Abstract
An in silico screen for myogenic long noncoding RNAs (lncRNAs) revealed nine lncRNAs that are upregulated more than 10-fold in myotubes versus levels in myoblasts. One of these lncRNAs, MyoD upstream noncoding (MUNC, also known as DRReRNA), is encoded 5 kb upstream of the transcription start site of MyoD, a myogenic transcription factor gene. MUNC is specifically expressed in skeletal muscle and exists as in unspliced and spliced isoforms, and its 5′ end overlaps with the cis-acting distal regulatory region (DRR) of MyoD. Small interfering RNA (siRNA) of MUNC reduced myoblast differentiation and specifically reduced the association of MyoD to the DRR enhancer and myogenin promoter but not to another MyoD-dependent enhancer. Stable overexpression of MUNC from a heterologous promoter increased endogenous MyoD, Myogenin, and Myh3 (myosin heavy chain, [MHC] gene) mRNAs but not the cognate proteins, suggesting that MUNC can act in trans to promote gene expression but that this activity does not require an induction of MyoD protein. MUNC also stimulates the transcription of other genes that are not recognized as MyoD-inducible genes. Knockdown of MUNC in vivo impaired murine muscle regeneration, implicating MUNC in primary satellite cell differentiation in the animal. We also discovered a human MUNC that is induced during differentiation of myoblasts and whose knockdown decreases differentiation, suggesting an evolutionarily conserved role of MUNC lncRNA in myogenesis. Although MUNC overlaps with the DRR enhancer, our results suggest that MUNC is not a classic cis-acting enhancer RNA (e-RNA) acting exclusively by stimulating the neighboring MyoD gene but more like a promyogenic lncRNA that acts directly or indirectly on multiple promoters to increase myogenic gene expression.
INTRODUCTION
Long noncoding RNAs (lncRNAs) are rapidly becoming recognized as important regulators of gene expression in development and disease (1, 2). Our genomes undergo widespread transcription (3), and lncRNA genes are in comparable abundance to protein-coding genes (4). While lncRNAs are not as well conserved between species as protein-coding genes (5), there have been a number of examples of lncRNAs that confer marked cellular and developmental phenotypes when their expression is altered by experiment or disease (6–9). Recently, a large class of lncRNAs named e-RNAs, for enhancer RNAs, has been described that are produced from known DNA enhancer elements and that activate transcription of neighboring genes by facilitating enhancer-promoter contacts and/or recruiting core transcription factors such as the mediator complex (reviewed in references 10 and 11).
Skeletal myogenesis is an ordered process where the activation of MyoD in Pax3/Pax7/MyoD-expressing skeletal myoblasts results in a cascade of gene expression changes that lead to terminal differentiation into multinucleated myotubes and myofibers (12, 13). The key transcriptional players in skeletal myogenesis are well known, but the mechanisms of their activation are not fully understood. MicroRNAs (miRNAs) play a significant role in myogenesis at many key points (14). In addition, it is becoming clear that lncRNAs are involved in the regulation of myogenesis. lncRNAs identified as being upregulated during muscle differentiation often overlap MyoD-binding sites across the genome and are transcribed in a MyoD-dependent manner (15). These lncRNAs are enriched in the enhancer regions of MyoD target genes and may play a role in facilitating expression of a neighboring cis-located gene, as reported for other e-RNAs that facilitate chromatin looping and recruitment of Mediator to transcriptional start sites (TSS) (8).
Categorizing lncRNAs into classes with common roles is not yet possible by sequence analysis. With the advent of widely available public databases of genome-wide histone modification, transcription factor binding, and RNA expression and the development of lncRNA prediction models (5), it is now possible to predict lncRNAs in a variety of developmental contexts in silico. Using some of these available data sets, we discovered a number of lncRNAs whose expression is activated during skeletal myogenesis and that are expressed in mature skeletal muscle. In this report we focus on one lncRNA located upstream to MyoD in the genome and demonstrate that it acts in trans to facilitate MyoD's role in initiating the myogenic cascade in vitro in mouse and human myoblasts, as well as in vivo during skeletal muscle regeneration in response to injury.
MATERIALS AND METHODS
Cell culture.
Under growth conditions C2C12 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS); when cells were differentiating, medium was switched to DMEM containing 2% FBS. C3H 10T1/2 cells were grown in Eagle's basal medium with 2-mM l-glutamine, 1.5 g/liter sodium bicarbonate, and Earle's balanced salt solution (BSS), with 10% FBS for growth conditions and 2% FBS for differentiation conditions. LHCN cells were cultured on gelatin-coated plates in DMEM-M199 medium (4:1, vol/vol), supplemented with 15% FBS, zinc sulfate (0.03 μg/ml), vitamin B12 (1.4 μg/ml), dexamethasone (0.055 μg/ml), hepatocyte growth factor (HGF; 2.5 ng/ml), basic fibroblast growth factor (bFGF; 10 ng/ml), and HEPES (0.02 M). Differentiation was performed in serum-free DMEM-M199 medium (4:1, vol/vol), supplemented with HEPES (0.02 M), zinc sulfate (0.03 μg/ml), vitamin B12 (1.4 μg/ml), and insulin (10 μg/ml).
lncRNA screen.
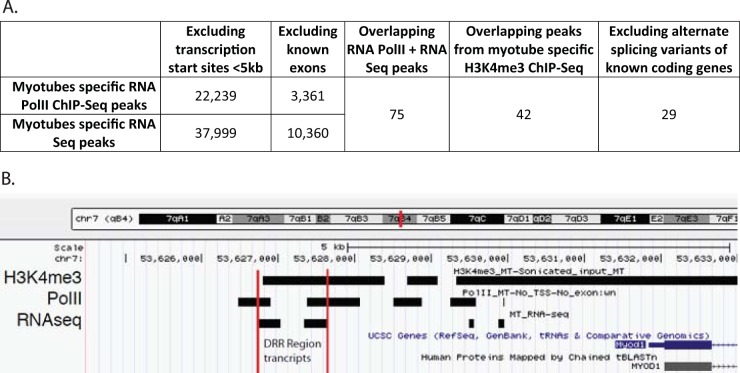
Myoblast sites were defined as the regions identified both by polymerase II (Pol II) chromatin immunoprecipitation with high-throughput sequencing (Chip-Seq) (Gene Expression Omnibus [GEO] accession number GSM721286) and high-throughput sequencing of RNA transcripts (RNA-Seq) (GSM521256) and by H3K4me3 (histone H3 trimethylated at lysine 4) Chip-Seq (GEO accession number GSM721292) experiments in myoblast cells. Myotube sites were defined by similar experiments in myotube cells (GEO accession numbers GSM721287, GSM521258, and GSM721293). Both sites were filtered for previously known transcripts, including protein-coding and non-protein-coding genes, known miRNAs, and transcription start sites. All of these filters were retrieved for the mm9 assembly on 12 June 2012 from the University of California Santa Cruz (UCSC) Genome Browser. Finally, myoblast sites were filtered for any sites common to myotubes, and myotube sites were filtered for any sites common to myoblasts (Fig. 1).
FIG 1.
(A) Workflow (left to right) of the computational screen that identified potential long noncoding RNAs induced during muscle differentiation. (B) UCSC Genome Browser screenshot showing the locations of Pol II ChIP-Seq, H3K4me3 ChIP-Seq, and RNA-Seq signals. Chr7, chromosome 7.
siRNA transfection of cells.
Cells were transfected either with Life Technologies Silencer Select small interfering RNAs (siRNAs) targeting MUNC (target sequence GGAUGAGCUGUGUGCUUCU or CGACCAAUGGGAGAGAGCA) or with commercial negative-control Silencer Select siRNAs. The Silencer Select RNAs have proprietary modifications that allow them to efficiently target nuclear RNAs. siRNA was transfected at a final concentration of 30 nM in growth medium with 3 μl/ml total medium volume of Lipofectamine RNAimax. siRNA and Lipofectamine were mixed in 2 ml/10 ml total medium volume Opti-Mem for 25 min prior to addition to cells seeded onto plates at 25% confluence.
Stable overexpression of MUNC in C2C12 cells.
Using amplified sequences of unspliced MUNC (PCR with C2C12 genomic DNA as a template), an insert was cloned into the pLPCX vector via the In-Fusion method (Clontech). The construct was linearized and introduced into the cells (XtremeGENE transfection reagent; Roche). After 24 h, pools of stably transfected cells were selected with puromycin.
RNA expression by quantitative reverse transcription-PCR (qRT-qPCR).
RNA was isolated by TRIzol extraction, and cDNA synthesis was performed using Life Technologies Superscript III RT cDNA synthesis kits using random hexamer priming. Prior to cDNA synthesis, RNA samples were treated with RQ1 RNase-free DNase to eliminate potential DNA contamination of samples. Quantitative PCR (qPCR) was performed with a Bio-Rad iCycler, using iQ SYBR green Supermix. Primers for the noncoding RNA (ncRNA) screen were designed using BatchPrimer, version 3. All primers used in this study are listed in Tables 1 and 2.
TABLE 1.
