Abstract
The cytoplasmic-element-binding (CPEB) protein is a sequence-specific RNA-binding protein that regulates cytoplasmic polyadenylation-induced translation. In mouse embryo fibroblasts (MEFs) lacking CPEB, many mRNAs encoding proteins involved in inflammation are misregulated. Correlated with this aberrant translation in MEFs, a macrophage cell line depleted of CPEB and treated with lipopolysaccharide (LPS) to stimulate the inflammatory immune response expresses high levels of interleukin-6 (IL-6), which is due to prolonged nuclear retention of NF-κB. Two proteins involved in NF-κB nuclear localization and IL-6 expression, IκBα and transforming growth factor beta-activated kinase 1 (TAK1), are present at excessively low and high steady-state levels, respectively, in LPS-treated CPEB-depleted macrophages. However, only TAK1 has an altered synthesis rate that is CPEB dependent and CPEB/TAK1 double depletion alleviates high IL-6 production. Peritoneal macrophages isolated from CPEB knockout (KO) mice treated with LPS in vitro also have prolonged NF-κB nuclear retention and produce high IL-6 levels. LPS-injected CPEB KO mice secrete prodigious amounts of IL-6 and other proinflammatory cytokines and exhibit hypersensitivity to endotoxic shock; these effects are mitigated when the animals are also injected with (5Z)-7-oxozeaenol, a potent and specific inhibitor of TAK1. These data show that CPEB control of TAK1 mRNA translation mediates the inflammatory immune response.
INTRODUCTION
Inflammation is triggered by bacterial pathogens, as well as lipopolysaccharide (LPS), a bacterial cell wall component that activates the transcription of multiple inflammatory response genes, including those for cytokines and chemokines (1, 2). Despite the importance of these inflammation mediators for host defense against infection, their excessive production can elicit organ failure and septic shock that results in lethality. Therefore, limitation of cytokine production is essential for the termination of inflammation and the prevention of endotoxic tissue damage (2).
The production of inflammatory mediators is controlled at multiple levels, including transcription, translation, and protein stability (3). LPS promotes the nuclear import of NF-κB (4) by indirectly regulating the activity of the IκB kinase (IKK) complex, which phosphorylates IκBα, a factor that normally retains NF-κB in the cytoplasm (5–7). Phosphorylated IκBα is rapidly destroyed, thereby releasing NF-κB to translocate to the nucleus and activate the transcription of target genes (8, 9). A proximal upstream LPS-activated factor is the Toll-like receptor, whose signaling pathway includes transforming growth factor beta-activated kinase 1 (TAK1), a mitogen-activated protein (MAP) kinase kinase kinase (10). TAK1 stimulation of p38 MAP kinase leads to IKK activity and NF-κB import. LPS stimulation of TAK1 also triggers the stabilization of many mRNAs, which is dependent upon the interplay of several 3′ untranslated region (UTR)-binding proteins such as those that associate with the AU-rich element (ARE) stabilization/destruction sequence (11, 12).
The cytoplasmic-element-binding (CPEB) protein is an mRNA-binding protein that interacts with the cytoplasmic polyadenylation element (CPE), a U-rich sequence in mRNA 3′ UTRs that controls poly(A) tail length and translation (13, 14). As a consequence of its regulation of mRNA expression, CPEB mediates germ cell development (15, 16), neuronal synaptic plasticity (17–20), and cellular senescence (21–24). Several observations suggest that CPEB might also be involved in the immune response. First, in mouse embryo fibroblasts (MEFs) derived from CPEB knockout (KO) mice, many mRNAs that encode proteins involved in inflammation and the NF-κB signaling pathway are aberrantly expressed (25). Indeed, NF-κB is hyperactivated and elevated amounts of interleukin-6 (IL-6) are produced in CPEB KO MEFs (24). Second, CPEB influences insulin signaling (25, 26), which is often coincident with inflammation. Third, the CPE resembles the ARE, a sequence often present in cytokine mRNA 3′ UTRs that has been implicated in their stability and/or translation (11, 27, 28).
In this report, we have investigated the involvement of CPEB in the inflammatory immune response. Following treatment with LPS in vitro, macrophages lacking CPEB secrete high levels of IL-6 and display prolonged nuclear retention of NF-κB. Two molecules that could mediate IL-6 production, IκBα and TAK1, have altered steady-state levels that correlate with cytokine secretion. However, only the synthesis of TAK1 is regulated by CPEB; it is elevated when CPEB is absent. IκBα, on the other hand, is controlled at the level of protein stability. The observation that double depletion of both CPEB and TAK1 mitigates elevated IL-6 production demonstrates the interplay between these two factors. CPEB KO mice exhibit strong endotoxic shock and prodigious proinflammatory cytokine production following LPS administration, in contrast to wild-type (WT) mice. Macrophages derived from CPEB KO mice treated with LPS in vitro display both elevated IL-6 production and enhanced TAK1 synthesis. Most importantly, the endotoxic shock and strong IL-6 secretion exhibited by CPEB KO mice treated with LPS are alleviated when the animals are injected with (5Z)-7-oxozeaenol, a specific inhibitor of TAK1. These data show that CPEB control of TAK1 synthesis mediates the inflammatory immune response.
MATERIALS AND METHODS
Animal studies.
