Abstract
Caspase-1 is activated by the inflammasome complex to process cytokines like interleukin-1β (IL-1β). Pro-caspase-1 consists of three domains, CARD, p20, and p10. Association of pro-caspase-1 with the inflammasome results in initiation of its autocatalytic activity, culminating in self-cleavage that generates catalytically active subunits (p10 and p20). In the current study, we show that Nedd8 is required for efficient self-cleavage of pro-caspase-1 to generate its catalytically active subunits. Nedd8 silencing or treating cells with the neddylation inhibitor MLN4924 led to diminished caspase-1 processing and reduced IL-1β maturation following inflammasome activation. Coimmunoprecipitation and mass spectrometric analysis of 293 cells overexpressing pro-caspase-1 (and CARD) and Nedd8 suggested possible neddylation of caspase-1 CARD. Following inflammasome activation in primary macrophages, we observed colocalization of endogenous Nedd8 with caspase-1. Similarly, interaction of endogenous Nedd8 with caspase-1 CARD was detected in inflammasome-activated macrophages. Furthermore, enhanced autocatalytic activity of pro-caspase-1 was observed following Nedd8 overexpression in 293 cells, and such activity in inflammasome-activated macrophages was drastically diminished upon treatment of cells with MLN4924. Thus, our studies demonstrate a role of Nedd8 in regulating caspase-1 activation following inflammasome activation, presumably via augmenting autoprocessing/cleavage of pro-caspase-1 into its corresponding catalytically active subunits.
INTRODUCTION
Caspase-1 is a critical enzyme required for processing of the 31-kDa pro-interleukin-1β (pro-IL-1β) protein to the 17-kDa active (secreted form) IL-1β, a key proinflammatory mediator that modulates the host response to disease and infection (1–7). Caspase-1 activity is also required for processing of cytokines like IL-18 and induction of the intracellular pathogen-induced cell death mechanism known as pyroptosis. Pro-caspase-1 is synthesized as an inactive precursor of 45 kDa consisting of three domains: CARD (9 kDa), p20 (20 kDa), and p10 (10 kDa). Pro-caspase-1, which possesses autocatalytic activity, cleaves itself at specific aspartic acid (Asp) residues to generate enzymatically active caspase-1, which is comprised of a tetramer of p10-p20 subunits. Activation of caspase-1 occurs following recruitment of pro-caspase-1 to a multiprotein complex known as the inflammasome (8–37). Various signals (e.g., from the pathogen, endogenous factors like ATP, cholesterol crystals, and monosodium urate) trigger formation of the inflammasome, comprised of a homo- and hetero-oligomeric complex of proteins, such as NLRP3, AIM2, NLRC4, NLRP1, NLRP6, NLRP7, and RIG-I. Among the different inflammasome complexes, the NLRP3/ASC inflammasome is best characterized. It is postulated that the inflammasome complex recruits pro-caspase-1 via a CARD-CARD interaction between pro-caspase-1 and ASC. A conformational change in pro-caspase-1 due to its recruitment to the inflammasome induces its autocatalytic activity, resulting in self-cleavage to yield p10 and p20 subunits, the functional subunits of active caspase-1.
Caspase-1-mediated production of active IL-1β is a critical regulator of the host response under various stress conditions. Thus, deregulated inflammasome/caspase-1 activation resulting in high or low active IL-1β secretion can manifest in various diseases, like pneumonia (viral or bacterial), diabetes, atherosclerosis, obesity, cancer, gout, Alzheimer's, arthritis, etc. (8–37). Moreover, IL-1β maturation disorders (inflammasomopathies) have been associated with various autoinflammatory diseases, such as familial cold autoinflammatory syndrome, Muckle-Wells syndrome, and familial Mediterranean fever (38, 39). Since the inflammasome plays a crucial role in health and disease, it is imperative to understand the cellular/molecular mechanism regulating its activation. Toward that end, we have uncovered that neddylation may play a role in inflammasome activation, by virtue of it enhancing cleavage and processing (autocatalytic activity) of pro-caspase-1 to yield active caspase-1.
We recently identified Nod2 and Nod-like receptor proteins (NLRPs) as important host factors controlling respiratory virus (respiratory syncytial virus [RSV] and influenza A virus [IAV]), specific innate immune responses following induction of type I interferon (40), and activation of NLRP3/ASC inflammasomes in RSV-infected cells (41). We also recently reported that RSV utilizes lipid rafts during virus budding (42), and since budding events of several viruses are regulated by Nedd proteins like Nedd4 (Nedd4 is a component of ESCRT [the endosomal sorting complex required for transport] pathway [43, 44] that utilizes L (late-assembly) domains during virus budding), we knocked down (via RNA interference [RNAi]) several Nedd proteins, including Nedd4 and Nedd8 (as a control). During those studies, we surprisingly uncovered that Nedd8 knockdown drastically reduced IL-1β production from RSV-infected cells. This observation led to studies focusing on the role of neddylation (45, 46) in regulating inflammasome activation. Silencing of Nedd8 in macrophages and epithelial cells led to reduced IL-1β secretion and maturation. This was due to diminished processing of pro-caspase-1 to yield functional subunits (i.e., the p10 subunit). The knockdown studies were further validated by using MLN4924, a neddylation inhibitor that blocks the activity of Nedd8-activating enzyme (NAE) (47–49). Further studies revealed colocalization and coimmunoprecipitation of endogenous Nedd8 with caspase-1 in inflammasome-activated macrophages. Biochemical analysis by overexpressing caspase-1 and Nedd8 in 293 cells showed that caspase-1 is neddylated and that CARD of pro-caspase-1 is the site of neddylation. Mechanistically, our studies also suggested that neddylation enhances autocleavage/autocatalytic activity of pro-caspase-1, since increased levels of cleaved caspase-1 products were detected in cells overexpressing Nedd8 and caspase-1. Finally, the physiological significance of neddylation was borne out by the observation that administration of MLN4924 to mice treated with lipopolysaccharide (LPS) and ATP (to trigger inflammasome-mediated production of mature IL-1β) led to significantly reduced IL-1β levels in serum. Thus, our studies have uncovered a role of neddylation during inflammasome-dependent activation of caspase-1 and subsequent release of mature IL-1β.
MATERIALS AND METHODS
Antibodies and reagents.
Dulbecco's modified Eagle's medium (DMEM), RPMI 1640, OPTI-MEM, fetal bovine serum (FBS), and penicillin-streptomycin were purchased from Life Technologies (Grand Island, NY). A bronchial epithelial cell growth medium (BEGM) bullet kit was purchased from Lonza. Bacterial lipopolysaccharide (LPS-EB; Escherichia coli O111:B4) was purchased from Invivogen. ATP and nigericin were purchased from Sigma-Aldrich (St. Louis, MO). Primary antibodies used included rabbit anti-human caspase-1 p10, rabbit anti-mouse caspase-1 p10, goat anti-mouse caspase-1 p20, mouse anti-green fluorescent protein (anti-GFP; Santa Cruz Biotechnology), rabbit anti-caspase-1 CARD (LSBio), rabbit anti-Nedd8 (Cell Signaling), goat anti-mouse IL-1β p17 (R&D Systems), mouse anti-human IL-1β 3ZD (NCI—Frederick Cancer Research and Development Center), mouse anti-FLAG, mouse anti-myc, and mouse antihemagglutinin (anti-HA; Sigma), and antiactin antibody (Bethyl Laboratories). Secondary antibodies included goat anti-mouse antibody–horseradish peroxidase (HRP), goat anti-rabbit antibody–HRP, and donkey anti-goat antibody–HRP conjugates (Jackson ImmunoResearch), and goat anti-rabbit antibody–fluorescein isothiocyanate (FITC), goat anti-rabbit antibody–Texas Red, goat anti-mouse antibody–FITC, goat anti-mouse antibody–Texas Red, and donkey anti-goat antibody-Texas Red conjugates (ProSci Inc.). Control small interfering RNA (siRNA) A and Nedd8 siRNA were purchased from Santa Cruz Biotechnology. Enzyme-linked immunosorbent assay (ELISA) kits for human IL-1β, mouse IL-1β, and tumor necrosis factor alpha (TNF-α) were purchased from eBioscience. A mouse IL-18 ELISA kit was purchased from MyBioSource.
Virus purification and cell culture.
RSV (A2 strain) was propagated in CV-1 cells (40, 50–52). RSV was purified by centrifugation (two times) on discontinuous sucrose gradients as described previously (40, 50–53). Influenza A virus [IAV; A/PR/8/34 (H1N1)] was grown in the allantoic cavities of 10-day-old embryonated eggs (40, 54). IAV was purified by centrifugation (two times) on discontinuous sucrose gradients (40, 54). 293 cells were maintained in DMEM supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin. Normal human bronchial epithelial (NHBE) cells were maintained in BEGM complete medium (Lonza) in accordance with the manufacturer's instructions, and cells used for the experiments were in their third passage or sooner. Immortalized mouse bone marrow-derived macrophages (NR-9456 cells) were maintained in DMEM supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin. THP-1 cells were maintained in RPMI 1640 medium supplemented with 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol. THP-1 differentiation was achieved by culturing 5 × 105 cells in complete medium containing 50 nM phorbol 12-myristate 13-acetate (PMA) for 24 h in 12-well cell culture plates. After 24 h, PMA-containing medium was replaced with fresh complete medium, and cells were maintained for 48 h before treatments. Bone marrow-derived macrophages (BMDMs) were obtained from femurs and tibias of wild-type C57BL/6 mice and were cultured for 6 to 8 days as described elsewhere (40, 54, 55). Cells were plated in 12-well cell culture plates containing RPMI 1640, 10% FBS, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 20 ng/ml granulocyte-macrophage colony-stimulating factor. 293 and THP-1 cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA). NHBE cells were obtained from Lonza. NR-9456 cells were a gift from Michael T. Berton (UT Health Science Center at San Antonio).
RNA isolation, PCR amplification, and RT-PCR.