Primers used for RT-qPCR confirmation of lncRNAs overexpressed in myotubes
| ncRNA no. | Direction of primera | Sequence |
|---|---|---|
| 1 | F | TTCGTGAGAGTATCCCACAGG |
| R | TGTGAAGAGGAGATGTCCAGAA | |
| 2 | F | AGCCTCAGGATGAGCTGTGT |
| R | CTCAATGCAGGGCCTCTTAG | |
| 3 | F | TTCCAAAAAGGAGGAAGCAA |
| R | ATGGATGTGGGGTTCATCAT | |
| 4 | F | CCAATGCTAAACAACCATCTGA |
| R | ATCCATTTGGAGGGCACTG | |
| 5 | F | CAGGACCTTTGCACATGTTT |
| R | GGATGAAGGGAGACAGAAGC | |
| 6 | F | TAAGGGTAAAGGCGGAGCTA |
| R | TTGCAGACTCCGCTCAGTAA | |
| 7 | F | TCGACATACCCTGTCTGCAA |
| R | CTTCCCATCTCCCAGTGTTG | |
| 8 | F | CCATGTGCAAGAACTCCAAA |
| R | TGGTACCCCTTCTCCAAATG | |
| 9 | F | GACCTTGACCTTTCCCCAGT |
| R | TTCCAGCTCTGTGTGGTCAG | |
| 10 | F | CACATGGATCCCTGGAGTG |
| R | AGAGCATGCCTTCATTCTCAA | |
| 11 | F | CAGCAGAGGTTGGTCCTCTT |
| R | GGAGGTGGGTATGCAGTGAG | |
| 12 | F | ACCATGGAGCCATTCACTTT |
| R | AGCTATTTTGGGAGCGCTTA | |
| 13 | F | GCTTGGTGTCCCTCAGTGAT |
| R | GTGCTCTCAGCCACACAGAA | |
| 14 | F | CCCGACTGGAGATCCTCATA |
| R | AGTAGGGGTTTGGGCAGAGT | |
| 15 | F | GACATAGGGAGGGTCCCAGT |
| R | AGGTAGTGTTCCTGGCTTGC | |
| 16 | F | TAGCGCCAGTCTTCTTCAGG |
| R | GTTAGAGCCAGGGCCTCAAT | |
| 17 | F | ATCTGACCTGCCAGGAAGC |
| R | CGTCTTTTCCTGTTCTCTTCCA | |
| 18 | F | CCCACAGGGACAGAGATAGG |
| R | TCTCTGTGACAGCTGGAGGA | |
| 19 | F | AGTCAGACCAGGCATCTTGC |
| R | ACAAGCCTTTCCCTTTCCTC | |
| 20 | F | GGGCACAGATGGTGAGTTG |
| R | CTGGAGTGTGGGCTGCTG | |
| 21 | F | AAATGTGTGTGTGGGTACGTG |
| R | GGGGGAATGTTCAAGACCTTA | |
| 22 | F | GGGGTTGGAACAGTGAAGAA |
| R | CATTAGCTCCAGCAGGCATT | |
| 23 | F | GCCTAGATGGTTGGCATTGT |
| R | TGAGTGGGTAAGGCACACAG | |
| 24 | F | TGTGCTTGCCCATACAACTC |
| R | TTGGGACACTGTGTGGGATA | |
| 25 | F | GCCACCCATCTACTTTTCCA |
| R | TCAGGTGCTTTCTGTGCATC | |
| 26 | F | CTGCAGGAAGTGCTGCTCTA |
| R | CAAGCACAGTGGCACAAGAT | |
| 27 | F | CGAAAGTGGACATGTTGTCG |
| R | AATCCTGTGGGGTGTAGCTG | |
| 28 | F | TCTCAGAGGCTCCCAAAGAA |
| R | GGCTTCCCCTTAATCTCCAC | |
| 29 | F | TGAGCTCTGGGGAGTCTCTG |
| R | GGTGGGAAAGAACAGCACAG |
F, forward; R, reverse.
TABLE 2.
Primers and siRNAs used in this study
| Primer name | Direction of primera | Sequence |
|---|---|---|
| qGAPDH F | F | GCACAGTCAAGGCCGAGAAT |
| qGAPDH R | R | GCCTTCTCCATGGTGGTGAA |
| qMHC F | F | TCCAAACCGTCTCTGCACTGTT |
| qMHC R | R | AGCGTACAAAGTGTGGGTGTGT |
| qMYOD F | F | CATCCGCTACATCGAAGGTC |
| qMYOD R | R | GTGGAGATGCGCTCCACTAT |
| qMYOGENIN F | F | AGCGCAGGCTCAAGAAAGTGAATG |
| qMYOGENIN R | R | CTGTAGGCGCTCAATGTACTGGAT |
| q human ACTIN β F | F | G GCACCAGATCATGTTTGAG |
| q human ACTIN β R | R | GAGTCCATCACGATGCCAGT |
| q human MHC F | F | CTTCCCTGCACCAGATTCTC |
| q human MHC R | R | GTATAAGCCCGAGGTGGTGA |
| q human MYOD F | F | GGGGCTAGGTTCAGCTTTCT |
| q human MYOD R | R | GCTCTGGCAAAGCAACTCTT |
| q human MYOGENIN F | F | GCCAGACTATCCCCTTCCTC |
| q human MYOGENIN R | R | GAGGCCGCGTTATGATAAAA |
| q human DRR F | F | CTGGGCAGAGCAGCCAAGGGAGCTG |
| q human DRR R | R | GAGGGGCTCATTTGGTGGGGAGTGGG |
| CER ChIP F | F | GGGCATTTATGGGTCTTCCT |
| CER ChIP R | R | CTCATGCCTGGTGTTTAGGG |
| DRR ChIP F | F | TCAGGCCAGGACCATGTCT |
| DRR ChIP R | R | CTGGACCTGTGGCCTCTTAC |
| Myogenin promoter ChIP F | F | GAATCACATGTAATCCACTGGA |
| Myogenin promoter ChIP R | R | ACGCCAACTGCTGGGTGCCA |
| 1F | F | AGCCTCAGGATGAGCTGTGT |
| 1R | R | CTCAATGCAGGGCCTCTTAG |
| 2F | F | TTCCAAAAAGGAGGAAGCAA |
| 2R | R | ATGGATGTGGGGTTCATCAT |
| Nested 5′ RACE primer outer | R | TCTCTCCCATTGGTCGGTTG |
| Nested 5′ RACE primer inner (R-B′) | R | GTTATTCACCGAGGGACACG |
| F-A | F | TAGCCAAGGGAGCTGAAATG |
| R-B | R | GTTATTCACCGAGGGACACG |
| siMUNC-1 | GGAUGAGCUGUGUGCUUCUTT | |
| siMUNC-2 | CGACCAAUGGGAGAGAGCATT | |
| si HUMAN MUNC | AGUCCUCCCCUCUCAGCUCCCtt | |
| si-MYOD | CCAAUGCGAUUUAUCAGGUGCUUUG | |
| pLPCX-MUNC F (in fusion cloning primer) | F | GCCTCGGCCAAACATCGATA |
| pLPCX-MUNC R (in fusion cloning primer) | R | GAGTCCGGTAGCGCTAGC |
| MUNC GT110AG F (MUNC splicing mutant cloning primer) | F | GAGCTGTGTGCTTCTCCAGAGCAGTGGGCCTACAGCCTAAG |
| MUNC GT110AG R (MUNC splicing mutant cloning primer) | R | CTTAGGCTGTAGGCCCACTGCTCTGGAGAAGCACACAGCTC |
F, forward; R, reverse.
Microarray analysis.
RNA was hybridized to Affymetrix Mouse Exon ST arrays and analyzed for gene expression using Affymetrix Expression Console software and Microsoft Excel. To design the heat map of gene expression, the microarray data were analyzed using Bioconductor. The top 400 genes which demonstrated the most variance between samples were used for unsupervised hierarchical clustering. Gene ontology (GO) analysis of the top gene clusters was performed using DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/home.jsp) (16).
Western blotting.
Cells were lysed in IPH buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% NP-40, 50 mM EDTA), and run on SDS–10% PAGE gels and transferred to nitrocellulose membranes. Membranes were blocked for 30 min in 5% milk containing phosphate-buffered saline–Tween 20 (PBS-T) and incubated overnight with primary antibody in 3% bovine serum albumin (BSA). Secondary antibody incubation was carried out for 1 h after cells were washed and at a 1:4,000 dilution before samples were washed and incubated with Millipore Immobilon horseradish peroxidase (HRP) substrate. Chemiluminescent images were captured on a G:BOX Geldoc system (Syngene). Antibodies and dilutions were used as follows: for MyoD, C-20 at 1:250 (Santa Cruz); for myogenin, 1:250 (Santa Cruz); for myosin heavy chain (MHC), 1:250 (Millipore); for tubulin 1:3,000 (Sigma).
ChIP studies.
Cells were washed in PBS and cross-linked with 1% formaldehyde for 10 min and then treated with 0.15 M glycine for 15 min. Cells were then washed twice in PBS and lysed in lysis buffer containing 1% SDS with 50 mM Tris, 10 mM EDTA, and protease and RNase inhibitors. Chromatin was sonicated to an average size of 300 bp and incubated overnight with protein G Dynabead-antibody complexes, with 2 μg of antibody per 1.5 × 106 cells. After overnight incubation, Dynabeads were washed with radioimmunoprecipitation assay (RIPA) buffer containing 150 mM NaCl, followed by one wash in 500 mM NaCl plus RIPA buffer containing 250 mM LiCl and two washes with Tris-EDTA. Beads were then de-cross-linked overnight at 65°C and treated with proteinase K, RNase A, and RNase H. DNA was then isolated by phenol-chloroform extraction and analyzed by qPCR.
Immunofluorescence.
Cells were plated on glass coverslips in the presence of 30 nM siRNA. Cells were collected in growth medium or after 24, 72, or 120 h in differentiation medium, fixed with 4% formaldehyde in PBS and permeabilized in 0.5% Triton X-100 in PBS. Coverslips were blocked in 5% goat serum. Coverslips were incubated at room temperature with primary antibody for 1 h and with Alexa Fluor 488- or 549-conjugated secondary antibody for 1 h, with three Tris-buffered saline (TBS) washes following each antibody incubation. Coverslips were then mounted with Vectashield mounting solution (Vector Laboratories). Antibodies used were anti-MyoD C-20 antibody (Santa Cruz Laboratories) and antimyosin heavy chain A4.1025 antibody (Millipore). Antibodies were diluted 1:200 in 5% goat serum containing PBS.
Microscopy.
Images were captured using a Nikon Microphot SA upright microscope equipped with a Nikon NFX35 camera using SPOT imaging software (Diagnostic Instruments, Inc.) and a Nikon Plan Apo 60×oil objective lens. Fluorescence images were acquired on the same day using the same exposure times, gamma, and gain between samples. Images were enhanced for brightness and contrast to the same extent within Photoshop.
Polyribosome fractionation and qRT-PCR.
A polysome fractionation assay was performed as described previously (17). The total RNAs from monoribosome and polyribosome fractionations were extracted separately and subjected to qRT-PCR analysis.
