Abstract
Macrophages play important roles in many diseases and are frequently found in hypoxic areas. A chronic hypoxic microenvironment alters global cellular protein expression, but molecular details remain poorly understood. Although hypoxia-inducible factor (HIF) is an established transcription factor allowing adaption to acute hypoxia, responses to chronic hypoxia are more complex. Based on a two-dimensional differential gel electrophoresis (2D-DIGE) approach, we aimed to identify proteins that are exclusively expressed under chronic but not acute hypoxia (1% O2). One of the identified proteins was cathepsin B (CTSB), and a knockdown of either HIF-1α or -2α in primary human macrophages pointed to an HIF-2α dependency. Although chromatin immunoprecipitation (ChIP) experiments confirmed HIF-2 binding to a CTSB enhancer in acute hypoxia, an increase of CTSB mRNA was evident only under chronic hypoxia. Along those lines, CTSB mRNA stability increased at 48 h but not at 8 h of hypoxia. However, RNA stability at 8 h of hypoxia was enhanced by a knockdown of tristetraprolin (TTP). Inactivation of TTP under prolonged hypoxia was facilitated by c-Jun N-terminal kinase (JNK), and inhibition of this kinase lowered CTSB mRNA levels and stability. We postulate a TTP-dependent mechanism to explain delayed expression of CTSB under chronic hypoxia.
INTRODUCTION
Chronic diseases such as diabetes, atherosclerosis, and cancer are characterized by hypoxic areas resulting, for example, from compromised perfusion of narrowed or leaky vessels. Cells of the immune system are involved in the outcome of these diseases. As part of the innate immune system, macrophages actively regulate inflammation but also the resolution of inflammation as well as tissue regeneration and remodeling. Macrophages invade hypoxic areas attracted by a number of cytokines produced by hypoxic cells. To survive and operate in a hypoxic environment, cells need a variety of adaptive mechanisms (1, 2).
Hypoxia-inducible factors (HIFs) are important to coordinate hypoxic responses and consist of a constitutively expressed β-subunit and an oxygen-regulated α-subunit. Both are members of the helix-loop-helix/Per, ARNT, and SIM (PAS) transcription factor family (1, 3). Among the α-subunits, HIF-1α and HIF-2α are best characterized. Both contain an oxygen-dependent degradation domain (ODD) with two conserved prolyl residues that are hydroxylated by prolyl hydroxylases (PHDs) 1 to 3 when sufficient oxygen is available, allowing their proteasomal degradation (4, 5). PHDs are impaired under hypoxia, which in turn causes accumulation and translocation of HIF-α into the nucleus. The α-subunit forms a heterodimer with the β-subunit and binds to hypoxia-responsive elements (HRE) in regulatory regions of target genes (6). By recruiting cofactors like p300 or CBP, the HIF proteins enhance transcription of about 400 target genes (7, 8). Although HIF abundance is mostly regulated by protein stability, regulation of HIF-1α mRNA via binding of tristetraprolin (TTP) to AU-rich elements (AREs) in the 3′ untranslated region (UTR) is established (9, 10).
TTP negatively regulates RNA stability, promoting RNA degradation. As a member of the TIS11 family of RNA-binding proteins, TTP binds to AREs located in the 3′ UTR of target mRNAs. About 5 to 8% of the transcriptome contains potential TTP binding sites (11–14). TTP's ability to bind to AREs is restricted by its phosphorylation by various kinases like c-Jun N-terminal kinase (JNK), p38 mitogen-activated protein kinase (p38 MAPK), or extracellular signal-regulated kinase (ERK), all of which are suggested to be activated by hypoxia (15, 16).
Recently, we defined chronic hypoxia and its consequences for macrophages by showing HIF-dependent and -independent adaptations using two-dimensional gel electrophoresis (2D-DIGE) coupled to tandem mass spectrometry (MS/MS) (17). In the proteomic approach, we identified proteins exclusively expressed under chronic hypoxia, including CTSB. Here, we describe an HIF-2α- and TTP-dependent regulatory mechanism to explain enhanced CTSB expression under chronic but not acute hypoxia.
MATERIALS AND METHODS
Cell culture.
Cell lines were purchased from ATCC (LGC Promochem, Wesel, Germany). The monocytic cell line THP-1 was incubated at 37°C with 5% CO2 in RPMI 1640 medium containing stable l-glutamine, 10% fetal calf serum (FCS), and 100 U/ml penicillin and 100 μg/ml streptomycin (P-S; PAA Laboratories, Cölbe, Germany). THP-1 cells were differentiated to macrophages with 10 nM 12-O-tetradecanoylphorbol-13-acetate (TPA) for 5 days, followed by 1-day incubations without TPA.
Isolation of primary human macrophages.
Primary human macrophages were isolated from buffy coats using Leucosept tubes and LSM 1077 medium (GE Healthcare, Munich, Germany). Cells were washed three times with phosphate-buffered saline (PBS) and were allowed to adhere to 6-well plates or 15-cm dishes (Cell+; Sarstedt, Nümbrecht, Germany) for 1 h at 37°C. Nonadherent cells were removed, and the remaining monocytes were incubated for at least 7 days with RPMI 1640 medium containing 5% human serum and P-S. Macrophages were used at a density of approximately 80%.
Transfection of primary human macrophages.
For small interfering RNA (siRNA) transfections, human macrophages were seeded on 6-well plates as described above. A total of 50 nM siRNA against HIF-1α (ON-TARGETplus SMART pool, human HIF1A; Thermo Scientific, Karlsruhe, Germany), HIF-2α (ON-TARGETplus SMART pool, human EPAS1; Thermo Scientific), or TTP (ON-TARGETplus SMART pool, human TTP; Thermo Scientific) was used with 16.8 μl HiPerFect (Qiagen, Hilden, Germany) in 500 μl medium with P-S for each well. After the mix was added, cells were incubated for 24 h. Then medium was removed and replaced by medium containing P-S and human serum to start experiments.
Experimental procedures.
Cells were incubated and harvested at different time points at 1% O2 in a hypoxic incubator (Invivo2 400; Ruskinn Technology, Leeds, United Kingdom). Fresh hypoxic medium was provided to cells after 24 h without reoxygenation. Proteins were harvested under hypoxic conditions, while RNA was harvested directly after cells were removed from the incubator. For RNA stability, primary human macrophages were incubated under hypoxia for 8 h or 48 h and treated with 2.5 μg/ml actinomycin D (ActD; Sigma, Steinheim, Germany) or dimethyl sulfoxide (DMSO; Sigma) as a control 4 h prior to harvesting cells. For kinase inhibition, cells were pretreated with 20 μM SP600125 (Sigma), 10 μM LY294002 (Sigma), or 10 μM SB203580 (Sigma) for 1 h followed by hypoxic incubations. DMSO-treated cells served as a control.
Western analysis.
Cells were resuspended in lysis buffer (6.65 M urea, 10% glycerol, 1% SDS, 10 mM Tris-HCl, pH 7.4) and sonified. After centrifugation (15,000 × g, 5 min), the protein content was determined in the supernatants by a Lowry protein assay kit (Bio-Rad, Munich, Germany). For immune detection of HIF-1α and HIF-2α, 100 μg protein was separated on 7.5% SDS gels, blotted on Immobilon-FL polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Schwalbach, Germany), and incubated with primary antibodies against human HIF-1α (polyclonal; Novus Biologicals, Hiddenhausen, Germany) or HIF-2α (polyclonal; R&D Systems, Abingdon, United Kingdom). For detection of CTSB and TTP, 12% SDS gels were used and incubated with an antibody against CTSB (polyclonal; Abcam, Cambridge, United Kingdom) or a rabbit antiserum against TTP (kindly provided by Pavel Kovarik, University of Vienna, Vienna, Austria). Membranes were incubated with the appropriate secondary antibodies conjugated with horseradish peroxidase, followed by detection using enhanced chemiluminescence (ECL solutions from GE Healthcare).
