Summary
Obesity and type 2 diabetes are strongly associated with abnormal lipid metabolism and accumulation of intramyocellular triacylglycerol, but the underlying cause of these perturbations are yet unknown. Herein, we show that the lipogenic gene, stearoyl-CoA desaturase 1 (SCD1), is robustly up-regulated in skeletal muscle from extremely obese humans. High expression and activity of SCD1, an enzyme that catalyzes the synthesis of monounsaturated fatty acids, corresponded with low rates of fatty acid oxidation, increased triacylglycerol synthesis and increased monounsaturation of muscle lipids. Elevated SCD1 expression and abnormal lipid partitioning were retained in primary skeletal myocytes derived from obese compared to lean donors, implying that these traits might be driven by epigenetic and/or heritable mechanisms. Overexpression of human SCD1 in myotubes from lean subjects was sufficient to mimic the obese phenotype. These results suggest that elevated expression of SCD1 in skeletal muscle contributes to abnormal lipid metabolism and progression of obesity.
Introduction
Obesity and type 2 diabetes are two closely connected metabolic diseases that are now invading Westernized societies at epidemic rates. The onset of insulin resistance, a hallmark pathophysiological feature of type 2 diabetes, usually occurs after a period of weight gain. The temporal relationship between these two diseases implies that increased adiposity and/or a high lipid environment somehow decay insulin’s ability to regulate glucose homeostasis. A unifying theme that has emerged during the past decade suggests that lipid oversupply to metabolic organs responsible for glucose regulation, such as muscle, liver, and pancreas, eventually leads to impaired function in those tissues. Fitting with this paradigm, we and others have shown that human obesity is associated with abnormal accumulation of neutral lipids within skeletal myofibers (Hulver et al., 2003; Malenfant et al., 2001), a phenomenon that occurs in concert with reduced insulin stimulated glucose transport (Dohm et al., 1988) and impaired insulin signal transduction (Cortright et al., 2000; Goodyear et al., 1995; Itani et al., 2000). Moreover, pharmacological and genetic manipulations that deplete intramyocellular triacylglycerol (IMTG) have been shown to concomitantly restore insulin sensitivity (Gavrilova et al., 2000; Kim et al., 2000; Schmitz-Peiffer et al., 1997; Ye et al., 2001; Zierath et al., 1998). These reports add to a growing body of evidence that links genetic and/or acquired defects in muscle lipid handling and IMTG accumulation to whole-body energy dysregulation and glucose intolerance.
The underlying mechanisms that mediate obesity-associated changes in muscle lipid metabolism are yet unknown, but could provide novel therapeutic targets for the development of antiobesity and/or antidiabetic drugs. In an effort to identify candidate genes that might contribute to elevated IMTG content we performed transcriptional profiling analyses on skeletal muscle from lean and obese humans using Affymetrix Gene-Chip microarray technology. Herein, we report a unique upregulation of a lipogenic gene, stearoyl-CoA desaturase-1 (SCD1), in skeletal muscle from obese humans. The SCD1 protein localizes to the endoplastic reticulum where it catalyzes the Δ9-cis desaturation of saturated FA, preferring palmitoyl-CoA (C16:0) and stearoyl-CoA (C18:0) as substrates and producing palmitoleoyl-CoA (C16:1) and oleoyl-CoA (C18:1), respectively, as products (Enoch et al., 1976; Ntambi, 1995). Oleoyl-CoA and palmitoleoyl-CoA are essential substrates for the synthesis of triacylglycerol (TAG), phospholipid (PL) and cholesterol esters and represent two major fatty acids (FA) found in IMTG (Enoch et al., 1976). In knockout mouse models, the absence of SCD1 lowers tissues TAG content and protects against both diet-induced and genetic forms of obesity (Cohen et al., 2002). Thus, evidence from animal studies point to a principal role for SCD1 in regulating tissue TAG levels as well as whole body energy homeostasis (Miyazaki et al., 2001; Ntambi et al., 2002). The present study now suggests that elevated expression of SCD1 in skeletal muscle contributes to perturbations in lipid partitioning and progression of the obesity syndrome in humans.
Results
Subject demographics
The lean (BMI ≤ 25 kg/m2) and obese (BMI ≥ 35 kg/m2) subjects that were studied in this investigation consisted of all females and were matched with respect to age and ethnicity (Caucasian and African-American). The BMI cutoff for the obese group was selected based on our previous report demonstrating that muscle lipid accumulation is most severe in extreme obesity (Hulver et al., 2003). In earlier studies we demonstrated that this extremely obese/nondiabetic population exhibits severe whole-body and muscle insulin resistance, assessed by both biochemical and molecular markers (Caro et al., 1989).
Skeletal muscle lipid accumulation
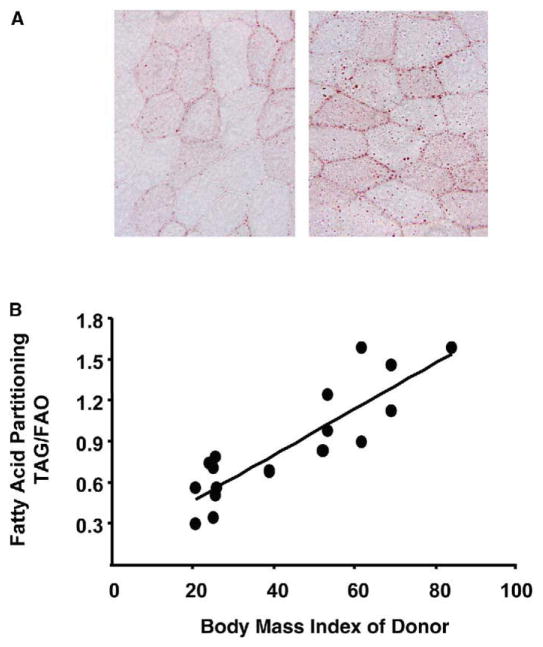
Figure 1A shows representative histological cross-sections of rectus abdominus muscle obtained from lean and obese humans. Consistent with several previous reports (Goodpaster et al., 2000; Malenfant et al., 2001), Oil Red O staining of neutral lipids revealed more intramuscular lipid droplets in obese compared to lean subjects. In a previous study we found that IMTG accumulation in obese subjects was associated with a 43% decrease in lipid oxidation (Hulver et al., 2003). Fatty acids entering the muscle are primarily utilized by the competing pathways of β-oxidation and TAG synthesis. Figure 1B shows that the amount of 14C-oleate incorporated in TAG, relative to the amount oxidized, was greater in muscle strips from obese subjects and correlated (r2 = 0.76, p < 0.01) with the BMI of the donor subject.
Figure 1. Abnormal lipid metabolism in muscle from obese subjects.
