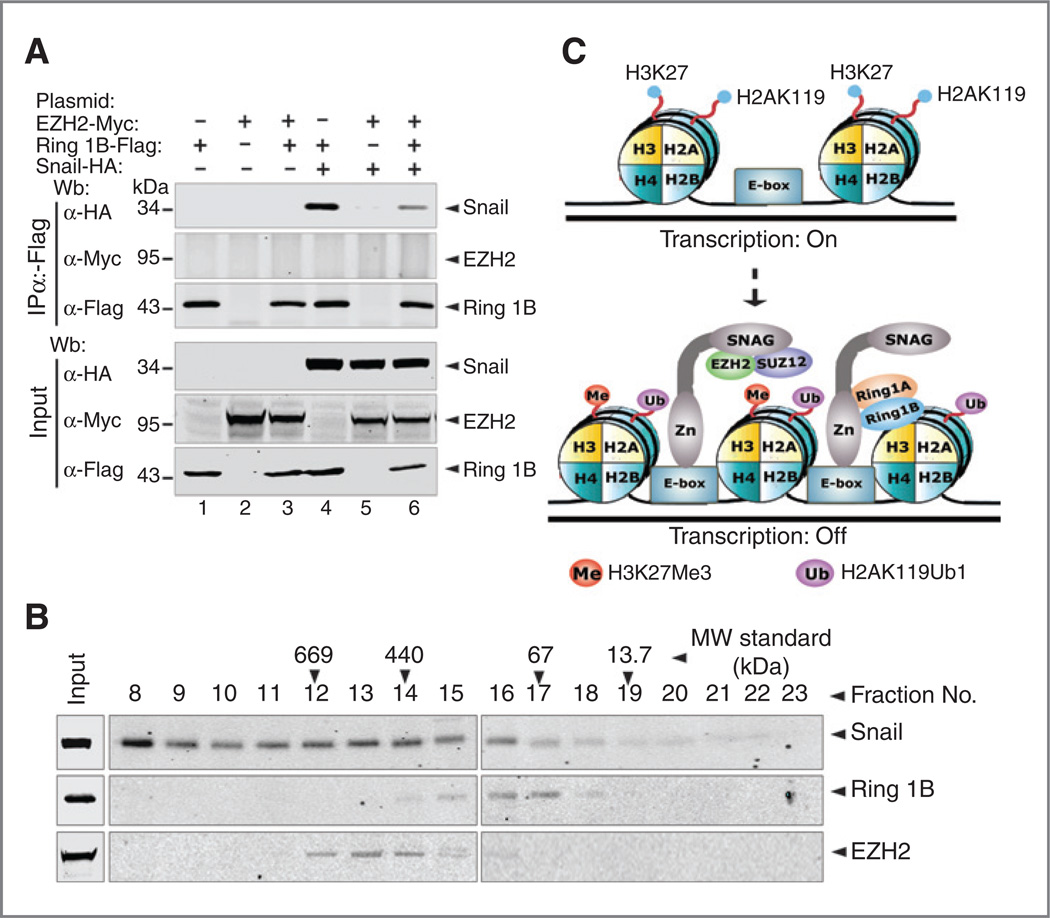
To determine if EZH2, Snail, and Ring1B form one functional complex, we coexpressed HA-Snail, Myc-EZH2, and Flag-Ring1B in HEK-293T cells and performed co-IP assays with Flag antibody. When these three proteins were coexpressed, Ring1B coimmunoprecipitated Snail, but not EZH2, indicating Snail, EZH2, and Ring1B do not coexist in the same complex (Fig. 7A). To further strengthen this observation, we performed size exclusion fractionation of whole-cell extracts prepared from HEK-293T cells expressing Snail, Ring1B, and EZH2. EZH2 was eluted in a single peak (fractions 12–14), Ring1B was detected in fraction 15 and was peaked at fractions 16 to 17; Snail was eluted in one peak (fractions 12– 16; Fig. 7B). These observations indicate that Snail forms two complexes, one with EZH2 and another one with Ring1B. Together, these data suggest that EZH2, Snail, and Ring1B do not coexist in the same protein complex; rather, EZH2 and Ring1B form two distinct protein complexes with Snail respectively (Fig. 7C).
Figure 7.
