Abstract
Measurements from small-angle X-ray scattering (SAXS) are highly informative to determine topological structures of bimolecular complexes in solution. Here, current and recent SAXS-driven developments are described, with an emphasis on computational modeling. In particular, accurate methods to computing one theoretical scattering profile from a given structure model are discussed, with a key focus on structure factor coarse-graining and hydration contribution. Methods for reconstructing topological structures from an experimental SAXS profile are currently under active development. We report on several modeling tools designed for conformation generation that make use of either atomic-level or coarse-grained representations. Furthermore, since large biomolecules can adopt multiple well-defined conformations, a traditional single-conformation SAXS analysis is inappropriate, so we also discuss recent methods that utilize the concept of ensemble optimization, weighing in on the SAXS contributions of a heterogeneous mixture of conformations. These tools will ultimately posit the usefulness of SAXS data beyond a simple space-filling approach by providing a reliable structural characterization of biomolecular complexes under physiological conditions.
Keywords: Computing, protein, DNA, RNA, ensemble optimization
1. Introduction
It is established that SAXS data can be highly informative in determining the topological structure of a biomolecule or a complex by characterizing how constituent parts are coherently organized[1, 2]. Structural information of SAXS data is encoded in a one-dimensional scattering profile determined from the spherical averaging of random orientations that a biomolecule can adopt in an aqueous solution. It is of technical and intellectual interest to achieve a reliable scattering measurement and understand the origins of this behavior, and it is also of practical importance to infer the fundamental information about biomolecular structures and dynamics from SAXS data[3, 4]. There are a fair number of excellent reviews discussing basic principles and general applications of SAXS[5, 6], and this short-review focuses on recent methodological developments aiming to better interpret SAXS data for topological structure characterization of large biomolecular complexes. One force driving the re-emergence of these developments is the increasing demand for using SAXS to study large, flexible, and often multimeric biomolecules that are recalcitrant to crystallographic or NMR structure determination.
A simple physical model for calculating the scattering intensity I (q) of a biomolecule can be given by the well-known Debye formula[7],
| (1) |
where q = 4π sin θ / λ is the scattering distance in reciprocal space or the amplitude of momentum transfer (2θ is the scattering angle and λ is the X-ray wavelength), fi(q) is the structure factor of atom i (i = 1,⋯,n and n is the total number of atoms) after excluded volume correction[8], and rij is the inter-particle distance between atom i and j. This Debye equation works under the approximation that each atom (or any coarse-grained residue/nucleotide) is spherical, and it is broadly applicable for theoretical scattering calculations. Once such a scattering profile is calculated, a macroscopic parameter regarding overall size, i.e. radius of gyration (Rg), can be derived via a Guinier analysis as an approximation to the Debye equation at the low-q limit,
| (2) |
The Rg -determining q-region is empirically defined by the criterion of q · Rg <1.3, which was empirically determined by Svergun and Feigin so that the deviation of IG (q) at the upper bound of q is within 10% of that from the Debye equation[9]. In addition, for a multimeric biomolecule, signature curvatures or "bumps" reflecting a collective spatial separation between two major structural groups can appear at higher-q regions of a scattering profile. These characteristic bumps are approximately located at
| (3) |
where Rcos, termed center-of-scattering distance, is related to a Bragg spacing between two large-group centers. Since the location of such a bump can be determined based on the first or second derivative of I (q), this Rcos analysis provides an estimate of the distance between these subunits. It can also add to the toolkit of macroscopic analyses based on, e.g., a Fourier-transformed pairwise distance distribution P(r), a Porod volume (Vp), a scattering cross-sectional Rg, and a volume of correlation (Vc)[10, 11] (which will not be further discussed here).
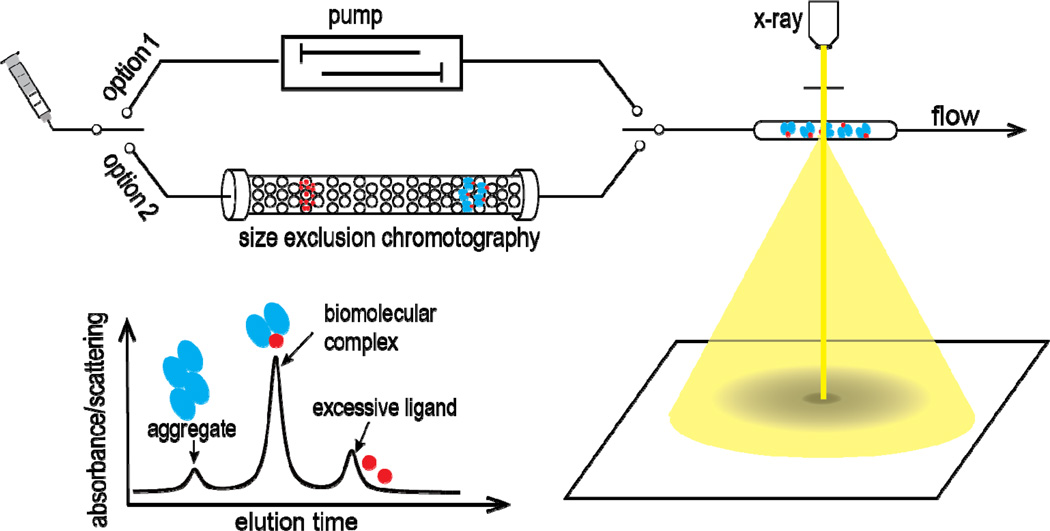
Acquisition of reliable SAXS data can be non-trivial. In fact, it is counter-intuitive that the sample preparation needed for a SAXS measurement could be stricter when compared to, e.g., crystallographic requirements given crystallization itself being a highly efficient purification process. For a well-behaved and non-aggregating sample, it is true that a simple injection – out of a single freeze-thaw cycle – into a flow-cell device could allow a successful data measurement. In particular, such a simple procedure can enable (i) the X-ray exposure to both a biomolecular sample and its corresponding buffer solution, (ii) the acquisition of their respective two-dimensional scattering images, and (iii) the reduction to a one-dimensional scattering profile of the biomolecule itself after buffer subtraction. Figure 1 illustrates a typical flow-cell setup via a programmable pump, which offers some key advantages over a static one, .e.g., keeping samples fresh during the exposure and minimizing radiation damage. For larger and/or flexible biomolecules that are prone to aggregation or form a large complex with a substrate, however, reliable data acquisition appears to require better sample handling, and a direct injection may be insufficient to achieve the desired homogeneity, especially when a molecule functions only in complex with its cognate ligand, such as chemical compound or DNA. Take a protein-DNA complex, for example – because DNA has a relatively stronger scattering than protein, excess DNA molecules in the buffer solution may contribute to the total scattering intensity and thus lead to unwanted data due to this sample heterogeneity.
Figure 1.
Schematic setup for SAXS data collection. There are two routine options available for SAXS data acquisition. A typical one is to use a programmable pump that allows a flow-cell setup for X-ray exposure and subsequent data acquisition for both a biological sample and its corresponding buffer. The other is a chromatography-coupled setup with a size exclusion column (SEC) that is designed to remove the unwanted species. The location of a homogeneous sample along the elution can be identified via a real-time IΩ (t) (Equation 4), together with chromatography light absorbance. A final one-dimensional scattering profile I (q) is obtained after buffer subtraction.