All experiments were conducted in accordance with approved NIH and institutional protocols for the treatment and handling of animals. Three-month-old randomly assigned CPEB WT and KO agouti males (13 matching littermates in each group) were injected intraperitoneally (i.p.) with LPS (20 mg/kg) or with normal saline alone. Some animals were also injected i.p. with LPS, followed by the TAK1 inhibitor (5Z)-7-oxozeaenol (5 mg/kg) or the vehicle (dimethyl sulfoxide), on day 1 and were monitored every 12 h for 7 days postinjection. One hundred microliters of blood was collected from the tail vein of each animal 4 h postinjection. The serum was used to assess IL-6 levels by enzyme-linked immunosorbent assay (ELISA; eBioscience) according to the manufacturer's instructions. The general condition and survival of animals were monitored every 12 h for 6 days. Animal survival rates were plotted by the Kaplan-Meier method (29), and statistical significance was scored by log rank test with a significance level of P < 0.05.
Histology.
For histological analysis, four WT and four CPEB KO mice were sacrificed 24 h after LPS injection. Liver and lung tissues were fixed overnight at 4°C in 10% buffered formaldehyde, paraffin embedded, sectioned, and stained with hematoxylin and eosin for morphological examination.
Measurements of cytokine levels.
A 100-μl volume of blood was collected from the tail of each animal (four animals per group) following phosphate-buffered saline (PBS) or LPS injection. All blood samples were clotted for 4 h at room temperature, centrifuged at 2,000 × g for 20 min, and subjected to cytokine ELISA array (RayBiotech) analysis with 96 different cytokines. The membranes were probed according to the manufacturer's protocol and quantified by scanning densitometry. IL-6 secretion from primary macrophages or the RAW 264.7 cell line was measured by mouse anti-IL-6 ELISA (BD Bioscience). Equal numbers of macrophages were plated in triplicate (5 × 105 cells/well) and treated with 100 μg/ml LPS (Sigma) for 0 to 4 h. In some cases, the cells were treated with the proteasome inhibitor MG132 or the NF-κB inhibitors JSH-23 and 5HPP-33 (5 mM; EMD Millipore) for 2 h; this was followed by the addition of LPS and incubation for an additional 4 h. The media were collected and assessed for IL-6 by ELISA according to the manufacturer's protocol.
Antibodies.
Immunoblotting was performed with purified polyclonal antibodies against NF-κB (p65) (Santa Cruz Biotechnology), TAK1, p38, IκBα, phospho-IκBα (Cell Signaling), and β-actin (Sigma).
Immunofluorescence.
Macrophages were grown in 12-well tissue culture plates on glass coverslips, washed with PBS, fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100, blocked with normal goat serum, and stained with NF-κB antibody (p65) (Santa Cruz) at 1:1,000 for 2 h and a secondary antibody conjugated with Alexa Fluor 594 (Molecular Probes). Images were acquired with a Zeiss Axiovert 200M microscope or a PerkinElmer Ultraview spinning-disc microscope with a Hamamatsu ORCA-ER camera with a 100× numerical aperture 1.4 Plan-Apochromat oil objective. Two-dimensional single-plane images (MetaMorph; Molecular Devices) were adjusted for fluorescence range intensity identically for each series of panels.
Lentivirus production and cell infection.
Viral stocks were prepared by calcium transfection of 293T cells with retroviral plasmid vectors containing CPEB short hairpin RNA (shRNA) or scrambled shRNA in a PLL.3 vector (30). The macrophages were used for experiments 5 days postinfection, and quasiquantitative reverse transcription (RT)-PCR was used to confirm CPEB knockdown.
Metabolic labeling of cells.
Cells were incubated in starvation medium (no methionine, 2 mM thymidine, and no serum) for 30 min and then labeled (150 μCi/ml Redivue [35S]methionine; Amersham Pharmacia) for 30 min. They were collected, washed three times in cold PBS, and lysed in radioimmunoprecipitation assay (RIPA) buffer. Immunoprecipitations were performed with anti-TAK1 or anti-IκBα polyclonal antibody overnight on ice. IgG served as a nonspecific control. LPS (1 mg/ml) was added to the cells for 4 h of incubation before starvation.
Cell cultures.
RAW 264.7 cells were purchased from ATCC and cultured in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and antibiotic-antimycotic solution (Gibco, Grand Island, NY) at 37°C in 5% CO2. The peritoneal cells were collected, centrifuged for 10 min at 1,500 rpm at 4°C, resuspended, plated in culture dishes, and allowed to adhere for 1 day. They were then washed twice with PBS to remove debris and contaminating cells and used for ELISA and immunofluorescence analysis.
RT-PCR.
RNA was extracted with TRIzol (Invitrogen); RT reactions with equal amounts of total RNA were performed by using Superscript II and oligo(dT) primer and mRNA-specific primers. The PCRs were titrated and thus are semiquantitative.
Immunoprecipitation.
RAW 264.7 macrophages infected with a virus expressing FLAG-CPEB or FLAG-CPEBΔZF (CPEB RNA-binding mutant protein lacking a zinc finger) were washed with PBS and lysed in RIPA buffer containing RNase inhibitors for 1 h on ice. The lysate was incubated with anti-FLAG antibody overnight at 4°C and then protein G-magnetic beads (Invitrogen) for an additional 6 h. The beads were washed four times with RIPA buffer, and the RNA was extracted with TRIzol reagent (Invitrogen).
Formaldehyde cross-linking and mRNA competition.