EZH2 and Ring1B form two distinct complexes with Snail. A, Snail, Ring1B, and EZH2 do not coexist in the same complex. Lysates from HEK-293T cells transfected with plasmids of HA-Snail, Flag-Ring1B, and Myc-EZH2 were incubated with anti-Flag M2 beads, and coeluted proteins were analyzed by HA and Myc antibodies. Co-IP showed that Ring1B does not interact with EZH2. B, size exclusion fractionation of whole-cell extracts prepared from HEK-293T cells expressing HA-Snail, Flag-Ring1B, and Myc-EZH2 was performed on Superose 6 10/300 GL column, and fractions of proteins were collected at 1 mL per fraction at 0.35 mL per minute in the cold room. Western bolt analysis of the proteins from each fraction with HA, Myc, or Flag antibodies showed that Ring1B and EZH2 form two distinct complexes with Snail. C, model for Snail-mediated repression. Snail recruits Ring1A and Ring1B via the carboxyl tandem zinc fingers to monoubiquitylate H2AK119, whereas Snail recruits EZH2 via SNAG domain to trimethylate H3K27, which may enhance Snail/Ring1A/B complex recruitment. Snail/Ring1A/B and Snail/EZH2 complexes may bind to different E-boxes or dynamically bind to the same E-boxes at the target promoters.
Discussion
PDAC is one of the most malignant human cancers, with a 5-year survival rate of only 5% and a median survival of less than 6 months (24, 25). Almost all patients with PDAC develop metastases due to its properties of rapid progression, potent invasiveness, and profound resistance to treatments. Identifying biomarkers for early diagnosis and prognosis is especially important to reduce the death rate of patients with PDAC.
H2AK119Ub1 modification is important during the embryonic development and organogenesis (12–15); however, the biologic significance of H2AK119Ub1 in tumor progression has not yet been elaborated. In this study, we found that H2AK119Ub1 is markedly elevated in tumor cells of PDAC, and the level of H2AK119Ub1 is positively associated with poor survival, indicating that H2AK119Ub1 is a potential epigenetic biomarker for diagnosis and prognosis of PDAC. However, we observed that levels of Snail, Ring1B, and H2AK119Ub1 are not correlated in some cases of PDAC specimens; for example, of the 60 cases,36 cases show Snail (H),37 cases show Ring1B (H), but only 28 cases show both Snail (H) and Ring1B (H; Fig. 1). The discrepancy between the level of Snail, Ring1B, and H2AK119Ub1 modification suggests additional mechanisms that are independent of the Snail/Ring1B complex and might be involved in Snail and Ring1B functions as well as H2AK119Ub1 modification in PDAC. To clarify this puzzle, ChIP-seq analyses of the occupancy of Snail and Ring1B, and the loci with high level of H2AK119Ub1 in the genome of PDAC specimens, will help to obtain global understanding of Snail, Ring1B, and H2AK119Ub1-dependent or -independent repressive pathways.
A number of the SNAG-associated histone modification complexes such as Sin3A-HDAC1/HDAC2, EZH2/SUZ12, LSD1-CoREST, and Ajuba-PRMT5 have been identified (6– 10), but how these complexes are orchestrated and assembled at the target chromatin is largely unknown. Here, we provided an example that how the SNAG-associated EZH2 and the zinc finger–associated Ring1A/B complexes cooperate at the target chromatin to regulate gene expression. EZH2 can trimethylate H3 at K27, which is required for Snail-dependent E-cadherin repression during cancer progression (22, 23). Consistently, we showed that depletion of EZH2 in PanC1 cells results in loss of Snail-mediated repression on E-cadherin, and a concomitant decrease in the binding of Snail, Ring1A, and Ring1B to the E-cadherin promoter, indicating that EZH2 functions upstream of Ring1A/B and promotes Snail to recruit Ring1A and Ring1B more efficiently at the E-cadherin promoter locus. This observation is well supported by previous findings that H3K27me3 is thought to involve in the initiation of gene repression and serve as a docking site for the recruitment of PRC1 proteins (26–29), whereby PRC1 is recruited to maintain the stable repression of genes. However, one remaining question is that how exactly Snail/EZH2 and Snail/Ring1A/B complexes are recruited to the target promoters because these two complexes do not coexist. Notably, we also observed that Snail occupancy at the target chromatin is affected by Ring1A and Ring1B, suggesting Ring1A and Ring1B may stabilize the Snail complex at the target chromatin. Although we showed that Snail binds to the RING domains of Ring1A and Ring1B, Snail is not a substrate of Ring1A and Ring1B (unpublished data). The detailed mechanism by which Ring1A and Ring1B regulate chromatin binding of Snail is not clear and needs to explore further. Moreover, we observed that depletion of Ring1A and Ring1B in PanC1 cells abolishes Ring1A and Ring1B binding and H2AK119Ub1 modification at the E-cadherin promoter, but expression of Snail in PanC1 cells still represses E-cadherin and induces cell migration in these cells (Fig. 4), indicating Snail may recruit additional repression complexes to repress transcription even in the absence of Ring1A and Ring1B. We postulate that the binding of Snail-associated complexes to Snail target chromatin may be dynamically and temporally regulated during the development and EMT events, which need to be dissected further in a system so that expression of Snail is finely controlled.