A) Histological cross-sections of rectus abdominus muscles obtained from lean and obese humans.
B) Fatty acid metabolism in muscle strips from lean (n = 8, BMI, 24.0 ± 0.79) and obese (n = 11, BMI, 58.0 ± 4.02) humans. The partitioning of FA between opposing metabolic pathways was evaluated by dividing the rate (nmol/g wet weight/h) of oleate esterified into TAG by the rate of oleate oxidized. Linear regression analysis revealed a strong correlation (r2 = 0.76, p = 0.01) between FA partitioning and body mass index of the donor subject.
Transcriptional profiling
Based on the results of our in vitro metabolic assays we suspected that muscles from the obese subjects might be transcriptionally programmed to promote lipid storage. In order to test this hypothesis we performed DNA microarray analyses using Affymetrix Human Genome chips U133A and U133B. Total RNA from lean (n = 8, BMI, 23.8 ± 0.58) and obese (n = 8, BMI, 53.8 ± 3.5) donors was extracted from a subset of the muscle specimens that were used for the in vitro muscle strip studies (Hulver et al., 2003). Table 1 shows relative mRNA expression levels of several candidate genes that could mediate obesity-associated changes in muscle fuel partitioning (see Supplemental Table 1 for normalized data). Surprisingly, mRNA expression of the lipogenic transcription factor, sterol regulatory element binding protein 1c (SREBP1c), as well as putative target genes involved in regulating glycerolipid synthesis; glycerol-3-phosphate acyltransferase (GPAT) and diacylglycerol acyltransferase (DGAT), were similar between the lean and obese subjects. Expression of acetyl-CoA carboxylase (ACC) β, an enzyme that acts as a negative regulator of mitochondrial FA uptake and oxidation, was increased 29% in the obese subjects. Genes encoding the lipid-activated transcription factors, PPARs α and δ (p < 0.01) were slightly suppressed in obese subjects, whereas PPARγ was elevated 57% (p < 0.05). PPAR-targeted FA oxidative genes, including pyruvate dehydrogenase kinase 4 (PDK4), carnitine palmitoyltransferase 1β (CPT1β) and malonyl-CoA decarboxylase (MCD) were either unaffected or only marginally down-regulated by obesity.
Table 1.
Transcriptional profiling of lipid-regulatory genes
| GeneChip (obese versus lean) | RTQ-PCR (obese versus lean) | |
|---|---|---|
| Lipid biosynthesis genes
| ||
| ACCα | 0.82 ± .17 | |
| ACCβ | 1.29 ± .09** | |
| DGAT | 1.04 ± 0.10 | 1.48 ± 0.26 |
| GPAT | 1.06 ± 0.02 | 1.38 ± 0.21 |
| SCD1 | 3.86 ± 1.24* | 3.13 ± 0.71* |
| SREBP1c | 1.08 ± .13 | 1.00 ± .08 |
|
| ||
| PPAR nuclear receptors
| ||
| PPARα | 0.86 ± 0.04 | 1.24 ± 0.10 |
| PPARδ | 0.62 ± 0.09* | 1.17 ± 0.10 |
| PPARγ | 1.57 ± 0.23* | 1.41 ± 0.20* |
|
| ||
| Fatty acid oxidative genes
| ||
| CPT1β | 0.97 ± 0.08 | 1.16 ± 0.20 |
| MCD | 0.81 ± 0.06** | 1.28 ± 0.13 |
| PDK4 | 0.89 ± 0.07 | 1.50 ± 0.20 |
|
| ||
| Adipocyte-enriched genes
| ||
| Adipose differentiation-related protein | 1.08 ± 0.06 | |
| Fatty acid binding protein 4 (AP2) | 1.12 ± 0.14 | |
| Adipsin | 0.89 ± 0.11 | |
| Adipose most abundant gene transcript 1 | 1.02 ± 0.07 | |
Total RNA was extracted from rectus abdominus muscles of obese (n = 8, BMI, 53.8 ± 3.5) and lean control subjects (n = 8, BMI, 23.8 ± 0.58) and transcriptional profiling was performed on individual samples using Affymetrix Human Genome chips (column 1) as described in Experimental Procedures. Microarray results were compared against results of RTQ-PCR analysis (column 2) using total RNA extracted from rectus abdominus muscles obtained from obese (n = 9, BMI, 51.1 ± 2.8) and lean subjects (n = 7, BMI, 24.0 ± 0.58). Gene expression data are presented as fold-change (mean ± SEM) in obese relative to lean controls and were analyzed as described in Experimental Procedures (*p < 0.05, **p < 0.01). ACC, acetyl CoA carboxylase; DGAT; diacylglycerol acyltransferase 1, GPAT; mitochondrial glycerol-3-phosphate acyltransferase, SREBP1c; sterol regulatory element binding protein 1c, CPT1β; carnitine palmitoyltransferase 1β, MCD; malonyl-CoA decarboxylase, PDK4; pyruvate dehydrogenase kinase 4, PPAR; peroxisome proliferator activated receptor.
Most remarkably however, obesity was associated with a robust (3-fold; p < 0.05) increase in transcript levels of SCD1, which ranked highest amongst the known metabolic genes that were differentially expressed between groups and represented the only lipid-related transcript that was regulated by more than 2-fold in either direction (Table S2). Notably absent from this list were adipocyte-enriched genes other than SCD1. The Affymetrix array includes several classic adipocyte markers, including adipsin, adiponectin, AP2 and leptin, that were equally low in muscle specimens obtained from obese compared to lean subjects. This finding implies that adipose tissue contamination was similarly low in both groups.