RAW 264.7 macrophages transfected with a plasmid encoding FLAG-CPEB pretreated with LPS for 4 h were treated with 1% formaldehyde in PBS for 15 min at room temperature. This chemical cross-linking was stopped by the addition of glycine to a final concentration of 0.2 M, followed by 5 min of incubation at room temperature. The collected cells were washed three times with PBS, and then a cell lysate was prepared in RIPA buffer. After overnight immunoprecipitation with FLAG antibody (Sigma-Aldrich) and Dynabeads (Invitrogen/Life Technologies), the precipitate was washed three times with RIPA buffer and digested with proteinase K for 1 h at 42°C. The cross-links were reversed by incubation at 65°C for 1 h. RNA was extracted with TRIzol, carrier glycogen was added, and RNA was precipitated with sodium acetate and ice-cold ethanol for 2 h at −80°C. The collected RNA pellet was washed with ice-cold 70% ethanol, air dried, suspended in RNase-free water, and subjected to an RT reaction for detection of TAK1 mRNA by PCR assay.
For the RNA competition assay, transfected RAW 264.7 macrophages were used for similar CPEB immunoprecipitations (no formaldehyde) but the Dynabeads containing CPEB were incubated for 1 h at 4°C with CPE-containing or CPE-lacking competitor RNAs derived from the cyclin B1 3′ UTR. The beads were then washed three times with RIPA buffer, and the RNA was extracted and assayed for TAK1 mRNA by RT-PCR.
RESULTS
CPEB mediates IL-6 production.
MEFs derived from CPEB KO mice do not senesce as do those derived from WT animals but instead are immortal (23, 24). Although aberrantly translated myc and p53 mRNAs in CPEB-lacking MEFs are responsible for this immortalization, we suspected that additional mRNAs are involved. Consequently, polysomal mRNA from WT and CPEB KO MEFs were used to screen microarrays, which revealed that, in addition to misregulated members of the insulin signaling pathway in the KO MEFs (25), more than 20% of the transcripts whose translation was affected by CPEB gene disruption were involved in the immune response and included a large number of cytokines, chemokines, and NF-κB-related molecules. To investigate whether these changes in translation could reflect alterations in the inflammatory immune response, mouse macrophages (RAW264.7 cell line) were depleted of CPEB by infection with a lentivirus expressing shRNA (Fig. 1A, top). This procedure had no detectable effect on IL-6 protein levels in unstimulated cells. However, when the cells were treated with LPS, IL-6 protein and RNA levels were dramatically increased, which was particularly evident in the CPEB-depleted cells (Fig. 1A and B; see Fig. S1 in the supplemental material). Because NF-κB is one of the major LPS-activated transcription factors that control cytokine production (31), we surmised that it might be responsible for the stimulation of IL-6. Indeed, Fig. 1B shows that two inhibitors of NF-κB activity (IN1, JSH-23; IN2, 5HPP-33) significantly mitigated the elevation of IL-6 secretion in CPEB-depleted cells. These inhibitors also had a similar effect on IL-6 secretion in WT cells.
FIG 1.
CPEB controls of NF-κB activity. (A) Lentivirus/shRNA knockdown (KD) of CPEB in RAW 264.7 macrophage cells (top) and knockdown followed by LPS treatment and RT-PCR for IL-6 (bottom). Cont or cont, control. (B) ELISA of IL-6 secretion in RAW macrophages following CPEB knockdown and treatment with LPS in the presence or absence of the NF-κB inhibitors JSH-23 (IN1) and 5HPP-33 (IN2). These experiments were performed four times. (C) NF-κB immunocytochemistry and 4′,6-diamidino-2-phenylindole (DAPI) counterstain in RAW cells following CPEB knockdown and treatment with LPS for 0 to 4 h. (D) Quantification of the extent of NF-κB nuclear localization. These experiments were performed three times, and approximately 250 to 300 cells of each sample were examined. Here and in all of the other figures, the bars on histograms refer to the standard error of the mean and one asterisk refers to a P < 0.05 significance level, two asterisks refer to a P < 0.01 significance level, and three asterisks refer to a P < 0.001 significance level (Student's t test).
NF-κB is tightly regulated at the level of nuclear entry. To determine whether the nuclear/cytoplasmic distribution of NF-κB is mediated by CPEB, control and CPEB-depleted RAW macrophages were treated with LPS and immunostained for the transcription factor (Fig. 1C). CPEB-depleted cells displayed prolonged nuclear NF-κB retention compared to WT cells after 0.5 to 4 h of LPS treatment, indicating that this strong nuclear localization is most likely responsible for the upregulation of IL-6 production in these cells (Fig. 1D).
Aberrant NF-κB signaling in CPEB-deficient macrophages.
IκBα is a cytoplasmic protein that controls the nuclear entry of NF-κB. In response to extracellular signals, IκBα is phosphorylated and destroyed in a ubiquitin-mediated manner, which liberates NF-κB to enter the nucleus and activate transcription. To investigate whether CPEB regulates IκBα destruction, control and CPEB-depleted cells were incubated with LPS with or without MG132, a proteasome inhibitor. Figure 2A shows that there were low levels of IκBα when CPEB knockdown cells were treated with LPS. In contrast, IκBα in CPEB-depleted cells incubated with the proteasome inhibitor MG132 was restored to control levels, demonstrating that CPEB mediates IκBα destruction.
FIG 2.