Total RNA isolation from cells was performed using a monophasic solution of phenol and guanidine thiocyanate (TRIzol; Sigma-Aldrich, St. Louis, MO), as recommended by the suppliers. Total cellular RNA (1 μg) was used to generate cDNA by using Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Carlsbad, CA). PCR was routinely performed using 0.625 units of Taq polymerase, 10 pmol of each oligonucleotide primer, 1 mM MgCl2, and 100 μM deoxynucleotide triphosphates in a final reaction volume of 25 μl. The amplification cycle was as follows: an initial denaturing step (95°C for 3 to 5 min) was followed by 28 cycles of denaturing (94°C for 30 s), annealing (60°C for 30 s), and extending (72°C for 30 s), followed by 10 min at 72°C for elongation. Following amplification, the PCR products were analyzed on 1.5% agarose gels, and band intensities were quantified by densitometry (Syngene gel documentation system). Equal loading in each well was confirmed by analyzing expression of the housekeeping glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene. The primers used to detect the genes for the indicated proteins by reverse transcription-PCR (RT-PCR) are as follows (forward and reverse sequences, respectively, are shown in the 5′-to-3′ direction): human pro-IL-1β, AAACAGATGAAGTGCTCCTTCCAG and TGGAGAACACCACTTGTTGCTCCA (product size, 391 bp); human GAPDH, GTCAGTGGTGGACCTGACCT and AGGGGTCTACATGGCAACTG (product size, 420 bp); human Nedd8, ATGCTAATTAAAGTGAAGACGCTGAC and TCACTGCCTAAGACCACCTCCTC (product size, 246 bp); mouse pro-IL-1β, GACAGTGATGAGAATGACCTGTTC and CCTGACCACTGTTGTTTCCC (product size, 730 bp); mouse GAPDH, GCCAAGGTCATCCATGACAACTTTGG and GCCTGCTTCACCACCTTCTTGATGTC (product size, 314 bp); mouse Nedd8, ATGCTAATTAAAGTGAAGACGCTGAC and TCACTGCCCAAGACCACCTCCTC (product size, 246 bp); mouse Ubc12, TTTGACCACCACGTGATTAT and CTTTAATAGCCTGGTGGGAC (product size, 211 bp); human pro-caspase-1, GGTCCTGAAGGAGAAGAGAA and AGGCCTGGATGATGATCACC (product size, 842 bp); human AS, GGACGCCTTGGCCCTCACC and GGCGCGGCTCCAGAGCCCTG (product size, 150 bp); human NLRP3, TTCTTTCTGTTTGCTGAGTTTTTG and TTCCTGGCATATCACAGTGG (product size, 467 bp).
Cell transfection and RNA interference.
293 cells were transfected with Lipofectamine 2000 (Invitrogen) at 80% confluence (40) in 12-well cell culture plates with expression plasmids carrying genes for human ASC (1.6 μg/well), human NLRP3 (1.6 μg/well), human pro-IL-1β (0.1 μg/well), myc-procaspase-1 or myc-procaspase-1–HA (1.6 μg/well), FLAG-Nedd8 or GFP-Nedd8 (1.6 μg/well), myc-tagged CARD or p30 domain proteins (1.6 μg/well), or empty pcDNA6.1 (1.6 μg/well) vector expressing FLAG, myc, HA, or GFP tags alone. At 24 h posttransfection, 293 cells were either treated with ATP (5 mM) or nigericin (15 μM) or infected with RSV at a multiplicity of infection (MOI) of 1 or with IAV (MOI, 1). BMDMs and NR-9456 cells were silenced by using Lipofectamine 2000 (40 pmol siRNA with 3 μl Lipofectamine per reaction mixture) for 24 h. NHBE cells were silenced by using Primefect and Primefect diluent (Lonza) according to the manufacturer's transfection protocol A for 24 h.
Treatments and infections.
Macrophages were treated with LPS for 4 h (BMDMs with 100 ng/ml, NR-9456 and THP-1 with 1 μg/ml) followed by ATP (5 mM) for 30 min and nigericin (15 μM) for 30 min. For MLN4924 experiments, macrophages were stimulated with LPS for 2 h, followed by addition of 250 nM MLN-containing medium with fresh LPS for 4 h, then ligand (nigericin or ATP) treatment for 30 min. 293 cells, NHBE cells, and BMDMs were infected with purified RSV or IAV at an MOI of 1 in serum-free antibiotic-free OPTI-MEM medium (Gibco). Virus adsorption was performed for 1.5 h at 37°C. Following adsorption, cells were washed twice with phosphate-buffered saline (PBS), and the infection was continued in the presence of complete medium for the specified time points. For the in vivo experiment, 6- to 8-week-old C57BL/6 male mice (obtained from Jackson Laboratories) were injected intraperitoneally (i.p.) with LPS (300 μg) in PBS for 3 h, then injected i.p. with MLN4924 (3 mg/kg of body weight, 6 mg/kg, or vehicle in PBS; 5 mice per group) for 4 h, then injected i.p. with ATP (100 μl of a 100 mM PBS solution) for 15 min. Mice were then bled via the retro-orbital method and sacrificed. Cytokine levels in serum were detected by an ELISA.
Immunofluorescence.
For immunofluorescence, 1 × 105 BMDMs were seeded onto 12-mm number 1.5 glass coverslips (Fisher) and cultured for 48 h. BMDMs were treated or infected following the protocol described above. BMDMs were then fixed with 10% stock formaldehyde in PBS overnight at 4°C, treated with 50 mM glycine for 1 h, permeabilized with 1% Triton X-100 in PBS for 1 h, blocked with 1% bovine serum albumin in PBS for 1 h, probed with goat anti-mouse caspase-1 p20 (1:300 for 1 h, washed 5 times [10 min per wash] with PBS, probed with rabbit anti-Nedd8 [1:300]) for 1 h, washed 5 times, probed with donkey anti-goat antibody–Texas Red conjugate (1:100); 30 min, washed 5 times, probed with goat anti-rabbit antibody–FITC conjugate for 30 min, washed 5 times, and finally mounted onto glass slides by using 5 μl SlowFade Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and clear nail polish. Slides were then analyzed on an Olympus FV-1000 confocal multiphoton spectral laser scanning microscope.
Western blotting and immunoprecipitation.
Primary antibodies for Western blotting included the following: rabbit anti-human caspase-1 p10 (1:500), rabbit anti-mouse caspase-1 p10 (1:500), goat anti-mouse caspase-1 p20 (1:500), rabbit anti-caspase-1 CARD (1:2,000), rabbit anti-Nedd8 (1:1,000), goat anti-mouse IL-1β p17 (1:2,000), mouse anti-human IL-1β (1:1,000), antiactin (1:5,000), mouse anti-GFP (1:2,000), mouse anti-myc (1:2,000), mouse anti-FLAG (1:2,000), and mouse anti-HA (1:2,000). Secondary antibodies for Western blotting included the following: goat anti-mouse antibody–HRP (1:5,000), goat anti-rabbit antibody–HRP (1:5,000), and donkey anti-goat antibody–HRP (1:5,000).
For immunoprecipitation, 293 lysate was harvested using 1% Triton X-100 in PBS with 5 mM Tris (pH 7.4) and 1× Roche Complete Mini EDTA-free protease inhibitor cocktail. Lysates were sonicated 3 times for 5 s under cold conditions. Lysates were then centrifuged at 13,000 rpm for 10 min, and clear lysate supernatant was transferred to a new tube. For 293 cells, lysates were mixed with 50 μl rabbit anti-c-myc–agarose (Sigma), 50 μl mouse anti-HA–agarose (Thermo Scientific), or 50 μl mouse anti-FLAG–agarose (Clontech) for 4 h at 4°C. For Western blotting with BMDMs, cell lysate was prepared as described above for 293 cells. The medium supernatant was precipitated with trichloroacetic acid (TCA) to analyze proteins in the medium by Western blotting. BMDM medium supernatant was collected and, following addition of TCA (15%), it was left on ice for 30 min. The medium was then centrifuged (at 14,000 rpm for 10 min at 4°C). Ice-cold acetone was added to the pellet, and following mixing of the pellet in acetone, it was once again centrifuged for 5 min (at 14,000 rpm at 4°C). Washing of the pellet with ice-cold acetone was repeated twice. After drying the pellet (to remove acetone), 2× sample buffer was directly added to the pellet for Western blot analysis.
For immunoprecipitation with BMDMs, cell lysate was mixed with 4 μg rabbit anti-Nedd8 IgG (Cell Signaling) for 1 h, followed by addition of 50 μl protein G-agarose (Thermo Scientific) for 3 h at 4°C. Samples were centrifuged at 1,000 rpm for 1 min, and the pellet was washed 4 times with PBS–5 mM Tris (pH 7.4) and protease inhibitor. For each sample, protein was eluted from the final pellet with 100 mM glycine (pH 2.9). Eluted protein was precipitated with 20% TCA overnight at 4°C. Precipitated protein was pelleted by centrifugation at 13,000 rpm for 10 min, washed twice with ice-cold acetone, resuspended in SDS dissolving buffer, and separated on a 12% SDS-PAGE gel. Protein was transferred onto a 0.2-μm nitrocellulose membrane (Bio-Rad) for Western blot analysis, or the SDS-PAGE gel was stained with Coomassie blue for mass spectrometry analysis. For analysis of cleaved pro-caspase-1 (p10) and pro-IL-1β (p17) levels (see Fig. 1B and E and 4B and C, below), we prepared cell lysate and TCA-precipitated (acetone-washed) pellet from the medium supernatant as described above. A portion of the cell lysate and TCA pellet (in SDS sample buffer) derived from medium supernatant was utilized for Western blotting with p10- and p17-specific antibodies. In order to detect total actin protein (loading control for the experiments shown in Fig. 1B and E and 4B and C), for each set an equal volume of cell lysate was added to an equal volume of SDS sample buffer containing the resuspended TCA pellet from the medium supernatant. This was followed by Western blotting with antiactin antibody.
FIG 1.