Alternatively, the implementation of a chromatography-coupled SAXS setup is becoming a standard option for SAXS data acquisition that allows the separation of different sample species of, e.g., monomer and dimer. Figure 1 illustrates such a chromatography-coupled setup equipped with a size exclusion column (SEC). When a mixture of molecules is injected into the column, each molecule of a different size moves along the column at a different speed. As a result, this implementation is able to improve sample homogeneity and remove the unwanted species, e.g., by separating complex-forming biomolecules from large aggregates or excess ligands, which is practically important for a reliable SAXS measurement. This "real-time" X-ray exposure is made possible by the availability of 3rd generation synchrotrons, where the exposure can occur on the second or even sub-second timescale, so a SAXS profile is determined along the chromatography elution[12, 13]. This chromatography-coupled setup has several advantages over a flow-cell setup. First, the target sample is rather fresh and presumably aggregate-free right out of the SEC. Second, the size separation can improve sample homogeneity, so excess ligands should not contribute to scattering measurements and only the biomolecular complex of interest is accounted for toward the final scattering profile. Finally, scattering intensity can be practically monitored in real time by a physical quality IΩ (t), the number of X-ray photons scattered per second into a detector that subtends a solid angle Ω,
| (4) |
Essentially, IΩ (t) is related to an effective cross-section of a biomolecule[7], which can be quantified by an integral over the entire q-range at each exposure time point of t (Equation 4). In addition, IΩ (t) can be correlated with chromatography light absorbance (Figure 1), which together provides an effective means to locate the specific scattering profile It (q) for the sample of interest. Alternatively, instead of using IΩ, a quality of either Rg or I (q = 0) can be used to monitor the scattering intensity along the elution[12]. In general, although it may require some modification for a high-throughput SAXS measurement[14], this chromatography-coupled setup can be particularly powerful for aggregation-prone samples or ligand-binding molecules to reach the needed homogeneity for an accurate and reliable SAXS measurement.
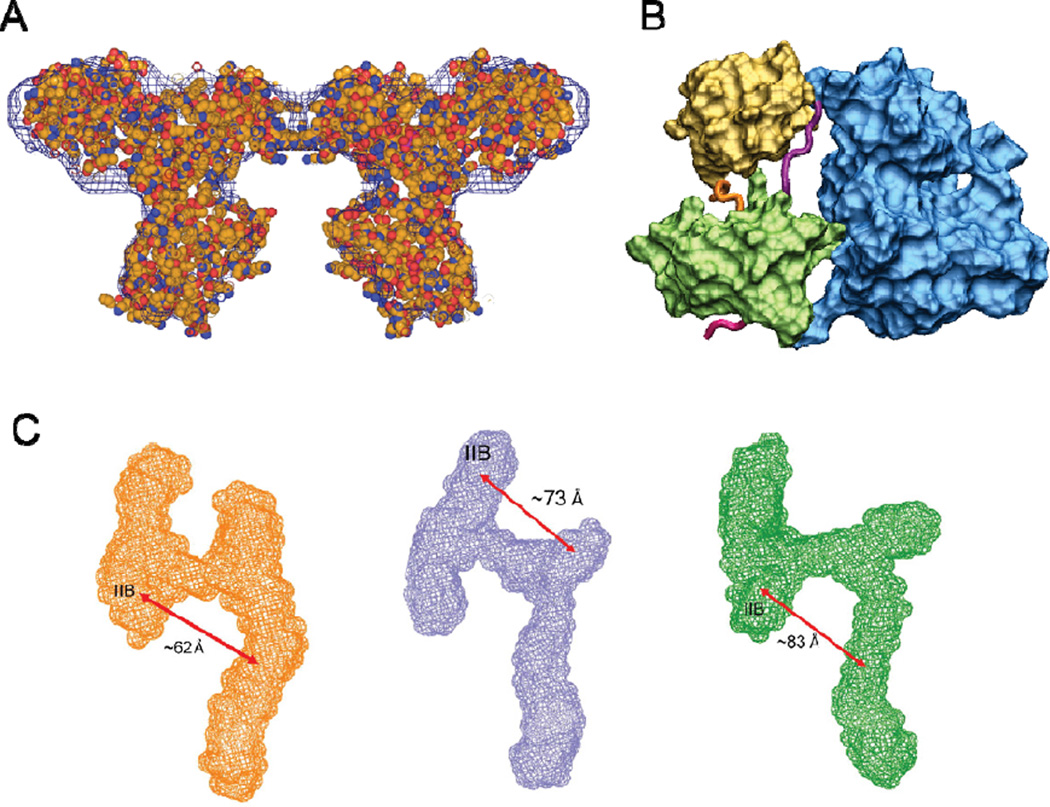
It is clear that topological structures can be derived from SAXS data. In fact, there are quite a few working examples of SAXS-derived structure models that are consistent with their corresponding high-resolution crystal structures. For example, one elegant proof-of-principle study on a motor protein p97 has shown that its SAXS-derived topology matches well with the crystal structure (Figure 2A)[15]. A similar match is also observed for a multidomain protein Src kinase where its crystal-like conformation (not shown but essentially identical) was found as a dominantly major species based on SAXS data representing its inactive state in solution[3, 16] (Figure 2B). More recently, a remarkable SAXS application has been demonstrated on an HIV viral RNA where each of three insertion mutants (plus the wild-type) can adopt a distinct "A"-like topological shape with notable repositioning of the legs and arms of the "A" (Figure 2C). This information about overall topology readily explains its specific recognition of a protein partner for optimal function of retroviral replication and translocation[17]. It is known that the ability to resolve competing structure models for a given SAXS measurement depends on the resolution of SAXS data itself and the overall scattering difference between the competing models. Nonetheless, these working examples emphasize that such topological structures can be inferred despite their low-resolution nature, somewhat similar to the early discovery of the low-resolution DNA double helix[2]. In the midst of broadened SAXS applications, the emerging potential of a SAXS analysis for visualizing the topology of large biomolecular complexes is apparent, especially when already known structures of individual components are productively used in theoretical and computational studies designed for SAXS data analysis.
Figure 2.
Topological structures derived using SAXS data. (A) The SAXS-derived shape (bush mesh) overlaps well with the crystal structure (colored balls) of a p97 ADP-AlFx complex (PDB entry 1OZ4)[15]. Reproduced with permission from Nagar and Kuriyan[19]. (B) The best-fit topology derived from SAXS data matches the crystal structure of a multidomain Src kinase (PDB entry 1QCF)[16, 20]. (C) Three SAXS-derived "A"-like topological structures of an HIV viral RNA each having a distinct distance between the two legs of the "A". Reproduced with permission from Fang et al[17].
2. Theoretical SAXS computing for protein, RNA/DNA, and their complexes
Typically, SAXS data analysis is performed in two directions. First, a one-dimensional SAXS profile I (q) alone can be used directly to derive macroscopic physical parameters such as Rg (Equation 2) and RCOS (Equation 3). Other attainable parameters include a Porod volume (Vp), a scattering cross-sectional Rg, and a volume of correlation (Vc)[10, 11]. The second direction is to use the information in SAXS data for shape reconstruction of biomolecules that are difficult to study with conventional biophysical techniques, which is becoming an area of focus in structural biology[18]. Key to these structure-driven SAXS data analyses are (i) an accurate method for theoretically determining a SAXS profile from a given structure and (ii) an ability to computationally generate a set of plausible structures that can be used for data interpretation. The former is essentially the theoretical foundation of most SAXS data analyses, while the latter is mainly driven by computation-intensive modeling (discussed next) that could ultimately convey the usefulness of SAXS data toward a topological structure characterization.