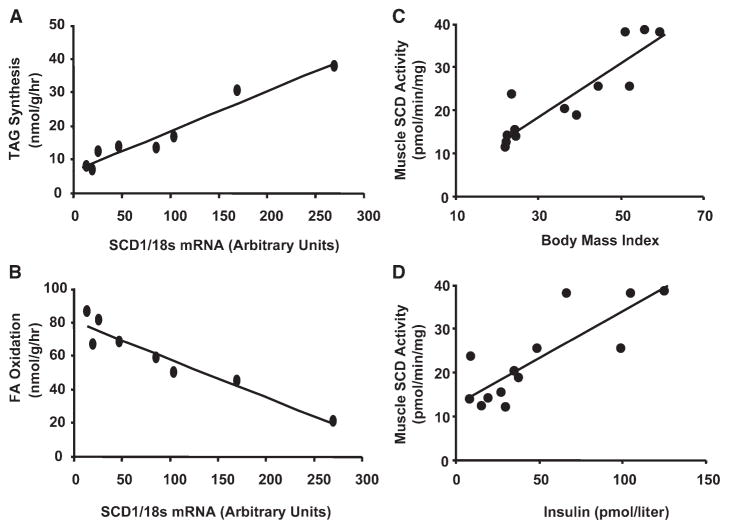
Expression profiling of a select set of candidate genes was performed concurrently using real-time quantitative PCR and muscle specimens obtained from an independent cohort of subjects (Table 1). Consistent with results from the GeneChip assay, as well as previous reports (Kruszynska et al., 1998; Park et al., 1997), muscle expression of PPARγ was increased in association with obesity. PPARs α and δ, along with several FA oxidative target genes, were not differentially regulated between the two groups. Again, elevated muscle SCD1 mRNA abundance (approximately 3-fold) emerged as a trait that corresponded with obesity, whereas expression of other lipogenic genes was unremarkable. Thus, data from two independent transcriptional profiling studies pointed to skeletal muscle SCD1 as a strong genomic marker of obesity. The gene expression data were then correlated with results from our in vitro metabolic studies. SCD1 mRNA levels exhibited a remarkably strong positive association with BMI (r2 = 0.90, p < 0.001) (Figure S1) and rates of IMTG synthesis (r2 = 0.95, p < 0.001) (Figure 2A), and an equally impressive negative association with rates of FA oxidation (r2 = 0.92, p < 0.001) (Figure 2B). Using specimens from a separate cohort of subjects we found that both BMI (r2 = 0.80, p < 0.001) and fasting insulin (r2 = 0.68, p < 0.01) correlated positively with muscle SCD1 enzyme activity (Figures 2C and 2D). Accordingly, mean SCD1 enzyme activity was 2.1-fold higher (p < 0.01) in muscle microsomes prepared from obese compared to lean donors. Similar to our previous reports, blood glucose values were not different between groups (5.2 ± 0.2 versus 4.8 ± 0.3 mmol/liter in lean and obese respectively), but fasting insulin levels were 5-fold higher in obese compared to leans subjects, consistent with insulin resistance.
Figure 2. Relationship between muscle fatty acid metabolism and SCD1 expression.
Muscle samples were obtained from lean and obese humans with a body mass index ranging from 22–60 kg/m2. Gene expression data from Table 1 were combined with results from in vitro metabolic assays (detailed in Hulver et al. [2003]) to show strong correlations between SCD1 mRNA levels (A) rates of intramuscular triacylglycerol (IMTG) synthesis and (B) fatty acid oxidation. The relationships between SCD1 activity and (C) body mass index and (D) serum insulin levels were evaluated using rectus abdominus specimens that were harvested from a second cohort of subjects.
Metabolic profiling
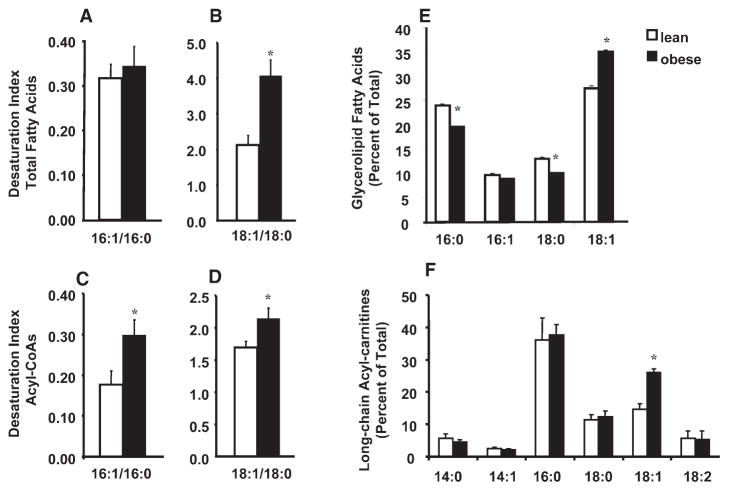
To determine whether increased SCD1 expression and activity corresponded with changes in muscle FA composition, we proceeded to measure the FA desaturation index (C16:1/C16:0 and C18:1/C18:0) of muscle lipid extracts from lean (n = 7, BMI, 23.6 ± 1.1 kg/m2) and obese (n = 7, BMI, 65.1 ± 7.4 kg/m2) human subjects. The desaturation index of total tissue lipids, 16:1/16:0 and 18:1/18:0 (Figures 3A and 3B), was 10% and 40% (Figure 3B) greater in muscle samples from obese compared to lean subjects, respectively. Likewise, the desaturation index of free acyl-CoAs, 16:1/16:0 and 18:1/8:0 (Figures 3C and 3D), was 40% and 25% greater in muscles from obese compared to lean subjects, respectively. The glycerolipid fraction from muscle of obese subjects also contained a higher percentage of C18:1 and lower percentages of both C18:0 and C16:0 (Figure 3E). Similarly, when we evaluated the composition of muscle acylcarnitines, reflective of mitochondrial FA uptake and metabolism (An et al., 2004), the oleoylcarnitine levels (expressed as a percent of total long-chain acylcarnitines) were increased from 15% in lean to 26% in the obese subjects (Figure 3F). Palmitoelyl carnitine esters (C16:1) were not detected. In summary, elevated mRNA expression of SCD1, an enzyme that catalyzes the synthesis of oleoyl-CoA, was indeed associated with increased C18:1 content of each of the lipid fractions that were examined.
Figure 3. Monounsaturated fatty acid content of muscle lipids is increased with obesity.
The desaturation index (16:1/16:0 and 18:1/18:0) was measured in various muscle lipid fractions. Total fatty acids (A) and (B) were extracted from rectus abdominus skeletal muscle obtained from lean (n = 7, BMI, 23.6 ± 1.1 kg/m2) and obese (n = 7, BMI, 65.1 ± 7.4 kg/m2). Fatty acyl-CoAs (C) and (D) were extracted from rectus abdominus muscle obtained from lean (n = 8, BMI, 23.8 ± 0.58) and obese (n = 8, BMI, 53.8 ± 3.5). The FA composition of (E) intra-myocellular glycerolipids and (F) mitochondrial-derived acylcarnitines are expressed as a percent of the total. Values represent the mean ± SEM, and differences between groups were analyzed by Student’s t test, *p < 0.05.