Isolation and growth of primary myoblasts.
Mice were genotyped and sacrificed at day 9. The procedure was performed according to the protocol of Springer et al. (18).
Mouse skeletal muscle regeneration following cardiotoxin injury.
Cardiotoxin regeneration experiments were performed as described in Dey et al. 19. Invitrogen in vivo silencers targeting the MUNC-1 sequence were injected on days 2 and 5 of regeneration using Invivofectamine (Invitrogen). Tissue samples from regenerating tibialis anterior (TA) muscle were collected on day 14 following cardiotoxin injection and analyzed by qRT-PCR and immunofluorescence. The use of animals in all of the studies was done following protocols approved by the Animal Care and Use Committee (ACUC) of the University of Virginia.
Microarray data accession number.
Microarray data were deposited in the Gene Expression Omnibus (GEO) database with accession number GSE63673.
RESULTS
lncRNAs induced during myogenesis.
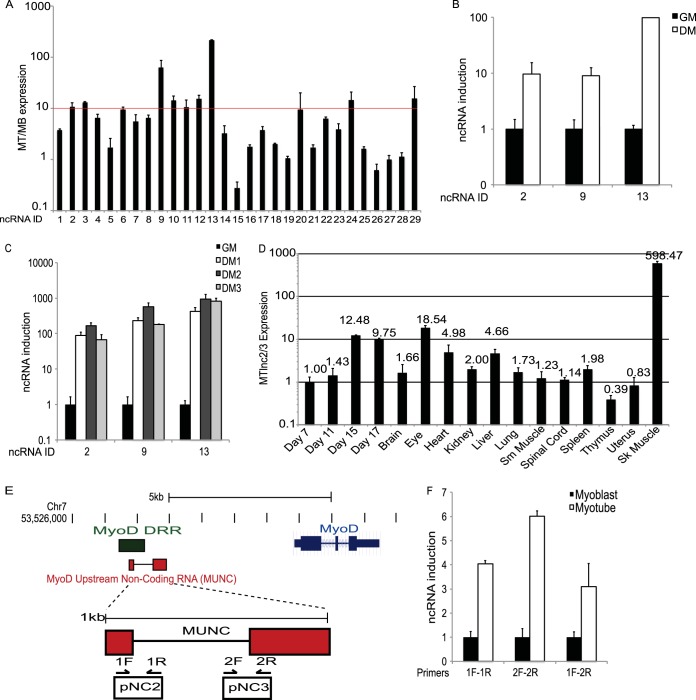
We accessed publicly available data showing where the mouse myoblast and myotube genomes are enriched in histone H3K4 trimethylation (a mark of actively transcribed chromatin), RNA polymerase II (20), and RNA transcripts (21) to identify transcripts that are enriched in myotubes over myoblasts (Fig. 1). From this set we removed all sequences containing known exons and any sequences within 2 kb from known transcriptional start sites (TSS) to predict 29 new potential myotube-specific genes which could be lncRNAs (Table 3). qRT-PCR of RNA from C2C12 myoblasts in growth medium (GM) and from myotubes obtained after 4 days in differentiation medium (DM) confirmed that 20 of these transcripts were induced >2-fold and nine were induced >10-fold in myotubes relative to myoblasts (Fig. 2A). Several of these RNAs were located in the genome proximal to protein-coding genes of interest in skeletal muscle differentiation. We focused on the two most upregulated RNAs, ncRNAs 9 and 13, and on two that were also upregulated >10-fold and encoded very close to each other, ncRNAs 2 and 3. Transdifferentiation of 10T1/2 fibroblasts by MyoD overexpression was accompanied by the induction of these RNAs (Fig. 2B, ncRNAs 2 and 3 show the same patterns; also data not shown). The same RNAs were upregulated on the first day of C2C12 differentiation, approaching peak expression between days 2 and 3 of differentiation (Fig. 2C), and they were present at a very high level specifically in skeletal muscle (Fig. 2D).
TABLE 3.
In silico predicted putative long noncoding RNAs induced during myogenesisa
| Chromosome | RNA-Seq start coordinate (nt)b | RNA-Seq stop coordinate (nt) | RNA-Seq IDc | ncRNA ID |
|---|---|---|---|---|
| 7 | 30766640 | 30766824 | MT_18580 | 1 |
| 7 | 53626843 | 53627031 | MT_19022 | 2 |
| 7 | 53627330 | 53627626 | MT_19023 | 3 |
| 7 | 53629487 | 53629561 | MT_19024 | 4 |
| 7 | 53629872 | 53629954 | MT_19025 | 5 |
| 14 | 22566952 | 22567040 | MT_141 | 6 |
| 14 | 32026105 | 32026292 | MT_248 | 7 |
| 19 | 3765132 | 3765533 | MT_20418 | 8 |
| 8 | 13201970 | 13202444 | MT_1230 | 9 |
| 8 | 13202768 | 13202922 | MT_1231 | 10 |
| 8 | 13203007 | 13203263 | MT_1232 | 11 |
| 8 | 126419999 | 126420073 | MT_2710 | 12 |
| 1 | 20612312 | 20612490 | MT_2953 | 13 |
| 6 | 29381128 | 29381504 | MT_24494 | 14 |
| 6 | 88850233 | 88850446 | MT_25164 | 15 |
| 6 | 149263245 | 149263339 | MT_25901 | 16 |
| 11 | 46206645 | 46206735 | MT_21873 | 17 |
| 11 | 48687223 | 48687425 | MT_21890 | 18 |
| 11 | 50026771 | 50027017 | MT_21919 | 19 |
| 11 | 58952620 | 58952746 | MT_22298 | 20 |
| 16 | 23989822 | 23989993 | MT_6896 | 21 |
| 3 | 14530455 | 14530643 | MT_8317 | 22 |
| 15 | 27958322 | 27958509 | MT_11215 | 23 |
| 4 | 119962964 | 119963170 | MT_26893 | 24 |
| 2 | 91789193 | 91789586 | MT_13961 | 25 |
| 9 | 21891831 | 21892015 | MT_28975 | 26 |
| 13 | 75846117 | 75846436 | MT_16126 | 27 |
| 13 | 75846651 | 75847469 | MT_16127 | 28 |
| 5 | 31912782 | 31912875 | MT_30734 | 29 |
Genomic locations and identifiers of transcripts of >200 bases that are induced in myotubes, overlap myotube-specific H3K4me3 and RNA Pol II occupancy sites, and are located outside of known protein-coding loci. Twenty-nine of these putative lncRNAs were identified in silico for experimental confirmation.
nt, nucleotide.
ID, identification number.
FIG 2.
Predicted muscle-specific noncoding RNAs are upregulated in myotubes. (A) qPCR confirmation of predicted noncoding RNAs induced in myotubes versus myoblasts. Nine predicted RNA-Seq fragments at seven independent genomic loci were >10-fold induced in myotubes (MT) versus levels in myoblasts (MB). (B) qPCR analysis of MyoD transfected transdifferentiating 10T1/2 fibroblasts for several predicted myogenically regulated lncRNAs when cells were transferred to low-serum differentiation medium (DM) versus growth medium (GM). (C) RT-qPCR of lncRNAs from screen in differentiating C2C12 myoblasts on the indicated days after the switch to low-serum differentiation medium. (D) RT-qPCR of lncRNAs 2 and 3 (or MUNC) from mouse embryos (embryonic days 7, 11, 15, and 17) and murine tissues showing high expression in skeletal muscle. (E) Schematic of MUNC genomic region upstream of the MyoD1 locus. MUNC overlaps the previously characterized distal regulatory region (DRR) enhancer and putative noncoding transcripts 2 and 3 (pNC2 and pNC3). (F) RT-qPCR analysis of MUNC expression in primary myoblasts and myotubes. MUNC 1F-1R and 2F-2R primers measure the primary unspliced MUNC while the 1F-2R primers measure spliced MUNC (Fig. 3E). Data represent means ± standard errors of the means (n = 3).
MUNC lncRNA.
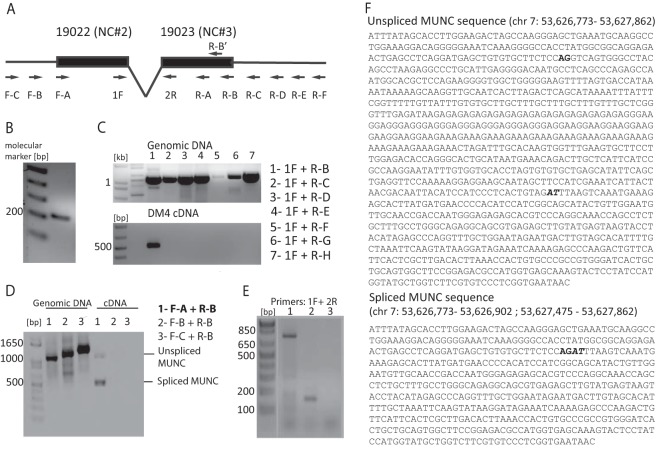
RNAs 2 and 3 were of particular interest because they were located upstream of the TSS of MyoD, a master regulator of skeletal muscle differentiation (12), with ncRNA 2 overlapping a previously known MyoD enhancer element, known as the distal regulatory region (DRR) (Fig. 2E). 5′ cap capture rapid amplification of cDNA ends (RACE) PCR and sequencing of the products confirmed ncRNAs 2 and 3 to be part of the same spliced transcript, with two exons each corresponding roughly to transcripts 2 and 3 (Fig. 3B and F). 3′ RACE PCR using oligo(dT) priming was unable to amplify a product at this locus, suggesting that the RNA was not polyadenylated. RT-PCR with a single forward-directed primer (1F) and 3′ primers located downstream of this locus suggested that the 3′ end of the transcript did not extend beyond primer R-B, at the 3′ end of RNA 3 (Fig. 3C). When we anchored the 3′ primer at R-B and used a series of 5′ primers, RT-PCR failed to give a product with primers upstream from the indicated TSS (Fig. 3D). Interestingly, the 1F and 2R primers revealed that there were two isoforms of the RNA (Fig. 3D, cDNA lane 1), and sequencing the two products showed that the smaller product was from a 518-base spliced RNA composed of the two exons separated by a 577-base intron, while the larger product was a 1,095-base genome-length, unspliced RNA (Fig. 3F). Primers were designed to distinguish the spliced RNA (primers 1F-2R) and unspliced RNA (primers 1F-1R or 2F-2R) (Fig. 2E), and, indeed, RT-PCR with primers 1F-2R selectively detected the spliced RNA (Fig. 3E). qRT-PCR with the three sets of primers showed that both spliced and unspliced isoforms are induced in primary myotubes relative to levels in myoblasts (Fig. 2F).