We organize this next section as follows. According to the source of scattering contribution to the total intensity, two main contributing components are discussed: one about a biomolecule itself; and the other about its surrounding hydration layer. For the former, several computing methods, either at an atomistic level or at a residue/nucleotide-simplified level, are briefly described. For the latter, the ability to effectively account for hydration contribution is an important aspect of theoretical SAXS calculations, which represents a major distinction among these currently available SAXS computing methods.
2.1. Scattering from a biomolecule itself
2.1.1. Atomic-level representation
One of the most influential SAXS computing methods is CRYSOL, a program that Svergun and colleagues developed[21], where a high-resolution structure is used as an input for scattering calculations. A similar atomic-level representation is also used in several other methods[22, 23]. While there may be some technical differences, one common focus is to improve the prediction accuracy of scattering profiles at high-q regions (approximately between q = 0.5–1.0Å−1), mostly via the use of structure coordinates and structure factors at the atomic level.
2.1.2. Residue/nucleotide-simplified representation
In parallel, a coarse-grained molecular representation has been developed for SAXS calculations, owing partially to the low-resolution nature of experimental SAXS data itself, which provides a fair standpoint for such a simplification. The need for a simplified approach also comes from the demand for analyzing large amounts of data from coarse-grained simulations, widely used for conformational sampling (discussed in the next section). In general, such coarse-grained SAXS computing methods are able to accurately calculate the scattering profiles approximately up to q = 0.5Å−1.
Several generalized residue/nucleotide-level methods are available for SAXS computing[24, 25]. Notably, we have simplified the atomic-level structure factors fi(q) (Equation 1) into residue-level and/or nucleotide-level structure factors Fi(q)[26–28], so the Debye equation is replaced by
| (5) |
where N is the number of residues or nucleotides (including coarse-grained water molecules). This speeds up significantly the scattering calculations for large biomolecular complexes, thereby enabling a more precise rendering of the scattering of the surrounding hydration layers (discussed next). Recently, this method has been implemented in the computer program Fast-SAXS-pro, allowing a user to compute the fit of an experimental SAXS profile to any complex formed by a mixture of proteins and/or RNA/DNA molecules[28]. This concept of coarse-graining is also adopted in several other methods, showing that satisfactory accuracy is achieved from the calculation[29, 30]. In addition, we note that a higher efficiency of computing can be further achieved via the use of a quasi-uniform spherical grid that gives rise to an effective orientation averaging, speeding up the calculation from a more costly O(N2) task to a much faster linear O(N) task[23]. This approximate treatment of orientation averaging does not appear to sacrifice the accuracy of the SAXS calculations. Indeed, as the simplification improves the performance of the calculations, we have recently implemented this O(N) scheme into our Fast-SAXS-pro algorithm. Overall, the Debye-based approach of using coarse-grained structure factors (Equation 5) can be broadly applicable due to its simplicity and speed of calculation, and can be especially useful when a large amount of structure models are to be scored for their fit to experimental data.
2.2. Scattering from a surrounding hydration layer
The hydration layer surrounding a biomolecule is known to contribute to the total scattering intensity. This hydration contribution can lead to excess electron density contributing to the intensity after buffer subtraction, partially due to specific hydrophobic or electrostatic features of biomolecular surfaces that lead to variation in water density or ionic characters in the surrounding hydration layer, compared to the rest of the bulk solution[31]. This hydration is a considerable contributing factor especially for those molecules with a large solvent-accessible surface area. For instance, it has reportedly contributed to an apparent swelling in overall shape when compared to the volume of a dry protein based on the low-resolution shape restoration using DAMMIN[32]. Modeling this hydration contribution can be achieved by placing water molecules, either implicitly[25] or explicitly via a coarse-grained structure factor as used in Fast-SAXS-pro[26] (see Figure 3A). One key point is to use these water molecules as a proxy to represent the density difference between the hydration layer and the bulk solvent. For example, CRYSOL accounts for this effect by assigning a default density that is 10% higher than that of the bulk solvent in a blind prediction mode. An exact hydration difference can be further refined via fitting to experimental data, as used in CRYSOL and more recently, in an updated version of FoXS[33]. It is a valid idea to refine such hydration parameters as the layer thickness and density against experimental data, although this parameter fitting can be done only if each structure would have its own hydration parameters. When a straight prediction is required to deal with a large ensemble of structure models, the extent to which this one-structure-one-parameter scenario can be applied remains to be seen. Finally, it should be noted that this hydration modeling is meant to account for the difference between the bulk solvent and the hydration layer, which should be distinguished from a non-homogeneous treatment within the hydration layer, as demonstrated on several high-resolution structures in a recent review by Rambo and Tainer[2]. This heterogeneity can be more pronounced when the hydration layer spreads across the surface of both a protein and a nucleic acid (RNA/DNA), which will be discussed next.
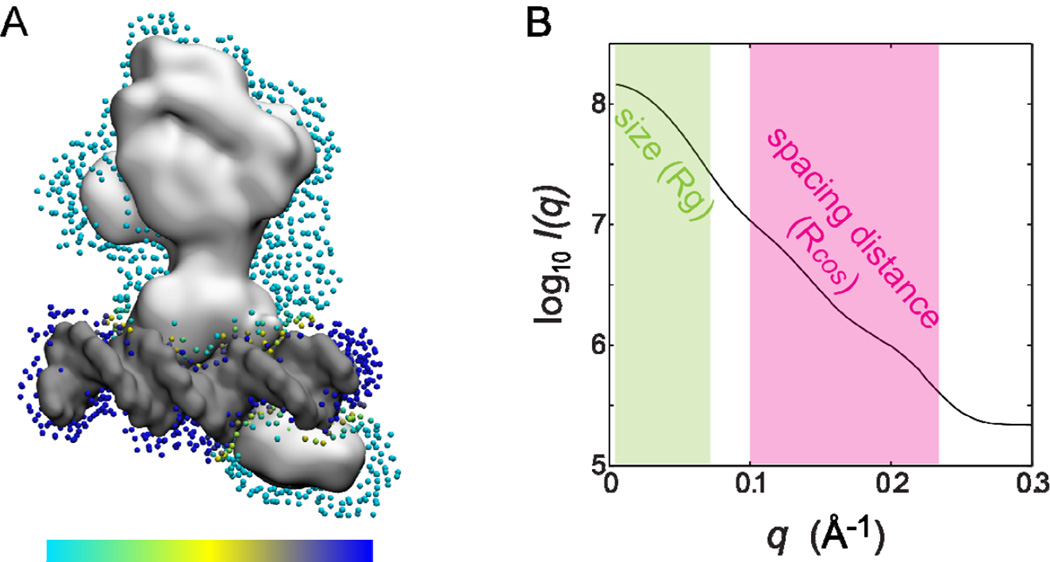
Figure 3.
Theoretical SAXS computing for biomolecular complexes of protein and DNA/RNA. (A) Hydration contribution is represented by dummy water molecules in the hydration layer surrounding a protein-DNA complex (e.g., a multi-domain nuclear receptor HNF-4α with PDB entry 4IQR[34] shown in white, a double-strand DNA in gray, and dummy water molecules as blue dots). The hydration contribution is different for the protein compared to the DNA (or RNA); this difference is modeled by assigning a different weighting factor (scaled by the underlying color bar). For example, a weighting factor of 4% is used for the protein (in light blue) and 7% for the DNA (in dark blue) (Equation 5). (B) A theoretical SAXS profile of this HNF-4α protein-DNA complex using Fast-SAXS-pro[28]. The scattering profile at low-q (in green) provides information about overall size such as Rg (Equation 2), while a mid-q region (in magenta) can reveal the information regarding internal structural spacing such as Rcos (Equation 3). Note that this Rcos analysis can be performed only if the q-range is higher than q = 2π / dmax where dmax is the maximum size of a molecule.