RNA isolation from human macrophages.
Total RNA was isolated from cell cultures using peqGold (Peqlab, Erlangen, Germany) by following the manufacturer's instructions and stored at −80°C. Concentrations were determined using a NanoDrop ND-1000 spectrophotometer (Peqlab). Reverse transcription (RT) was performed with 1 μg RNA using the Maxima first-strand cDNA synthesis kit for RT-PCR (Thermo Scientific), and cDNA was stored at −20°C.
Quantitative PCR (qPCR).
Gene expression was analyzed using SYBR green fluorescent mix (Thermo Scientific) on a CFX96 real-time PCR detection system (Bio-Rad). Primer sequences are given in Table 1. Analysis of data was performed using Bio-Rad software. mRNA data were normalized to TATA box binding protein (TBP) except for RNA stability measurements, which were normalized to ribosomal 18S RNA showing a higher stability than TBP.
TABLE 1.
Primers used for qPCR
| Target | Primer sequence (5′→3′) |
|
|---|---|---|
| Forward | Reverse | |
| TBP | GGGCCGCCGGCTGTTTAACT | GGGCCGCCGGCTGTTTAACT |
| 18S rRNA | GTAACCCGTTGAACCCCATT | CCATCCAATCGGTAGTAGCG |
| CTSB qPCR | AACACGTCACCGGAGAGATGA | CCCAGTCAGTGTTCCAGGAGTT |
| CTSB ChIP | TGTCATTCAAGAGCCAGGTG | AGCTCATCATGCTTGGCTTT |
| CTSB ChIP start | GCCCTGTTTCCAACTGGTTA | GCTGGGGAAAGAAAAGAACC |
| CTSB ChIP middle | GGGCTGCGTCCTATGTATGT | CAGGGCCATTTTTCAACAGT |
| CTSB ChIP end | GTCTCGGTCTCAGCCAGTTC | CCAGTAGGGTGTGCCATTCT |
| TTP | CAAGTAGCCAAAGCCGTTGCCAAA | ATACAAGGGAAGCAGACGACCCAA |
| SNX5 | GCAGAAACTGGGAGAAGGTG | GAGAAGAAAGCCGCTGAAGA |
| OSTF1 | TTGCATGAAGCAGCAAAAAG | TAAGGCAGTGCTTCCAGCTT |
| H4 | AAGCGCATTTCTGGTCTCAT | AAGGGCCTTTTGGAGTCTGT |
2D-DIGE coupled with MS/MS.
Experimental procedures for 2D-DIGE and MS/MS, including evaluation of results, were performed as described by Fuhrmann et al. (17).
ChIP.
For chromatin immunoprecipitation (ChIP) analysis, 15-cm dishes with primary human macrophages were harvested after 4 h (HIF-ChIPs) or 8 h and 48 h (Pol II-ChIPs) under hypoxia. ChIP experiments were carried out as previously described (18). Briefly, cross-linking was performed in hypoxia using 1% formaldehyde for 10 min at 37°C and quenched by 0.125 M glycine. Cells were then collected after 2 washing steps in PBS and centrifuged at 500 × g for 5 min. Pellets were frozen at −80°C. Cells were resuspended in 1.5 ml L1A buffer (10 mM HEPES-KOH [pH 7.9], 85 mM KCl, 1 mM EDTA [pH 8.0], 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM dithiothreitol [DTT], and protease inhibitor cocktail from Roche, Mannheim, Germany) and lysed in 250 μl of L1B buffer (L1A and 1% Nonidet P-40) for 15 min on a roller mixer at 4°C. Cross-linked chromatin was sheared to an average DNA fragment size around 200 to 500 bp using a Branson Sonifier 250 (Dietzenbach-Steinberg, Germany). DNA quality was estimated by electrophoresis. After centrifugation, 5% of the supernatant was used as an input. After preclearing with Sepharose CL-4B beads for 2 h, equal amounts of chromatin were immunoprecipitated overnight with 4 μg of rabbit polyclonal antibody anti-HIF-1α (Novus Biologicals), 8 μl anti-HIF-2α (the antibody was a kind gift from Chris Pugh, Oxford, United Kingdom), 10 μl anti-Pol II (Cell Signaling), anti-H3 (Merck Millipore), or anti-rabbit IgG (Merck Millipore). Immune complexes were recovered by 2-h incubations with protein A–Sepharose CL-4 beads at 4°C. After reverse cross-linking, DNA was purified using QIAquick PCR purification kits (Qiagen) according to instructions provided. Enrichment of specific DNA fragments in the immunoprecipitated material was determined by qPCR on a CFX96 (Bio-Rad). Primers are listed in Table 1.
Statistics.
Experiments were repeated at least 3 times. Data are expressed as means ± standard errors of the means (SEM). Statistically significant differences were calculated after analysis of variance (ANOVA) and Bonferroni's test or Student's t test; P values of <0.05 were considered significant.
RESULTS
2D-DIGE identifies proteins that are exclusively expressed under chronic hypoxia.
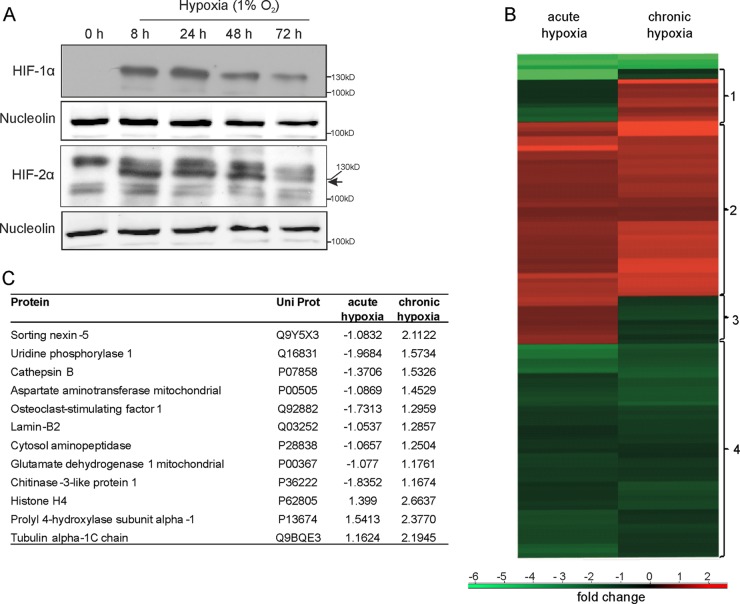
Primary human macrophages were incubated for 8 to 72 h under hypoxia (1% O2) and analyzed for HIF-1α and HIF-2α protein expression (Fig. 1A). HIF-1α expression increased from 8 to 24 h of hypoxia, but protein levels declined after 48 h, while HIF-2α remained stable for 48 h and disappeared after 72 h, possibly by a PHD-dependent destabilization of the HIF proteins. A previous proteome approach in THP-1 macrophages showed effects of chronic hypoxia on several metabolic pathways and cellular responses such as glycolysis, mitochondrial protein regulation, autophagy, and protein folding (17). In order to enhance the reproducibility of the experiments and to reduce the variability naturally occurring in human blood donor samples, proteome analysis was performed with THP-1 cells. To distinguish between acute and chronic hypoxia, we compared protein expression in THP-1 cells after 8 and 72 h of hypoxia using 2D-DIGE followed by MS/MS (Fig. 1B). Proteins were selected for MS by comparing normoxic controls to acute hypoxic and to chronic hypoxic samples, which gave an impression of proteins generally upregulated under each condition (17). To distinguish between chronic and acute hypoxia, the normalized expression levels of both hypoxic groups were compared to each other. We identified four clusters representing distinct expression patterns visualized by the heat map, with responses to either acute or chronic hypoxia being normalized to normoxic controls. Cluster 1 shows proteins exclusively upregulated under chronic hypoxia. All 12 proteins identified in this cluster are shown in Fig. 1C. In contrast, cluster 3 contains proteins only increased in acute hypoxia, including ELAV-like protein 1 or spliceosome RNA helicase DDX39B. Proteins in cluster 2 are upregulated in both acute and chronic hypoxia. Interestingly, enrichment analysis coupled with gene ontology reveals a significant enrichment of proteins involved in glycolysis (P = 3.7883 × 10−5), like glucose-6-phosphate isomerase, phosphoglycerate kinase 1, or glyceraldehyde-3-phosphate dehydrogenase. Cluster 4 contains proteins decreased in both conditions, including mitochondrial proteins (P = 0.02) and proteins located in the endoplasmic reticulum (P = 0.03). Within this cluster, we found, e.g., calreticulin and the protein disulfide isomerases A3, A4, and A6. Since we were specifically interested in proteins that are differently expressed in chronic hypoxia, we further concentrated on proteins appearing in cluster 1.