FIG 3.
Characterization of MyoD upstream noncoding transcript. (A) Schematic of primers used for 5′ RACE PCR and PCR primer walking to determine ends of MUNC transcript. (B) 5′ end mapping of MUNC: PCR product generated by 5′ RACE PCR on cap-captured DM4 C2C12 RNA with the R-B′ (nested) primer. (C) 3′ end mapping of MUNC: PCR products with 1F and indicated R primers on genomic DNA (positive control) and DM4 C2C12 cDNA. Only the R-B primer gave a product on the cDNA, putting it at the 3′ end of MUNC. (D) PCR on genomic DNA (positive control) and cDNA from DM4 C2C12 cells confirms the 5′ end of MUNC and the presence of unspliced and spliced isoforms. Primers F-A plus R-B produced two products on cDNA: genome-length unspliced (∼1,000 bases) and spliced (∼500 bases) cDNAs. (E) PCR products amplified by 1F-2R primers. Lane 1, genomic DNA from C2C12 with extension time of 60 s; lane 2, cDNA from DM3 C2C12 with an extension time of 20 s; lane 3, negative control (no DNA) with an extension time of 20 s. (F) Sequences of unspliced and spliced MUNC forms amplified by F-A and R-B primers. Bold represents the 5′ splice site, and bold italic represents the 3′ splice site. Coordinates are according to the UCSC Genome Browser (assembly of July 2007).
The Coding Potential Calculator (22) tool predicted the unspliced and spliced transcripts to be noncoding RNAs with low coding potential and low evolutionary conservation in any of their short open reading frames (Fig. 4A). Polysome fractionation (Fig. 4B) found very little of the spliced or unspliced transcript in the polyribosome-containing fraction compared to mRNA for the protein MyoD or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Fig. 4C). Thus, these transcripts are noncoding, and we have named them unspliced and spliced MUNC, for MyoD upstream noncoding. While this work was in progress, Mousavi and coworkers independently discovered the unspliced MUNC, naming it DRReRNA (23).
FIG 4.
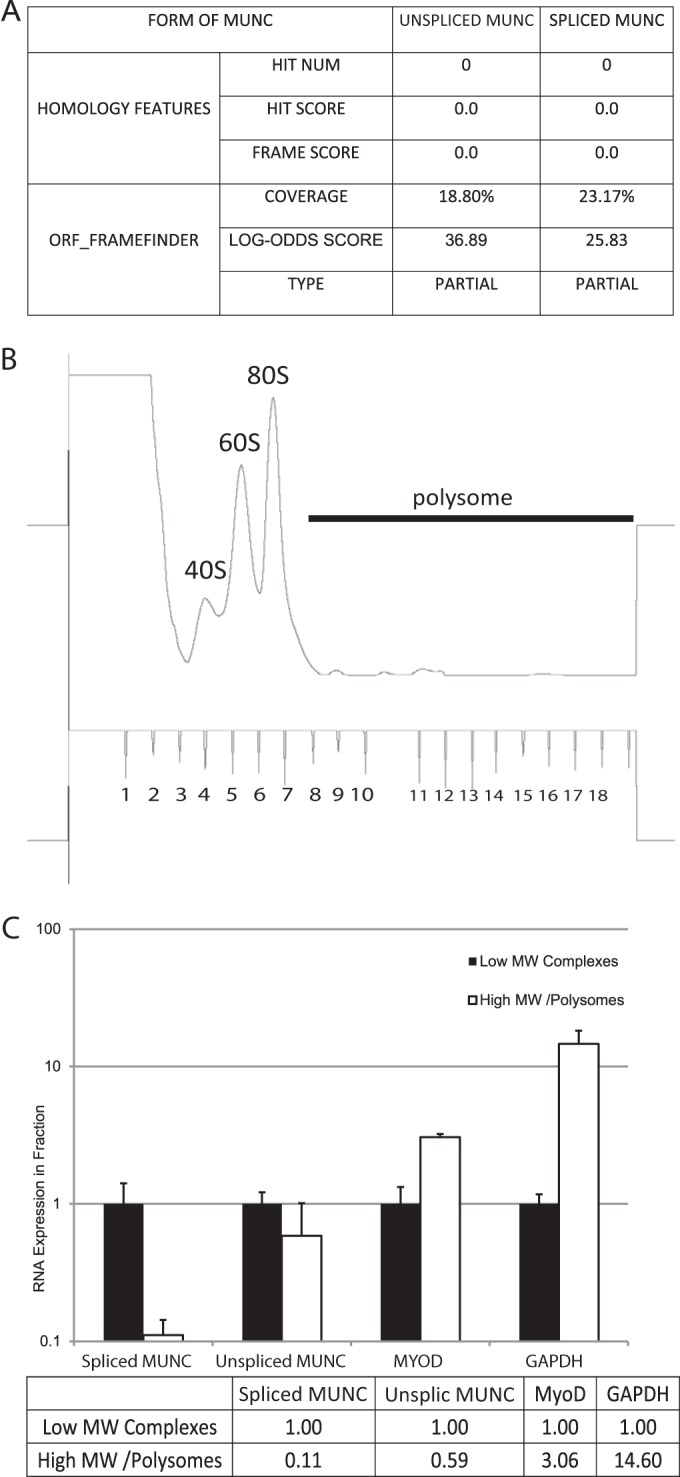
Unspliced and spliced MUNC forms are predicted to have low coding potential and are not associated with polysomes during differentiation. (A) Analysis obtained from the Coding Potential Calculator based on evolutionary conservation and open reading frame (ORF) attributes (http://cpc.cbi.pku.edu.cn/). Both forms of MUNC are likely to be noncoding transcripts. (B) Polysome fractionation profile of differentiating C2C12 cells. Monosomes, fractions 3 to 8; polysomes, fractions 9 to 18. (C) qRT-PCR of polysome fractions and monosome fractions. Spliced and unspliced (Unsplic) MUNCs are depleted from the polysome fraction while the mRNAs for MyoD and GAPDH are enriched in the polysome fraction. Data represent means ± standard errors of the means (n = 3). MW, molecular weight.
Distribution of MUNC in tissues and cells.
lncRNA MUNC, 9, and 13 levels were high in mouse skeletal muscle, but MUNC had the greatest specificity of expression for skeletal muscle (Fig. 2D and 5A and B).
FIG 5.
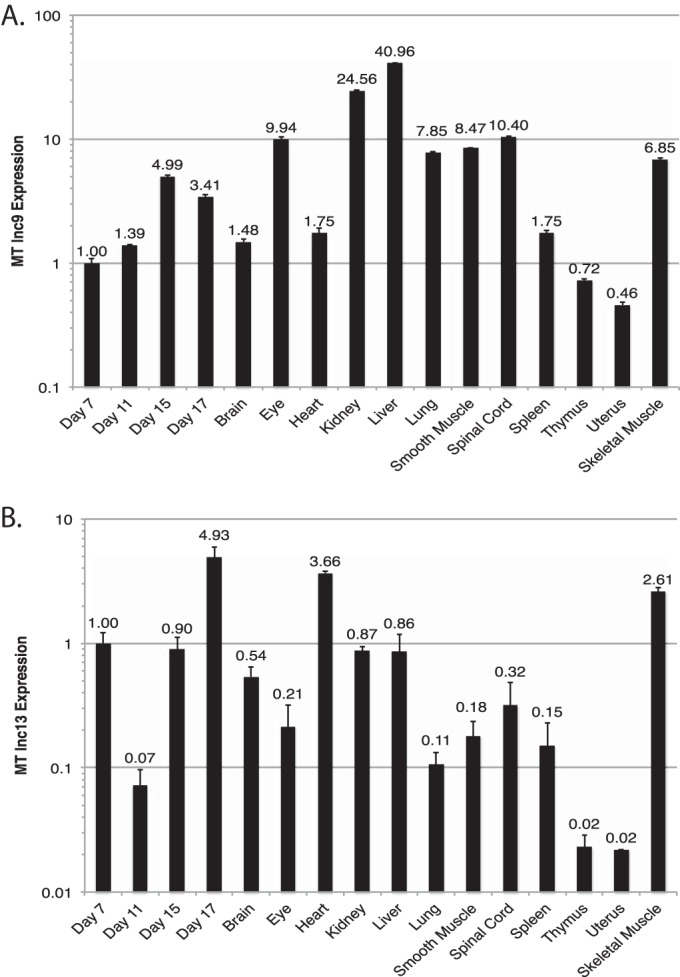
Tissue expression of two other myogenically upregulated long noncoding RNA transcripts, 9 and 13 (lnc9 and lnc13, respectively). Panels A and B show qRT-PCR results of the indicated transcripts across a panel of embryonic and adult mouse tissue samples. Values are normalized to expression of RPS13, a housekeeping gene with low tissue variability, and plotted relative to expression in day 7 embryonic tissue. Data represents means ± standard errors of the means (n = 3). MT, myotube.
Knockdown of MUNC decreases muscle differentiation.