To account for a non-homogeneous distribution within the hydration layer, a different approach has been developed for an explicit treatment of a complex of protein and nucleic acid (DNA/RNA). In Fast-SAXS-pro[28], this heterogeneity is explicitly taken into account by assigning a different scaling factor for dummy water molecules according to their proximity to protein and DNA/RNA (Figure 3A). Based on several model systems we have tested, a general trend is observed that an RNA has more excess electron density in its corresponding hydration layer than a DNA, and a DNA has more than a protein[28]. Compared to a homogeneous hydration layer, e.g., used in CRYSOL, this approach of treating each hydration layer differently with a non-homogeneous distribution – implemented in Fast-SAXS-pro – has a pronounced effect on the theoretical scattering profile of large macromolecules such as a protein-RNA complex.
3. Computational modeling for topological structures
Topological structure determination from a given SAXS observation is challenging but currently under active development. In the context of large multimeric complexes, there are quite a few computing methods to address this issue. Often, SAXS data is supplemented by imposing known knowledge of structural information on individual subunits, as well as substantial assistance from computational modeling. Here, we focus on the tool development for modeling the molecules that can adopt a single conformation or exist in a mixture of multiple conformations.
3.1. Space-filling bead modeling
There has been remarkable progress in using SAXS data to reconstruct a 3D topological shape[25, 32, 35]. One of the most popular methods is DAMMIN[32], which takes a space-filling strategy and utilizes a bead-like molecular representation to model the shape of the scattered volume. Despite the assumption of the lack of physical connectivity between the beads, it is still possible for a space-filling approach to build a packed topological shape representing the most probable structure. Figure 4A illustrates an example of a DAMMIN-built shape for a protein-DNA complex, using a theoretical SAXS profile calculated from its known crystal structure. Despite some discrepancy, a reasonable overlap between the crystal structure and its corresponding SAXS-built topological shape suggests that the information encoded in SAXS data is able to outline the structural organization of its subunits. It should be noted that this illustration is mainly for reconstructing a single conformation; it may only represent a somewhat "averaged" shape if multiple conformations co-exist. Since this space-filling method uses only prior knowledge of a Porod volume that encloses all the dummy atoms within a molecule, but without any knowledge of, e.g., protein sequence or atomic coordinate, it provides an effective ab initio shape reconstruction.
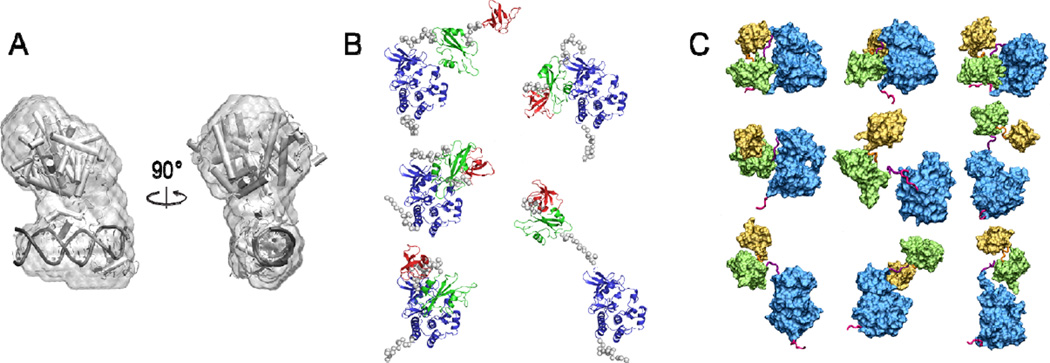
Figure 4.
SAXS-based topological shape reconstruction. (A) Space-filling using DAMMIN[32]. A bead-like shape model is illustrated and reconstructed from a theoretical "synthetic" SAXS profile of the protein-DNA complex (shown in Figure 3). A total of 20 independent DAMMIN runs were performed to generate structure candidates for the most probable shape model[6, 32]. (B) Conformations generated by a rigid-body docking program BUNCH[37] for a multidomain c-Src protein kinase (PDB entry 2SRC[41]). (C) A set of conformations of Hck kinase generated after clustering the data of residue-simplified simulations[16].
3.2. Rigid-body docking
As a straightforward choice, rigid-body modeling can be used to dock high-resolution structures available for individual subunits into a multimeric complex for the fitting of its theoretical SAXS profile against experimental data. There are several docking-based methods specially designed for SAXS data interpretation[36–38]. Figure 4B illustrates a small set of conformers for a multidomain c-Src kinase generated by the program BUNCH[37]. These docked conformations can be used in two ways: one is to rank the most-probable conformations using a direct fitting to experimental data; and the other is to serve as a candidate pool of conformers for an ensemble optimization (discussed in the next section). It should be noted that there are various docking programs available from the broader field of macromolecular docking[39, 40], which could be adopted for SAXS data analyses as well. Since such a docking approach simplifies the conformational search within a simple six-degree-of-freedom space, it can be remarkably suitable when the intrinsic flexibility of individual subunits is negligible so the rigid-body assumption would hold upon the complex formation.
3.3. Flexible-docking simulations
Conformation generation via molecular dynamics (MD) simulations is known to recognize the intrinsic structure flexibility that can be displayed in an aqueous environment. More importantly, it is designed to allow induced-fit and even large-scale conformational changes to occur, which is often required for biomolecules in order to function. Typical MD simulations are performed at either an atomic or a residue-simplified level[30, 42, 43, 44, 45]. For example, Pelikan and coworkers have used all-atom MD simulations at a high temperature to generate a large pool of structure ensembles for SAXS[42]. Alternatively, given the low-resolution nature of SAXS data, coarse-grained (CG) modeling can be introduced to reduce the number of degrees of freedom and thus enables the generation of a diverse set of conformers, as demonstrated for a multidomain Hck kinase[16]. Figure 4C illustrates a minimum basis-set of Hck conformations generated from coarse-grained simulations, ranging from compacted to extended shapes and from assembled to fully disassembled. More recently, the predictive power of coarse-grained modeling is being enabled for the study of protein-protein interactions[46, 47], which is expected to significantly enhance the ability of simulating a multi-component complex. In fact, a recent proof-of-principle study shows a simple CG model is able to correctly predict the conformation transition from an inactive to an active state of an estrogen-binding domain[48]. Overall, these studies provide exemplary applications of using either all-atom or CG simulations as a flexible-docking tool for SAXS data interpretation of intrinsically flexible biomolecular complexes.
4. Optimization of conformational ensembles against experimental SAXS data
The usefulness of SAXS data for topological structure characterization is arguably determined by the ability to explore a set of structure models in conformation space. From a computation standpoint, how to generate as many conformations as possible is becoming a central piece of a reliable SAXS shape reconstruction. While it may be prohibitive to obtain a comprehensive sampling in a high-dimensional configuration space such as protein folding, the exhaustiveness of a conformational search is largely achievable in the context of protein-protein interactions, in part due to a reduced number of degrees of freedom involved. This conformation generation in an exhaustive fashion is poised to provide the technical feasibility of an effective and robust SAXS data analysis.