CPEB control of the NF-κB signaling pathway. (A) Western blot analysis of IκBα, phospho-S32/S36-IκBα, phospho-S176/S180-IKKα/β, phospho-T180/Y180-p38 MAP kinase, TAK1, NF-κB, and actin in control (cont) and CPEB-depleted RAW macrophages treated with LPS or the proteasomal inhibitor MG132 for the times indicated. All blot assays were performed with at least three independent samples; all produced similar results. KD, knockdown. (B) Control and CPEB-depleted cells were treated with LPS for 4 h; this was followed by 40 min of incubation with [35S]methionine (35S-met), subsequent immunoprecipitation (IP) of IκBα and TAK1, and analysis by SDS-PAGE and autoradiography. Also shown are the [35S]methionine-labeled nonprecipitated whole-cell extracts, as wells as IκBα. The amount of immunoprecipitated TAK1 was quantified by scanning densitometry from three independent experiments (histogram). ns, nonspecific band. (C) Lysates from FLAG-CPEB-expressing RAW macrophages were supplemented with RNA containing or lacking three CPEs. CPEB was then FLAG immunoprecipitated, and the relative amounts of TAK1 mRNA remaining bound to CPEB were assessed by RT-PCR. Nonspecific IgG and FLAG-CPEBΔZF, an mRNA-binding mutant protein lacking two zinc fingers, served as controls for immunoprecipitation (top). In other experiments, RAW macrophages infected with lentivirus expressing FLAG-CPEB or FLAG-CPEBΔZF (dZn) were treated with formaldehyde to covalently cross-link RNA and protein in vivo. CPEB was then immunoprecipitated with FLAG antibody, the precipitate was heated to reverse the cross-links, and the RNA was extracted and subjected to analysis of TAK1 mRNA by semiquantitative RT-PCR. The input represents 10% of the total (bottom). CPE+, CPE-containing RNA; CPE−, CPE-lacking RNA. (D) Macrophages infected with lentiviruses expressing scrambled (control) or CPEB-targeting (KD) shRNAs were incubated with TAK1 siRNA for 3 days; this was followed by LPS stimulation for 4 h and semiquantitative RT-PCR detection of TAK1 and tubulin mRNAs. Western blot analysis of TAK1 protein after knockdown of TAK1 RNA by siRNA (left). Tubulin was used as a loading control. Semiquantitative RT-PCR detection of IL-6 mRNA in macrophages following CPEB and TAK1 depletion. Tubulin served as a control (right). (E) Control (CONT), CPEB-depleted (KD), or CPEB/TAK1 double-depleted (KD) macrophages were incubated with LPS for 4 h. The culture medium was assessed for IL-6 by ELISA.
We next assessed the phosphorylation of IκBα at serines 32 and 36, which is required for its ubiquitination and destruction (32). In CPEB-depleted cells treated with LPS and MG132, serines 32 and 36 were hyperphosphorylated (Fig. 2A), which demonstrates the involvement of CPEB in this signaling pathway. We therefore examined the activation of upstream kinases known to initiate NF-κB nuclear import via IκBα phosphorylation. In contrast to WT cells, CPEB-depleted cells showed strong phosphorylation of IKK (S180/181) and p38 (T180/Y182), indicating a robust activation of these kinases that mediate NF-κB activation (Fig. 2A). Steady-state NF-κB levels were unaffected by CPE depletion or LPS treatment.
Upstream of p38 MAP kinase is TAK1, a mitogen-activated protein kinase kinase kinase (10, 32, 33). We suspected that TAK1 could be an essential upstream kinase controlled by CPEB because its mRNA has numerous CPEs in its >2-kb 3′ UTR. Western blotting of LPS-stimulated WT and CPEB-depleted cells for TAK1 shows that, indeed, the level of this kinase was elevated in the absence of CPEB (Fig. 2A), although TAK1 mRNA levels were unaffected (see Fig. S1 in the supplemental material). To determine whether CPEB mediates the synthesis of TAK1, we pulse-labeled LPS-treated WT and CPEB knockdown cells with [35S]methionine for 30 min and then immunoprecipitated TAK1 and analyzed it by SDS-PAGE (Fig. 2B). This procedure measures the instantaneous synthesis of a protein without significant influence of protein destruction. For comparison, we performed the same experiment with IκBα. Knockdown of CPEB resulted in an ∼2.2-fold increase in the synthesis of TAK1 but not of IκBα, suggesting that CPEB controls TAK1 mRNA translation.
Next, we expressed FLAG-CPEB or FLAG-CPEBΔZF, which lacks a zinc finger that is important for optimal RNA binding (30, 34), in RAW 264.7 macrophages. Lysates prepared from these cells were supplemented with two concentrations of competing RNAs that lacked or contained three CPEs. After 30 min of incubation, CPEB (the WT, as well as the ΔZF form) was immunoprecipitated (IgG served as a second control) and TAK1 mRNA in the RNP complex was detected by RT-PCR. Figure 2C (top) demonstrates that the CPE-containing RNA was an effective competitor for TAK1 mRNA binding while the CPE-lacking RNA elicited no diminution of CPEB binding to TAK1 mRNA. We also treated other CPEB-expressing cells with formaldehyde to covalently cross-link RNAs and proteins in vivo. The CPEB-RNP complexes were then immunoprecipitated with FLAG antibody and washed extensively, the precipitate was heated to reverse the cross-link, and the RNA was extracted and analyzed for TAK1 mRNA by quasiquantitative RT-PCR. Figure 2C (bottom) shows that TAK1 mRNA was strongly immunoprecipitated with CPEB, in contrast to the mutant form of the protein. Taken together, the data thus far presented in Fig. 2 indicate that CPEB directly modulates the synthesis of TAK1 to control downstream signaling to NF-κB and IL-6 production.