Human skeletal muscle cell culture
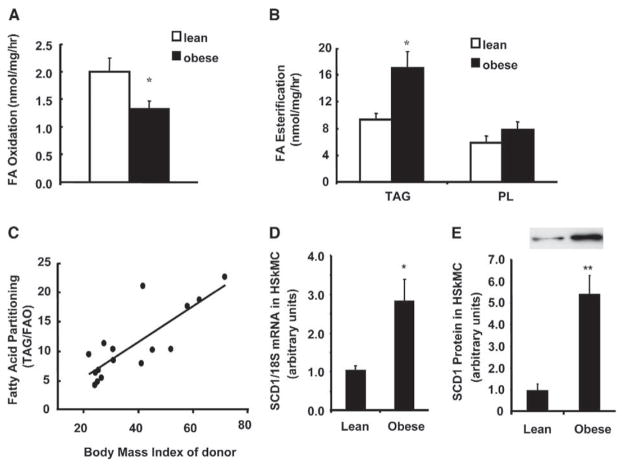
To address the possibility that obesity-associated perturbations in lipid balance might reflect heritable and/or imprinted metabolic defects, we next evaluated FA utilization in primary human skeletal myocytes (HSKMC) that originated from lean (n = 7, BMI = 23.5 ± 0.77) or obese (n = 9, BMI = 45.1 ± 3.89) donors. Fatty acid oxidation was higher (p < 0.05) in myocytes from lean (1.99 ± 0.27 nmol/mg/hr) compared to obese (1.33 ± 0.15 nmol/mg/hr) subjects (Figure 4A). In contrast, FA incorporation into TAG was 83% greater (p < 0.02) in myocytes from obese compared to lean subjects, whereas label incorporation into PL was not different between groups (Figure 4B). Only small amounts of FA (<10% of the total) were incorporated into DAG and other minor lipid species (not shown). The partitioning of FA between opposing metabolic pathways was then evaluated by dividing the rate (nmol/mg/hr) of oleate esterified into TAG by the rate of oleate oxidized, thereby providing an index of FA utilization. Obesity affected oxidation and esterification in opposite directions, resulting in a 227% increase in the TAG:oxidation partitioning index, from approximately 5:1 in myocytes from lean subjects to 11:1 in those from obese subjects. Linear regression analysis revealed a strong correlation (r2 = 0.66, p < 0.001) between the FA partitioning index of primary myocytes and BMI of the donor subject (Figure 4C). Notably, the results presented in Figure 4 have been reproduced using donors from multiple cohorts that were recruited from the Greenville, NC and Baton Rouge, LA geographic regions.
Figure 4. Fatty acid metabolism and SCD1 expression in cultured HSKMC from lean and obese subjects.
Myoblasts were isolated from human rectus abdominus muscle obtained from lean or obese subjects. Day 8 myotubes were incubated 3 hr at 37°C in DFM with 100 μM [14C]oleate and 0.25% BSA.
A) Fatty acid oxidation was determined by measuring 14C-label incorporation into CO2 and acid soluble metabolites.
B) Glycerolipid synthesis was determined by measuring 14C-label incorporation into triacylglycerol (TAG) and phospholipid (PL).
C) Relationship between fatty acid partitioning ([14C]-oleate incorporation into TAG (nmol/mg protein/h) divided by the amount oxidized] and body mass index of the donor subject. Metabolic assays were performed in triplicate and values represent the mean ± SEM from 7–9 subjects.
D) Total RNA was isolated and SCD1 mRNA expression was quantified by RTQ-PCR. Relative expression levels were normalized to 18S mRNA and data are presented as means ± SEM of cells from 7 subjects per group.
E) Cell extracts were prepared from day 8 myotubes and used for Western blot analysis of SCD1 protein abundance. Differences between groups were analyzed by Student’s t test. *p < 0.05, **p < 0.01.
Because the expression profiling studies (Table 1) implicated SCD1 as a potential mediator of aberrant muscle lipid metabolism, we next sought to evaluate this same parameter in the primary myocytes. SCD1 again emerged as a strong marker of abnormal muscle lipid metabolism, as mRNA levels were increased 2.8-fold (Figure 4D) and protein levels were increased 5.7-fold (Figure 4E) in myotubes that originated from obese compared to lean subjects. Predictably, SCD1 transcript levels in muscle were approximately 10- to 20-fold lower than those in adipose tissue (Table S3). However, the CT values for SCD1 (obtained by RTQ-PCR) using muscle RNA were comparable to other lipid-related metabolic transcripts, such as CPT1β and MCD; suggesting that the gene is expressed at moderately abundant levels in both muscle tissue and skeletal myotubes. We also found that SCD1 mRNA levels were similarly abundant in human fibroblasts, preconfluent myoblasts, confluent myoblasts, and mature myotubes (Table S3); thus, gene expression is not enriched in myoblasts and does not appear to be regulated during myocyte differentiation. In contrast, SCD1 was expressed at low levels in human pre-adipocytes and robustly induced upon adipocyte differentiation. This finding is consistent with results from mouse 3T3-L1 adipocytes (Ntambi et al., 1988) and indicates that contamination by pre-adipocytes would not cause an artifactual increase in the SCD1 mRNA levels measured in human myotubes.
To further address the possibility of adipocyte contamination we carefully evaluated cellular morphology and performed immunocytochemical staining of the cultures (Figure 5). Similar to our previous report (Muoio et al., 2002b), we found that 80%–90% of HSKMC exhibited an elongated, multinucleated morphology within 6–8 days after switching to differentiation media. Formation of multinucleated myotubes and expression of the hallmark muscle-specific protein, myosin, were similar between cells from lean and obese donors (Figures 5A and 5B). Consistent with the metabolic assays, Oil Red O staining of neutral lipid droplets following an overnight FA exposure was more intense in cells harvested from obese subjects. Importantly, lipid droplets were seen only in mature myotubes (Figures 5C–5F). Lipid-loaded adipocytes are characterized by large triacylglycerol droplets that occupy the majority of the cytoplasmic space. This distinct morphology was absent from our cultures. Finally, the presence of SCD1 in mature myotubes was confirmed by immunocytochemistry (Figure 5H). Western blotting of cell and tissue lysates detected only a single 37 kDa band representative of the SCD1 protein (data not shown), thus demonstrating high specificity of the antibody. Immunostaining of cultured myotubes from lean subjects revealed a diffuse signal that was co-expressed with sarcomeric myosin (Figure 5I), again implying that the SCD1 detected in muscle tissue and/or myotubes was not an artifact of adipocyte contamination.
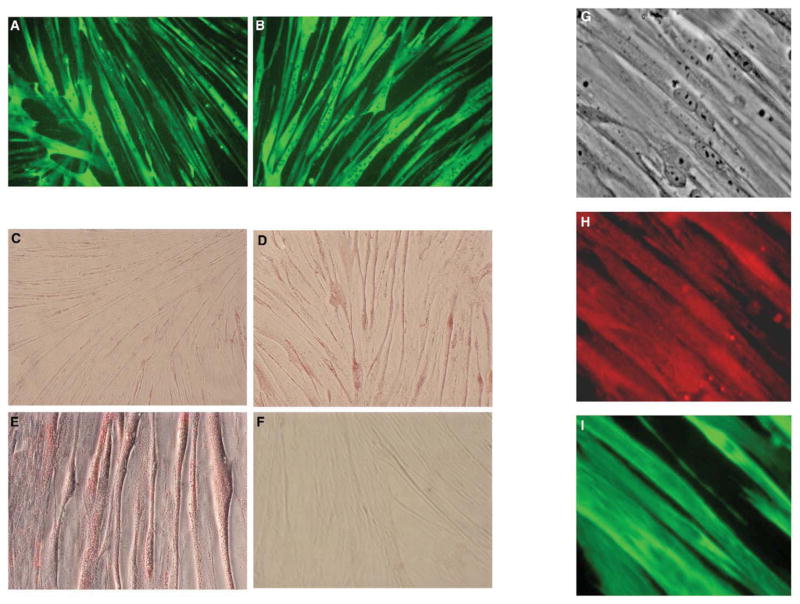
Figure 5. Characterization of primary human myotubes.