FIG 1.
Protein expression in response to acute versus chronic hypoxia. (A) Western analysis of HIF-1α and HIF-2α in primary human macrophages incubated for up to 72 h under hypoxia. Expression of nucleolin serves as a loading control. (B) Heat map showing protein expression of 2D-DIGE experiments performed with THP-1 macrophages incubated in acute (8 h) or chronic (72 h) hypoxia. (C) List of the 12 proteins identified by MS/MS from cluster 1 of panel B that are expressed under chronic hypoxia. Expression values are ratios between control conditions and acute or chronic hypoxia.
Chronic hypoxia provokes mRNA changes associated with late protein expression.
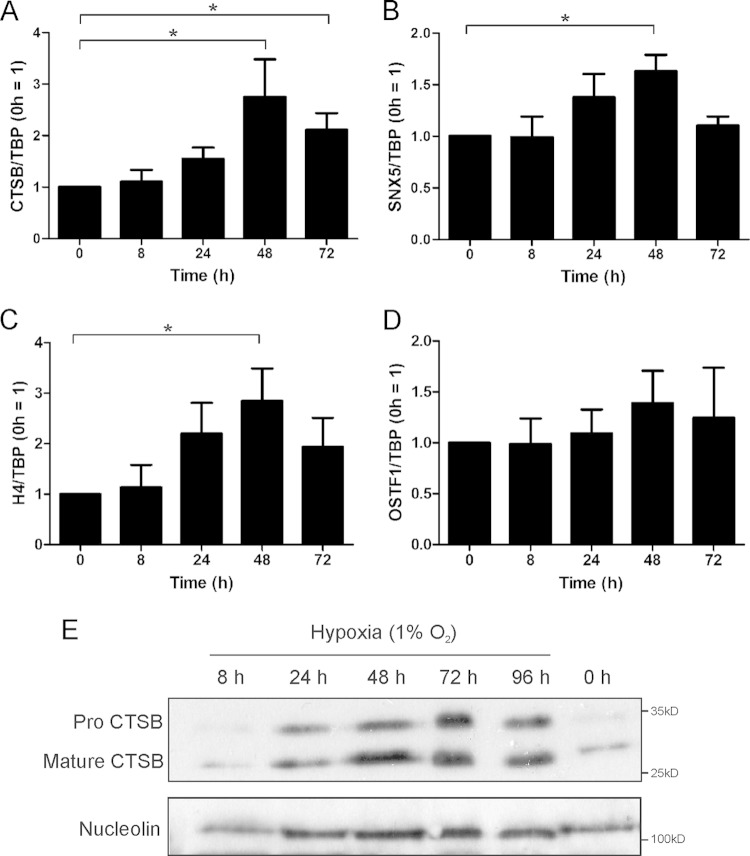
Regulatory mechanisms controlling protein expression under chronic hypoxia, as indicated by cluster 1, are poorly understood. Therefore, we analyzed mRNA expression of 10 of these proteins in primary human macrophages (excluding tubulin and lamin), detecting distinct expression patterns. Three proteins showed no mRNA regulation, suggesting a regulatory mechanism at the protein level. Two mRNAs decreased in chronic hypoxia, and one increased even in acute hypoxia. Interestingly, four proteins out of 10 showed similar mRNA expression patterns. The CTSB, sorting nexin 5 (SNX5), and histone H4 mRNAs did not respond to immediate hypoxia (8 h) but were slightly upregulated after 24 h and significantly increased after 48 h (Fig. 2A to C), while osteoclast stimulating factor 1 showed the same pattern without reaching significance (Fig. 2D). Since the CTSB mRNA increase was significant both at 48 and 72 h of hypoxia and showed the most obvious difference between acute and chronic hypoxia, we decided to concentrate on CTSB for further mechanistic studies. Western analysis of CTSB expression in primary human macrophages confirmed DIGE results with weak expression at 0 and 8 h of hypoxia but a strong response at 24 to 72 h both in pro-CTSB and mature CTSB (Fig. 2E). As CTSB mRNA and protein expression occurred relatively late, an immediate transcriptional regulation under hypoxia appeared unlikely and raised the question about the involvement of the HIF proteins in chronic hypoxic protein expression.
FIG 2.
Chronic hypoxia-regulated proteins. Primary human macrophages were incubated for 0 to 72 h under hypoxia. RNA expression of cathepsin B (CTSB) (A), sorting nexin 5 (SNX5) (B), histone H4 (H4) (C), and osteoclast stimulating factor 1 (OSTF1) (D) was measured by qPCR and normalized to the expression of TATA box binding protein (TBP). The normoxic control was set to 1. *, P < 0.05.(E) CTSB protein was analyzed by Western analysis; nucleolin serves as a loading control. Data are expressed as means ± SEM (n ≥ 3 experiments).
CTSB is an HIF-2α target.