Nedd8 silencing in BMDMs reduces caspase-1 activation and IL-1β processing during inflammasome activation. (A) RT-PCR analysis of Nedd8 expression in BMDMs transfected with control siRNA or Nedd8 siRNA. (B) Western blotting results for caspase-1 p10 and mature IL-1β p17 in supernatants (supn) of Nedd8-silenced BMDMs treated with LPS followed by ATP or nigericin treatment. Actin served as a loading control (i.e., control for cell numbers). (C) RT-PCR analysis of Nedd8 expression in NR-9456 cells transfected with control siRNA or Nedd8 siRNA. (D) ELISA results for IL-1β in supernatants of Nedd8-silenced NR-9456 cells treated with LPS and nigericin. The values represent the means ± standard deviations from three independent experiments performed in triplicate. *, P < 0.05 using Student's t test. (E) Western blotting results for caspase-1 p10 in supernatants of Nedd8-silenced NR-9456 cells treated with LPS and nigericin. Actin served as a loading control (i.e., control for cell numbers). RT-PCR data (A and C) and Western blotting data (B and E) are representative of three independent experiments with similar results.
FIG 4.
Inhibition of neddylation by MLN4924 (MLN) diminishes caspase-1 activation and IL-1β maturation. (A) ELISA results for IL-1β in supernatants of MLN-treated BMDMs treated with LPS and ATP or nigericin. The values represent the means ± standard deviations from three independent experiments performed in triplicate. *, P < 0.05 using Student's t test. (B) Western blotting results for caspase-1 p10 and mature IL-1β p17 in supernatants (supn) of MLN-treated BMDMs treated with LPS and ATP or nigericin. Actin served as a loading control (i.e., control for cell numbers). (C) Western blotting results for caspase-1 p10 and IL-1β p17 in the supernatants of MLN-treated THP-1 cells incubated with LPS and nigericin in the presence of DMSO (vehicle control) or MLN. Actin served as a loading control (i.e., control for cell numbers). Western blotting data (B and C) are representative of three independent experiments with similar results.
Mass spectrometry.
Following immunoprecipitation, proteins were separated by one-dimensional (1-D) SDS-PAGE and stained with Coomassie blue. Another SDS-PAGE gel was used for Western blotting with specific antibodies. Bands of interest (as deduced by comparing the Western blot bands with the corresponding bands in the Coomassie blue-stained gel) were excised, and the proteins were digested in situ with trypsin (Promega). The digests were analyzed by capillary high-performance liquid chromatography–electrospray ionization-tandem mass spectrometry (HPLC-ESI-MS/MS) on a Thermo Fisher LTQ Orbitrap Velos mass spectrometer fitted with a New Objective Digital PicoView 550 NanoESI source. Online HPLC separation of the digests was accomplished with an Eksigent/AB Sciex NanoLC-Ultra 2-D HPLC system: PicoFrit column (New Objective; 75-μm inner diameter; New Objective) packed to 15 cm with C18 adsorbent (218MS; 5 μm, 300 Å; Vydac). Precursor ions were acquired in the Orbitrap in centroid mode at 60,000 resolution (m/z 400); data-dependent collision-induced dissociation (CID) spectra of the six most intense ions in the precursor scan above a set threshold were acquired at the same time in the linear trap. Mascot (Matrix Science) was used to search the spectra against a database containing sequences of locally generated recombinant proteins and constructs concatenated to the Swiss-Prot database. GlyGly-lysine, methionine oxidation, and deamidation of glutamine and asparagine were considered variable modifications; trypsin was specified as the proteolytic enzyme, with two missed cleavages allowed. Subset search of the identified proteins by X! Tandem, cross-correlation with the Mascot results, and determinations of protein and peptide identity probabilities were accomplished by using Scaffold (Proteome Software). The thresholds for acceptance of peptide and protein assignments in Scaffold were 95% and 99.9%, respectively.
RESULTS
Diminished IL-1β and pro-caspase-1 processing in Nedd8-silenced macrophages.
In order to evaluate the role of Nedd8 in IL-1β maturation, we silenced Nedd8 in primary BMDMs (Fig. 1A). The Nedd8-lacking cells were treated with LPS and inflammasome activators (nigericin and ATP) (Fig. 1B). IL-1β maturation was evaluated by Western blotting of medium supernatant with an antibody specific for mature IL-1β (p17) (Fig. 1B). Western blotting of medium supernatant with a caspase-1 p10 subunit-specific antibody was also performed to detect the levels of processed/cleaved caspase-1 (Fig. 1B). Nedd8 is essential for IL-1β maturation, since the levels of mature IL-1β were reduced in Nedd8-silenced cells (Fig. 1B). The loss of IL-1β maturation was a result of inefficient processing of pro-caspase-1, which was evident from the reduced levels of caspase-1 p10 subunit in Nedd8-silenced cells (Fig. 1B).
Similar results were obtained by using immortalized BMDMs (NR-9456 cells) silenced for Nedd8 expression (Fig. 1C). ELISA analysis of medium supernatant demonstrated loss of IL-1β production following inflammasome activation (LPS plus nigericin treatment) of NR-9456 cells treated with Nedd8 siRNA (Fig. 1D). Diminished IL-1β production was due to inefficient processing of pro-caspase-1, which was evident from the reduced levels of caspase-1 p10 subunit in Nedd8-lacking NR-9456 cells (Fig. 1E). Lack of mature IL-1β is not due to differences in pro-IL-1β levels, since expression of pro-IL-1β was similar in control siRNA and Nedd8 siRNA-treated BMDMs and NR-9456 cells (see Fig. S1A and B in the supplemental material). Similarly, pro-caspase-1 expression levels were unaltered in Nedd8-silenced cells compared to control cells (data not shown). During the time frame of our experiment, loss of Nedd8 had no effect on NF-κB activation, since LPS treatment of Nedd8-silenced cells did not alter TNF-α production (see Fig. S1C). These studies demonstrated that Nedd8 is an important regulator of IL-1β maturation and pro-caspase-1 processing, following inflammasome activation in macrophages.
Nedd8 is required for optimal IL-1β maturation/production following infection of lung epithelial cells with respiratory syncytial virus.
Recently, we showed that RSV activates the NLRP3/ASC inflammasome in macrophages (41). Similar activation of the NLRP3 inflammasome was noted in RSV-infected primary lung epithelial cells (56). Thus, we evaluated whether the Nedd8 requirement is restricted to only myeloid cells (i.e., macrophages) or if it can also regulate IL-1β production from nonmyeloid cells (i.e., epithelial cells). Since epithelial cells do not activate inflammasomes following treatment with LPS and inflammasome-activating agents (e.g., nigericin, ATP), we infected primary NHBE cells with RSV. NHBE cells silenced for Nedd8 expression (Fig. 2A) were infected with RSV, and IL-1β production was assessed in these cells. The requirement of Nedd8 is not only limited to myeloid cells, since IL-1β production from RSV-infected NHBE cells was significantly diminished following Nedd8 silencing (Fig. 2B). Western blot analysis of medium supernatant with antibody specific for mature IL-1β (p17) confirmed the loss of mature IL-1β (p17) in RSV-infected Nedd8-silenced cells (see Fig. S1D in the supplemental material). No change in expression of pro-IL-1β (Fig. 2A) was noted in Nedd8-lacking NHBE cells. Furthermore, the RSV infectious titer was comparable in control siRNA-treated cells versus Nedd8 siRNA-treated cells (data not shown). Thus, Nedd8 regulates IL-1β maturation/production in both nonmyeloid (NHBE) and myeloid (macrophage) cells.
FIG 2.
Nedd8 is required for optimal IL-1β maturation/release from RSV-infected NHBE cells, and silencing of Ubc12 in NR-9456 cells reduces IL-1β production. (A) RT-PCR analysis of Nedd8 and pro-IL-1β expression in RSV-infected NHBE cells transfected with control siRNA or Nedd8 siRNA. (B) ELISA results for IL-1β in supernatants of Nedd8-silenced NHBE cells infected with RSV for 16 h. (C) RT-PCR analysis of Ubc12 and pro-IL-1β expression in untreated (UT) and LPS-treated NR-9456 cells transfected with control siRNA or Ubc12 siRNA. (D) ELISA results for IL-1β in supernatants of Ubc12-silenced NR-9456 cells treated with LPS and ATP. The values shown in panels B and D represent the mean ± standard deviation from three independent experiments performed in triplicate. *, P < 0.05 using Student's t test. RT-PCR data (A and C) are representative of three independent experiments with similar results.
Loss of IL-1β production from Ubc12-silenced macrophages.
Ubc12 functions as the E2 conjugating enzyme for Nedd8 (57). Ubc12 is required for shuttling activated Nedd8 to the E3 ligase, which then conjugates Nedd8 to specific substrates. Ubc12 is the only E2 conjugating enzyme of the Nedd8 pathway. Thus, we silenced Ubc12 in macrophages to further provide evidence that the Nedd8 pathway regulates IL-1β maturation/production following inflammasome activation. For these studies, we silenced Ubc12 in NR-9456 cells (Fig. 2C). These cells were then treated with LPS and ATP to activate inflammasomes. Expression of Ubc12 is critical for IL-1β production, since diminished IL-1β production was noted following Ubc12 silencing (Fig. 2D). Loss of IL-1β production was not due to reduced pro-IL-1β (Fig. 2C) or pro-caspase-1 (data not shown) expression in Ubc12-silenced cells. These studies validated an essential role of Nedd8 in regulating IL-1β production following inflammasome activation.
Enhanced IL-1β production from 293 cells overexpressing Nedd8.