If this scenario is correct, then IL-6 expression should be reduced by TAK1 depletion in cells lacking CPEB. Figure 2D shows that transduction of CPEB-depleted cells with TAK1 small interfering RNA (siRNA) significantly reduced TAK1. As a result, cells stimulated with LPS had smaller amounts of IL-6 mRNA, as measured by RT-PCR and secreted diminished levels of IL-6 protein, as determined by ELISA (Fig. 2E). On the basis of these data, we propose that CPEB knockdown elicits IκBα instability by activating the IKKα/β complex via elevated synthesis of the upstream TAK1 kinase, resulting in NF-κB nuclear retention and IL-6 transcription.
TAK1 mediates the inflammatory response in CPEB KO mice.
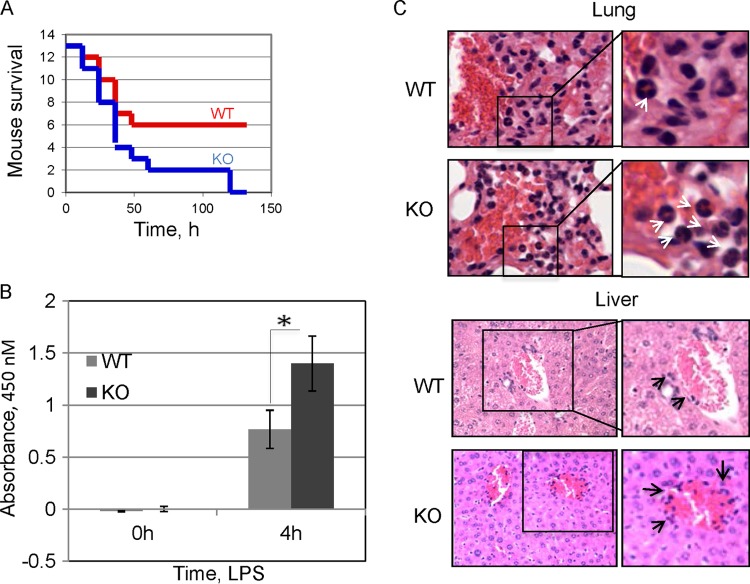
Peritoneal macrophages obtained from WT and CPEB KO mice treated with LPS in vitro secrete substantial amounts of IL-6, but the amount secreted by the KO macrophages is almost twice as large (Fig. 3A). Moreover, the KO macrophages displayed prolonged nuclear NF-κB retention (see Fig. S2 in the supplemental material), similar to that observed in a macrophage cell line depleted of CPEB (Fig. 1C), and had reduced TAK1 protein levels (see Fig. S3 in the supplemental material). These data suggest that CPEB KO mice might be particularly susceptible to endotoxic shock. Under normal conditions, CPEB KO mice show no obvious signs of chronic inflammation; however, when fed a high-fat diet, they exhibit liver insulin resistance (25), a phenotype that is often related to inflammation. To assess whether the animals have a strong inflammatory response under stress conditions, we injected the peritoneal cavities of WT and CPEB KO mice with a 50% lethal dose (LD50) of LPS, which stimulates the immune response and triggers septic shock (32). LPS-induced lethality for KO mice was substantially elevated relative to that of WT animals and reached 100% by day 6 (Fig. 3B). Both WT and CPEB KO mice had nearly identical lymphocyte profiles (see Fig. S4 in the supplemental material) and total blood cell counts (see Table S1 in the supplemental material), indicating no obvious predisposition to inflammation. Histological analysis of liver and lung tissue samples taken 24 h after LPS injection revealed a substantial increase in the number of neutrophils in both WT and CPEB KO animals. However, the levels of neutrophils in the KO samples were significantly higher (Fig. 3C), indicating an especially strong inflammatory response.
FIG 3.
Enhancement of the LPS-induced inflammatory response of CPEB KO mice. (A) WT and CPEB KO mice were injected i.p. with an LD50 of LPS, and a Kaplan-Meier (29) survival curve was determined. The KO mice exhibited statistically significantly decreased survival (13 mice per group, P < 0.05, Student's t test). (B) Peritoneal macrophages from WT and CPEB KO mice were treated with LPS (100 mg/ml) for 0 or 4 h, after which time the media were analyzed for IL-6 by ELISA (n = 3; *, P < 0.05). (C) Representative hematoxylin-and-eosin-stained sections of lung and liver tissues obtained from control and LPS-injected WT and KO mice. White arrows indicate increased neutrophil infiltration in KO tissue following LPS injection, and black arrows indicate inflammatory cell infiltration around portal areas.
Next, we used a cytokine/chemokine array to analyze the levels of blood serum cytokines of WT and CPEB KO animals after control PBS or LPS injection by performing an ELISA for 96 different mediators of inflammation (see Fig. S5 in the supplemental material). In addition to the expected elevation of IL-6, six other cytokines (IGF-bp3, IGF-1, IL-6, IL-12, and IL-8 [KC]) showed a statistically significant change, although they were little altered upon PBS injection, irrespective of the genotype. l-Selectin was elevated in the serum of both PBS- and LPS-injected KO mice. ELISA analysis showed that LPS injection elicited significantly higher levels of IL-6, IL-8, IL-12p40, IGF-1, and IGF-bp3 in the KO animals (Fig. 4), which generally reflected the changes in the levels of their corresponding mRNAs (see Fig. S1 in the supplemental material). Thus, elevation of proinflammatory cytokines IL-6, IL-8, and IL-12p40, the major markers of lethal sepsis, correlate with the increased morbidity of the LPS-injected CPEB KO mice.