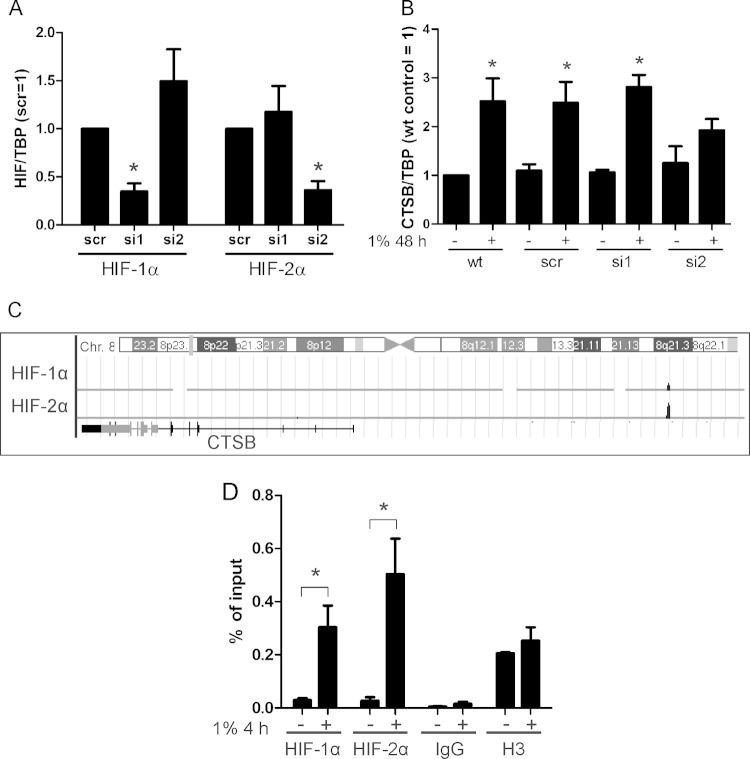
Since CTSB is regulated by hypoxia, the impact of HIF-1α and HIF-2α was determined by transfecting primary human macrophages with either siRNA against HIF-1α (si1), HIF-2α (si2), or a scrambled control (scr) (Fig. 3A). Based on the most obvious increase of CTSB mRNA at 48 h, we decided to focus on this time point of hypoxic incubation for further experiments. CTSB mRNA increased at 48 h of hypoxia in wild-type (wt), scr, and si1 cells but not in si2 cells, pointing to CTSB as an HIF-2 target gene (Fig. 3B). ChIP experiments coupled with sequencing showed HIF-1 and HIF-2 binding to the CTSB gene, visualized by the human genome browser (Fig. 3C) (19). Data were confirmed by ChIP, with subsequent qPCR analysis performed after 4 h of incubation under hypoxia (Fig. 3D). HIF-2α and HIF-1α, albeit to a lower extent, bound to the CTSB gene, while the negative control with IgG revealed no binding and the positive control using a histone H3 antibody showed no significant difference between both groups. These experiments confirmed binding of HIF-2α to the CTSB enhancer after 4 h of hypoxia, although HIF-2α-dependent regulation of the CTSB mRNA occurred after 48 h only, again pointing to an RNA-regulating mechanism.
FIG 3.
Identification of CTSB as an HIF-2 target. (A) Primary human macrophages were transfected with siRNA (50 nM) against HIF-1α (si1), HIF-2α (si2), or a scrambled control (scr). Cells were incubated for 0 and 48 h under hypoxia. (B) mRNA was isolated from wild-type (wt), scrambled (scr), HIF-1α (si1), and HIF-2α (si2) knockdown macrophages to analyze CTSB mRNA expression by qPCR. Data are normalized to TBP expression, and the wt control was set to 1. (C) Results of ChIP-Seq data visualizing HIF-1α and -2α binding to the CTSB enhancer region using the UCSC Genome Browser. (D) HIF binding to the CTSB enhancer was analyzed after 4 h of hypoxia by ChIP, using HIF-1α- or HIF-2α-specific antibodies. Enrichment is shown as the percentage of input. Data are expressed as means ± SEM (n ≥ 3 experiments). *, P < 0.05.
Delayed CTSB induction is not regulated at the transcriptional level.
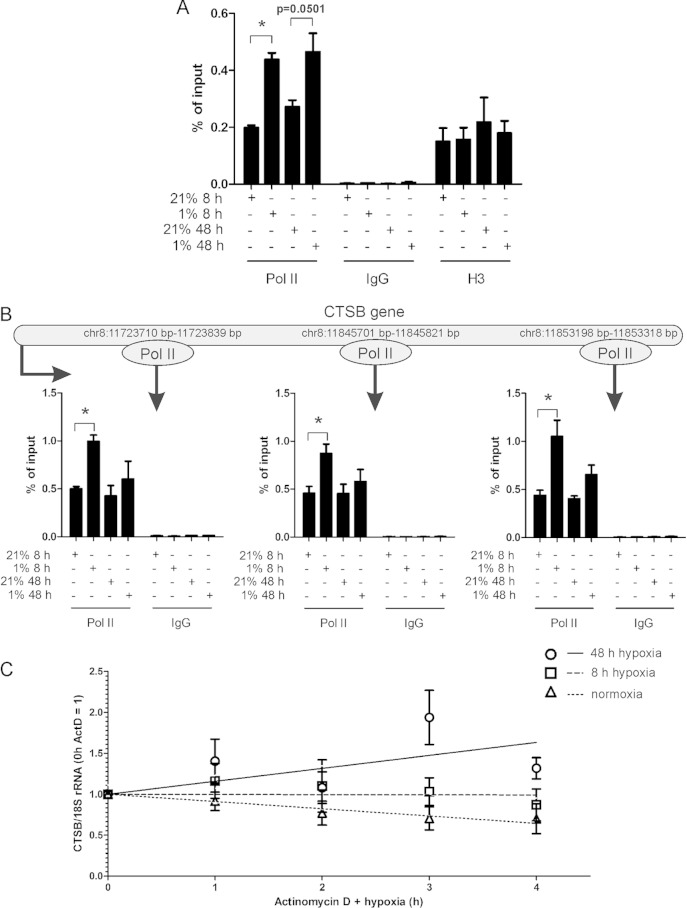
To distinguish whether delayed upregulation of CTSB mRNA depends on transcriptional regulation or an altered RNA stability, we performed ChIP analysis using a polymerase II (Pol II) antibody (Fig. 4A). Pol II binding to the CTSB enhancer significantly increased in cells exposed to hypoxia for 8 or 48 h. There was no significant difference in binding of Pol II comparing short-term and long-term exposures to hypoxia. Additionally, we probed different sections of the CTSB gene for Pol II binding and found a strong binding of Pol II at the start as well as in the middle and the end of the gene after 8 h of hypoxic incubation (Fig. 4B). After 48 h of hypoxia, Pol II binds to a smaller extent to the CTSB gene, which might result in a decreased transcription compared to that of 8 h of hypoxia but may go together with a slightly enhanced transcription compared to the normoxic control. These findings suggest that Pol II is active and CTSB is transcribed after 8 h of hypoxia, initiated by HIF-2 even if an increase in mRNA is not detectable. As the mRNA increase occurred much later and transcription decreases over time, enhanced RNA stability at late time points appeared as a potential explanation.
FIG 4.
Regulation and stability of CTSB mRNA. (A) Primary human macrophages were incubated for 8 or 48 h under hypoxia followed by Pol II-ChIP at the CTSB promoter. (B) Pol II-ChIP samples were used to probe different sections of the CTSB gene for Pol II binding. Enrichment is shown as the percentage of input. (C) Primary human macrophages were incubated under hypoxic conditions for 4 or 44 h and treated with actinomycin D (ActD; 2.5 μg/ml) for an additional 4 h. DMSO-treated controls were set to 1. CTSB mRNA was analyzed by qPCR and normalized to 18S rRNA. Data are expressed as means ± SEM (n ≥ 3 experiments). *, P < 0.05.
CTSB mRNA stability increases in chronic hypoxia.