4.1. Exhaustive conformational search
There has been considerable interest in developing new sampling techniques aiming to generate structurally diverse conformations, because brute-force simulations using either all-atom or CG approximations may be easily trapped in local minima. In fact, several advanced sampling techniques have been developed in the past to address this hurdle, including umbrella sampling and replica exchange[49]. Recently, a specific push-pull-release (PPR) method was designed to enhance the simulations of protein-protein interactions by repeating the PPR cycles to facilitate the encounter formation[47]. Figure 5A illustrates a recent example of using PPR-enhanced simulations to identify the top conformations of an estrogenreceptor/ DNA complex[50]. To achieve an exhaustive search, we have further extended this PPR sampling with a rotation-based pose generation that uniformly covers the conformational space of five rotational degrees of freedom. This rotation-enhanced PPR is demonstrated to be effective for an exhaustive search for different modes of protein-protein interactions in a receptor-ligand complex TGFb-FKBP12[51]. A similar comprehensive conformational search can be achieved by a grid-based docking of, e.g., protein and RNA (see an example in Figure 5B). Overall, these advanced sampling methods enable the generation of a large set of structure models required for a comprehensive SAXS data analysis.
Figure 5.
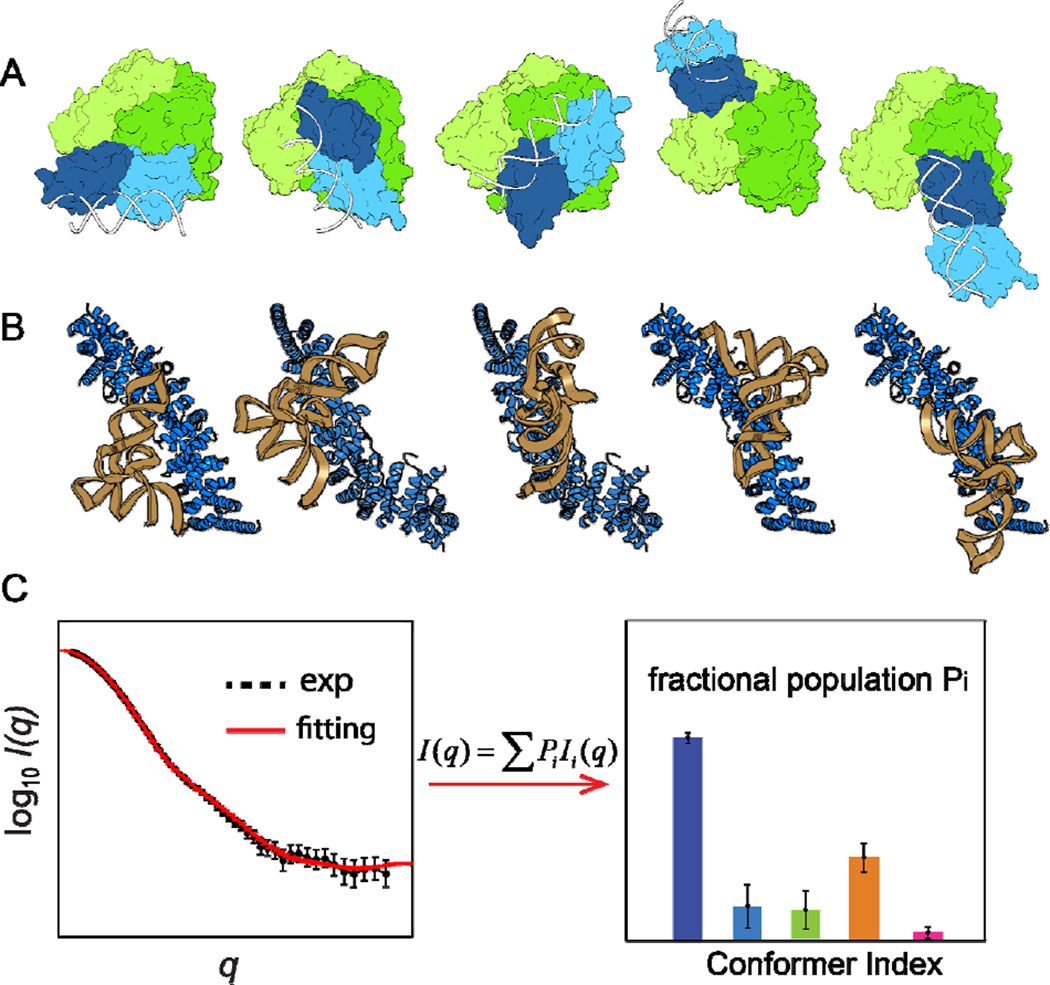
Computational modeling and SAXS-driven ensemble optimization. (A) Flexible-docking simulations predict the structures of a multidomain estrogen receptor (in surface plots) in complex with a DNA duplex (in gray ribbon). Modified from Ref.[50]. (B) Conformations of a protein-RNA complex generated from an exhaustive grid-based docking[39, 55], where a tRNA is shown in magenta (PDB entry 4TRA[56]) and a multirepeat protein in blue (PDB entry 2GL7[57]). (C) A cartoon illustrating the optimization of a theoretical SAXS profile against experimental data, which infers the most-probable fractional population for each conformer (Equation 6)[16].
4.2. SAXS data inference for topological structure determination
Based on the computation-generated conformations, structural interpretation of SAXS data often proceeds in two steps. First, theoretical SAXS profiles can be calculated (described in Section 2 above). Then, optimization of these theoretical profiles against experimental data is performed to infer the best-fit conformational ensembles. A pioneering work for such an ensemble-based analysis is the ensemble optimization method (EOM), where SAXS fitting is based on a pool of randomly generated models in which protein domain are treated as rigid bodies and connected by self-avoiding linkers of dihedral angles complying with a quasi-Ramachandran plot[52]. A similar strategy is also adopted in several other studies utilizing this concept of ensemble optimization[37, 42, 45, 53]. It should be noted that each conformation in the EOM-optimized ensemble contributes equally to the scattering averaging; this equality in the scattering of each conformation is different from the way that was used in the program OLIGOMER[54], which is designed for a completely different purpose of separating oligomeric species of, e.g., monomer and dimer.
A different strategy of using a non-equal weight for each conformation is emerging in SAXS-driven ensemble optimization. This was first attempted in the minimal ensemble method (MES) that was initially used to distinguish disordered systems from those adopting well-defined conformations[42]. This weighted scheme became more pronounced when the basis-set supported SAXS (BSS-SAXS) approach was introduced by assigning a fractional population Pi to each conformation member of the basis-set, each with a distinct theoretical SAXS profile of Ii(q)[16]. Both MES and BSS-SAXS yield a small number of conformers that best-fit experimental SAXS data. The key difference is that the former method relies on a full optimization over a large pool of structures (e.g., in the order of 10,000) and the latter instead uses a two-step clustering algorithm (based on the similarity in both structures and SAXS profiles) to reduce the conformer pool to a basis-set of conformations (in the order of 10) each with a distinguishable scattering profile. Heuristically, BSS-SAXS can be equivalent to EOM and MES in terms of best-fitting observed SAXS data. Nonetheless, the final theoretical SAXS profile for the entire basis-set in BSS-SAXS is given by
| (6) |
where Ns is the total of conformers used in the basis-set. Clearly, this implementation is designed to account for the differential co-existence of multiple well-defined conformations. It has successfully explained ligand-induced conformational changes of a multidomain protein Hck kinase[16] and can be broadly applied to probe any large-scale change of conformational equilibrium[3]. This weighted scheme is further enhanced and adopted in the method of ensemble-refinement of SAXS (EROS)[30], which has been applied to study the salt-induced conformational transition of an endosome-associated ESCRT-III domain and the conformation of a tyrosine phosphatase[30, 58]. More recently, the use of such a weighted ensemble optimization has led to a successful shape reconstruction for an HIV-1 viral RNA using SAXS data (shown in Figure 2C).