Differentiated HSKMC exhibit multi-nucleated (arrows) morphology that is characteristic of mature myotubes. Fluorescent photomicrographs (200×) show that immuno-detection of the sarcomeric protein, myosin (green) was similar between cells from (A) lean and (B) obese subjects. Myotubes were incubated overnight with 500 μM fatty acid (oleate:palmitate 2:1) bound to 0.5% BSA and then fixed and stained for neutral lipids. Phase contrast photomicrographs (100×) revealed less intense staining in myotubes that originated from (C) lean compared to (D) obese donors. (E) Higher magnification (200×) showing lipid droplets in mature myotubes. (F) Myotubes from obese subjects that were incubated overnight with 0.5% BSA minus fatty acid displayed few lipid droplets. (G) Phase contrast photomicrographs (40×) of day 8 myotubes that were incubated with antibodies against human SCD1 and mouse sarcomeric myosin. Proteins were visualized using secondary antibodies conjugated to distinct fluorophores. Fluorescent photomicrographs show co-expression of (H) SCD1 (red) and (I) sarcomeric myosin (green) in multinucleated myotubes. Negative controls that were performed without addition of primary antibody did not present a fluorescent signal (data not shown).
Metabolic impact of SCD1 overexpression
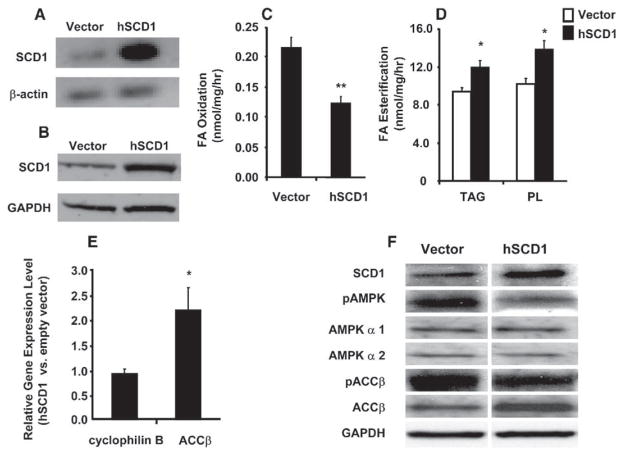
To establish a causal relationship between SCD1 and lipid imbalance, we proceeded to increase expression of the enzyme in primary myocytes from lean subjects by transient transfection. We expected this maneuver to produce a metabolic phenotype resembling that displayed by myocytes from the obese subjects. In preliminary experiments, we used a green fluorescent protein expression vector to confirm reasonably high efficiency transfection (40%–50%) of primary human myotubes (not shown). Likewise, using the hSCD1 expression vector we were able to achieve a robust increase in SCD1 mRNA (>10-fold, Figure 6A) with an approximate 2-fold increase in protein when compared against control cells transfected with the empty parent vector (Figure 6B). Transfected myotubes were then subjected to acute (3 hr) incubations with radiolabeled oleic acid to assess changes in FA metabolism. SCD1 overexpression decreased oleate oxidation by 45% (Figure 6C) and increased incorporation into myocellular TAG and PL by 22% and 27%, respectively (Figure 6D). The net impact on cellular lipid utilization was a 2.1-fold shift that favored the partitioning of FA toward storage and away from oxidation.
Figure 6. hSCD1 overexpression in primary HSKMC alters FA partitioning.
Preconfluent myoblasts were transiently transfected with pcDNA3.0-hSCD1 or the empty parent vector. After 7 days in DFM, cells were harvested to obtain total RNA. Overexpression of SCD1 was confirmed in differentiated myotubes by (A) semiquantitative PCR of mRNA and (B) Western blot analysis of protein. β-actin mRNA and GAPDH protein were used as loading controls. Metabolic assays were performed following a 3 hr incubation with 100 μM [14C]oleate and 0.25% BSA. (C) Fatty acid oxidation was determined by measuring 14C-label incorporation into CO2. (D) Glycerolipid synthesis was determined by measuring 14C-label incorporation into triacylglycerol (TAG) and phospholipid (PL). All assays were performed in triplicate and values represent the mean ± SEM from four separate experiments. Differences between groups were analyzed by Student’s t test, *p < 0.05, **p < 0.01. (E) ACCβ mRNA levels were quantified by RTQ-PCR and are expressed as relative units in cells transfected with SCD1 versus empty vector controls. Cyclophilin B was measured as an endogenous control. Data are representative of five experiments and differences between groups were analyzed by Student’s t test, *p < 0.05. (F) Total and phosphorylated AMPK and ACCβ were analyzed by immunoblotting with antibodies against the AMPKα 1 and 2 catalytic subunits, ACCβ, AMPK-thr172, and ACCβ-ser79. GAPDH protein expression was not different between groups.
Disruption of SCD1 in mice results in decreased hepatic expression and activity of ACCα and ACCβ (Dobrzyn et al., 2004b), enzymes that promote lipogenesis and inhibit FA oxidation. Both enzymes are inactivated via phosphorylation by their upstream kinase, AMP-activated kinase (AMPK) (Dobrzyn et al., 2004b). Interestingly, our microarray study indicated that high muscle SCD1 expression in obese subjects was accompanied by an increase in ACCβ, the major isoform expressed in muscle (Table 1). In light of the animal data, we reasoned that the metabolic consequences of SCD1 overexpression might be at least partly mediated by the induction and/or activation of ACCβ. Indeed, transcript levels of ACCβ were increased 2.2-fold in HSKMC transfected with hSCD1 compared to the empty vector controls (Figure 6E). Additionally, SCD1 overexpression caused a decrease in the phosphorylated levels of both AMPK and ACCβ (Figure 6F), which in turn favors increased ACCβ activity and lower rates of FA oxidation (Mills et al., 1983). Total protein levels of the AMPK α 1 and 2 catalytic subuntis were unchanged whereas ACCβ protein expression increased in alignment with changes in the transcript. In the aggregate, these results demonstrate that high expression of SCD1 alters muscle lipid balance in a manner that is likely to contribute to the development of obesity.