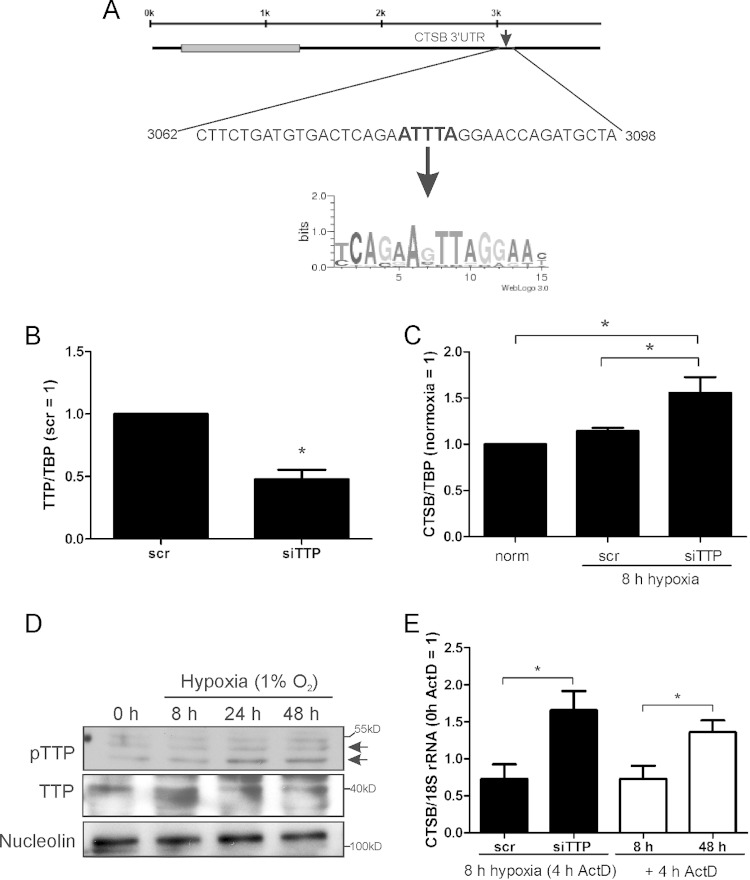
Following our hypothesis, we analyzed RNA stability using actinomycin D (ActD) to inhibit transcription in primary human macrophages exposed to hypoxia for 8 or 48 h. ActD was added for the last 4-h period. After 8 h of hypoxia, CTSB mRNA showed no change in stability compared to that of the appropriate controls, while after 48 h of hypoxia, CTSB mRNA stability significantly increased (Fig. 4C). Since binding of proteins influencing mRNA stability in the 3′ UTR of mRNAs is well established, we screened for factors, potentially influencing RNA stability, which bind to AU-rich elements (AREs) within the 3′ UTR of CTSB, using AREsite (http://rna.tbi.univie.ac.at/cgi-bin/AREsite.cgi). We identified one potential ARE in the 3′ UTR at base positions 3080 to 3084 (Fig. 5A). Additionally, CTSB mRNA was characterized by Emmons et al. as a target of TTP (20). Thus, TTP may account for CTSB mRNA degradation after 8 h of hypoxic incubation.
FIG 5.
TTP regulates CTSB mRNA. (A) AREsite (http://rna.tbi.univie.ac.at/cgi-bin/AREsite.cgi) was used to screen the 3′ UTR of CTSB for AU-rich elements, and one site was predicted for TTP binding. (B) Primary human macrophages were transfected with siRNA against TTP (siTTP). The knockdown was validated by analyzing mRNA levels of TTP normalized to TBP and compared to the scrambled control. (C) Macrophages were transfected with siRNA (50 nM) against TTP (siTTP) or a scrambled control siRNA (scr) and incubated for 8 h under hypoxia. CTSB mRNA was measured using qPCR. (D) Primary human macrophages were time-dependently exposed to hypoxia. TTP, phospho-TTP (pTTP), and nucleolin were analyzed by Western blotting. Different exposure times were used to obtain appropriate pTTP or TTP signals. (E) Macrophages with a knockdown of TTP were incubated for 8 h in hypoxia, with ActD (2.5 μg/ml) added for the last 4 h. mRNA of CTSB was measured and normalized to 18S rRNA (black bars). Macrophages were incubated for 8 or 48 h under hypoxia, with ActD added for the last 4 h. These graphs (white bars) were created from data shown in Fig. 4C. Data are expressed as means ± SEM (n ≥ 3 experiments). *, P < 0.05.
TTP destabilizes CTSB mRNA.
To examine the role of TTP in CTSB regulation, we transfected primary human macrophages with either a scrambled control (scr) or siRNA against TTP (siTTP), reaching a knockdown of roughly 50% (Fig. 5B). Cells were then incubated for 8 or 48 h under hypoxia, and CTSB mRNA was measured (Fig. 5C). As expected, cells transfected with scrambled siRNA showed no difference in CTSB mRNA level at 8 h of hypoxic incubation compared to normoxic controls (norm). CTSB mRNA in TTP knockdown cells significantly increased in acute hypoxia (8 h) compared to controls (norm, scr). These data suggest that TTP keeps CTSB mRNA levels low under acute hypoxia, while chronic hypoxia inactivates TTP, which restricts RNA degradation of CTSB. Since TTP is inactivated by phosphorylation, we exposed macrophages for 0 to 48 h to hypoxia and performed Western analysis to analyze phosphorylated TTP (pTTP) (Fig. 5D). Western analysis revealed bands at different sizes, indicating distinct stages of TTP phosphorylation. At normoxia and acute hypoxia, only low levels of pTTP were detectable, while phosphorylation increases at 24 to 48 h. The increase in pTTP points to its inactivation under chronic hypoxia. Experiments using an siRNA-induced TTP knockdown were used to validate TTP and pTTP bands (data not shown). To establish whether TTP really affects CTSB mRNA stability in acute hypoxia, we exposed TTP knockdown cells for 8 h to hypoxia and treated them for the last 4 h with ActD (Fig. 5E, black bars). As expected, siTTP-treated cells showed an increase in CTSB mRNA stability. This effect was very similar to the mRNA expression after 48 h of hypoxia, which supports our hypothesis that TTP is responsible for the degradation of CTSB mRNA in acute hypoxia (Fig. 5E, white bars were adapted from Fig. 4C). Our data suggest an HIF-2-induced transcriptional induction of CTSB mRNA, which is degraded in a TTP-dependent manner in acute hypoxia but is stabilized in chronic hypoxia due to TTP inactivation.
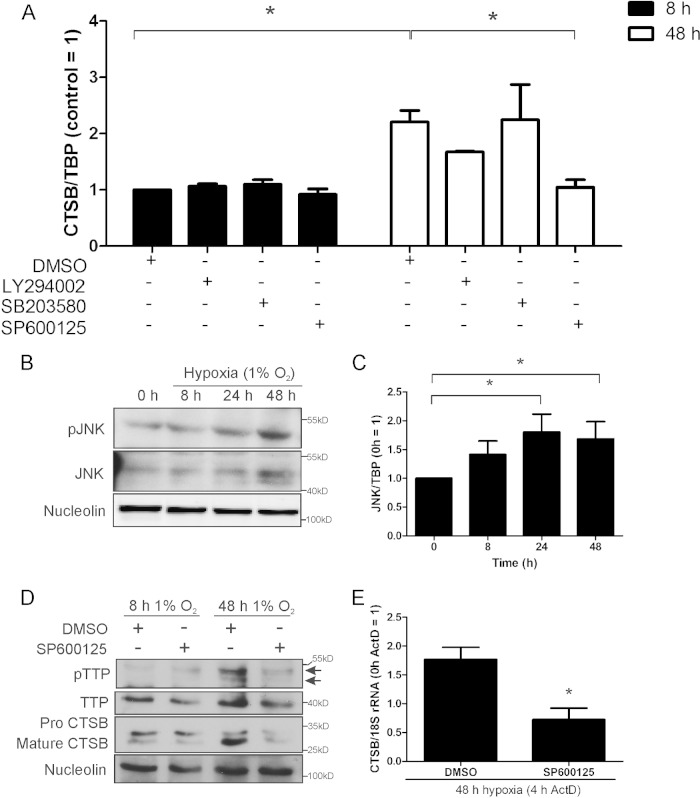
Inhibition of JNK attenuates CTSB mRNA under chronic hypoxia.