FIG 4.
Serum levels of selected cytokines from WT and CPEB KO mice. Six of the proteins (IGF-bp3, IGF1, l-selectin, IL-6, IL-12, and KC) were consistently different among the four groups. The histograms represent the relative densitometric values of the proteins that were significantly different among the groups. P values of <0.05 are indicated by asterisks (n = 4).
To determine whether TAK1 mediates the inflammatory immune response in vivo in a CPEB-dependent manner, we used (5Z)-7-oxozeaenol, a resocyclin acid lactone that specifically inhibits TAK1 activity in vitro and in vivo (35, 36). Figure 5A shows a Kaplan-Meier curve (29) demonstrating that, as depicted earlier, CPEB KO mice succumb more readily than WT mice to LPS administration. However, injection of (5Z)-7-oxozeaenol into LPS-injected KO mice caused a nearly 50% increase in their survival, which was similar to that seen in WT animals injected with LPS. The serum IL-6 levels of similarly injected animals are more reflective of the interplay between CPEB and TAK1 than the animal mortality rate. Figure 5B demonstrates that LPS induced a much stronger IL-6 response in the CPEB KO mice than in WT animals and that the TAK1 inhibitor abrogated this response such that it was similar to WT levels (WT LPS). Thus, CPEB and TAK1 control cytokine production and inflammation in vivo, as well as in vitro.
FIG 5.
CPEB regulation of TAK1 in vivo. (A) Kaplan-Meier analysis of WT and CPEB KO mouse survival following LPS and (5Z)-7-oxozeaenol injection (10 animals per group). DMSO, dimethyl sulfoxide; Cont, control; inh, inhibitor. (B) Serum from animals treated as described for panel A was analyzed for IL-6 by ELISA. (C) Schematic of CPEB-dependent NF-κB nuclear translocation. CPEB binds and represses TAK1 mRNA; in the absence of CPEB, an excess of TAK1 elicits prolonged nuclear retention of NF-κB via activation of IKK, p38, and IκBα. In the absence of LPS, NF-κB is retained in the cytoplasm through interaction with IκBα. Upon LPS stimulation, IκBα is phosphorylated and subsequently degraded, thereby releasing NF-κB for translocation to the nucleus and stimulation of IL-6 transcription.
On the basis of the evidence presented above, we propose that CPEB inhibits the synthesis of TAK1, probably by binding 3′ UTR CPEs of TAK1 mRNA. LPS promotes TAK1 mRNA unmasking and induces TAK1 synthesis, which in turn activates the p38 MAP kinase signaling pathway that leads to IκBα destruction, NF-κB nuclear import, and IL-6 production. In the absence of CPEB, all of these events proceed at an accelerated pace (Fig. 5C). However, TAK1 is unlikely to be the only mRNA that CPEB regulates to influence the immune response, which is indicated by the observation that (5Z)-7-oxozeaenol significantly, but not completely, rescued IL-6 levels and mouse survival. Thus, we surmise that perhaps a network of mRNAs under CPEB regulatory control also contributes, perhaps substantially, to inflammatory activity as a result of bacterial invasion.
DISCUSSION
This study demonstrates that CPEB regulates the inflammatory response and does so by modulating the expression of TAK1, which, through stimulation of the p38 MAP kinase signaling cascade, promotes NF-κB-mediated transcription of chemokine and cytokine genes (33). Although CPEB KO mice have a normal life span and display no overt immunological disorders, stimulation with LPS results in excessive serum proinflammatory cytokine levels and increased morbidity. Only upon LPS treatment of animals or cultured cells is CPEB deficiency manifest by elevated cytokine production. Thus, in the nonstressed state, CPEB depletion has little detectable effect on inflammation and only upon the application of stress (i.e., LPS) does CPEB deficiency allow certain signaling events to occur without their normal constraints, leading to an inflammatory response. Thus, CPEB behaves like a stress response protein. Consider that CPEB-deficient primary mouse or human cells do not senesce as do WT cells but are immortal (22, 23). In the brain, CPEB deficiency results in aberrant synaptic function, which is induced by another type of stress, electrical stimulation (17, 20). Various signaling events may converge on CPEB or CPEB-bound mRNAs that are normally tightly controlled; when CPEB is absent, unrestricted signaling may produce a range of cell or tissue phenotypes.
In primary macrophages, as well as in a macrophage cell line treated with LPS, ablation of CPEB results in prolonged IκBα destabilization, which leads to increased NF-κB nuclear retention and IL-6 transcription. The observation that IL-6 transcription is also elevated in later passages of MEFs derived from CPEB KO mice because of phosphorylation of p65, the 65-kDa subunit of NF-κB (24), suggests that IκBα destruction is the result but not the cause of the activated NF-κB signaling pathway. Indeed, CPEB has no effect on IκBα synthesis but instead affects its destabilization upon the activation of upstream signaling via p38 and TAK1 kinases. TAK1 mRNA, in particular, contains multiple 3′ UTR CPEs (which also resemble AREs), is bound by CPEB, and is translated at an excessive level when CPEB is absent. Thus, CPEB, in this case, likely acts as a repressor protein to mediate the inflammatory immune response via TAK1 mRNA expression. It is important to point out that TAK1 is not the only CPEB-regulated mRNA that influences inflammation. Indeed, in MEFs and very likely in macrophages as well, a large number of mRNAs linked to inflammation are regulated at the translational level by CPEB (25). However, the data presented here certainly indicate that TAK1 mRNA is among the most important CPEB target mRNAs.