In summary, these observations demonstrate that the repressive activity of Snail is contributed not only by SNAG domain but also by the carboxyl zinc fingers; the histone ubiquitination also plays an important role in Snail-mediated transcriptional repression. Together, this sheds new light on the epigenetic machinery in Snail-induced EMT and cancer metastasis. These results provide unequivocal evidence to support that Ring1A and Ring1B could be explored for new therapeutics and that H2AK119Ub1 could be a potential bio-marker of pancreatic cancer prognosis.
Supplementary Material
Acknowledgments
The authors thank Guiping Li, Han Qin, Xuemei Yang, and Minmin Shi for technical help on histology and imaging and Yanyan Song for statistical analysis.
Grant Support
This work was supported by the Ministry of Sciences and Technology of China (2013CB 910900), the National Science Foundation of China (grant Nos. 81172028 and 81372309), and the Shanghai Committee of Science and Technology (13JC1401302). Z. Hou is a Pujiang Scholar of Shanghai metropolitan. F.J. Rauscher is supported by NIH grants CA129833, CA010815, CA163761,CA167151, DOD-BCRP W81XWH-11-1-0494, The Samuel Waxman Cancer Research Foundation, Susan G. Komen for the Cure, The Noreen O‧Neill Foundation for Melanoma Research, and the Ovarian Cancer Research Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors‧ Contributions
Conception and design: J. Chen, H. Xu, F.J. Rauscher, III, C. Peng, Z. Hou
Development of methodology: J. Chen, H. Xu, Z. Hou
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): J. Chen, H. Xu, X. Zou, J. Wang, A. Zhou, E.Y. Chin, Z. Hou
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J. Chen, H. Xu, B. Shen Z. Hou
Writing, review, and/or revision of the manuscript: J. Chen, H. Xu, F.J. Rauscher, III, C. Peng, Z. Hou
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): H. Xu, Y. Zhu, H. Chen, C. Peng, Z. Hou Study supervision: JH. Xu, X. DengC. Peng, Z. Hou
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Nieto MA. Epithelial-mesenchymal transitions in development and disease: old views and new perspectives. Int J Dev Biol. 2009;53:1541–1547. doi: 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- 2.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- 3.Moody SE, Perez D, Pan T-c, Sarkisian CJ, Portocarrero CP, Sterner CJ, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, et al. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 5.Batlle E, Sancho E, Franci C, Domínguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 6.Lin Y, Wu Y, Li J, Dong C, Ye X, Chi Y-I, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. EMBO J. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hector Peinado, Esteban Ballestar, Manel Esteller, Cano aA. Snail mediates E-cadherin repression by the recruitment of the Sin3A–histone deacethylase1-2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou Z, Peng H, Ayyanathan K, Yan KP, Langer EM, Longmore GD, et al. The LIM protein AJUBA recruits protein arginine methyltransferase 5 to mediate SNAIL-dependent transcriptional repression. Mol Cell Biol. 2008;28:3198–3207. doi: 10.1128/MCB.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong C, Wu Y, Yao J, Wang Y, Yu Y, Rychahou PG, et al. G9a interacts with Snail and is critical for Snail-mediated E-cadherin repression in human breast cancer. J clin invest. 2012;122:1469–1486. doi: 10.1172/JCI57349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tong Z, Cai M, Wang X, Kong L, Mai S, Liu Y, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2011;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 11.Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int J Dev Biol. 2009;53:355–370. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 13.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol cell. 2005;20:845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 14.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A. 2003;100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alberga A, Boulay J-L, Kempe E, Dennefeld C, Haenlin M. The snai gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 17.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Mol Cell Biol. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Romero C, Rooman I, Skoudy A, Guerra C, Molero X, Gonzalez A, et al. The epigenetic regulators Bmi1 and Ring1B are differentially regulated in pancreatitis and pancreatic ductal adenocarcinoma. J pathol. 2009;219:205–213. doi: 10.1002/path.2585. [DOI] [PubMed] [Google Scholar]
- 19.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Der Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 20.Maacke H, Opitz S, Jost K, Hamdorf W, Henning W, Krüger S, et al. Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int J Cancer. 2000;88:907–913. doi: 10.1002/1097-0215(20001215)88:6<907::aid-ijc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Dancey CP, Reidy J. Estatística sem matemática para psicologia: usando SPSS para Windows. Brazil: Artmed/Bookman. 2006 [Google Scholar]
- 22.Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani R-S, Tomlins SA, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 25.Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057. doi: 10.1016/S0140-6736(04)15841-8. [DOI] [PubMed] [Google Scholar]
- 26.Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a Histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 27.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 28.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Min J, Zhang Y, Xu R-M. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.