Key to the ensemble optimization is minimizing the difference between theoretical results against experimental data via an error-weighted score,
| (7) |
where qmin and qmax are the lower and upper bounds of the observed q-range, respectively, σ(q) are the measurement errors of experimental data log Iexp (q) (in a logarithmic scale), and Ical(q) is the weighted average calculated from an ensemble of Ns conformations (Equation 6). The offset constant Δ can be calculated by the intensity difference at qmin. Due to a large fluctuation around the beam stop, however, we also found that the value of Δ can be better optimized by minimizing the difference between log Ical(q) and log Iexp (q) over the Rg-determining q-region (Equation 2 and Figure 3B). Note that a slightly different variation of can be defined using a linear scale of I(q)[3, 11, 30, 53, 59]; however, the use of a logarithmic scale log I (q) is not merely for a mathematical convenience but for a physical consideration. More recently, a new parameter has been introduced in an attempt to reduce overfitting when the "noise" level is high, but reportedly gives a similar performance when the noise level is low[11]. Nonetheless, a -based scheme can yield a quantitative optimization of fractional populations Pi (see an illustration in Figure 5C). In practice, a set of optimal Pi values can be achieved via a maximum likelihood method or a Monte Carlo (MC) algorithm. For example, in a Bayesian-like MC algorithm based on the Metropolis criterion of [16], any MC move attempt is accepted with a probability[60],
| (8) |
where is the difference between two adjacent MC steps (from i to j). It has been demonstrated that this MC approach is able to find an optimal solution in a rapid fashion and further estimate the uncertainties of Pi values (Figure 5C). It should be noted that this error estimation provides an effective assessment regarding the robustness of SAXS-inferred conformational ensembles[16]. Another assessment about the goodness of fit can be achieved by examining the score distribution ; specifically, the dependence of on the size (Ns) of the conformational basis-set can be examined for a self-consistent completeness check[16]. Additional cross-validation analyses can be performed, but are not discussed here. Overall, this approach of MC-assisted BSS-SAXS shape reconstruction provides an alternative means to infer the best-fit conformational ensembles from SAXS data.
5. Conclusions and Perspectives
While SAXS data alone is almost never the primary source of information for high-resolution structure determination, it is now known that topological structures, albeit at low-resolution, can be derived to provide a highly informative, often much-needed structural knowledge. For large, flexible, multimeric biomolecules, e.g., at the range of 50–150 kDa, SAXS remains one of very few biophysical techniques available for an effective structural characterization with regard to overall shape and topology. This topological characterization has often benefited from known information of individual subcomponents within a complex, and of course can be supplemented by attainable information from other biophysical techniques including, but not limited to, NMR and chemical cross-linking[61]. Nonetheless, it is worthwhile to restate that SAXS is a warranted technique for biomolecules not amenable for NMR or crystallographic studies.
Recent advances in SAXS-based topological structure determination include the technical improvement of a chromatography-coupled SAXS experimentation, but also new SAXS computing methods that can recognize the scattering difference between proteins and nucleic acids with regard to hydration contribution. From the perspective of SAXS data analyses, the interplay of computation-intensive simulations and experimental SAXS measurements is becoming apparent. On one hand, conformation generation from large-scale computations provides a solid theoretical foundation for SAXS data interpretation. It should be noted that the experimental technique itself has also undergone active developments in various directions, e.g., at wide angles[62], in a high-throughput or time-resolved fashion[14, 63], and more recently, in the context of utilizing X-ray free-electron lasers[64]. The sophistication of SAXS data acquisition has helped improve the accuracy of theoretical prediction itself[38, 44, 65]. It also presents new opportunities of developing novel computational algorithms to better interpret SAXS data for topological structure determination, and it is almost certain that such a development will benefit from the ever-increasing power of computational modeling[66]. Retrospectively, the wide use of synchrotron sources worldwide – that are either under operation or in the near future – may push for a new wave of computational tool developments appropriate for SAXS-based topological studies of large, flexible, multimeric biomolecules.
Acknowledgements
Technical support with figure re/production from Marc Parisien, Wei Huang, Krishna M. Ravikumar, and Marc Parisien is greatly appreciated. Stimulating discussions with Benoît Roux, Herbert Levine, and José N. Onuchic over the years have been instrumental. This work also benefited from fruitful interactions with Lee Makowski, Lin Yang, Rita Graceffa, Sanghyun Park, and Srinivas Chakravarthy at the Argonne National Lab, the Advanced Photon Source (APS), and the National Synchrotron Light Source (NSLS) as well as from the participation and lecture contribution at SAXS workshops organized at APS, NSLS, and ESRF. Comments from an anonymous reviewer have been very constructive for revision. The work of SY was supported in part by CWRU, the Cleveland Foundation, the American Cancer Society (ACS IRG-91-022-15), and the Department of Defense Breast Cancer Research Program (W81XWH-11-1033). Beamtime access was supported by the U.S. Department of Energy (DE-AC02-06CH11357 to APS and DE-AC02-98CH10886 to NSLS) and by the National Institutes of Health (9P41GM103622-18 to APS-BioCAT and P41RR012408 and P41GM103473 to NSLS-X9). Computational support was provided by the Ohio Supercomputer Center and the CWRU high-performance computing cluster.
References
- 1.Svergun DI, Koch MHJ, Timmins PA, May RP. Small angle x-ray and neutron scattering from solutions of biological macromolecules. USA: Oxford University Press; 2013. [Google Scholar]; Trewhella J, Hendrickson WA, Kleywegt GJ, Sali A, Sato M, Schwede T, Svergun DI, Tainer JA, Westbrook J, Berman HM. Structure. 2013;21:875. doi: 10.1016/j.str.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 2.Rambo RP, Tainer JA. Annu Rev Biophys. 2013;42:415. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 3.Bernado P, Blackledge M. Nature. 2010;468:1046. doi: 10.1038/4681046a. [DOI] [PubMed] [Google Scholar]
- 4.Yang S, Roux B. Structure. 2011;19:3. doi: 10.1016/j.str.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Graewert MA, Svergun DI. Curr Opin Struc Biol. 2013;23:748. doi: 10.1016/j.sbi.2013.06.007. [DOI] [PubMed] [Google Scholar]; Putnam CD, Hammel M, Hura GL, Tainer JA. Q Rev Biophys. 2007;40:191. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]; Perez J, Nishino Y. Curr Opin Struct Biol. 2012;22:670. doi: 10.1016/j.sbi.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 6.Blanchet CE, Svergun DI. Annu Rev Phys Chem. 2013;64:37. doi: 10.1146/annurev-physchem-040412-110132. [DOI] [PubMed] [Google Scholar]