In addition to CPEB, other RNA-binding proteins regulate the inflammatory response as well (37, 38). TTP (tristetraproline), AUF1 (AU-rich binding factor 1), HuR (also known as ELAV; embryonic lethal abnormal vision), TIA (T-cell intracellular antigen), and KSRP (K homology-type splicing regulatory protein) bind AREs in cytokine 3′ UTRs that mainly mediate their stability but can also regulate their translation (3). Chemokine mRNAs generally do not contain AREs but instead harbor 3′ UTR GAIT (IFN-activated inhibitor of translation) elements, stem-loop structures bound by GAIT complexes that mediate their translation (39). In several cases, the ablation of these RNA-binding proteins is linked to immune system impairment, increased sensitivity to LPS, and misregulation of cytokine production. For example, mice lacking AUF1 succumb to endotoxic shock because of increased stability of the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and IL-1β (40) and also have severe dermatitis (41). Moreover, depletion of AUF1 from a leukemic cell line (THP-1) (42) somewhat increases TAK1 levels and stimulates NF-κB-mediated gene transcription. In addition, a recent report demonstrated that depletion of hnRNP K also increases TAK1 levels (43). Because these observations are similar to those obtained with CPEB KO mice and cultured cells lacking CPEB, we hypothesize that there might be a causal link between CPEB, AUF1, hnRNP K, and TAK1-dependent inflammation. Therefore, an investigation of the interplay between CPEB and other RNA-binding proteins to control the inflammatory response is warranted. In this vein, the complex relationship among ARE-binding proteins is illustrated by TTP and TIA-1; mice lacking the gene for either of these proteins develop arthritis and have high levels of TNF-α, but animals lacking both genes have low levels of TNF-α and have very severe arthritis (44). Thus, ARE-binding proteins are promiscuous and can compete with or complement one another, as well as regulate the levels of other RNA-binding proteins.
Regulatory cascades that cross talk via multiple RNA-binding proteins are common in developing systems such as oocytes and embryos and may be just as prevalent in the immune system (3, 37). Understanding how the myriad of RNA-binding proteins coordinates the expression of cytokine/chemokine mRNAs is a daunting task, especially given that many of the proteins bind the same or nearly the same sequence. Nonetheless, it is abundantly evident that tuning of the translational apparatus is an essential regulatory feature of the inflammatory immune response.
Supplementary Material
ACKNOWLEDGMENTS
We thank Leslie Berg and Catherine Yin for help with fluorescence-activated cell sorter analysis. We thank Tsuyoshi Udagawa for shRNA-CPEB constructs.
M.I. was supported by institutional postdoctoral training grant T32 HD07312, and I.M.A. was supported by institutional postdoctoral training grant T32 HD07349. This work was funded by NIH grant GM46779.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00800-14.
REFERENCES
- 1.Bezbradica JS, Medzhitov R. 2009. Integration of cytokine and heterologous receptor signaling pathways. Nat Immunol 10:333–339. doi: 10.1038/ni.1713. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R, Horng T. 2009. Transcriptional control of the inflammatory response. Nat Rev Immunol 9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P. 2010. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 4.Xiao C, Ghosh S. 2005. NF-kappaB, an evolutionarily conserved mediator of immune and inflammatory responses. Adv Exp Med Biol 560:41–45. doi: 10.1007/0-387-24180-9_5. [DOI] [PubMed] [Google Scholar]
- 5.Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. 1997. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science 278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 6.Stancovski I, Baltimore D. 1997. NF-kappaB activation: the I kappaB kinase revealed? Cell 91:299–302. doi: 10.1016/S0092-8674(00)80413-4. [DOI] [PubMed] [Google Scholar]
- 7.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. 1997. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKalpha and IKKbeta, necessary for IkappaB phosphorylation and NF-kappaB activation. Cell 91:243–252. doi: 10.1016/S0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann A, Baltimore D. 2006. Circuitry of nuclear factor kappaB signaling. Immunol Rev 210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 9.Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. 1995. Rel/NF-kappa B/I kappa B family: intimate tales of association and dissociation. Genes Dev 9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 10.Sakurai H. 2012. Targeting of TAK1 in inflammatory disorders and cancer. Trends Pharmacol Sci 33:522–530. doi: 10.1016/j.tips.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Hao S, Baltimore D. 2009. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T, Kruys V, Huez G, Gueydan C. 2002. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem Soc Trans 30:952–958. doi: 10.1042/bst0300952. [DOI] [PubMed] [Google Scholar]
- 13.Hake LE, Richter JD. 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 14.Richter JD. 2007. CPEB: a life in translation. Trends Biochem Sci 32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Tay J, Hodgman R, Sarkissian M, Richter JD. 2003. Regulated CPEB phosphorylation during meiotic progression suggests a mechanism for temporal control of maternal mRNA translation. Genes Dev 17:1457–1462. doi: 10.1101/gad.1071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tay J, Richter JD. 2001. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell 1:201–213. doi: 10.1016/S1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- 17.Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD. 2004. Selective modulation of some forms of Schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Mem 11:318–327. doi: 10.1101/lm.72704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richter JD, Klann E. 2009. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev 23:1–11. doi: 10.1101/gad.1735809. [DOI] [PubMed] [Google Scholar]