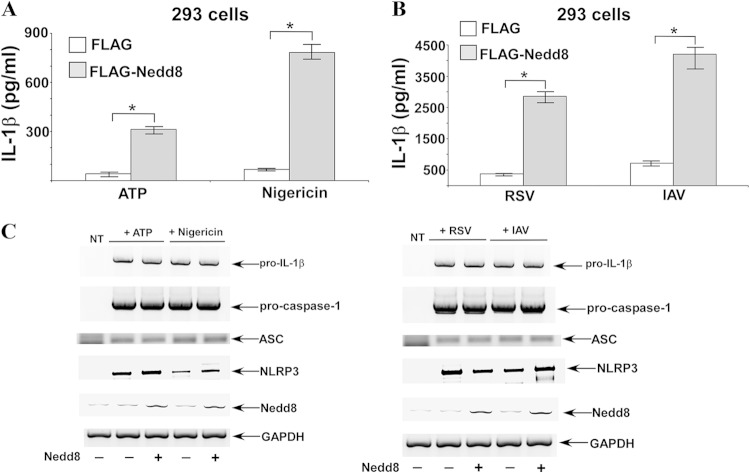
In order to further confirm Nedd8's role as a positive regulator of IL-1β production, we overexpressed Nedd8 in 293 cells along with pro-caspase-1, pro-IL-1β, ASC, and NLRP3. The 293 cell reconstitution system has been utilized successfully to analyze NLRP3 inflammasome-mediated IL-1β production. Cells overexpressing pro-IL-1β and inflammasome components (in the presence or absence of Nedd8 expression) were stimulated with NLRP3 inflammasome-activating agents, such as ATP, nigericin, and viruses (RSV and IAV). All of the inflammasome-activating agents drastically enhanced IL-1β production only in the presence of Nedd8 (Fig. 3A and B). The levels of pro-caspase-1, pro-IL-1β, ASC, and NLRP3 expression were similar in control cells and cells expressing Nedd8 (Fig. 3C). Note that although RT-PCR detected endogenous Nedd8 in Nedd8-nontransfected cells, enhanced Nedd8 expression was observed in cells transfected with FLAG-Nedd8. These studies demonstrated a role of Nedd8 as a positive regulator of IL-1β production.
FIG 3.
Enhanced IL-1β secretion from Nedd8-expressing 293 cells. (A and B) 293 cells were transfected with pro-IL-1β, pro-caspase-1, ASC, NLRP3, and FLAG-Nedd8 or empty FLAG vector (control). These cells were then either treated with ATP or nigericin (A) or infected with human RSV or IAV (B). IL-1β in supernatants was measured by ELISA. The values represent the means ± standard deviations from three independent experiments performed in triplicate. *, P < 0.05 using Student's t test. (C) RT-PCR analysis of gene expression in inflammasome-activated (treated with ATP or nigericin and infected with RSV or IAV) 293 cells transfected with pro-IL-1β, pro-caspase-1, ASC, NLRP3, or Nedd8. NT, nontransfected.
Neddylation inhibitor MLN4924 blocks IL-1β maturation/production in vitro and in vivo.
Since Nedd8 is embryonic lethal, Nedd8 knockout (KO) mice do not exist. Nedd8 conditional KO also did not yield a productive model, since Nedd8 is required for cellular homeostasis. Therefore, in order to validate the results obtained with RNAi technology and establish the in vivo physiological relevance of neddylation during IL-1β production, we utilized an inhibitor of neddylation called MLN4924, which blocks the activity of Nedd8-activating enzyme (47–49). MLN4924 is currently being developed as an anticancer drug during tumorogenesis, since neddylation of the tumor suppressor protein p53 results in loss of its activity. Several clinical trials conducted with human cancer patients have concluded that MLN4924 is a safe and highly efficacious anticancer drug. Since neddylation could also be involved in IL-1β maturation/production, we next assessed the effects of MLN4924 on pro-caspase-1 processing and IL-1β maturation/production. Both murine (BMDM) and human (THP-1) macrophages were treated with LPS and inflammasome-activating agents (ATP and nigericin) either in the presence or absence of MLN4924. Treatment of BMDMs with MLN4924 resulted in significantly reduced IL-1β production (Fig. 4A). Loss of IL-1β production was due to diminished IL-1β maturation and reduced pro-caspase-1 processing in BMDMs, since Western blot analysis of medium supernatant revealed loss of mature IL-1β (p17) (Fig. 4B) and processed p10 subunit of pro-caspase-1 (Fig. 4B) following MLN4924 treatment. Similar to BMDMs, MLN4924 treatment of THP-1 cells also led to loss of IL-1β maturation and reduced pro-caspase-1 processing (Fig. 4C). MLN4924 treatment did not significantly alter the expression of pro-IL-1β in BMDMs (see Fig. S2A in the supplemental material) and THP-1 cells (see Fig. S2B, upper panel). The levels of pro-caspase-1 were unaltered in MLN4924-treated THP-1 cells (see Fig. S2B, lower panel) or BMDMs (data not shown). The loss of mature IL-1β (p17 subunit) and processed caspase-1 (p10 subunit) in the medium supernatant of Nedd8-silenced (data not shown) and neddylation-inactivated MLN4924-treated cells (see Fig. S2D) was not due to a problem in secretion of these cleaved fragments, since we failed to detect any p17 and p10 in the cell lysate following inflammasome triggering by nigericin (see Fig. S2C). Our results corroborated results from previous studies (58) that demonstrated NLRP3 inflammasome activation in BMDMs (by ATP or nigericin) triggering rapid release of p10 and p17 in the medium supernatant following cleavage of pro-caspase-1 and pro-IL-1β, respectively. Therefore, p10 and p17 can only be detected in the medium supernatant and not in the cell lysate of ATP- and nigericin-treated cells (58). Although we detected p10 and p17 in the medium supernatant of inflammasome-activated BMDMs (Fig. 4B), albeit at reduced levels in MLN4924-treated cells (Fig. 4B), we failed to detect any p10 and p17 in the cell lysate of inflammasome-activated (activated by nigericin) control or MLN4924-treated cells (see Fig. S2C), thus precluding any possibility of interference with secretion of p10 and p17 in neddylation-inhibited cells. In accordance with previous studies (58), we also observed that inflammasome activation resulted in loss of intracellular pro-caspase-1 and pro-IL-1β proteins due to their release into the medium supernatant (see Fig. S2C). Diminished levels of intracellular caspase-1 and complete loss of intracellular pro-IL-1β protein in inflammasome-activated control and MLN4924-treated cells (see Fig. S2C) indicated that reduced levels of p10 and p17 in the medium supernatant of MLN4924-treated cells (Fig. 4B) were not due to a secretion problem, but rather due to diminished processing of pro-caspase-1 and pro-IL-1β. We observed similar results in inflammasome-activated Nedd8-silenced BMDMs (data not shown). In addition, note that nigericin (and ATP) treatment of BMDMs causes release of pro-caspase-1 and pro-IL-1β in the medium supernatant independent of inflammasome activation (58). Therefore, although MLN4924 diminished caspase-1 activation (and pro-IL-1β cleavage), low levels of intracellular pro-caspase-1 and pro-IL-1β were detected in control and MLN4924-treated cells activated with inflammasome-triggering agent nigericin (see Fig. S2C).
Similar to Nedd8-silenced cells, MLN4924 treatment during the time frame of our experiment did not alter NF-κB activation status, since LPS triggered similar levels of TNF-α production from control and MLN4924-treated cells (see Fig. S2D in the supplemental material). Interestingly, Nedd8 was required for both canonical NLRP3 inflammasome-mediated and noncanonical NLRC4 inflammasome-mediated IL-1β release. Previous studies have shown that 2-h infection of BMDMs with flagellin-deleted Salmonella enterica serovar Typhimurium (ΔfliC ΔfliB) results in NLRC4-mediated IL-1β release (59). We therefore analyzed IL-1β release from control and MLN4924-treated BMDMs infected with ΔfliC ΔfliB S. Typhimurium for 2 h. We observed a significant loss of IL-1β release from MLN4924-treated Salmonella (ΔfliC ΔfliB)-infected cells (see Fig. S2E). Thus, our result indicated that neddylation can also regulate NLRC4 inflammasome-dependent IL-1β release.
Since MLN4924 inhibited IL-1β production/maturation, we next administered MLN4924 to mice to investigate the in vivo role of neddylation during IL-1β production. In order to assess the involvement of Nedd8, IL-1β levels were measured in the serum of mice treated (via the intraperitoneal route) with LPS and ATP (in the absence or presence of MLN4924). A similar procedure was utilized previously to investigate inflammasome activation in mice (32). Neddylation is also required in vivo for IL-1β production, since significantly reduced levels of IL-1β were detected in the serum of MLN4924-treated animals (Fig. 5A). IL-18 is also released following caspase-1 activation. We observed diminished IL-18 levels in the serum of MLN4924-treated mice (Fig. 5B). The effect of MLN4924 was specific for IL-1β inhibition during the time frame of our experiment, because levels of serum TNF-α were unaltered in MLN4924-treated and untreated mice (Fig. 5C). These results demonstrated that IL-1β and IL-18 production in vivo is positively regulated by neddylation. Furthermore, our in vivo studies suggest the potential use of MLN4924 as a specific anti-IL-1β (and anti-inflammasome) therapeutic agent to control inflammasome-dependent IL-1β production during various disease processes.
FIG 5.
MLN4924 (MLN) administration reduces IL-1β and IL-18 production in mice. (A and B) C57BL/6 mice (n = 5 per group) were injected (i.p. route) with LPS (for 3 h), followed by i.p. injection of MLN (for 4 h) and finally i.p. administration of ATP (for 15 min). Serum collected from the mice was analyzed for IL-1β (A) and IL-18 (B) by ELISA. The values represent the means ± standard deviations. *, P < 0.05 using Student's t test. (C) C57BL/6 mice (n = 5 per group) were injected (i.p. route) with LPS (for 3 h), followed by i.p. injection of MLN (for 4 h). Serum collected from the mice was analyzed for TNF-α by ELISA. The values represent the means ± standard deviations.
Nedd8 enhances the autocatalytic activity of caspase-1.