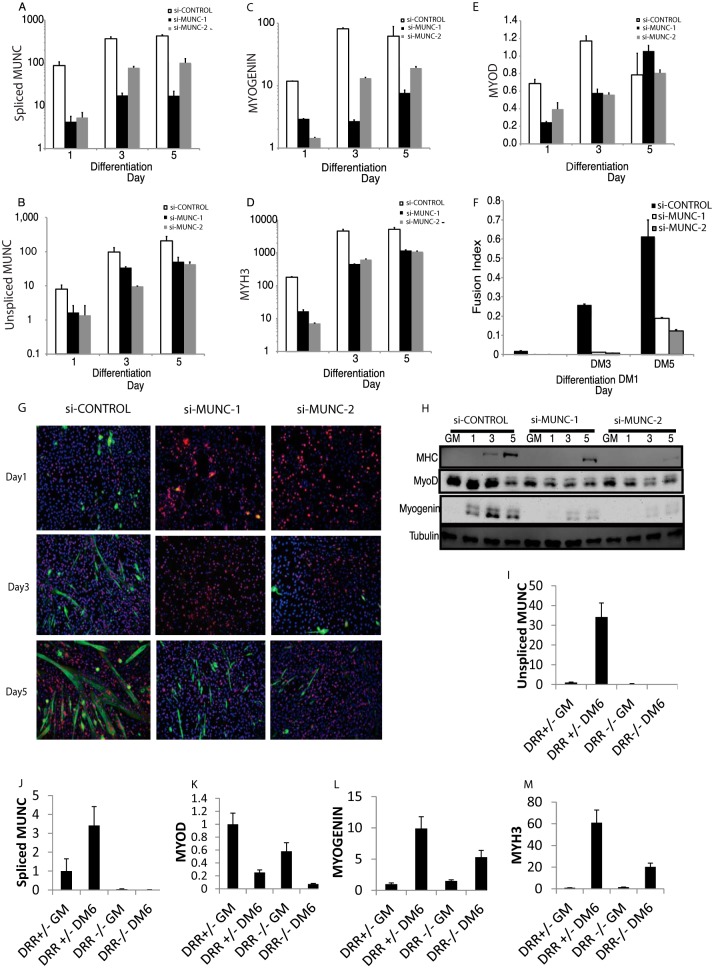
Two independent siRNA duplexes targeting MUNC suppressed the upregulation of spliced and unspliced MUNC over the course of differentiation (Fig. 6A and B). MUNC knockdown significantly repressed the mRNAs of myogenic markers upregulated during differentiation. The mRNA for Myogenin, encoding an early marker of muscle differentiation and a transcription factor essential for muscle differentiation, and for Myh3 (encoding myosin heavy chain [MHC]), a marker of terminal differentiation, were both decreased by nearly 10-fold on days 1, 3, and 5 of differentiation (Fig. 6C and D). MyoD mRNA was repressed 2-fold on days 1 and 3 and not repressed on day 5 of differentiation (Fig. 6E). MHC and myogenin proteins were significantly repressed upon MUNC knockdown (Fig. 6H). MyoD protein was reduced at most 2-fold on days 1 and 3 of differentiation, with no repression on day 5 (Fig. 6H). Immunofluorescence for MHC showed fewer MHC-positive multinucleated myotubes when MUNC was decreased (Fig. 6G), and the fusion index was repressed by at least 5-fold (Fig. 6F). To confirm whether MUNC was equally important for differentiation of primary myoblasts, we isolated primary myoblasts from mice lacking the MUNC locus (DRR−/−) (24) and compared their differentiation ability (after 6 days of differentiation) in vitro with that of primary myoblasts which were heterozygotes for the MUNC locus (DRR+/−). Cells lacking MUNC (Fig. 6I and J) showed a 2-fold decrease of MyoD RNA level in both growth medium and differentiation medium (Fig. 6K). During differentiation the myogenin RNA level was decreased 2-fold (Fig. 6L), and the Myh3 RNA level was diminished 3-fold (Fig. 6M), showing that primary myoblasts lacking MUNC are impaired in differentiation.
FIG 6.
MUNC knockdown represses the induction of myogenic differentiation markers and impairs myotube formation in culture. (A to E) qRT-PCR measuring induction of unspliced and spliced MUNC (I and J) and myogenic markers MyoD, myogenin, and Myh3 (MHC) during differentiation of C2C12 cells incubated with either control siRNA or siRNA targeting the 5′ or 3′ end of MUNC. MUNC levels were normalized to those of GAPDH. Data represent means ± standard errors of the means (n = 3). Note log scale of the y axis in panels A to D. (F) Fusion index of differentiating C2C12 cells shows that MUNC knockdown impairs myotube formation. The fusion index was calculated by dividing the number of nuclei contained within multinucleated cells by the number of total nuclei in a field. (G) Immunofluorescence of MHC (green) and MyoD (blue) in differentiating C2C12 cells. C2C12 cells incubated with control siRNA show much greater formation of MHC-positive, multinucleated cells on differentiation days 3 and 5 than cells incubated with siRNA targeting MUNC. Data represent means ± standard errors of the means (n = 3). (H) Western blot analysis of MHC, myogenin, and MyoD in differentiating C2C12 cells. Independent siRNA targeting MUNC reduces expression of these myogenic proteins. (I to M) qRT-PCR measuring induction of unspliced and spliced forms of MUNC (I and J) and myogenic markers MyoD, myogenin, and Myh3 (K to M) during differentiation of primary murine myoblasts derived from DRR+/− and DRR−/− mice. Expression levels are normalized to those of GAPDH. Data represent means ± standard errors of the means (n = 3).
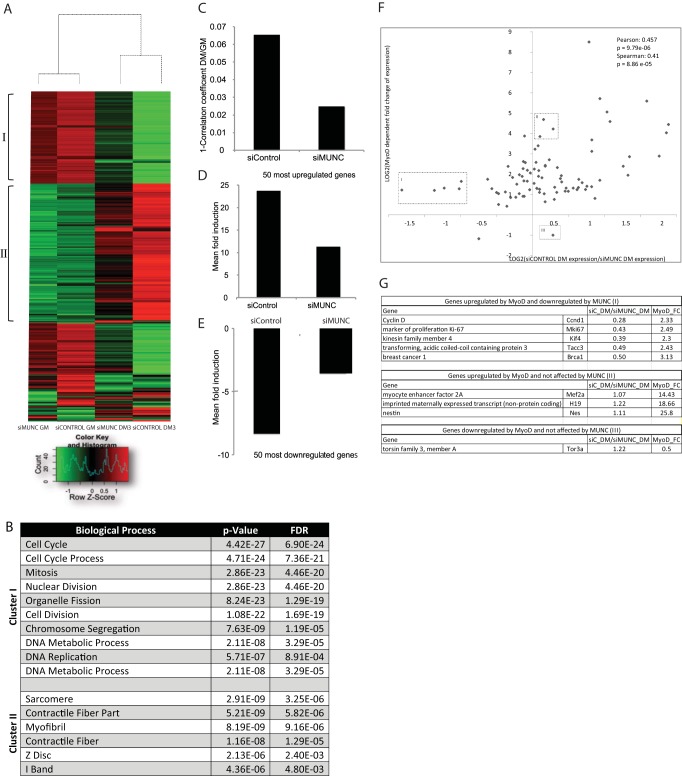
A global analysis of gene expression in C2C12 cells during differentiation was carried out by hybridization of cDNAs to Affymetrix microarrays. Unsupervised hierarchical clustering of the gene expression patterns shows two groups of genes, I and II, that are repressed and induced during differentiation, respectively (Fig. 7A, compare siControl GM versus DM3). Gene ontology enrichment analysis shows that the repressed genes are enriched in genes related to cell proliferation and that the induced genes are enriched in genes related to muscle function (Fig. 7B). Knockdown of MUNC clearly attenuated the extent of repression of cluster I genes related to cell proliferation or induction of cluster II genes related to muscle function (Fig. 7A). The vast majority of genes are correlated in expression between cells in GM and DM, but the deviation from a perfect correlation coefficient of 1 is a measure of the extent of gene expression change during differentiation (Fig. 7C). Knockdown of MUNC decreased this deviation from a perfect correlation coefficient from 0.065 to 0.025. Thus, there is less change in gene expression when differentiation is induced after MUNC depletion. Consistent with this, genes that are most upregulated or downregulated during differentiation in cells transfected with an siRNA control (siControl) show a significant attenuation in the fold change in the cells where MUNC is knocked down (Fig. 7D and E). Thus, the global gene expression patterns confirm that MUNC is required for the changes in gene expression that accompany differentiation, probably not because MUNC acts directly on all these genes but because it is indirectly affecting a few key factors during initial steps of myogenesis.
FIG 7.
Global gene expression changes that occur during skeletal myogenesis are inhibited by MUNC depletion. (A) Hierarchical clustering of the 400 genes that varied the most upon differentiation of C2C12 cells. The cells were treated with either MUNC or control silencer RNAs in both GM and DM. Signals are scaled to Z scores of the rows. I and II, clusters I and II, respectively. (B) Clusters I and II represent genes that are enriched in GO terms associated with cell cycle and growth (I) and with muscle-specific processes (II). (C) Deviation from a perfect correlation coefficient of 1 of gene expression profiles of GM versus DM3 cells with either siControl or siMUNC as measured by the Affymetrix exon array analysis. (D and E) Mean expression fold change of the 50 most upregulated or downregulated genes in control differentiating cells. This fold change is suppressed in siMUNC cells. Values are means ± standard errors of the means (n = 5). (F) Values on the y axis represent fold change of genes known to be induced (top) or repressed (bottom) by MyoD (25). Values on the x axis represent the fold change of the same genes after knockdown of MUNC (left, repressed by MUNC; right, induced by MUNC). Boxes represent the following: I, genes induced by MyoD but downregulated by MUNC; II, genes induced by MyoD but not regulated by MUNC; III, genes downregulated by MyoD but not affected by MUNC. (G) Description of the genes belonging to the three subclasses (boxes I to III) identified in panel F. FDR, false discovery rate.
As shown in Fig. 7F, we compared the fold change in expression of genes affected by knockdown of MUNC (left quadrants, genes repressed by MUNC and so induced by siMUNC; right quadrants, genes induced by MUNC; top quadrants, genes induced by MyoD; bottom quadrants, genes repressed by MyoD) with genes that are known to be induced by MyoD (25). There was a statistically very significant positive correlation between the effects of MyoD and MUNC, suggesting that MUNC and MyoD stimulate similar promyogenic genes. However, there were notable exceptions. In Fig. 7F and G, we highlight in clusters I to III a few genes upregulated by MyoD that were not similarly regulated by MUNC and one gene that was repressed by MyoD but induced or not affected by MUNC.