- 7.Als-Nielsen J, McMorrow D. Elements of modern X-ray physics. Hoboken: Wiley; 2011. [Google Scholar]
- 8.Fraser RDB, MacRae TP, Suzuki E. J Appl Crystallogr. 1978;11:693. [Google Scholar]
- 9.Svergun DI, Feigin LA, Taylor GW. Structure analysis by small-angle x-ray and neutron scattering. New York: Plenum Press; 1987. [Google Scholar]
- 10.Glatter O, Kratky O. Small angle x-ray scattering. London ; New York: Academic Press; 1982. [Google Scholar]
- 11.Rambo RP, Tainer JA. Nature. 2013;496:477. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathew E, Mirza A, Menhart N. J Synchrotron Radiat. 2004;11:314. doi: 10.1107/S0909049504014086. [DOI] [PubMed] [Google Scholar]; Berthaud A, Manzi J, Perez J, Mangenot S. J Am Chem Soc. 2012;134:10080. doi: 10.1021/ja301667n. [DOI] [PubMed] [Google Scholar]
- 13.David G, Perez J. J Appl Crystallogr. 2009;42:892. [Google Scholar]; Wright GS, Hasnain SS, Grossmann JG. Biochem J. 2011;439:39. doi: 10.1042/BJ20110948. [DOI] [PubMed] [Google Scholar]; Watanabe Y, Inoko Y. J Chromatogr A. 2013;1303:100. doi: 10.1016/j.chroma.2013.06.048. [DOI] [PubMed] [Google Scholar]
- 14.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, 2nd, Tsutakawa SE, Jenney FE, Jr, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MW, Tainer JA. Nat Methods. 2009;6:606. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies JM, Brunger AT, Weis WI. Structure. 2008;16:715. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]; Davies JM, Tsuruta H, May AP, Weis WI. Structure. 2005;13:183. doi: 10.1016/j.str.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 16.Yang SC, Blachowicz L, Makowski L, Roux B. P Natl Acad Sci USA. 2010;107:15757. doi: 10.1073/pnas.1004569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang XY, Wang JB, O'Carroll IP, Mitchell M, Zuo XB, Wang Y, Yu P, Liu Y, Rausch JW, Dyba MA, Kjems J, Schwieters CD, Seifert S, Winans RE, Watts NR, Stahl SJ, Wingfield PT, Byrd RA, Le Grice SFJ, Rein A, Wang YX. Cell. 2013;155:594. doi: 10.1016/j.cell.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levitt M. Proc Natl Acad Sci U S A. 2009;106:11079. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagar B, Kuriyan J. Structure. 2005;13:169. doi: 10.1016/j.str.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Schindler T, Sicheri F, Pico A, Gazit A, Levitzki A, Kuriyan J. Molecular Cell. 1999;3:639. doi: 10.1016/s1097-2765(00)80357-3. [DOI] [PubMed] [Google Scholar]
- 21.Svergun D, Barberato C, Koch MHJ. J Appl Crystallogr. 1995;28:768. [Google Scholar]
- 22.Zuo X, Cui G, Merz KM, Jr, Zhang L, Lewis FD, Tiede DM. Proc Natl Acad Sci U S A. 2006;103:3534. doi: 10.1073/pnas.0600022103. [DOI] [PMC free article] [PubMed] [Google Scholar]; Schneidman-Duhovny D, Hammel M, Sali A. Nucleic Acids Res. 2010;38:W540. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]; Grishaev A, Guo L, Irving T, Bax A. J Am Chem Soc. 2010;132:15484. doi: 10.1021/ja106173n. [DOI] [PMC free article] [PubMed] [Google Scholar]; Poitevin F, Orland H, Doniach S, Koehl P, Delarue M. Nucleic Acids Res. 2011;39:W184. doi: 10.1093/nar/gkr430. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bardhan J, Park S, Makowski L. J Appl Crystallogr. 2009;42:932. doi: 10.1107/S0021889809032919. [DOI] [PMC free article] [PubMed] [Google Scholar]; Park S, Bardhan JP, Roux B, Makowski L. J Chem Phys. 2009;130:134114. doi: 10.1063/1.3099611. [DOI] [PMC free article] [PubMed] [Google Scholar]; Virtanen JJ, Makowski L, Sosnick TR, Freed KF. Biophys J. 2010;99:1611. doi: 10.1016/j.bpj.2010.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]; Liu H, Morris RJ, Hexemer A, Grandison S, Zwart PH. Acta Crystallogr A. 2012;68:278. doi: 10.1107/S010876731104788X. [DOI] [PubMed] [Google Scholar]; Kofinger J, Hummer G. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87:052712. doi: 10.1103/PhysRevE.87.052712. [DOI] [PubMed] [Google Scholar]; Oroguchi T, Ikeguchi M. Chem Phys Lett. 2012;541:117. [Google Scholar]
- 23.Watson MC, Curtis JE. J Appl Crystallogr. 2013;46:1171. [Google Scholar]
- 24.Walther D, Cohen FE, Doniach S. J Appl Crystallogr. 2000;33:350. [Google Scholar]; Guo DY, Blessing RH, Langs DA, Smith GD. Acta Crystallographica Section D. 1999;55:230. doi: 10.1107/S0907444998008208. [DOI] [PubMed] [Google Scholar]
- 25.Svergun DI, Petoukhov MV, Koch MH. Biophys J. 2001;80:2946. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang S, Park S, Makowski L, Roux B. Biophys J. 2009;96:4449. doi: 10.1016/j.bpj.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S, Parisien M, Major F, Roux B. J Phys Chem B. 2010;114:10039. doi: 10.1021/jp1057308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravikumar KM, Huang W, Yang S. J Chem Phys. 2013;138:024112. doi: 10.1063/1.4774148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stovgaard K, Andreetta C, Ferkinghoff-Borg J, Hamelryck T. BMC Bioinformatics. 2010;11:429. doi: 10.1186/1471-2105-11-429. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zheng W, Tekpinar M. Biophys J. 2011;101:2981. doi: 10.1016/j.bpj.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; dos Reis MA, Aparicio R, Zhang Y. Biophys J. 2011;101:2770. doi: 10.1016/j.bpj.2011.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rozycki B, Kim YC, Hummer G. Structure. 2011;19:109. doi: 10.1016/j.str.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheatham TE, 3rd, Kollman PA. Annu Rev Phys Chem. 2000;51:435. doi: 10.1146/annurev.physchem.51.1.435. [DOI] [PubMed] [Google Scholar]; Makarov V, Pettitt BM, Feig M. Acc Chem Res. 2002;35:376. doi: 10.1021/ar0100273. [DOI] [PubMed] [Google Scholar]; Bragg L, Perutz MF. Proceedings of the Royal Society of London. Series A. Mathematical and Physical Sciences. 1952;213:425. [Google Scholar]
- 32.Svergun DI. Biophys J. 1999;76:2879. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Biophys J. 2013;105:962. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandra V, Huang P, Potluri N, Wu D, Kim Y, Rastinejad F. Nature. 2013;495:394. doi: 10.1038/nature11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chacon P, Moran F, Diaz JF, Pantos E, Andreu JM. Biophys J. 1998;74:2760. doi: 10.1016/S0006-3495(98)77984-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konarev PV, Petoukhov MV, Svergun DI. J Appl Crystallogr. 2001;34:527. doi: 10.1107/S1600576716005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petoukhov MV, Svergun DI. Biophys J. 2005;89:1237. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneidman-Duhovny D, Hammel M, Sali A. J Struct Biol. 2011;173:461. doi: 10.1016/j.jsb.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabb HA, Jackson RM, Sternberg MJE. Journal of Molecular Biology. 1997;272:106. doi: 10.1006/jmbi.1997.1203. [DOI] [PubMed] [Google Scholar]