Next, we embarked on a series of studies focusing on the mechanism by which Nedd8 regulates IL-1β maturation/production. First, we performed coimmunoprecipitation (co-IP) studies with cell lysates obtained from 293 cells expressing GFP-tagged Nedd8 (GFP-Nedd8) along with various inflammasome-associated proteins. We did not detect any interaction of Nedd8 with NLRP3 or ASC (data not shown). However, co-IP analysis of cells coexpressing GFP-Nedd8 and myc-pro-caspase-1 revealed two bands (indicated by red arrows) corresponding to 45 kDa and 80 kDa (Fig. 6A). Based on the molecular mass of GFP-Nedd8 (8-kDa Nedd8 plus 27-kDa GFP = 35 kDa) and pro-caspase-1 (45 kDa), the 80-kDa band suggested possible covalent linkage (since the protein samples were boiled with SDS reducing buffer) of GFP-Nedd8 monomer with pro-caspase-1 (i.e., 35-kDa GFP-Nedd8 conjugated to 45-kDa caspase-1 = 80 kDa). Since the CARD of caspase-1 could be neddylated (see below) (Fig. 6B), the 45-kDa band may represent neddylated CARD (i.e., 35-kDa GFP-Nedd8 conjugated to 9-kDa CARD = ∼45 kDa). Mass spectrometry analysis revealed the presence of caspase-1 and Nedd8 in the 80-kDa and 45-kDa bands (see Fig. S3 in the supplemental material). The observed effect was not due to nonspecific binding of GFP to pro-caspase-1, since expression of GFP alone did not result in its interaction with pro-caspase-1 (Fig. 6A). Moreover, Western blotting of the cell lysate showed similar levels of GFP and GFP-Nedd8 expression (Fig. 6A). However, similar analysis showed reduced pro-caspase-1 levels in GFP-Nedd8-expressing cells, which could be attributed to the ability of Nedd8 to augment the autoprocessing activity of pro-caspase-1 (see Fig. 7). Furthermore, we could observe interaction of pro-caspase-1 with Nedd8 by using Nedd8 fused to a smaller tag-like FLAG tag (see below) (see also Fig. S5B in the supplemental material). The rationale for using GFP-Nedd8 was based on the ability to detect appreciable shift in the molecular mass of pro-caspase-1 if GFP-Nedd8 (especially with mononeddylation of pro-caspase-1) were covalently linked to it. Use of a smaller tagged Nedd8 protein may not show an apparent difference in pro-caspase-1 molecular mass, if it is mononeddylated. Thus, our studies demonstrated possible neddylation of caspase-1 following overexpression of pro-caspase-1 and Nedd8 in 293 cells.
FIG 6.
Interaction of Nedd8 with caspase-1. (A) Cell lysates were collected from 293 cells expressing GFP-Nedd8 and myc-procaspase1 and immunoprecipitated (IP) with myc antibody, followed by immunoblotting (IB) with GFP antibody. Total lysate was also blotted with GFP and myc antibodies. The lower panel shows the membrane with a longer exposure. The 45-kDa and 80-kDa bands observed in Nedd8-expressing cells are indicated with arrows. (B) Cell lysates collected from 293 cells expressing GFP-Nedd8 and myc-CARD (pro-caspase-1 CARD) and immunoprecipitated with myc antibody, followed by blotting with GFP antibody. The lower panel shows the gel with a longer exposure. The 80-kDa band observed in Nedd8-expressing cells is indicated by an arrow. The data are representative of three independent experiments with similar results.
FIG 7.
Nedd8-dependent enhanced processing of pro-caspase-1. (A) Cell lysate collected from 293 cells expressing FLAG-Nedd8 and double-tagged pro-caspase-1 (myc–caspase-1–HA; myc tag on the N terminal and HA tag on the C terminal) were immunoprecipitated (IP) with myc antibody, followed by immunoblotting (IB) with myc antibody. (B) Cell lysate collected from 293 cells expressing FLAG-Nedd8 and myc–caspase-1–HA were immunoprecipitated with HA antibody, followed by blotting with HA antibody. (C) A longer-exposed version of the blot shown in panel B. The higher-molecular-mass (approximately 53-kDa) caspase-1 was visible in Nedd8-expressing cells. (D) Western blotting results for the total lysate corresponding to the experiment shown in panels A to C. Cell lysates collected from 293 cells expressing FLAG-Nedd8 and myc–caspase-1–HA were subjected to Western blotting with either HA or FLAG antibody. The data are representative of three independent experiments with similar results.
Next, we investigated which domain of pro-caspase-1 interacts with Nedd8. For these studies, we expressed a myc-tagged N-terminal CARD (myc-CARD) or C-terminal p30 (myc-p30) domain (which is cleaved to generate p10 and p20, the functional subunits of active caspase-1) in 293 cells along with GFP-Nedd8. Co-IP analysis revealed interaction of GFP-Nedd8 with the caspase-1 CARD (i.e., with myc-CARD) (Fig. 6B; see also Fig. S4A and B in the supplemental material). In contrast, the p30 domain of caspase-1 did not interact with Nedd8 (see Fig. S5A in the supplemental material). The CARD was also covalently linked to Nedd8, since immunoprecipitation with myc antibody and blotting with GFP antibody yielded two bands (Fig. 6B): (i) a 45-kDa band which might correspond to mononeddylated CARD (35-kDa GFP-Nedd8 conjugated to 9-kDa CARD = ∼45 kDa), and (ii) an 80-kDa band which might represent dineddylated CARD. These studies demonstrated that the CARD of pro-caspase-1 interacts with Nedd8, possibly via covalent attachment.
To gain further mechanistic insights, we expressed doubly tagged pro-caspase-1 (myc-caspase-1-HA tagged with myc and HA at the N terminal and C terminal, respectively) in 293 cells along with FLAG-Nedd8. The rationale for using myc-caspase-1-HA was to track processing (cleavage) of pro-caspase-1 into various subunits. As expected, immunoprecipitation with HA antibody followed by blotting with FLAG antibody yielded a band of 53 kDa, which may correspond to mononeddylated pro-caspase-1 (45-kDa pro-caspase-1 plus 8-kDa Nedd8 = 53 kDa) (see Fig. S5B in the supplemental material). Mass spectrometry analysis confirmed that the 53-kDa band observed in Nedd8-expressing cells was indeed caspase-1 (see Fig. S6 in the supplemental material). Moreover, data from Nedd8-expressing cells revealed a Gly-Gly modification (this modification on the lysine residue serves as a signature for neddylation [60, 61]) in two lysine residues (residues K37 and K53) of caspase-1 CARD. Thus, K37 and K53 may serve as neddylation sites of caspase-1.
In order to examine the processing (cleavage) status of pro-caspase-1 in the presence or absence of Nedd8, we next performed immunoprecipitation with myc antibody followed by blotting with myc antibody to detect the levels of cleaved CARD fragment. Similarly, to observe the status of the cleaved p10 subunit, we also immunoprecipitated with HA antibody followed by anti-HA blotting. Surprisingly, we detected enhanced processing of pro-caspase-1 in the presence of Nedd8 (Fig. 7A, B, and C). Increased levels of cleaved fragments of the p10 subunit (10 kDa) and p30 subunit (30-kDa subunit generated after cleavage of the CARD) were also detected in the presence of Nedd8 (Fig. 7A). Furthermore, greater quantities of the CARD fragment were also detected in cells expressing Nedd8 (Fig. 7B and C). It is important to note that we did not detect any unneddylated CARD (9 kDa). Instead, a 17-kDa band, which may represent mononeddylated CARD (9-kDa CARD plus 8-kDa Nedd8 = 17 kDa), along with additional processing intermediates were detected in the presence of Nedd8. It is important to mention that a longer exposure was required to visualize 53-kDa neddylated caspase-1 (Fig. 7C), as deduced from our co-IP studies shown in Fig. S5B in th supplemental material. Interestingly, the 53-kDa caspase-1 band was only observed in cells expressing Nedd8 (Fig. 7C). The increased apparent molecular mass of caspase-1 and its enhanced processing were also obvious by Western blotting of a total cell lysate with HA antibody (to detect pro-caspase-1 and C-terminal processing) (Fig. 7D). Mass spectrometry analysis confirmed that the cleavage products were, in fact, processed caspase-1 (see Fig. S7 in the supplemental material). These results suggested probable neddylation of caspase-1 (CARD) and, furthermore, demonstrated that neddylation could enhance the autocatalytic activity of pro-caspase-1, resulting in efficient self-cleavage to yield functional p10 and p20 subunits. Involvement of Nedd8 is consistent with diminished caspase-1 p10 levels following inflammasome activation in Nedd8-silenced or NAE-inhibited (MLN4924-treated cells) cells (Fig. 1B and E and 4B and C).
Endogenous interaction of Nedd8 with caspase-1 following inflammasome activation.
Next, we investigated the possible interaction of endogenous Nedd8 with caspase-1 and whether this occurs following inflammasome activation. Activation of inflammasome by ATP, nigericin, and viruses (RSV and IAV) led to colocalization (following coimmunofluorescence) of Nedd8 with caspase-1 (Fig. 8). Interestingly, Nedd8 was detected in the “speck” that represents the inflammasome complex (19, 62, 63). Thus, endogenous Nedd8 colocalizes with caspase-1, which is most probably bound to or in close proximity to the inflammasome complex (the speck).
FIG 8.
Colocalization of endogenous Nedd8 with caspase-1 following inflammasome activation. (A) Coimmunofluorescence analysis of Nedd8 (red) and caspase-1 (green) in BMDMs following LPS treatment in the absence or presence of ATP or nigericin. (B) Coimmunofluorescence analysis of Nedd8 and caspase-1 in BMDMs following infection with RSV or IAV. Merged images (yellow) show colocalization of Nedd8 with caspase-1. Magnified versions of the merged “speck” are shown on the extreme right sides of the two panels. The images are representative of 30 viewing fields from two independent experiments with similar results.
The effect of the neddylation inhibitor MLN4924 on the interaction of Nedd8 with caspase-1 upon inflammasome activation was investigated by examining colocalization of Nedd8 with caspase-1 in the presence of MLN4924. As expected, activation of inflammasome by LPS and nigericin treatment triggered colocalization of Nedd8 with caspase-1 in cells treated with dimethyl sulfoxide (DMSO; vehicle control) (see Fig. S8 in the supplemental material). However, in MLN4924-treated cells, although the caspase-1 “speck” was visible in inflammasome-activated cells, we failed to detect any colocalization of Nedd8 with caspase-1 (see Fig. S8).