Knockdown of MUNC decreases MyoD binding to the DRR but not as much to the CER.
To gain insight into the mechanism by which MUNC facilitates muscle differentiation, we examined by ChIP the binding of MyoD and myogenin to the regulatory sites known to bind the two transcription factors, the core enhancer region (CER) at kb −20 relative to MyoD TSS and the DRR (at kb −5 relative to MyoD TSS), and to the sites in the kb −0.5 region of the Myogenin promoter (Fig. 8). These results are expressed as the amount of DNA (relative to input) detected in each ChIP assay (Fig. 8A, B, and C). Under control siRNA conditions, after 3 days in differentiation medium, all three loci were enriched in the MyoD ChIP over IgG ChIP. With MUNC knockdown, however, MyoD binding was decreased to <5% at the DRR, to 40% at the Myogenin promoter, and to 60% at the CER (Fig. 8A to C). This variability between sites could indicate that knockdown of MUNC does not inhibit MyoD binding to all sites simply by repressing MyoD protein levels. A less likely possibility is that the DRR has such a low affinity for MyoD (compared to the other sites) that MyoD binding to the DRR is more severely affected by the 50% decrease of MyoD protein seen after MUNC knockdown. We consider the latter explanation unlikely, given that in control cells >1% of the input DRR locus is precipitated with MyoD while a much smaller proportion of the two other sites (0.05 to 0.12%) is precipitated with MyoD, suggesting that the DRR is preferentially occupied by MyoD in control cells.
FIG 8.
MUNC is required for MyoD binding at certain target sites and not others. MUNC expression is dependent on MyoD. Graphs represent results of MyoD and myogenin (MyoG) ChIP at the MyoD distal regulatory region (A) Myogenin promoter region (B), and MyoD core enhancer region (C). Cells were treated with either control siRNA or siMUNC and incubated in differentiation medium for 72 h. Data represent means ± standard errors of the means (n = 3). (D to F) MUNC expression level in proliferating C2C12 cells (GM) and in differentiating cells (differentiation day 4, DM4) under control conditions (siRNA targeting luciferase GL2 [siGL2]) and after MyoD knockdown (siMyoD). Cells were transfected with siRNA and harvested 48 h later (GM) or retransfected (at DM0 and DM2) and incubated in differentiation medium for 4 days (DM4). Data represent means ± standard errors of the means (n = 3).
Myogenin is known to facilitate chromatin remodeling in a MyoD-dependent manner (26) and binds to the DRR, CER, and its own promoter during myoblast differentiation (27). Knockdown of MUNC decreased the binding of myogenin at the DRR to 60% (Fig. 8A) without any effect at the Myogenin promoter or at the CER (Fig. 8B and C). The milder impairment of myogenin binding to the DRR compared to MyoD binding suggests that there is some specificity of the action of MUNC on MyoD binding to the DRR that is unlikely to be explained by MUNC expression simply opening the local chromatin at the DRR to make it more accessible to all transcription factors.
MUNC expression is dependent on MyoD presence.
MyoD ChIP analysis showed that MUNC is required for MyoD binding to the DRR, which overlaps the 5′ end of the MUNC locus. To test whether MUNC induction during differentiation is itself dependent on MyoD, we checked the MUNC expression level after MyoD knockdown. MyoD depletion in differentiating cells downregulated expression of both unspliced and spliced MUNC (Fig. 8D to F). Given that MUNC is required for MyoD expression and binding to the DRR, these results suggest a positive feedback loop between MUNC and MyoD, where both myogenic factors promote the expression of the other.
Overexpression of MUNC stimulates MyoD, Myogenin, and Myh3 mRNA levels but not the proteins.
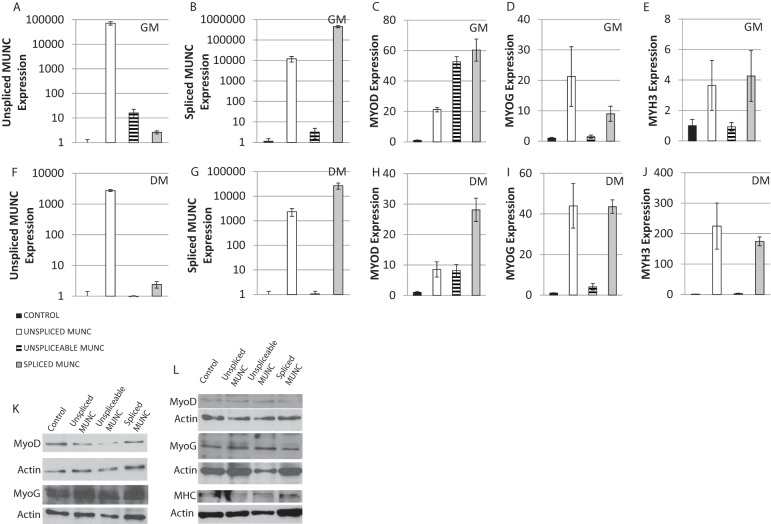
To assay whether expression of MUNC from a heterologous site activates MyoD in trans, we examined the effect of stable overexpression of exogenous MUNC on C2C12 cells. Overexpression of all three forms of MUNC, i.e., MUNC wild type (WT; expressing both unspliced and spliced MUNC), unspliceable MUNC (unspliced MUNC with a point mutation in the splice donor sequence [AG→AT], interfering with splicing), and spliced MUNC. The levels of MUNC, MyoD, myogenin, and Myh3 were measured in GM (Fig. 9A to E) and after 3 days in DM (Fig. 9F to J).
FIG 9.
Stable overexpression of MUNC enhances RNA of myogenic markers but not that of the corresponding proteins. qRT-PCR expression of MUNC isoforms and myogenic markers following C2C12 transfection with linearized vectors encoding the WT unspliced form of MUNC, an unspliceable form of MUNC with a point mutation preventing RNA splicing, and a spliced form of MUNC. Measurements were performed on proliferating cells (GM) and differentiating cells after 3 days in differentiation medium (DM3). Expression data of unspliced MUNC (A and F), spliced MUNC (B and G), MyoD (C and H), myogenin (D and I), and Myh3 (E and J) RNAs are shown. Data are normalized to GAPDH expression and then normalized again in each panel to the level in vector-transfected cells in GM or DM. Data represent means ± standard errors of the means (n = 3). (K) Western blot analysis showing level of MyoD and MyoG proteins in C2C12 cells overexpressing MUNC in GM. Actin was used as a loading control. (L) The same experiment as shown in panel K except performed in cells after 3 days in DM.
In GM overexpression of WT unspliced MUNC (105-fold increase) (Fig. 9A) caused a 20-fold increase of MyoD RNA (Fig. 9C) and 15-fold increase of Myogenin RNA (Fig. 9D). There was a mild induction of Myh3 RNA compared to levels in control cells (Fig. 9E). Overexpression of WT unspliced MUNC also induced expression of spliced MUNC (104-fold) (Fig. 9B), but we could not distinguish whether the spliced MUNC was from the exogenously derived transcript or from an endogenous MUNC induced by the exogenous unspliced form. In cells at day 3 in differentiation medium (DM3), overexpression of MUNC WT led to a 1,000-fold induction of both unspliced and spliced isoforms of MUNC (Fig. 9F and G). This was accompanied by a 10-fold increase of MyoD, 40-fold increase of myogenin, and 200-fold increase of Myh3 transcripts (Fig. 9H to J). The lower fold induction of MyoD and myogenin in DM3 is, of course, explained by the induction of the transcripts in the control cells simply by their transfer to differentiation medium.
Overexpression of the unspliceable form of MUNC was not very efficient, with only a 10-fold increase of unspliced MUNC and no increase of spliced MUNC in GM (Fig. 9A and B). Despite this, there was a 50-fold induction of MyoD (Fig. 9C) but no induction of myogenin or Myh3 transcript (Fig. 9D and E). In DM, unspliceable MUNC did not show a marked induction of MUNC RNA (Fig. 9F) but still led to significant induction of MyoD RNA compared to the control (Fig. 9H). Thus, the induction of MyoD RNA does not need supraphysiological levels of MUNC.
Overexpression of spliced MUNC did not increase unspliced MUNC by much but had approximately the same effects on the RNAs of MyoD, myogenin, and Myh3 as WT MUNC in both GM and DM (Fig. 9C to J).
Although we show results with single pairs of primers, to ensure that the full-length MyoD, MyoG, and Myh3 transcripts were induced, we used primers distributed all along the lengths of the RNAs and obtained the same results. Collectively, these results lead us to suggest that spliced and unspliced MUNC are both stimulators of MyoD RNA, while myogenin and Myh3 RNAs could be induced more by spliced MUNC than by unspliceable MUNC. However, the caveat is that the unspliceable MUNC was not overexpressed sufficiently, perhaps accounting for the failure to stimulate myogenin and Myh3 RNAs.
A very interesting result was obtained when we examined the MyoD, myogenin, and Myh3 proteins in the cells overexpressing WT or spliced MUNC. Although there was robust induction of the three RNAs after overexpression of MUNC, the levels of the three proteins were not induced in GM (Fig. 9K) or in DM (Fig. 9L).
This is a very exciting result on two counts. First, MUNC overexpression dissociates the induction of MyoD, myogenin, and Myh3 transcripts from the general differentiation program, perhaps because of the absence of parallel signals that emerge during normal differentiation to stimulate the translation or stability of these proteins. Second, the induction of myogenin and Myh3 RNAs by MUNC, without inducing the MyoD protein, definitively suggests that MUNC's transcription stimulatory function is not secondary to the induction of MyoD protein.
Murine MUNC (mMUNC) is required for skeletal muscle regeneration in vivo.