Discussion
Mounting evidence suggests that disordered muscle lipid metabolism could play a central role in the development of obesity and type 2 diabetes. Herein and in a previous report (Hulver et al., 2003) we demonstrated that skeletal muscle strips from extremely obese humans partition FA substrate preferentially toward esterification pathways and away from mitochondrial β-oxidation. Because these studies were performed using an in vitro muscle system, the results imply that muscles from the obese subjects are inherently programmed to store high levels of IMTG. High IMTG are proposed to provide a surplus of endogenous FA substrate that can be used to synthesize insulin desensitizing lipid metabolites such as fatty acyl CoAs, diacylglycerols, and ceramides. Whereas intense efforts have focused on identifying the precise lipid mediators that impair insulin signaling, relatively few studies have addressed the underlying mechanisms that cause IMTG to accumulate.
Using cDNA microarray analysis we discovered that obesity was associated with a 3-fold increase in muscle mRNA levels of the lipogenic enzyme, SCD1. In contrast, muscle expression of other lipid biosynthetic and/or adipocyte-enriched genes was similar between the lean and obese groups. SCD1 expression levels correlated strongly with BMI as well as the obesity-related changes in muscle FA partitioning. Compelling evidence from animal studies has implicated SCD1 as an important regulator of tissue lipid balance (Cohen et al., 2002; Dobrzyn et al., 2004b). SCD1 catalyzes the synthesis of oleate and palmitoleate from saturated FA, and therefore plays a crucial role in supplying the monounsaturated FA that are required for de novo synthesis of TAG and PL (Enoch et al., 1976). Targeted disruption of the SCD1 gene in knockout mice reduces TAG levels and increases FA catabolism in liver (Ntambi and Miyazaki, 2004; Ntambi et al., 2002) and muscle (Dobrzyn et al., 2004a). In comparison, we found that human muscle strips and primary HSKMC that express high levels of SCD1 displayed increased TAG synthesis and decreased FA oxidation.
Despite a convincing body of evidence that links defects in muscle lipid metabolism to obesity and insulin resistance, it remains uncertain whether these perturbations represent a contributing factor or an adaptive consequence of the disease process. In this regard, the HSKMC culture system provides an attractive model in which muscle fuel metabolism can be evaluated apart from the confounding systemic influences of the obese environment. Similar to previous reports (Gaster et al., 2004; Henry et al., 1995, 1996), we found that cultured HSKMC express metabolic characteristics that approximate the in vivo phenotype of the donor subject. Others have shown that muscle cells from diabetes-prone Pima Indians exhibit impaired insulin responsiveness when compared against cells from control subjects (Henry et al., 1996). Additionally, perturbations in palmitate oxidation (Gaster et al., 2004) and insulin-stimulated glycogen synthase activity (Nikoulina et al., 2002) were evident in HSKMC obtained from a more heterogenous pool of type 2 diabetics. We evaluated fuel metabolism in HSKMC from lean compared to extremely obese (but nondiabetic) subjects. In vivo assessments indicated that the obese condition features abnormal accumulation of IMTG droplets and reduced FA catabolism. These same traits persisted in the cultured myocytes that had been removed from the hyperinsulinemic, dyslipidemic in vivo environment for approximately 30 days. Moreover, HSKMC from obese compared to lean subjects expressed 2.8-fold higher mRNA levels of SCD1, an outcome nearly identical to that obtained using biopsied muscle specimens.
We also demonstrated that specific overexpression of SCD1 in myotubes from lean subjects altered FA partitioning in a manner that resembled the high rates of muscle TAG synthesis and low rates of FA oxidation observed with obesity. Thus, SCD1 overexpression in our cell system provoked metabolic adjustments that were reciprocal to those displayed when the gene was disrupted in knockout mice. Notably, the metabolic impact of SCD1 overexpression was not limited to effects on the acyl-CoA substrates of SCD1. Rather, elevated SCD1 activity caused robust changes in the metabolic fate of oleoyl-CoA, the major monounsaturated product of the enzyme. This result suggests that the alterations in FA partitioning were not simply due to a mass action effect, but instead were conferred by a more global level of regulation. The notion that changes in SCD1 activity can impact multiple lipid-handling pathways is also supported by results from the loss-of-function studies. In liver and adipose tissue from SCD1 null mice, absence of the gene not only decreases TAG synthesis (Miyazaki et al., 2000, 2001), but also increases metabolic rate and FA catabolism in association with marked induction of several PPAR target genes (Cohen et al., 2002; Ntambi et al., 2002). These observations imply that SCD1 might impart transcriptional control by altering intracellular levels of lipid ligands that activate the PPARs or other nuclear hormone receptors.
More recent investigations have shown that the SCD1 null mice display enhanced AMPK activity, decreased expression and activity of ACCα and ACCβ and lower levels of the ACC product, malonyl-CoA (Dobrzyn et al., 2004a, 2004b). Similarly, we found that overexpression of SCD1 in human myocytes resulted in decreased phosphorylation of both AMPK and ACCβ, and an approximate 2-fold induction of ACCβ mRNA and protein levels. Consistent with our cell-based studies, muscle ACCβ mRNA expression was also up-regulated in the obese subjects. Taken together with the animal data, our results suggest that high SCD1 activity augments the production of malo-nyl-CoA. Since malonyl-CoA functions as a potent inhibitor of CPT1, the rate limiting enzyme of β-oxidation, changes in this metabolite are likely to play an important role in conferring SCD1-mediated changes in cellular lipid balance.
In addition to its impact on fuel metabolism, SCD1 also regulates the FA composition of neutral lipids as well as membrane PL (Enoch et al., 1976). Thus, SCD1-induced modifications in lipid signaling molecules and/or membrane FA composition might alter the activities of multiple regulatory enzymes and/or proteins. The absence of SCD1 in knockout mice lowers the oleate and palmityloleate content of various lipid fractions and elevates palmitate and stearate levels (Ntambi et al., 2002). In comparison, we found that high SCD1 activity in human muscle corresponded with increases in the monounsaturated FA composition of esterified lipids, free acyl-CoAs as well as mito-chondrial-derived acylcarnitines. Others have suggested that changes in the intracellular ratio of stearic to oleic acid may impact skeletal muscle insulin sensitivity (Storlien et al., 1996). Indeed, disruption of SCD1 appears to promote glucose tolerance in mice (Rahman et al., 2003). We found a positive correlation between SCD1 activity and fasting insulin levels, but further studies are required to determine whether or not SCD1 directly opposes insulin action in human muscle.