Based on our results with 293 cells, one may envision possible neddylation of endogenous caspase-1 following inflammasome activation. If indeed pro-caspase-1 were neddylated following inflammasome activation, one would expect to observe a high-molecular-mass pro-caspase-1 protein, but the size of pro-caspase-1 does not change after inflammasome activation, as demonstrated previously by others and us (data not shown). We postulate that this could be due to the fact that once pro-caspase-1 is neddylated, it is rapidly cleaved and, therefore, it is difficult to detect steady-state levels of neddylated pro-caspase-1. We speculate that neddylation of endogenous pro-caspase-1 (i.e., neddylation of pro-caspase-1 CARD) following inflammasome activation results in rapid cleavage of the neddylated CARD to release the p30 domain (which is then autocleaved into functional p10 and p20 subunits). In order to further probe into the possible interaction of endogenous Nedd8 with caspase-1 (specifically the CARD), we performed co-IP studies with anti-Nedd8 antibody and antibody specific for caspase-1 CARD (CARD antibody). Western blotting with CARD antibody detected pro-caspase-1 (45 kDa) in wild-type (WT) BMDMs, while the band corresponding to pro-caspase-1 was not detected in caspase-1 KO BMDMs (see Fig. S9A in the supplemental material). Furthermore, CARD antibody can only detect caspase-1 CARD (by Western blotting) following transfection of 293 cells with the CARD portion of caspase-1 (data not shown). These results showed that the CARD antibody is specific for caspase-1. For co-IP studies, we used cell lysates, since we failed to detect CARD in the medium supernatant of inflammasome-activated (i.e., LPS- and nigericin-treated) BMDMs (see Fig. S9B). Cell lysates from either LPS-treated or LPS- and nigericin-treated BMDMs were immunoprecipitated with Nedd8 antibody, followed by blotting with caspase-1 CARD-specific antibody. Co-IP analysis revealed a band of 18-kDa in inflammasome-activated BMDMs (Fig. 9A). Based on the apparent molecular mass, the band may represent the cleaved mononeddylated CARD portion of pro-caspase-1 (9-kDa CARD plus 8-kDa Nedd8 = 17 kDa). These results demonstrated an endogenous interaction of Nedd8 with pro-caspase-1 following inflammasome activation.
FIG 9.
Endogenous interaction of Nedd8 with caspase-1 CARD following inflammasome activation. (A) Cell lysates collected from BMDMs treated with LPS alone (4 h) or LPS and nigericin (LPS treatment for 4 h followed by nigericin treatment for 30 min) were immunoprecipitated (IP) with Nedd8 antibody and immunoblotted (IB) with caspase-1 CARD-specific antibody. (B) BMDMs were either untreated (UT; lane 1) or treated with LPS for 2 h (lane 2), followed by addition of MLN4924 (lane 4) or DMSO (control; lane 3) along with LPS. After 4 h, cells treated with LPS plus MLN4924 (lane 4) or LPS plus DMSO (lane 3) (total LPS treatment, 6 h) were treated with nigericin for 30 min. The cell lysates were subjected to Western blotting with anti-CARD antibody. The 17-kDa cleaved CARD fragment observed following inflammasome activation (i.e., after nigericin treatment) is indicated by a red arrow. Actin served as a loading control (i.e. control for cell numbers).
Neddylation is required for optimal cleavage of CARD from endogenous pro-caspase-1 following inflammasome activation.
Our studies with 293 cells suggested that neddylation could function as an essential cellular factor regulating efficient processing/cleavage of pro-caspase-1, which is necessary to yield the functional p20 and p10 subunits. Furthermore, we observed an endogenous interaction of Nedd8 with caspase-1 (CARD) following inflammasome activation. Based on these studies, we speculated that inhibiting neddylation in BMDMs results in diminished cleavage of CARD from endogenous pro-caspase-1 following inflammasome activation. In order to elucidate a role of neddylation in pro-caspase-1 processing in BMDMs, we treated these cells with LPS for 2 h, followed by addition of vehicle or MLN4924 (with LPS). After MLN4924 (with LPS) treatment for 4 h, nigericin was added to BMDMs for 30 min. The cell lysates were subjected to Western blotting with CARD antibody. As expected, pro-caspase-1 processing was evident in vehicle-treated cells. A band of 17 kDa (red arrow) was detected in vehicle-treated, inflammasome-activated cells (Fig. 9B, lane 3). This band may correspond to mononeddylated CARD (9-kDa CARD plus 8-kDa Nedd8 = 17 kDa), as discussed above. We failed to visualize a 9-kDa band, which should have represented the 9-kDa CARD fragment (Fig. 9B, blot with longer exposure). Interestingly, MLN4924 treatment almost completely abolished the 17-kDa band (Fig. 9B, lane 4). As mentioned above, we performed Western blotting with cell lysate, since we failed to detect cleaved CARD in the supernatant of inflammasome-activated cells (see Fig. S9B in the supplemental material).
Furthermore, blocking neddylation by MLN4924 did not prevent formation of ASC “specks” (see Fig. S10 in the supplemental material), suggesting that neddylation does not play a role in ASC oligomerization during inflammasome complex formation. This result suggested that neddylation could be involved in efficient processing of endogenous pro-caspase-1 following inflammasome activation, and especially that cleavage of CARD from pro-caspase-1 could be dependent on neddylation.
DISCUSSION
Inflammasome-mediated caspase-1 activation leading to maturation and production of IL-1β is a critical host response that dictates health and disease. This pathway has been implicated in a wide spectrum of diseases, including pneumonia (viral or bacterial), diabetes, atherosclerosis, obesity, cancer, gout, Alzheimer's, arthritis, etc. (8–37). Furthermore, it also constitutes an important host defense mechanism against various pathogens (viruses, bacteria, fungi, and parasites). Caspase-1 is also required for induction of pyroptosis, a specialized cell death pathway which is primarily upregulated during bacterial infection. During IL-1β maturation, active caspase-1 cleaves 31-kDa inactive pro-IL-1β to 17-kDa active (secreted) IL-1β, which is then secreted from the cells to activate (via paracrine and autocrine mechanisms) its cognate IL-1R. This results in amplification of the proinflammatory response, which shapes the host immune response during various stresses. Inactive pro-caspase-1 (45 kDa) consists of three domains: CARD (9 kDa), p20 (20 kDa), and p10 (10 kDa). Recruitment of pro-caspase-1 to the inflammasome complex triggers its autocleavage activity, most probably due to conformational alteration of the inflammasome-bound pro-caspase-1. As a consequence of self-cleavage, p10 and p20 subunits are generated. A homo-hetero-tetramer of p10-p20 constitutes the enzymatically active form of caspase-1. Surprisingly, our studies demonstrated an involvement of Nedd8 in inflammasome-dependent caspase-1 activation and IL-1β maturation. We postulate that interaction of Nedd8 with the CARD of pro-caspase-1 is essential for optimal autocatalytic processing of pro-caspase-1 into its cleaved active subunits.
Covalent conjugation of Nedd8 to proteins (the process known as neddylation) via an isopeptide linkage regulates the biochemical (and biological) properties of the target protein (45, 46). Neddylation can act as a posttranslational modification to either commit target proteins to proteasomal degradation or alter the biological activity of the target protein. The latter event occurs due to conformational alteration of the neddylated target protein, which triggers either its activation or deactivation in terms of its cellular, molecular, biochemical, and biological activity/function. Only a few Nedd8 target proteins have been identified so far, and there has been no report of neddylation regulation of inflammasome activity. Neddylation of the cullin family of proteins is well characterized (64, 65). Cullins act as scaffold proteins during assembly of ubiquitin E3 enzymes. Neddylation of cullin activates ubiquitin ligase activity, resulting in ubiquitination of target proteins. Neddylation can also affect protein functions directly by modulating their biological activity/function. Neddylation of p53 and p77 proteins suppresses their transcriptional activities (66, 67). Neddylation of pVHL promotes its interaction with fibronectin, resulting in assembly of extracellular matrix (68). BCA3 is neddylated to inhibit NF-κB-mediated transcription by virtue of an interaction of neddylated BCA3 with SIRT1 (a histone deacetylase) (69).
A recent study reported that neddylation of an apoptosis-associated caspase, caspase-7, inhibited its proteolytic activity, thus inhibiting induction of apoptosis (70). A separate study failed to observe neddylated caspase-7 (71). Thus, further studies are required to establish whether caspase-7 is posttranslationally modified by neddylation. However, our studies suggest that an apoptosis-unrelated caspase, caspase-1, is modified by neddylation. Contrary to caspase-7 neddylation, caspase-1 neddylation augments its autocatalytic activity, leading to generation of enzymatically active caspase-1, which cleaves pro-IL-1β into mature IL-1β. Although we have demonstrated that overexpressed pro-caspase-1 is neddylated in 293 cells by mass spectrometry, whether this modification occurs in macrophages is still unknown. Since our study suggested that neddylated pro-caspase-1 could be cleaved rapidly, it may be technically challenging to acquire enough endogenous caspase-1 protein for mass spectrometric analysis. Nevertheless, the neddylation inhibitor MLN4924 reduced the processing of endogenous pro-caspase-1 into its cleaved fragments comprised of CARDs and p10 domains following inflammasome activation, indicating that neddylation has a biological role in caspase-1 processing. In the future, we plan to further expand our studies to explore the role of neddylation in IL-1β maturation and whether endogenous pro-caspase-1 is neddylated after inflammasome activation.