- 19.Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. 2012. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell 47:253–266. doi: 10.1016/j.molcel.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zearfoss NR, Alarcon JM, Trifilieff P, Kandel E, Richter JD. 2008. A molecular circuit composed of CPEB-1 and c-Jun controls growth hormone-mediated synaptic plasticity in the mouse hippocampus. J Neurosci 28:8502–8509. doi: 10.1523/JNEUROSCI.1756-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burns DM, D'Ambrogio A, Nottrott S, Richter JD. 2011. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns DM, Richter JD. 2008. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev 22:3449–3460. doi: 10.1101/gad.1697808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. 2006. Control of cellular senescence by CPEB. Genes Dev 20:2701–2712. doi: 10.1101/gad.1438906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Groppo R, Richter JD. 2011. CPEB control of NF-kappaB nuclear localization and interleukin-6 production mediates cellular senescence. Mol Cell Biol 31:2707–2714. doi: 10.1128/MCB.05133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexandrov IM, Ivshina M, Jung DY, Friedline R, Ko HJ, Xu M, O'Sullivan-Murphy B, Bortell R, Huang YT, Urano F, Kim JK, Richter JD. 2012. Cytoplasmic polyadenylation element binding protein deficiency stimulates PTEN and Stat3 mRNA translation and induces hepatic insulin resistance. PLoS Genet 8:e1002457. doi: 10.1371/journal.pgen.1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkissian M, Mendez R, Richter JD. 2004. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes Dev 18:48–61. doi: 10.1101/gad.1136004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gratacós FM, Brewer G. 2010. The role of AUF1 in regulated mRNA decay. Wiley Interdiscip Rev RNA 1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schott J, Stoecklin G. 2010. Networks controlling mRNA decay in the immune system. Wiley Interdiscip Rev RNA 1:432–456. doi: 10.1002/wrna.13. [DOI] [PubMed] [Google Scholar]
- 29.Kaplan EL, Meier P. 1958. Nonparametric estimation from incomplete observations. J Am Stat Assoc 53:457–481. doi: 10.1080/01621459.1958.10501452. [DOI] [Google Scholar]
- 30.Nagaoka K, Udagawa T, Richter JD. 2012. CPEB-mediated ZO-1 mRNA localization is required for epithelial tight-junction assembly and cell polarity. Nat Commun 3:675. doi: 10.1038/ncomms1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pahl HL. 1999. Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 32.Skaug B, Jiang X, Chen ZJ. 2009. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem 78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 33.Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. 1999. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398:252–256. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 34.Hake LE, Mendez R, Richter JD. 1998. Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol 18:685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ninomiya-Tsuji J, Kajino T, Ono K, Ohtomo T, Matsumoto M, Shiina M, Mihara M, Tsuchiya M, Matsumoto K. 2003. A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem 278:18485–18490. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang D, Hu Y, Sun Q, Zhao J, Cong Z, Liu H, Zhou M, Li K, Hang C. 2013. Inhibition of transforming growth factor beta-activated kinase 1 confers neuroprotection after traumatic brain injury in rats. Neuroscience 238:209–217. doi: 10.1016/j.neuroscience.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 37.Anderson P. 2008. Post-transcriptional control of cytokine production. Nat Immunol 9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 38.Anderson P. 2009. Intrinsic mRNA stability helps compose the inflammatory symphony. Nat Immunol 10:233–234. doi: 10.1038/ni0309-233. [DOI] [PubMed] [Google Scholar]
- 39.Yao P, Potdar AA, Arif A, Ray PS, Mukhopadhyay R, Willard B, Xu Y, Yan J, Saidel GM, Fox PL. 2012. Coding region polyadenylation generates a truncated tRNA synthetase that counters translation repression. Cell 149:88–100. doi: 10.1016/j.cell.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu JY, Sadri N, Schneider RJ. 2006. Endotoxic shock in AUF1 knockout mice mediated by failure to degrade proinflammatory cytokine mRNAs. Genes Dev 20:3174–3184. doi: 10.1101/gad.1467606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadri N, Schneider RJ. 2009. AUF1/HNRNPD-deficient mice develop pruritic inflammatory skin disease. J Investig Dermatol 29:557–570. doi: 10.1038/jid.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar S, Han J, Sinsimer KS, Liao B, Foster RL, Brewer G, Pestka S. 2011. RNA-binding protein AUF1 regulates lipopolysaccharide-induced IL10 expression by activating IkappaB kinase complex in monocytes. Mol Cell Biol 31:602–615. doi: 10.1128/MCB.00835-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liepelt A, Mossanen JC, Denecke B, Heymann F, De Santis R, Tacke F, Marx G, Ostareck DH, Ostareck-Lederer A. 2014. Translation control of TAK1 mRNA by hnRNP K modulates LPS-induced macrophage activation. RNA 20:899–911. doi: 10.1261/rna.042788.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips K, Kedersha N, Shen L, Blackshear PJ, Anderson P. 2004. Arthritis suppressor genes YIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci U S A 101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.