Next, we aimed at identifying kinases involved in TTP inactivation under chronic hypoxia. We treated primary human macrophages with compounds to inhibit phosphoinositide 3-kinase (LY294002; 10 μM), p38 MAPK (SB203580; 10 μM), or JNK (SP600125; 20 μM) for either 8 or 48 h under hypoxia. DMSO was used as a control (Fig. 6A). CTSB mRNA remained unaffected by the inhibitors when incubated for 8 h under hypoxia, while a CTSB mRNA increase was seen in the DMSO controls, when exposed to hypoxia for 48 h. Inhibition of phosphoinositide 3-kinase with LY294002 or p38 MAPK with SB203580 left CTSB mRNA levels unaffected, while blocking JNK with SP600125 significantly reduced the CTSB mRNA amount. Thus, JNK was considered to be involved in inactivating TTP and thereby increasing the CTSB mRNA amount under chronic hypoxia. To follow JNK activity, we performed Western analysis to detect the phosphorylated and thus active form of the kinase. As seen in Fig. 6B and C, expression of JNK increased over time at the protein and mRNA levels, with prominent expression at 48 h of hypoxia. This was paralleled by an increased protein phosphorylation, which also culminated at 48 h of hypoxia. Thus, JNK appeared to be active under chronic hypoxia. To verify that TTP phosphorylation is causatively linked to JNK activity, we cultured cells for 8 or 48 h with SP600125 or DMSO as a control under hypoxia followed by Western analysis (Fig. 6D). As expected, TTP was not affected by SP600125 after 8 h. However, the increase of pTTP seen at 48 h was diminished by the inhibitor. Additionally, we stained the blot for CTSB. The protein was upregulated after 48 h, while treatment with SP600125 reduced CTSB levels to control values. To explore the role of JNK in CTSB mRNA stabilization, we exposed macrophages for 48 h to hypoxia with SP600125 or DMSO as a control, adding ActD for the last 4 h (Fig. 6E). Substantiating our hypothesis, inhibition of JNK significantly destabilized CTSB mRNA under chronic hypoxic conditions. Apparently, JNK is activated under chronic hypoxia and phosphorylates TTP. Inhibition of JNK reduces pTTP, followed by degradation of CTSB mRNA and thus reduced protein expression.
FIG 6.
JNK influences CTSB mRNA expression and stability. Primary human macrophages were incubated with DMSO, LY294002 (10 μM), SB203580 (10 μM), or SP600125 (20 μM) under hypoxia for 8 or 48 h. (A) CTSB mRNA was analyzed using qPCR. (B) Macrophages were time-dependently exposed to hypoxia, and protein amount of c-Jun N-terminal kinase (JNK) and phosphorylated JNK was measured by Western analysis, while nucleolin served as a loading control. (C) Macrophages were incubated for 0 to 48 h under hypoxic conditions, and JNK mRNA was analyzed by qPCR. Data were normalized against TBP. (D) Primary human macrophages were incubated for the indicated time periods under hypoxia and treated with either SP600125 (20 μM) or DMSO. Western analysis was performed using an antibody against TTP or nucleolin (loading control). Blots were exposed to films for variable times to obtain appropriate pTTP or TTP signals. (E) Macrophages were incubated with SP600125 (20 μM) under hypoxia for 48 h, and transcription was blocked by adding actinomycin D (ActD; 2.5 μg/ml) during the last 4 h. CTSB mRNA was measured and normalized to 18S rRNA (0-h ActD control = 1). Data are expressed as means ± SEM (n ≥ 3 experiments). *, P < 0.05.
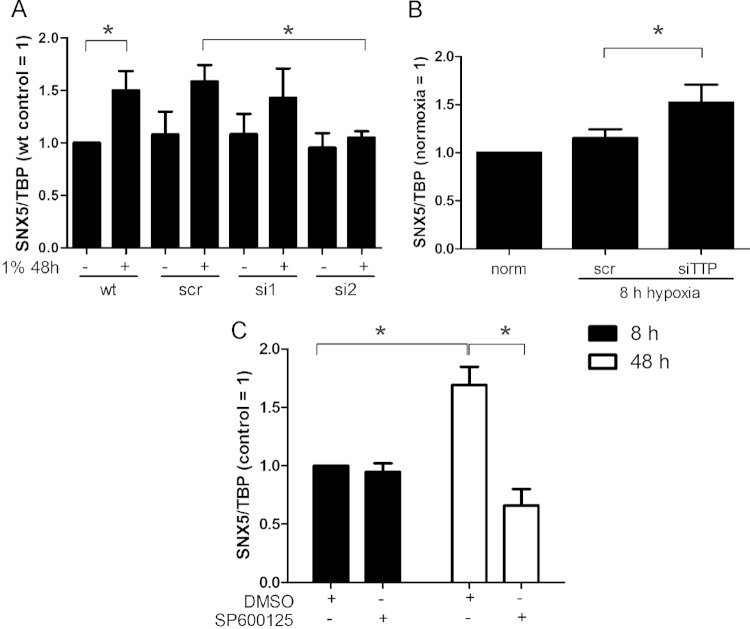
SNX5.
To generalize our finding of how chronic hypoxia increases CTSB expression, we tested whether other proteins of cluster 1 (Fig. 1B) behaved similarly. SNX5 mRNA expression did not change at 8 h of hypoxia but significantly increased during chronic hypoxia (48 h) (Fig. 2B). siRNA knockdown of HIF-2α prevented upregulation of SNX5 mRNA, as seen for CTSB (Fig. 7A). To proof the involvement of TTP, we analyzed SNX5 mRNA levels in TTP knockdown cells (Fig. 7B). The TTP knockdown provoked an SNX5 mRNA increase under acute hypoxia (8 h) compared to normoxic controls. Additionally, inhibition of JNK with SP600125 significantly decreased SNX5 mRNA in response to 48 h of hypoxia (Fig. 7C).
FIG 7.
Regulation of SNX5. (A) RNA was isolated from wild-type (wt), scrambled (scr), HIF-1α (si1), and HIF-2α (si2) knockdown macrophages, and SNX5 expression was analyzed by qPCR. (B) Primary human macrophages were transfected with siRNA (50 nM) against TTP (siTTP) or a scrambled control siRNA (scr) for 8 or 48 h in hypoxia, and SNX5 mRNA was measured using qPCR. (C) Primary human macrophages were incubated with DMSO or SP600125 (20 μM) and incubated for 8 or 48 h under hypoxia. SNX5 mRNA was analyzed using qPCR and normalized to TBP; 8-h DMSO control was set to 1. Data are expressed as means ± SEM (n ≥ 3). *, P < 0.05.
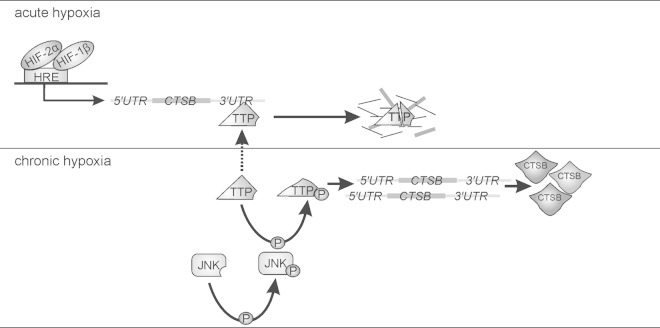
Taken together, CTSB and SNX5 mRNAs are induced in an HIF-2-dependent manner and destabilized by TTP under acute hypoxia. Chronic hypoxia inactivates TTP in a JNK-dependent manner, which in turn allows the expression of distinct TTP target proteins (Fig. 8).