It was recently shown that cullin neddylation affects NF-κB activity in human (THP-1) and mouse (RAW 264.7) macrophages (72). Treatment of these cells with MLN4924 led to reduced NF-κB activity and diminished production of NF-κB-dependent cytokines (TNF-α and IL-6). However, in our current studies conducted with primary BMDMs, we did not observe a discernible difference in LPS-mediated pro-IL-1β protein expression and TNF-α production following Nedd8 silencing or MLN4924 treatment. Moreover, we added MLN4924 to the cells after NF-κB induction (i.e., following LPS treatment). Cullins are well-characterized neddylated proteins (64, 65), and neddylated cullins (cullins 1, 2, and 3 are parts of SCF, SOCS, and BTB E3 ubiquitin ligase, respectively) are involved in ubiquitination of target proteins. In that regard, deubiquitinase enzymes (DUBs) have been demonstrated to regulate inflammasome activity (73–75). Previous studies have shown that DUB inhibitors reduces ASC oligomerization and IL-1β release (74, 75). Ubiquitinated NLRP3 was detected when cells were treated with DUB inhibitors, and ASC ubiquitination was observed during AIM2-mediated inflammasome activation (74–76). In that scenario, it is possible that silencing of Nedd8 or inhibiting neddylation prevents ubiquitination/deubiquitination, which will lead to diminished caspase-1 activation and IL-1β maturation. So far, no studies have directly shown that inhibiting ubiquitination adversely affects inflammasome activation and IL-1β release, although a study demonstrated that the cIAP (cIAP 1 and 2) E3 ubiquitin ligase is involved in caspase-1 ubiquitination and that lack of cIAP results in loss of caspase-1 activation (77). Moreover, proteasomal inhibitors did not have an effect on inflammasome activation and IL-1β release from both human and murine macrophages (75). In that regard, a recent study detected caspase-1 ubiquitination following inflammasome activation (78). However, such posttranslational modifications only occurred after caspase-1 activation and, thus, it was postulated that caspase-1 is ubiquitinated following its activation so that caspase-1 can undergo proteasomal degradation (78). Based on our studies and previous reports, it is possible that neddylation and ubiquitination represent two distinct mechanisms regulating caspase-1 activation and IL-1β release, since (i) proteasomal inhibitors have no effect on caspase-1 activation and IL-1β production (75), (ii) caspase-1 ubiquitination occurs following its activation (78), whereas neddylation is required for caspase-1 activation, (iii) it is postulated that ubiquitinated caspase-1 (and other inflammasome components) undergo proteasomal (and autophagosomal) degradation to ensure that inflammasome activation is regulated, and (iv) although cIAP E3 ligase is involved in caspase-1 ubiquitination (77), cIAP E3 ligases are distinct from the other E3 ligases (i.e., SCF, SOCS, and BTB E3 ubiquitin ligases) that require neddylated cullins, and therefore it is highly unlikely that loss of Nedd8 (and inhibition of neddylation activity) will also result in reduced ubiquitination of caspase-1 by cIAP.
Nevertheless, we are very much aware that the role of Nedd8 in caspase-1 activation could be indirect, either being regulated via the ubiquitination pathway or additional cellular factors. Therefore, we will conduct further studies to evaluate the ubiquitination status of inflammasome-associated proteins (including caspase-1) in Nedd8-silenced cells and cells treated with MLN4924.
In summary, our studies have demonstrated a role of Nedd8 in regulating inflammasome-dependent caspase-1 activation and IL-1β maturation/production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) grant AI083387 (to S.B.). J.A.S. was supported by NIH/NIDCR grant DE14318 for the COSTAR program and Translational Science Training (TST) program at the University of Texas Health Science Center at San Antonio, with funding provided by CTSA grant 8UL1 TR000149. Mass spectrometry analyses were conducted in the UTHSCSA Institutional Mass Spectrometry Laboratory, with support from UTHSCSA and NIH grant 7P30CA54174-14 (CTRC MS Shared Resource). Images were generated in the Core Optical Imaging Facility, which is supported by UTHSCSA, NIH-NCI P30 CA54174 (San Antonio Cancer Institute), NIH-NIA P30 AG013319 (Nathan Shock Center), and NIH-NIA P01AG19316.
We thank Leigh Knodler (Washington State University) for providing the Salmonella strain. We also thank Devendra Shah (Washington State University) and Narayan Paul (Washington State University) for suggestions. We thank Stephanie Baker for technical assistance.
We declare that we have no conflict of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00775-14.
REFERENCES
- 1.Kostura MJ, Tocci MJ, Limjuco G, Chin J, Cameron P, Hillman AG, Chartrain NA, Schmidt JA. 1989. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc Natl Acad Sci U S A 86:5227–5231. doi: 10.1073/pnas.86.14.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thornberry NA, Bull HG, Calaycay JR, Chapman KT, Howard AD, Kostura MJ, Miller DK, Molineaux SM, Weidner JR, Aunins J, Elliston KO, Ayala JM, Casano FJ, Chin J, Ding GJ-F, Egger LA, Gaffney EP, Limjuco G, Palyha OC, Raju SM, Rolando AM, Salley JP, Yamin T-T, Lee TD, Shively JE, MacCross M, Mumford RA, Schmidt JA, Tocci MJ. 1992. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356:768–774. doi: 10.1038/356768a0. [DOI] [PubMed] [Google Scholar]
- 3.Cerretti DP, Kozlosky CJ, Mosley B, Nelson N, Van Ness K, Greenstreet TA, March CJ, Kronheim SR, Druck T, Cannizzaro LA, et al. 1992. Molecular cloning of the interleukin-1 beta converting enzyme. Science 256:97–100. doi: 10.1126/science.1373520. [DOI] [PubMed] [Google Scholar]
- 4.Wilson KP, Black JA, Thomson JA, Kim EE, Griffith JP, Navia MA, Murcko MA, Chambers SP, Aldape RA, Raybuck SA, Livingston DJ. 1994. Structure and mechanism of interleukin-1 beta converting enzyme. Nature 370:270–275. doi: 10.1038/370270a0. [DOI] [PubMed] [Google Scholar]
- 5.Walker NP, Talanian RV, Brady KD, Dang LC, Bump NJ, Ferenz CR, Franklin S, Ghayur T, Hackett MC, Hammill LD, et al. 1994. Crystal structure of the cysteine protease interleukin-1 beta-converting enzyme: a (p20/p10)2 homodimer. Cell 78:343–352. doi: 10.1016/0092-8674(94)90303-4. [DOI] [PubMed] [Google Scholar]
- 6.Denes A, Lopez-Castejon G, Brough D. 2012. Caspase-1: is IL-1 just the tip of the ICEberg? Cell Death Dis 3:e338. doi: 10.1038/cddis.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dinarello CA, Simon A, van der Meer JW. 2012. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov 11:633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis BK, Wen H, Ting JP. 2011. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol 29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant C, Fitzgerald KA. 2009. Molecular mechanisms involved in inflammasome activation. Trends Cell Biol 19:455–464. doi: 10.1016/j.tcb.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. 2009. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol 10:241–247. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamkanfi M, Dixit VM. 2009. The inflammasomes. PLoS Pathog 5:e1000510. doi: 10.1371/journal.ppat.1000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroder K, Tschopp J. 2010. The inflammasomes. Cell 140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 13.Martinon F, Mayor A, Tschopp J. 2009. The inflammasomes: guardians of the body. Annu Rev Immunol 27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- 14.Jha S, Ting JP. 2009. Inflammasome-associated nucleotide-binding domain, leucine-rich repeat proteins and inflammatory diseases. J Immunol 183:7623–7629. doi: 10.4049/jimmunol.0902425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye Z, Ting JP. 2008. NLR, the nucleotide-binding domain leucine-rich repeat containing gene family. Curr Opin Immunol 20:3–9. doi: 10.1016/j.coi.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Franchi L, Warner N, Viani K, Nunez G. 2009. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev 227:106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tannahill GM, O'Neill LA. 2011. The emerging role of metabolic regulation in the functioning of Toll-like receptors and the NOD-like receptor Nlrp3. FEBS Lett 585:1568–1572. doi: 10.1016/j.febslet.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Masters SL, Latz E, O'Neill LA. 2011. The inflammasome in atherosclerosis and type 2 diabetes. Sci Transl Med 3:81ps17. doi: 10.1125/scitranslmed.3001902. [DOI] [PubMed] [Google Scholar]
- 19.Masters SL, Dunne A, Subramanian SL, Hull RL, Tannahill GM, Sharp FA, Becker C, Franchi L, Yoshihara E, Chen Z, Mullooly N, Mielke LA, Harris J, Coll RC, Mills KH, Mok KH, Newsholme P, Nunez G, Yodoi J, Kahn SE, Lavelle EC, O'Neill LA. 2010. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat Immunol 11:897–904. doi: 10.1038/ni.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franchi L, Munoz-Planillo R, Reimer T, Eigenbrod T, Nunez G. 2010. Inflammasomes as microbial sensors. Eur J Immunol 40:611–615. doi: 10.1002/eji.200940180. [DOI] [PubMed] [Google Scholar]
- 21.Schroder K, Zhou R, Tschopp J. 2010. The NLRP3 inflammasome: a sensor for metabolic danger? Science 327:296–300. doi: 10.1126/science.1184003. [DOI] [PubMed] [Google Scholar]
- 22.Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, Ravussin E, Stephens JM, Dixit VD. 2011. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med 17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin C, Flavell RA. 2010. Molecular mechanism of NLRP3 inflammasome activation. J Clin Immunol 30:628–631. doi: 10.1007/s10875-010-9440-3. [DOI] [PubMed] [Google Scholar]
- 24.Thomas PG, Dash P, Aldridge JR Jr, Ellebedy AH, Reynolds C, Funk AJ, Martin WJ, Lamkanfi M, Webby RJ, Boyd KL, Doherty PC, Kanneganti TD. 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30:566–575. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichinohe T, Lee HK, Ogura Y, Flavell R, Iwasaki A. 2009. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J Exp Med 206:79–87. doi: 10.1084/jem.20081667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. 2006. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem 281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 28.Kanneganti TD. 2010. Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol 10:688–698. doi: 10.1038/nri2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajamaki K, Lappalainen J, Oorni K, Valimaki E, Matikainen S, Kovanen PT, Eklund KK. 2010. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: a novel link between cholesterol metabolism and inflammation. PLoS One 5:e11765. doi: 10.1371/journal.pone.0011765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen H, Gris D, Lei Y, Jha S, Zhang L, Huang MT, Brickey WJ, Ting JP. 2011. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat Immunol 12:408–415. doi: 10.1038/ni.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, Abela GS, Franchi L, Nunez G, Schnurr M, Espevik T, Lien E, Fitzgerald KA, Rock KL, Moore KJ, Wright SD, Hornung V, Latz E. 2010. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 464:1357–1361. doi: 10.1038/nature08938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- 33.Lamkanfi M, Dixit VM. 2011. Modulation of inflammasome pathways by bacterial and viral pathogens. J Immunol 187:597–602. doi: 10.4049/jimmunol.1100229. [DOI] [PubMed] [Google Scholar]