Results from our investigation prompt questions regarding the mechanism(s) that drive muscle SCD1 induction in the setting of obesity. Persistence of this feature in the cell culture system implicates factors that may link to a genetic predisposition. However, because our study was executed with three relatively heterogenous cohorts of subjects, the obesity-associated increases in SCD1 abundance are unlikely to stem from a single inherited trait, but rather, might represent a common manifestation of multiple susceptibility genes. Alternatively, acquired defects in fuel metabolism may become imprinted into the metabolic memory of myogenic satellite cells. Compelling evidence from both animal and epidemiological studies indicates that deviant nutrition occurring during critical developmental periods can impose imprinted metabolic adaptations that persist into adulthood (Waterland and Jirtle, 2004). These observations suggest that transient environmental stresses can trigger permanent alterations in metabolic control. The mechanistic basis of “imprinting” is encompassed by the study of epigenetics, a term referring to the stable propagation of gene activity states that is not conferred by variations in DNA sequence (Egger et al., 2004). Thus, our findings could reflect an epigenetic phenomenon in which obesity-induced changes in DNA methylation and/or histone modification provoke irreversible perturbations in SCD1 gene regulation.
The SCD1 gene is transcriptionally regulated by many developmental, nutritional and hormonal factors (Ntambi, 1995). In liver and adipose tissue, SCD1 gene expression is repressed by leptin and induced by glucose and insulin (Ntambi and Miyazaki, 2004). Glucose oversupply has also been shown to increase expression and activity of SCD1 in rat skeletal muscle (Houdali et al., 2003). Both glucose and insulin-mediated stimulation of SCD1 occurs through the action of the lipogenic transcription factor, SREBP1c. Some studies also suggest that the SCD1 gene might be controlled by the PPARs (Singh Ahuja et al., 2001; Way et al., 2001). Whereas our transcriptional profiling studies did not detect changes in SREBP1c, we did observe an obesity-associated increase in muscle mRNA levels of PPARγ. Interestingly, in rodents, synthetic PPARγ ligands have been shown to induce SCD1 gene expression in both liver (Singh Ahuja et al., 2001) and skeletal muscle (Way et al., 2001). The potential roles of these and other molecular pathways in regulating SCD1 expression in the HSKMC system now awaits further investigation.
In summary, animal studies indicate that suppression of SCD1 confers protection against obesity and insulin resistance. Our studies now reveal a novel link between high muscle SCD1 activity and severe obesity in humans. We propose that elevated expression of SCD1 in skeletal muscle may represent a core mechanism contributing to reduced FA oxidation, increased IMTG synthesis and progression of the metabolic syndrome. The obese state is characterized by a constellation of metabolic abnormalities, including hyperglycemia, hyperinsulinemia and leptin resistance, which may converge to augment SCD1 expression. Pharmacological targeting of muscle SCD1 and/or its upstream regulators could provide new opportunities for preventing and/or treating obesity and its related co-morbidities.
Experimental procedures
Human subjects
The lean (BMI ≤ 25 kg/m2) and obese (BMI ≥ 35 kg/m2) groups that were studied in this investigation consisted of all females and were matched with respect to age and ethnicity (Caucasian and African-American). Their medical charts were reviewed to ensure participants were not diabetic. Subjects were excluded if fasting plasma glucose levels were >/= 6.9 pmole/L (126 mg/dL). The experimental protocol was approved by the Internal Review Boards for Human Research at East Carolina University and the Pennington Biomedical Research Center. Informed consent was obtained from all patients.
Lipid and immunostaining
Neutral lipid content was determined using the Oil Red O staining procedure, modified from Goodpaster et al. (Goodpaster et al., 2000) as previously described (Hulver et al., 2003). Staining of day 8 myotubes was performed as described by (Muoio et al., 2002a). Myotubes were rinsed three times with PBS, followed by fixing and permeablization in methanol at −20°C for 20 min, and then incubated with the primary antibodies, mouse anti-sarcomere myosin (MF20, Developmental Studies Hybridoma bank University of Iowa, Iowa City, IA) and rabbit anti-Human SCD1(SCD11-A, Alpha diagnostic Intl, Inc, San Antonio Texas), for 1 hr at room temperature. Following a 3× wash in PBS plus 0.05% tween 20, cells were incubated for 1 hr at room temperature with secondary antibodies; goat anti-mouse IgG conjugated to FITC (cat#115-095-166 Jackson Immuno Research Laboratories, West Grove, PA) and goat anti-rabbit IgG (H+L) conjugated to Alexa Fluor 594 (cat# A11012, Invitrogen Molecular probes, Carlsbad, CA) for sarcomeric myosin and human SCD1, respectively. Proteins were visualized by fluorescence microscopy using Nikon model eclipse TS 100 and appropriate filters. Images were captured using the Metamorph software. Negative controls that were performed without addition of primary antibody did not present a fluorescent signal.
Lipid metabolism in intact skeletal muscle strips
Rectus abdominus muscle strips were harvested and incubated for 1 hr at 30°C in Krebs-Henseleit buffer with the addition of 0.75 μCi [1-14C]palmitate (New England Nuclear, Boston, MA) and oxidation rates were determined as previously described (Hulver et al., 2003). Neutral lipids were extracted and quantified as previously described (Hulver et al., 2003).
Affymetrix microarray complementary RNA synthesis and labeling, hybridization, and expression profiling
Total RNA was extracted from skeletal muscle samples using the TRIzol reagent (Invitrogen Life Technologies) and RNeasy Mini kit (Qiagen). Double-stranded cDNA was synthesized from 6 μg of total RNA using the SuperScript Choice system (Invitrogen) and the T7-Oligo(dT) promoter primer kit (Affymetrix). cDNA was purified using Phase Lock Gels (Eppendorf 5-prime) and used to synthesize biotin-labeled cRNA. Fifteen micrograms of biotin-labeled cRNA was hybridized onto Human Genome U133A and U133B GeneChips (Affymetrix). Procedures for cRNA preparation and GeneChip processing were performed as previously described (Chen et al., 2002), following quality control procedures that have been published (Hoffman et al., 2004). A detailed description of all quality control parameters as well as all image files (.dat) corresponding to this work is available http://microarray.cnmcresearch.org.
Microarray data analysis
Data analysis was performed using Affymetrix Microarray Suite 5.0 software to generate an absolute analysis for each chip. Each chip was scaled globally to a target intensity value of 800 to allow for inter-array comparisons. All image files (.DAT) and cell intensity files (.CEL) corresponding to each expression profile are available at http://microarray.cnmcresearch.org/pgadatatable.asp. Further details of the analysis are provided in Supplemental Methods. Results of the microarray analysis were validated by performing RTQ-PCR using rectus abdominus muscle specimens that were obtained from an independent cohort of obese (n = 9, BMI, 51.1 ± 2.8) and lean (n = 7, BMI, 24.0 ± 0.58) subjects.