Finally, we wanted to determine whether MUNC had a physiological role in vivo during skeletal myogenesis. An early surprising finding in the field of skeletal myogenesis was that MyoD is dispensable for embryonic muscle development, with MyoD knockout mice developing skeletal muscle normally and reaching adulthood with minimal defects. This is explained by MyoD's close homolog, Myf5, compensating for the loss of MyoD during embryonic development (28, 29). However, the MyoD−/− mice are impaired in skeletal muscle regeneration following injury, indicating a critical role of MyoD in skeletal muscle satellite cells that have to proliferate and differentiate for successful regeneration of adult skeletal muscle (30). This role of MyoD is mirrored by the requirement of the DRR DNA locus for MyoD expression in adult satellite cells: the DRR is not required for MyoD expression during embryogenesis but must be intact for MyoD to be expressed in adult skeletal myoblasts (31).
To examine whether MUNC (which initiates in the DRR locus) is important for adult skeletal muscle regeneration, we knocked down MUNC in the tibialis anterior (TA) muscle by injection of an siRNA to MUNC (siMUNC) in an emulsion of Invivofectamine (Invitrogen). The 1F-1R and 2F-2R pairs of primers measure unspliced MUNC (Fig. 2E), while the 1F-2R pair of primers measures spliced MUNC (Fig. 3E). The steady-state levels of unspliced MUNC (1F-1R and 2F-2R) and spliced MUNC (1F-2R) are decreased in adult mouse skeletal muscle after 5 days of siMUNC injection compared to the level with siControl injection (Fig. 10A). This was accompanied by a significant decrease in the expression of MyoD and Myogenin, which are normally induced during adult muscle regeneration (32, 33), and Myh3 mRNAs (Fig. 10B), suggesting a role of MUNC in maintaining expression of these RNAs in adult skeletal muscle. Note that although myogenin protein is not seen in adult muscle, the mRNA is normally still detectable at low levels (33).
FIG 10.
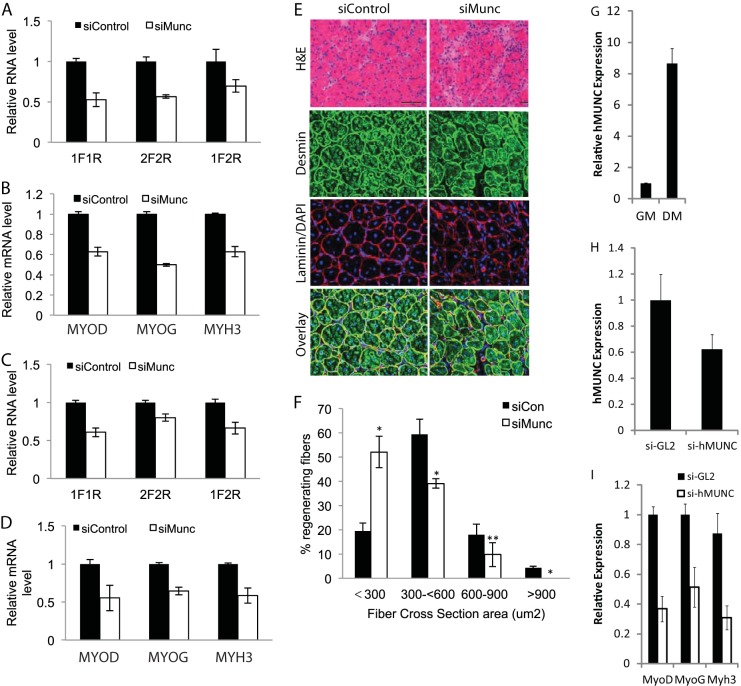
MUNC knockdown reduces myogenic marker expression during skeletal muscle regeneration in adult mice and impairs regeneration. MUNC is conserved between humans and mice. (A and B) qRT-PCR expression of MUNC isoforms (A) and myogenic markers (B) following knockdown of MUNC in three adult mouse TA muscles. Mice were injected twice with siMUNC (Invitrogen in vivo Silencer/Invivofectamine complexes), and RNA was harvested 5 days following the first injection. Control siRNA was injected into the contralateral leg. 1F-1R and 2F-2R measure the 5′ and 3′ regions of the unspliced MUNC, while 1F-2R measure spliced MUNC. Data represent means ± standard errors of the means (n = 3). (C and D) qRT-PCR expression of MUNC isoforms (C) and myogenic markers (D) after knockdown in adult mouse TA muscle 14 days after injury with cardiotoxin. Mice were injected with cardiotoxin and with then siMUNC (Invitrogen in vivo Silencer/Invivofectamine complexes) twice, on days 2 and 5 following injury. RNA was harvested 14 days following the injury. Control siRNA was injected into the contralateral leg. Data represent means ± standard errors of the means (n = 4). (E) Representative hematoxylin and eosin (H&E)-, desmin-, and laminin-stained sections of regenerating mouse TA muscle 14 days after cardiotoxin injection and control or MUNC in vivo siRNA knockdown. DAPI, 4′,6′-diamidino-2-phenylindole. (F) Quantitation of myofiber cross-sectional area. Data represents means ± standard errors of the means (n = 4). For statistical analysis, Student's t test was used. *, P < 0.05 (significant); **, P > 0.05 (not significant). (G) Induction of expression of human MUNC (hMUNC) RNA during differentiation of LHCN cells. (H and I) qRT-PCR analysis of human MUNC RNA (H) and myogenic markers (I) in LHCN human myoblasts after transfection with siRNA targeting human MUNC (si-hMUNC) and 7 days of in low-serum differentiation medium. Data are normalized to beta-actin expression. Data represent means ± standard errors of the means (n = 3).
We next injured the TA muscle with cardiotoxin and followed its regeneration by measuring the appearance of MyoD, Myogenin, and Myh3 RNAs on day 14 after injury. All three RNAs are normally induced during regeneration (31, 32). MUNC was depleted during the 2 weeks of skeletal muscle regeneration by siRNA injection on days 3 and 5 following cardiotoxin injury (Fig. 10C). There was a reduction in MyoD, Myogenin, and Myh3 mRNAs in the regenerated muscle after MUNC knockdown (Fig. 10D). There was a significant reduction in myofiber diameter and an increase in inflammatory infiltrates in the regenerating muscle after MUNC knockdown on day 14 after injury (Fig. 10E and F). This suggests that MUNC has a role in facilitating myogenic gene expression and skeletal muscle regeneration in adult skeletal muscle in vivo.
MUNC is conserved in humans.
lncRNAs are not well conserved in sequence compared to protein-coding RNAs, which prompted us to experimentally determine whether MUNC has a role in human cells as well. Alignment of human DRR sequence with mouse MUNC revealed striking conservation of sequence with the first exon of MUNC after allowing for a 36-base insertion in human DRR (Fig. 11). The sequence conservation extended upstream from the MUNC transcription start site (TSS), but, as shown in Fig. 3C, we did not see any evidence of transcription in mouse myoblasts upstream of the TSS. The conservation also extended to the 5′ region of the intron retained in unspliced mouse MUNC. The region in human DRR that matched mouse MUNC exon 1 is transcribed in human LHCN myoblasts, and the transcript (human MUNC) is significantly upregulated during differentiation (Fig. 10G). siRNA-mediated knockdown of the human MUNC transcript in LHCN cells was not very efficient (Fig. 10H), but despite this, the expression of MyoD, Myogenin, and Myh3 mRNAs was repressed upon differentiation (Fig. 10I). Thus, human myoblasts contain human MUNC RNA similar to mouse MUNC and with similar promyogenic function.
FIG 11.
Alignment between the human DRR sequence plus downstream 1,000 bp (chromosome 11, positions 17714232 to 17716026) and the mouse MUNC locus from 400 bp upstream of the TSS to end of MUNC (chromosome 7, positions 46371003 to 46372492). Blue in the human sequence indicates DRR; red in the mouse sequence indicates MUNC exon 1 and MUNC exon 2.
DISCUSSION
We identified several RNAs whose expression is upregulated during myogenesis. Within this set we discovered a noncoding RNA MUNC produced from a previously characterized enhancer upstream from the MyoD gene, required for MyoD expression in adult skeletal muscle satellite cells (24, 30). We fully characterized its transcriptional isoforms, unspliced and spliced, and showed that both forms can promote muscle differentiation. Previous studies found that this enhancer element facilitates muscle-specific transcription (34). Our findings suggest that this is likely due to the fact that this enhancer encodes an lncRNA which facilitates MyoD binding to the DRR and to a lesser extent to the Myogenin promoter, thereby promoting the expression of MyoD targets, i.e., Myogenin and both MyoD and MUNC itself.
There has been much recent focus on the rapidly expanding roles of lncRNAs in mammalian genomes. Noncoding RNAs have been implicated in regulation of gene expression by facilitating gene and chromosome silencing through the recruitment of PRC2 complexes to chromatin (35). Numerous examples have been discovered suggesting that lncRNAs play a common role in gene silencing in higher eukaryotes (1, 2, 36–38). There have also been discoveries of lncRNAs activating gene expression (11). Most enhancer RNAs (e-RNAs) facilitate expression of a gene neighboring the enhancer in cis by recruiting the transcriptional protein Mediator to chromatin (8). Some e-RNAs can function by facilitating looping of chromatin, bringing regulatory elements on the same chromosome into proximity with each other. MUNC differs from a classic e-RNA (i) because it is required for MyoD binding to chromatin at the myogenin locus (which is not in cis with MUNC or MyoD) and (ii) because it stimulates the expression of MyoD and Myogenin even when it is expressed from a heterologous locus in trans.
Mousavi and coworkers recently reported ncRNA products of the MyoD DRR and CER loci, finding them to facilitate myogenesis (23). In contrast to our study, knockdown of MUNC (which they call DRReRNA) in their study had no effect on MyoD expression, but, as in our study, there was repression of myogenin. We find that MUNC is required for MyoD localization to the DRR of the MyoD promoter and to the Myogenin promoter, while they report that it is required for directing MyoD only to the Myogenin promoter. Additionally, we notice that there is a differential requirement of MUNC for MyoD binding to different genomic targets; core enhancer region binding is not affected as much by siMUNC. In addition we saw only a slight effect in binding of myogenin to the DRR and no effect in binding to the myogenin promoter or CER. The differential effect of MUNC on MyoD and myogenin binding to the DRR suggests that MUNC is unlikely to act at this locus by opening up the chromatin to make it more accessible to all transcription factors. In the future we plan to do MyoD ChIP-Seq and MyoG ChIP-Seq experiments after knockdown of MUNC in differentiating C2C12 cells and compare the results with published data sets (20, 23) to find more binding sites for MyoD and MyoG that are affected by MUNC.