- 34.Dowling JK, O'Neill LA. 2012. Biochemical regulation of the inflammasome. Crit Rev Biochem Mol Biol 47:424–443. doi: 10.3109/10409238.2012.694844. [DOI] [PubMed] [Google Scholar]
- 35.Chae JJ, Cho YH, Lee GS, Cheng J, Liu PP, Feigenbaum L, Katz SI, Kastner DL. 2011. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity 34:755–768. doi: 10.1016/j.immuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee GS, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, Germain RN, Kastner DL, Chae JJ. 2012. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rathinam VA, Vanaja SK, Fitzgerald KA. 2012. Regulation of inflammasome signaling. Nat Immunol 13:333–342. doi: 10.1038/ni.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masters SL, Simon A, Aksentijevich I, Kastner DL. 2009. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease. Annu Rev Immunol 27:621–668. doi: 10.1146/annurev.immunol.25.022106.141627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryan JG, Kastner DL. 2008. Fevers, genes, and innate immunity. Curr Top Microbiol Immunol 321:169–184. doi: 10.1007/978-3-540-75203-5_8. [DOI] [PubMed] [Google Scholar]
- 40.Sabbah A, Chang TH, Harnack R, Frohlich V, Tominaga K, Dube PH, Xiang Y, Bose S. 2009. Activation of innate immune antiviral responses by Nod2. Nat Immunol 10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segovia J, Sabbah A, Mgbemena V, Tsai SY, Chang TH, Berton MT, Morris IR, Allen IC, Ting JP, Bose S. 2012. TLR2/MyD88/NF-κB pathway, reactive oxygen species, potassium efflux activates NLRP3/ASC inflammasome during respiratory syncytial virus infection. PLoS One 7:e29695. doi: 10.1371/journal.pone.0029695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang TH, Segovia J, Sabbah A, Mgbemena V, Bose S. 2012. Cholesterol-rich lipid rafts are required for release of infectious human respiratory syncytial virus particles. Virology 422:205–213. doi: 10.1016/j.virol.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol 82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhadina M, Bieniasz PD. 2010. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog 6:e1001153. doi: 10.1371/journal.ppat.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabut G, Peter M. 2008. Function and regulation of protein neddylation. “Protein modifications: beyond the usual suspects” review series. EMBO Rep 9:969–976. doi: 10.1038/embor.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson IR, Irwin MS, Ohh M. 2011. NEDD8 pathways in cancer, Sine Quibus Non. Cancer Cell 19:168–176. doi: 10.1016/j.ccr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Blank JL, Liu XJ, Cosmopoulos K, Bouck DC, Garcia K, Bernard H, Tayber O, Hather G, Liu R, Narayanan U, Milhollen MA, Lightcap ES. 2013. Novel DNA damage checkpoints mediating cell death induced by the NEDD8-activating enzyme inhibitor MLN4924. Cancer Res 73:225–234. doi: 10.1158/0008-5472.CAN-12-1729. [DOI] [PubMed] [Google Scholar]
- 48.Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J, Dick LR, Loke HK, McDonald AA, Ma J, Manfredi MG, Sells TB, Sintchak MD, Yang X, Xu Q, Koenig EM, Gavin JM, Smith PG. 2012. Treatment-emergent mutations in NAEβ confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell 21:388–401. doi: 10.1016/j.ccr.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 49.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 50.Bose S, Kar N, Maitra R, DiDonato JA, Banerjee AK. 2003. Temporal activation of NF-κB regulates an interferon-independent innate antiviral response against cytoplasmic RNA viruses. Proc Natl Acad Sci U S A 100:10890–10895. doi: 10.1073/pnas.1832775100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Echchgadda I, Kota S, DeLa Cruz I, Sabbah A, Chang T, Harnack R, Mgbemena V, Chatterjee B, Bose S. 2009. Anticancer oncolytic activity of respiratory syncytial virus. Cancer Gene Ther 16:923–935. doi: 10.1038/cgt.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kota S, Sabbah A, Chang TH, Harnack R, Xiang Y, Meng X, Bose S. 2008. Role of human beta-defensin-2 during tumor necrosis factor-alpha/NF-κB-mediated innate antiviral response against human respiratory syncytial virus. J Biol Chem 283:22417–22429. doi: 10.1074/jbc.M710415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueba O. 1978. Respiratory syncytial virus. I. Concentration and purification of the infectious virus. Acta Med Okayama 32:265–272. [PubMed] [Google Scholar]
- 54.Mgbemena V, Segovia JA, Chang TH, Tsai SY, Cole GT, Hung CY, Bose S. 2012. Transactivation of inducible nitric oxide synthase gene by Kruppel-like factor 6 regulates apoptosis during influenza A virus infection. J Immunol 189:606–615. doi: 10.4049/jimmunol.1102742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Racoosin EL, Swanson JA. 1989. Macrophage colony-stimulating factor (rM-CSF) stimulates pinocytosis in bone marrow-derived macrophages. J Exp Med 170:1635–1648. doi: 10.1084/jem.170.5.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Triantafilou K, Kar S, Vakakis E, Kotecha S, Triantafilou M. 2013. Human respiratory syncytial virus viroporin SH: a viral recognition pathway used by the host to signal inflammasome activation. Thorax 68:66–75. doi: 10.1136/thoraxjnl-2012-202182. [DOI] [PubMed] [Google Scholar]
- 57.Gong L, Yeh ET. 1999. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem 274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 58.Baroja-Mazo A, Martín-Sánchez F, Gomez AI, Martínez CM, Amores-Iniesta J, Compan V, Barberà-Cremades M, Yagüe J, Ruiz-Ortiz E, Antón J, Buján S, Couillin I, Brough D, Arostegui JI, Pelegrín P. 2014. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol 15:738. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- 59.Man SM, Hopkins LJ, Nugent E, Cox S, Glück IM, Tourlomousis P, Wright JA, Cicuta P, Monie TP, Bryant CE. 2014. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc Natl Acad Sci U S A 111:740. doi: 10.1073/pnas.1315179111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Witze ES, Old WM, Resing KA, Ahn NG. 2007. Mapping protein post-translational modifications with mass spectrometry. Nat Methods 4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 61.Jones J, Wu K, Yang Y, Guerrero C, Nillegoda N, Pan ZQ, Huang L. 2008. A targeted proteomic analysis of the ubiquitin-like modifier nedd8 and associated proteins. J Proteome Res 7:1274–1287. doi: 10.1021/pr700749v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qu Y, Misaghi S, Izrael-Tomasevic A, Newton K, Gilmour LL, Lamkanfi M, Louie S, Kayagaki N, Liu J, Komuves L, Cupp JE, Arnott D, Monack D, Dixit VM. 2012. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490:539–542. doi: 10.1038/nature11429. [DOI] [PubMed] [Google Scholar]
- 63.Embry CA, Franchi L, Nunez G, Mitchell TC. 2011. Mechanism of impaired NLRP3 inflammasome priming by monophosphoryl lipid A. Sci Signal 4:ra28. doi: 10.1126/scisignal.2001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. 2008. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bosu DR, Kipreos ET. 2008. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 3:7. doi: 10.1186/1747-1028-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. 2004. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 67.Watson IR, Blanch A, Lin DC, Ohh M, Irwin MS. 2006. Mdm2-mediated NEDD8 modification of TAp73 regulates its transactivation function. J Biol Chem 281:34096–34103. doi: 10.1074/jbc.M603654200. [DOI] [PubMed] [Google Scholar]
- 68.Russell RC, Ohh M. 2008. NEDD8 acts as a ‘molecular switch’ defining the functional selectivity of VHL. EMBO Rep 9:486–491. doi: 10.1038/embor.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao F, Cheng J, Shi T, Yeh ET. 2006. Neddylation of a breast cancer-associated protein recruits a class III histone deacetylase that represses NFκB-dependent transcription. Nat Cell Biol 8:1171–1177. doi: 10.1038/ncb1483. [DOI] [PubMed] [Google Scholar]
- 70.Broemer M, Tenev T, Rigbolt KT, Hempel S, Blagoev B, Silke J, Ditzel M, Meier P. 2010. Systematic in vivo RNAi analysis identifies IAPs as NEDD8-E3 ligases. Mol Cell 40:810–822. doi: 10.1016/j.molcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 71.Nagano T, Hashimoto T, Nakashima A, Kikkawa U, Kamada S. 2012. X-linked inhibitor of apoptosis protein mediates neddylation by itself but does not function as a NEDD8-E3 ligase for caspase-7. FEBS Lett 586:1612–1616. doi: 10.1016/j.febslet.2012.04.056. [DOI] [PubMed] [Google Scholar]
- 72.Chang FM, Reyna SM, Granados JC, Wei SJ, Innis-Whitehouse W, Maffi SK, Rodriguez E, Slaga TJ, Short JD. 2012. Inhibition of neddylation represses lipopolysaccharide-induced proinflammatory cytokine production in macrophage cells. J Biol Chem 287:35756–35767. doi: 10.1074/jbc.M112.397703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haneklaus M, O'Neill LA, Coll RC. 2013. Modulatory mechanisms controlling the NLRP3 inflammasome in inflammation: recent developments. Curr Opin Immunol 25:40–45. doi: 10.1016/j.coi.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 74.Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. 2013. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell 49:331–338. doi: 10.1016/j.molcel.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Lopez-Castejon G, Luheshi NM, Compan V, High S, Whitehead RC, Flitsch S, Kirov A, Prudovsky I, Swanton E, Brough D. 2013. Deubiquitinases regulate the activity of caspase-1 and interleukin-1β secretion via assembly of the inflammasome. J Biol Chem 288:2721–2733. doi: 10.1074/jbc.M112.422238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. 2012. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Labbe K, McIntire CR, Doiron K, Leblanc PM, Saleh M. 2011. Cellular inhibitors of apoptosis proteins cIAP1 and cIAP2 are required for efficient caspase-1 activation by the inflammasome. Immunity 35:897–907. doi: 10.1016/j.immuni.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 78.Van Opdenbosch N, Gurung P, Vande Walle L, Fossoul A, Kanneganti TD, Lamkanfi M. 2014. Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat Commun 5:3209. doi: 10.1038/ncomms4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.