- 40.Chen R, Li L, Weng Z. Proteins. 2003;52:80. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]; Dominguez C, Boelens R, Bonvin AM. J Am Chem Soc. 2003;125:1731. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]; Tovchigrechko A, Vakser IA. Nucleic Acids Res. 2006;34:W310. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Mol Cell. 1999;3:629. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 42.Pelikan M, Hura GL, Hammel M. Gen Physiol Biophys. 2009;28:174. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsutakawa SE, Van Wynsberghe AW, Freudenthal BD, Weinacht CP, Gakhar L, Washington MT, Zhuang Z, Tainer JA, Ivanov I. Proc Natl Acad Sci U S A. 2011;108:17672. doi: 10.1073/pnas.1110480108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Vigil D, Gallagher SC, Trewhella J, Garcia AE. Biophys J. 2001;80:2082. doi: 10.1016/S0006-3495(01)76182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Baaden M, Marrink SJ. Curr Opin Struct Biol. 2013;23:878. doi: 10.1016/j.sbi.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Xia Z, Bell DR, Shi Y, Ren P. J Phys Chem B. 2013;117:3135. doi: 10.1021/jp400751w. [DOI] [PubMed] [Google Scholar]
- 45.Jamros MA, Oliveira LC, Whitford PC, Onuchic JN, Adams JA, Blumenthal DK, Jennings PA. J Biol Chem. 2010;285:36121. doi: 10.1074/jbc.M110.116947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim YC, Hummer G. Journal of Molecular Biology. 2008;375:1416. doi: 10.1016/j.jmb.2007.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zheng W, Schafer NP, Davtyan A, Papoian GA, Wolynes PG. Proc Natl Acad Sci U S A. 2012;109:19244. doi: 10.1073/pnas.1216215109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang Q, Zhang PZ, Hoffman L, Tripathi S, Homouz D, Liu Y, Waxham MN, Cheung MS. P Natl Acad Sci USA. 2013;110:20545. doi: 10.1073/pnas.1312788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ravikumar KM, Huang W, Yang SC. Biophys J. 2012;103:837. doi: 10.1016/j.bpj.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang W, Ravikumar KM, Greene GL, Yang S. 2014 Submitted. [Google Scholar]
- 49.Torrie GM, Valleau JP. J Comput Phys. 1977;23:187. [Google Scholar]; Roux B. Comput Phys Commun. 1995;91:275. [Google Scholar]; Sanbonmatsu KY, Garcia AE. Proteins-Structure Function and Genetics. 2002;46:225. doi: 10.1002/prot.1167. [DOI] [PubMed] [Google Scholar]; Sugita Y, Okamoto Y. Chem Phys Lett. 1999;314:141. [Google Scholar]
- 50.Huang W, Greene GL, Ravikumar KM, Yang S. Proteins. 2013;81:1900. doi: 10.1002/prot.24331. [DOI] [PubMed] [Google Scholar]
- 51.Ravikumar KM, Huang W, Chin C, Yang SC. 2014 Submitted. [Google Scholar]
- 52.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Journal of the American Chemical Society. 2007;129:5656. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 53.Forster F, Webb B, Krukenberg KA, Tsuruta H, Agard DA, Sali A. Journal of Molecular Biology. 2008;382:1089. doi: 10.1016/j.jmb.2008.07.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. J Appl Crystallogr. 2003;36:1277. [Google Scholar]
- 55.Parisien M, Wang XY, Perdrizet G, Lamphear C, Fierke CA, Maheshwari KC, Wilde MJ, Sosnick TR, Pan T. Cell Rep. 2013;3:1703. doi: 10.1016/j.celrep.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Katchalskikatzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. P Natl Acad Sci USA. 1992;89:2195. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westhof E, Dumas P, Moras D. Acta Crystallogr A. 1988;44(Pt 2):112. [PubMed] [Google Scholar]
- 57.Sampietro J, Dahlberg CL, Cho US, Hinds TR, Kimelman D, Xu W. Mol Cell. 2006;24:293. doi: 10.1016/j.molcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 58.Francis DM, Rozycki B, Koveal D, Hummer G, Page R, Peti W. Nat Chem Biol. 2011;7:916. doi: 10.1038/nchembio.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jamros MA, Oliveira LC, Whitford PC, Onuchic JN, Adams JA, Jennings PA. PLoS Comput Biol. 2012;8:e1002695. doi: 10.1371/journal.pcbi.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Allen MP, Tildesley DJ. Computer simulation of liquids. Oxford England New York: Clarendon Press ; Oxford University Press; 1987. [Google Scholar]
- 61.Hennig J, Wang I, Sonntag M, Gabel F, Sattler M. J Biomol NMR. 2013;56:17. doi: 10.1007/s10858-013-9719-9. [DOI] [PubMed] [Google Scholar]; Grishaev A, Wu J, Trewhella J, Bax A. J Am Chem Soc. 2005;127:16621. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]; Hutin M, Sprafke JK, Odell B, Anderson HL, Claridge TD. J Am Chem Soc. 2013;135:12798. doi: 10.1021/ja406015r. [DOI] [PubMed] [Google Scholar]; Wang J, Zuo X, Yu P, Byeon IJ, Jung J, Wang X, Dyba M, Seifert S, Schwieters CD, Qin J, Gronenborn AM, Wang YX. J Am Chem Soc. 2009;131:10507. doi: 10.1021/ja902528f. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boura E, Rozycki B, Herrick DZ, Chung HS, Vecer J, Eaton WA, Cafiso DS, Hummer G, Hurley JH. Proc Natl Acad Sci U S A. 2011;108:9437. doi: 10.1073/pnas.1101763108. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mertens HD, Piljic A, Schultz C, Svergun DI. Biophys J. 2012;102:2866. doi: 10.1016/j.bpj.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]; Karaca E, Bonvin AM. Acta Crystallogr D Biol Crystallogr. 2013;69:683. doi: 10.1107/S0907444913007063. [DOI] [PubMed] [Google Scholar]; Russel D, Lasker K, Webb B, Velazquez-Muriel J, Tjioe E, Schneidman-Duhovny D, Peterson B, Sali A. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wang YX, Zuo X, Wang J, Yu P, Butcher SE. Methods. 2010;52:180. doi: 10.1016/j.ymeth.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischetti RF, Rodi DJ, Gore DB, Makowski L. Chem Biol. 2004;11:1431. doi: 10.1016/j.chembiol.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 63.Chen L, Wildegger G, Kiefhaber T, Hodgson KO, Doniach S. J Mol Biol. 1998;276:225. doi: 10.1006/jmbi.1997.1514. [DOI] [PubMed] [Google Scholar]; Lamb J, Kwok L, Qiu X, Andresen K, Park HY, Pollack L. J Appl Crystallogr. 2008;41:1046. doi: 10.1107/S0021889808028264. [DOI] [PMC free article] [PubMed] [Google Scholar]; Roh JH, Guo L, Kilburn JD, Briber RM, Irving T, Woodson SA. J Am Chem Soc. 2010;132:10148. doi: 10.1021/ja103867p. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tsuruta H, Irving TC. Curr Opin Struct Biol. 2008;18:601. doi: 10.1016/j.sbi.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spence JCH, Weierstall U, Chapman HN. Rep Prog Phys. 2012;75 doi: 10.1088/0034-4885/75/10/102601. [DOI] [PubMed] [Google Scholar]
- 65.Zheng W, Doniach S. J Mol Biol. 2002;316:173. doi: 10.1006/jmbi.2001.5324. [DOI] [PubMed] [Google Scholar]; Parisien M, Major F. J Struct Biol. 2012;179:252. doi: 10.1016/j.jsb.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]; Wu Y, Tian X, Lu M, Chen M, Wang Q, Ma J. Structure. 2005;13:1587. doi: 10.1016/j.str.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 66.Smith JC, Roux B. Structure. 2013;21:2102. doi: 10.1016/j.str.2013.11.005. [DOI] [PubMed] [Google Scholar]; Karplus M, McCammon JA. Nat Struct Biol. 2002;9:646. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]; Duan Y, Kollman PA. Science. 1998;282:740. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]; Freddolino PL, Liu F, Gruebele M, Schulten K. Biophys J. 2008;94:L75. doi: 10.1529/biophysj.108.131565. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. Science. 2011;334:517. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]