In addition, we find that MUNC expressed from a heterologous artificial locus in trans stimulates both MyoD and Myogenin promoters and not Myogenin alone. Despite these small differences, we agree that such trans stimulation of transcription by MUNC from a heterologous locus eliminates models where transcription stimulation involves specific three-dimensional interactions bringing the MUNC locus in proximity to a neighboring target promoter. We also suggest that an important function of MUNC is to stimulate the auto-activation of the MyoD promoter by facilitating the binding of MyoD to the DRR.
We provide the first evidence of MUNC function in vivo. Depletion of MUNC in regenerating mouse skeletal muscle in vivo impaired expression of myogenic markers and impeded regeneration, suggesting that its function is important for repair of damage to skeletal muscle in adults. Here, too, we saw a decrease in MyoD expression when MUNC was depleted.
One mechanism by which e-RNAs stimulate genes in cis is the transcription of the e-RNA that promotes the local opening of chromatin in cis. Although MUNC induces MyoD transcript, its role in myogenesis does not appear to be limited to MyoD RNA induction alone. MUNC knockdown reduced MyoD binding to target sites in the MyoD and Myogenin promoters. The reduction in MyoD binding at the DRR was much greater than the reduction in expression of MyoD itself and was not uniform at all MyoD binding sites, suggesting that MUNC facilitates MyoD binding to specific target sites, including to MyoD's auto-stimulatory DRR enhancer. Most strikingly, overexpression of MUNC stimulated Myogenin and Myh3 transcripts without inducing MyoD protein. Thus, while MUNC is required for MyoD transcription, increasing the level of MyoD protein is not necessary for MUNC to stimulate the transcription of Myogenin or Myh3, clearly suggesting that the promyogenic function of MUNC is not simply on account of its role as an e-RNA that induces MyoD. The global analysis of gene expression changes after MUNC knockdown reinforces this message by highlighting that (i) not all MUNC-regulated genes are necessarily MyoD dependent and (ii) not all MyoD-induced genes require MUNC.
Many uncertainties remain about which e-RNAs are conserved across species. Sequence conservation of lncRNAs is often quite poor between species. Not enough RNA secondary structures have been solved, so it is also difficult to identify lncRNAs conserved by structure alone. Hence, we are excited to show that a MUNC-like transcript is preserved in humans and that it may be equally important for differentiation. Future experiments will test whether human MUNC can complement the loss of mouse MUNC. The overexpression of unspliceable MUNC hints that spliced and unspliced MUNC may have different target specificities, but this conclusion needs to be corroborated by future experiments where we carefully regulate the levels of expression of spliced and unspliceable MUNC so that they are expressed at approximately equal levels.
Our results are in broad agreement with those of Mousavi and coworkers (23), and we conclude by summarizing the new findings in this report. (i) MUNC knockdown downregulates MyoD transcript and MyoD protein. (ii) MUNC is required for expression of muscle differentiation marker genes in primary myoblasts. (iii) An unexpected spliced isoform of MUNC suggests that MUNC is an RNA polymerase II-driven transcript and that not all the transcribed MUNC is simply left on chromatin, like an e-RNA. (iv) The differential requirement of MUNC for MyoD binding to different E boxes suggests a function in addition to regulating MyoD expression. (v) The differential requirement of MUNC for binding of MyoD and myogenin to the DRR site suggests that the role of MUNC is more specific than simply opening up the chromatin at the DRR to give access to all transcriptional factors. (vi) MUNC is required for skeletal muscle regeneration in adult mice. (vii) A human homolog of MUNC suggests evolutionary conservation of MUNC function. (viii) A positive feedback loop between MyoD and MUNC suggests how the two could be involved in turning on a switch toward differentiation. (ix) Induction of MyoD, Myogenin, or Myh3 transcripts by overexpressed MUNC is not sufficient to induce the corresponding proteins, thus dissociating the transcriptional induction program from other aspects of differentiation. (x) The induction of Myogenin and Myh3 RNAs by MUNC does not require an induction of MyoD protein, suggesting that MUNC has actions beyond simply acting as an e-RNA that induces MyoD. (xi) The global analysis of gene expression changes following MUNC knockdown shows that although many MUNC-induced genes are also induced by MyoD, there are clear examples of genes induced by MyoD alone or MUNC alone.
ACKNOWLEDGMENTS
We thank Woodring E. Wright, University of Texas Southwestern Medical Center, for providing us LHCN-M2 cell line. We thank David J. Goldhamer, University of Connecticut, for providing us o C57Bl/6J DRR loxP/loxP mice.
The study was supported by R01 AR053948 and P01 CA104106.
REFERENCES
- 1.Wang KC, Chang HY. 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wapinski O, Chang HY. 2011. Long noncoding RNAs and human disease. Trends Cell Biol 21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. 2011. The reality of pervasive transcription. PLoS Biol 9:e1000625. doi: 10.1371/journal.pbio.1000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. 2012. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A. 2010. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, Triller A, Spector DL, Bessis A. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M-C, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. 2010. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. 2013. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maass PG, Rump A, Schulz H, Stricker S, Schulze L, Platzer K, Aydin A, Tinschert S, Goldring MB, Luft FC, Bähring S. 2012. A misplaced IncRNA causes brachydactyly in humans. J Clin Invest 122:3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mousavi K, Zare H, Koulnis M, Sartorelli V. 2014. The emerging roles of eRNAs in transcriptional regulatory networks. RNA Biol 11:106–110. doi: 10.4161/rna.27950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orom UA, Shiekhattar R. 2013. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkes CA, Tapscott SJ. 2005. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama S, Asahara H. 2011. The myogenic transcriptional network. Cell Mol Life Sci 68:1843–1849. doi: 10.1007/s00018-011-0629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gagan J, Dey BK, Dutta A. 2012. MicroRNAs regulate and provide robustness to the myogenic transcriptional network. Curr Opin Pharmacol 12:383–388. doi: 10.1016/j.coph.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. 2012. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev 26:2763–2779. doi: 10.1101/gad.200113.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Bor Y-C, Misawa Y, Xue Y, Rekosh D, Hammarskjöld M-L. 2006. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature 443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- 18.Springer ML, Rando TA, Blau HM. 2002. Gene delivery to muscle. Curr Protoc Hum Genet Chapter 13:Unit13.4. doi: 10.1002/0471142905.hg1304s31. [DOI] [PubMed] [Google Scholar]
- 19.Dey BK, Gagan J, Yan Z, Dutta A. 2012. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev 26:2180–2191. doi: 10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asp P, Blum R, Vethantham V, Parisi F, Micsinai M, Cheng J, Bowman C, Kluger Y, Dynlacht BD. 2011. Genome-wide remodeling of the epigenetic landscape during myogenic differentiation. Proc Natl Acad Sci U S A 108:E149–E158. doi: 10.1073/pnas.1102223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35(Suppl 2):W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mousavi K, Zare H, Dell'Orso S, Grontved L, Gutierrez-Cruz G, Derfoul A, Hager G, Sartorelli V. 2013. ERNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen JCJ, Ramachandran R, Goldhamer DJ. 2002. Essential and redundant functions of the MyoD distal regulatory region revealed by targeted mutagenesis. Dev Biol 245:213–223. doi: 10.1006/dbio.2002.0638. [DOI] [PubMed] [Google Scholar]
- 25.Bergstrom DA, Penn BH, Strand A, Perry RLS, Rudnicki MA, Tapscott SJ. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell 9:587–600. doi: 10.1016/S1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- 26.Du C, Jin YQ, Qi JJ, Ji ZX, Li SY, An GS, Jia HT, Ni JH. 2012. Effects of myogenin on expression of late muscle genes through myoD-Dependent chromatin remodeling ability of myogenin. Mol Cells 34:133–142. doi: 10.1007/s10059-012-2286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson DS, Mortazavi A, Myers RM, Wold B. 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 28.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. 1992. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell 71:383–390. doi: 10.1016/0092-8674(92)90508-A. [DOI] [PubMed] [Google Scholar]
- 29.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. 1993. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell 75:1351–1359. doi: 10.1016/0092-8674(93)90621-V. [DOI] [PubMed] [Google Scholar]
- 30.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev 10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 31.Chen JCJ, Love CM, Goldhamer DJ. 2001. Two upstream enhancers collaborate to regulate the spatial patterning and timing of MyoD transcription during mouse development. Dev Dyn 221:274–288. doi: 10.1002/dvdy.1138. [DOI] [PubMed] [Google Scholar]
- 32.Zhao P, Hoffman EP. 2004. Embryonic myogenesis pathways in muscle regeneration. Dev Dyn 229:380–392. doi: 10.1002/dvdy.10457. [DOI] [PubMed] [Google Scholar]
- 33.Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS. 2003. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J Biol Chem 278:8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 34.Tapscott SJ, Lassar AB, Weintraub H. 1992. A novel myoblast enhancer element mediates MyoD transcription. Mol Cell Biol 12:4994–5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brockdorff N. 2013. Noncoding RNA and Polycomb recruitment. RNA 19:429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aguilo F, Zhou MM, Walsh MJ. 2011. Long noncoding RNA, polycomb, and the ghosts haunting INK4b-ARF-INK4a expression. Cancer Res 71:5365–5369. doi: 10.1158/0008-5472.CAN-10-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marín-Béjar O, Marchese FP, Athie A, Sánchez Y, González J, Segura V, Huang L, Moreno I, Navarro A, Monzó M, García-Foncillas J, Rinn JL, Guo S, Huarte M. 2013. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. 2013. Binding interactions between long noncoding RNA HOTAIR and PRC2 proteins. Biochemistry 52:9519–9527. doi: 10.1021/bi401085h. [DOI] [PMC free article] [PubMed] [Google Scholar]