FIG 8.
Scheme to illustrate CTSB expression under chronic hypoxia. For details, see the text.
DISCUSSION
Protein expression in response to acute hypoxia is well defined and linked to HIF-dependent transcriptional activation. Studies dealing with chronic hypoxia refer to physiological and pathological consequences of chronic hypoxia, e.g., during pregnancy (21, 22) or cancer (23, 24). While several screening approaches determining changes in the proteome or transcriptome during chronic hypoxia have recently been reported (25, 26), studies dealing with mechanistic questions are rare. To analyze differential protein expression in acute versus chronic hypoxia, we performed 2D-DIGE experiments coupled to MS/MS and focused on the regulatory mechanism provoking delayed CTSB protein expression. Induction of CTSB under chronic hypoxia was shown before, but it was not compared to acute hypoxia (27). Our experiments suggest that CTSB is a target of HIF-2, which binds to an enhancer region of CTSB, confirming earlier data that postulated a crucial role of HIF-2 in chronic hypoxic protein expression (27–29). Furthermore, binding of Pol II to different regions of the CTSB gene after 8 h of hypoxia provides evidence that CTSB is transcribed at early time points, but no mRNA or protein is detectable. These data suggest that the CTSB mRNA and protein increase under chronic hypoxia results from enhanced RNA stability. In fact, we noticed inactivation of TTP after 24 to 48 h of hypoxia. In addition, the knockdown of TTP confirmed its role in reducing target mRNAs such as CTSB in acute but not chronic hypoxia. Moreover, blocking JNK, a kinase known to phosphorylate and thus inactivate TTP under chronic hypoxia, attenuated stabilization of CTSB mRNA by decreasing TTP activity (16). Activation of JNK under prolonged hypoxia, as shown in our study, corroborates previous findings by Jin et al. (15). JNK activation is associated with an enhanced metastatic ability of cancer cells and linked to increased levels of urokinase-type plasminogen activator and matrix metalloprotease 9 (MMP-9) (30). Interestingly, MMP-9 is defined as a TTP target, suggesting that JNK-mediated TTP phosphorylation may stabilize MMP-9 mRNA (31). Also, overexpression of CTSB in various cancers is linked to malignant progression, favoring migration and invasion (27, 32–36). Additionally, we found both pro-CTSB and mature CTSB to be upregulated in human macrophages under chronic hypoxia, which suggests an enhanced activity of the enzyme under these conditions. Consistently, TTP is ubiquitously expressed in malignant tumors and dominantly detected in its phosphorylated and thus inactive form (37, 38). We provide a mechanism for protein regulation under chronic hypoxia, taking regulatory features of TTP and tumor-specific expression of TTP into consideration. Expression patterns of CTSB are shared by SNX5, osteoclast-stimulating factor 1, and histone H4 mRNA, with the further information that SNX5 is regulated by TTP as well. We suggest that an HIF-triggered expression in combination with TTP-dependent RNA decay is an important and more general regulatory mechanism for protein expression under chronic hypoxia.
Nakayama reported activation of nuclear factor kappa light-chain enhancer of activated B cells and cyclic AMP response element-binding protein during chronic hypoxia to induce MMP-1 in an HIF-2α-dependent manner (39). In light of our studies, we speculate that an additional mechanism might explain delayed MMP-1 expression, since the MMP-1 3′ UTR also contains several AREs as potential TTP binding sites.
In conclusion, we provide evidence that transcription of CTSB mRNA under hypoxia is HIF-2α dependent. The delay in protein expression is explained by mRNA destabilization, which is facilitated by TTP (Fig. 8). Inactivation of TTP due to phosphorylation by JNK under periods of chronic hypoxia stabilizes target mRNAs of, e.g., CTSB or SNX5, which results in protein expression as a late hypoxic event. Our work provides a molecular explanation for protein expression under periods of long-lasting hypoxia, as found in various diseases, which differs clearly from mechanisms under acute hypoxia.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 815, project Z1 [I.W.] and project A8 [B.B.]).
We declare that we have no conflicts of interest.
REFERENCES
- 1.Dehne N, Brune B. 2014. Sensors, transmitters, and targets in mitochondrial oxygen shortage—a hypoxia-inducible factor relay story. Antioxid Redox Signal 20:339–352. doi: 10.1089/ars.2012.4776. [DOI] [PubMed] [Google Scholar]
- 2.Brune B, Dehne N, Grossmann N, Jung M, Namgaladze D, Schmid T, von Knethen A, Weigert A. 2013. Redox control of inflammation in macrophages. Antioxid Redox Signal 19:595–637. doi: 10.1089/ars.2012.4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaelin WG Jr, Ratcliffe PJ. 2008. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Wang GL, Semenza GL. 1993. General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A 90:4304–4308. doi: 10.1073/pnas.90.9.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh MY, Powis G. 2012. Passing the baton: the HIF switch. Trends Biochem Sci 37:364–372. doi: 10.1016/j.tibs.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wenger RH, Stiehl DP, Camenisch G. 2005. Integration of oxygen signaling at the consensus HRE. Sci STKE 2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 7.Schodel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, Mole DR. 2011. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117:e207–e217. doi: 10.1182/blood-2010-10-314427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. 2009. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A 106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahling M, Persson AB, Klinger B, Benko E, Steege A, Kasim M, Patzak A, Persson PB, Wolf G, Bluthgen N, Mrowka R. 2012. Multilevel regulation of HIF-1 signaling by TTP. Mol Biol Cell 23:4129–4141. doi: 10.1091/mbc.E11-11-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim TW, Yim S, Choi BJ, Jang Y, Lee JJ, Sohn BH, Yoo HS, Yeom YI, Park KC. 2010. Tristetraprolin regulates the stability of HIF-1alpha mRNA during prolonged hypoxia. Biochem Biophys Res Commun 391:963–968. doi: 10.1016/j.bbrc.2009.11.174. [DOI] [PubMed] [Google Scholar]
- 11.Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 12.DuBois RN, McLane MW, Ryder K, Lau LF, Nathans D. 1990. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J Biol Chem 265:19185–19191. [PubMed] [Google Scholar]
- 13.Lai WS, Stumpo DJ, Blackshear PJ. 1990. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem 265:16556–16563. [PubMed] [Google Scholar]
- 14.Varnum BC, Ma QF, Chi TH, Fletcher B, Herschman HR. 1991. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol Cell Biol 11:1754–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin N, Hatton N, Swartz DR, Xia X, Harrington MA, Larsen SH, Rhoades RA. 2000. Hypoxia activates jun-N-terminal kinase, extracellular signal-regulated protein kinase, and p38 kinase in pulmonary arteries. Am J Respir Cell Mol Biol 23:593–601. doi: 10.1165/ajrcmb.23.5.3921. [DOI] [PubMed] [Google Scholar]
- 16.Cao H, Lin R. 2008. Phosphorylation of recombinant tristetraprolin in vitro. Protein J 27:163–169. doi: 10.1007/s10930-007-9119-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuhrmann DC, Wittig I, Heide H, Dehne N, Brune B. 2013. Chronic hypoxia alters mitochondrial composition in human macrophages. Biochim Biophys Acta 1834:2750–2760. doi: 10.1016/j.bbapap.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 18.Pham TH, Langmann S, Schwarzfischer L, El Chartouni C, Lichtinger M, Klug M, Krause SW, Rehli M. 2007. CCAAT enhancer-binding protein beta regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J Biol Chem 282:21924–21933. doi: 10.1074/jbc.M611618200. [DOI] [PubMed] [Google Scholar]
- 19.Tausendschön M, Rehli M, Dehne N, Schmidl C, Döring C, Hansmann M-L, Brüne B. 2015. Genome-wide identification of hypoxia-inducible factor-1 and -2 binding sites in hypoxic human macrophages alternatively activated by IL-10. Biochim Biophys Acta 1849:10–22. doi: 10.1016/j.bbagrm.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Emmons J, Townley-Tilson WH, Deleault KM, Skinner SJ, Gross RH, Whitfield ML, Brooks SA. 2008. Identification of TTP mRNA targets in human dendritic cells reveals TTP as a critical regulator of dendritic cell maturation. RNA 14:888–902. doi: 10.1261/rna.748408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Julian CG, Yang IV, Browne VA, Vargas E, Rodriguez C, Pedersen BS, Moore LG, Schwartz DA. 2014. Inhibition of peroxisome proliferator-activated receptor gamma: a potential link between chronic maternal hypoxia and impaired fetal growth. FASEB J 28:1268–1279. doi: 10.1096/fj.13-239749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanek J. 2013. Hypoxic patterns of placental injury: a review. Arch Pathol Lab Med 137:706–720. doi: 10.5858/arpa.2011-0645-RA. [DOI] [PubMed] [Google Scholar]
- 23.Hirai K, Nomura T, Yamasaki M, Inoue T, Narimatsu T, Chisato Nakada PD, Yoshiyuki Tsukamoto PD, Matsuura K, Sato F, Moriyama M, Mimata H. 2014. The Vav3 oncogene enhances the malignant potential of prostate cancer cells under chronic hypoxia. Urol Oncol 32:101–109. doi: 10.1016/j.urolonc.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 24.Danza G, Di Serio C, Ambrosio MR, Sturli N, Lonetto G, Rosati F, Rocca BJ, Ventimiglia G, del Vecchio MT, Prudovsky I, Marchionni N, Tarantini F. 2013. Notch3 is activated by chronic hypoxia and contributes to the progression of human prostate cancer. Int J Cancer 133:2577–2586. doi: 10.1002/ijc.28293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosqueira M, Willmann G, Zeiger U, Khurana TS. 2012. Expression profiling reveals novel hypoxic biomarkers in peripheral blood of adult mice exposed to chronic hypoxia. PLoS One 7:e37497. doi: 10.1371/journal.pone.0037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vigano A, Vasso M, Caretti A, Bravata V, Terraneo L, Fania C, Capitanio D, Samaja M, Gelfi C. 2011. Protein modulation in mouse heart under acute and chronic hypoxia. Proteomics 11:4202–4217. doi: 10.1002/pmic.201000804. [DOI] [PubMed] [Google Scholar]
- 27.Wickramasinghe NS, Banerjee K, Nagaraj NS, Vigneswaran N, Zacharias W. 2005. Hypoxia alters cathepsin B/inhibitor profiles in oral carcinoma cell lines. Anticancer Res 25:2841–2849. [PubMed] [Google Scholar]
- 28.Stiehl DP, Bordoli MR, Abreu-Rodriguez I, Wollenick K, Schraml P, Gradin K, Poellinger L, Kristiansen G, Wenger RH. 2012. Non-canonical HIF-2alpha function drives autonomous breast cancer cell growth via an AREG-EGFR/ErbB4 autocrine loop. Oncogene 31:2283–2297. doi: 10.1038/onc.2011.417. [DOI] [PubMed] [Google Scholar]
- 29.Qing G, Simon MC. 2009. Hypoxia inducible factor-2alpha: a critical mediator of aggressive tumor phenotypes. Curr Opin Genet Dev 19:60–66. doi: 10.1016/j.gde.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, Sun L, Zhao P, Yao L, Jin H, Liang S, Wang Y, Zhang D, Pang Y, Shi Y, Chai N, Zhang H. 2010. Hypoxia promotes metastasis in human gastric cancer by up-regulating the 67-kDa laminin receptor. Cancer Sci 101:1653–1660. doi: 10.1111/j.1349-7006.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Tubergen EA, Banerjee R, Liu M, Vander Broek R, Light E, Kuo S, Feinberg SE, Willis AL, Wolf G, Carey T, Bradford C, Prince M, Worden FP, Kirkwood KL, D'Silva NJ. 2013. Inactivation or loss of TTP promotes invasion in head and neck cancer via transcript stabilization and secretion of MMP9, MMP2, and IL-6. Clin Cancer Res 19:1169–1179. doi: 10.1158/1078-0432.CCR-12-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Withana NP, Blum G, Sameni M, Slaney C, Anbalagan A, Olive MB, Bidwell BN, Edgington L, Wang L, Moin K, Sloane BF, Anderson RL, Bogyo MS, Parker BS. 2012. Cathepsin B inhibition limits bone metastasis in breast cancer. Cancer Res 72:1199–1209. doi: 10.1158/0008-5472.CAN-11-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konduri S, Lakka SS, Tasiou A, Yanamandra N, Gondi CS, Dinh DH, Olivero WC, Gujrati M, Rao JS. 2001. Elevated levels of cathepsin B in human glioblastoma cell lines. Int J Oncol 19:519–524. doi: 10.3892/ijo.19.3.519. [DOI] [PubMed] [Google Scholar]
- 34.Yan S, Sloane BF. 2003. Molecular regulation of human cathepsin B: implication in pathologies. Biol Chem 384:845–854. doi: 10.1515/BC.2003.095. [DOI] [PubMed] [Google Scholar]
- 35.Sevenich L, Schurigt U, Sachse K, Gajda M, Werner F, Muller S, Vasiljeva O, Schwinde A, Klemm N, Deussing J, Peters C, Reinheckel T. 2010. Synergistic antitumor effects of combined cathepsin B and cathepsin Z deficiencies on breast cancer progression and metastasis in mice. Proc Natl Acad Sci U S A 107:2497–2502. doi: 10.1073/pnas.0907240107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sevenich L, Werner F, Gajda M, Schurigt U, Sieber C, Muller S, Follo M, Peters C, Reinheckel T. 2011. Transgenic expression of human cathepsin B promotes progression and metastasis of polyoma-middle-T-induced breast cancer in mice. Oncogene 30:54–64. doi: 10.1038/onc.2010.387. [DOI] [PubMed] [Google Scholar]
- 37.Suswam E, Li Y, Zhang X, Gillespie GY, Li X, Shacka JJ, Lu L, Zheng L, King PH. 2008. Tristetraprolin down-regulates interleukin-8 and vascular endothelial growth factor in malignant glioma cells. Cancer Res 68:674–682. doi: 10.1158/0008-5472.CAN-07-2751. [DOI] [PubMed] [Google Scholar]
- 38.Suswam EA, Shacka JJ, Walker K, Lu L, Li X, Si Y, Zhang X, Zheng L, Nabors LB, Cao H, King PH. 2013. Mutant tristetraprolin: a potent inhibitor of malignant glioma cell growth. J Neurooncol 113:195–205. doi: 10.1007/s11060-013-1112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakayama K. 2013. cAMP-response element-binding protein (CREB) and NF-kappaB transcription factors are activated during prolonged hypoxia and cooperatively regulate the induction of matrix metalloproteinase MMP1. J Biol Chem 288:22584–22595. doi: 10.1074/jbc.M112.421636. [DOI] [PMC free article] [PubMed] [Google Scholar]