Real-time quantitative PCR
RTQ-PCR was performed using an ABI PRISM 7000 Sequence Detection System instrument and TaqMan Universal PCR Master Mix used according to manufactures specifications (Applied Biosystems, Inc., Foster City, CA). Reactions were performed in triplicate and contained 20 ng cDNA as template, 900 nM forward and reverse primers and 200 nM probe. Target gene expression was normalized to 18S rRNA levels, which were assayed by multiplexing with the manufactures 5′VIC-labeled, primer-limited 18S endogenous control premix. Primers and 5′FAM-labeled Taqman probes were purchased as pre-validated assays (ABI) or designed using Primer Express software and sequences available in GenBank. Relative quantification of target genes was calculated using the 2−ΔΔCT method, which was validated for each primer/probe set using a 6 point serial standard curve as described previously (Muoio et al., 2002a). Derivation of the 2−ΔΔCT equation has been described in Applied Biosystems User Bulletin No. 2 (P/N 4303859).
Fatty acid analysis/SCD activity index
Total lipids were extracted from skeletal muscle obtained from lean (n = 7, BMI, 23.6 ± 1.1 kg/m2) and obese (n = 7, BMI, 65.1 ± 7.4 kg/m2) human subjects according to the methods of Bligh and Dryer (Bligh and Dryer, 1959). Fatty acids were methylated and analyzed by gas-liquid chromatography as previously described (Barakat et al., 1976). Heptadecanoic acid (Sigma, St. Louis, MO) was added as an internal standard for the quantitation of FA. The FA species palmitate (16:0), palmitoleate (16:1), stearate (18:0) and oleate (18:1) were identified by comparison of retention time with authentic standards (Nu-Chek Prep, Inc, Elysian, MN). The desaturation index was determined by calculating product:substrate ratios (16:1/16:0 and 18:1/18:0) using the quantitated values for palmitoleate, palmitate, oleate and stearate. Acylcarnitines were extracted from 50 mg rectus abdominus muscle as previous described (An et al., 2004). Acylcarnitines were quantified by direct injection electrospray tandem mass spectrometry (Millington et al., 1990), using a Quattro Micro LC-MS system (Waters-Micromass, Mil-ford, MA) equipped with a model HTS-PAL autosampler (Leap Technologies, Carrboro, NC), a model 1100 HPLC solvent delivery system (Agilent Technologies) and a data system running MassLynx software. Measurement of fatty acyl CoAs was made as previously described (Hulver et al., 2003).
SCD1 activity
Microsomal preparations were made from rectus abdominus muscle samples from lean (n = 6, BMI, 23.3 ± 0.42) and obese (n = 7, BMI, 48.4 ± 3.23) humans and SCD1 activity was measured as previously described (Miyazaki et al., 2000). Serum insulin was measured in blood samples obtained from these research participants as previously described (Hulver et al., 2002).
Cultures of primary human skeletal muscle cells
Rectus abdominus muscle samples weighing ~ 50 mg were obtained from lean (BMI = 23.5 ± 0.77, n = 7) and obese (BMI = 45.1 ± 3.89, n = 9) subjects. Satellite cells were cultured and differentiated in multinucleated myotubes as previously described (Muoio et al., 2002b). Fatty acid metabolism assays were performed in triplicate as detailed in (Muoio et al., 2002b).
Construction of human SCD1 expression plasmids
The full coding region of human SCD1 (hSCD1) was generated by PCR using human liver cDNA as the template and the following primers: 5′ primer, ttgaattcaccatgtacccatatgacgtcccggactacgccatgccggcccacttgctgc, which contains a sequence of N-terminal hemagglutinin epitope (HA) tag and a EcoRI restriction enzyme site; and 3′ primer tgctcgagtcagccactcttg tagtttcc, which contains a XhoI restriction enzyme site. The resulting PCR product was cloned into pcDNA3 (Invitrogen, Carlsbad, CA) and designated pcDNA3-HA-SCD1. The integrity of the PCR product was confirmed by DNA sequencing.
Overexpression of SCD1 in HSKMC
Myoblasts were subcultured onto 6-well type I collagen-coated plates at a density of ~ 40 × 103 cells per well. Upon reaching ~ 60% confluence, myocytes were transiently transfected with either hSCD1 or empty vector (pcDNA3.1) using GenePORTER transfection reagents (Gene therapy Systems, Inc, San Diego, CA) according to manufacturer’s specifications. Twenty-four hours posttransfection, medium was replaced with growth medium. When myocytes reached 90% confluence, growth media was replaced with differentiation media (DFM). Cell harvest and metabolic experiments were performed on differentiation day 7.
SCD1 mRNA and protein overexpression was confirmed by semi-quantitative PCR and Western blotting, respectively. cDNA was synthesized (iScript cDNa kit, Bio-Rad, Hercules, Ca) from total RNA and amplified using primer specific for hSCD1 (Forward: 5′-GCAGGACGATATCTCTAGCT-3′; Reverse: 5′-GTCTCCAACTTATCTCCTCCATTC-3′) and β-actin (Forward: 5′-ACAGGATGCAGAAGGAGATTACT-3′; Reverse: 5′-TGATCCACATCTGC TGGAAGGT-3′). PCR products were separated on a 2% agarose gel and results of semiquantitative analysis were confirmed by RTQ-PCR. Western analysis was performed using cell lysates harvested in 50 mM Hepes, (pH 7.5), 15 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 2 mM EDTA, 10% glycerol, 1% Triton x-100, 10 mM NaP2O7, 100 mM NaFl, 10 mM PMSF, and 10 μg/mL aprotinin. Proteins (30 μg) were separated using a 10% Criterion-Tris-HCl gel (Bio-Rad, Hercules, CA) and subsequently transferred to PVDF membrane (Bio-Rad, Hercules, CA). Blots were probed with primary antibodies against hSCD1 (4 μg/mL, Alpha Diagnostics International, San Anto-nio, TX) and GAPDH (1:100 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) followed by anti-rabbit and anti-goat secondary antibodies respectively (1:8000 and 1:10000 dilutions, respectively; Santa Cruz Biotechnology, Santa Cruz, CA). Proteins were visualized using Super-Signal Chemoluminescent Substrate (Pierce, Rockville, Il).
Supplementary Material
Acknowledgments
We thank Adrienne Cain for diligent work in the laboratory, Steven Smith, M.D., for assistance with transient transfection experiments, and both Dorothy Slentz and Julie Marchand for assistance with immunocytochemistry. We also thank Dr. Chris Newgard for his critical review of the manuscript. This research was supported by funding from the National Institute of Health Diabetes and Digestive and Kidney Diseases (NRSA F32-DK-6260501; M.W.H, F32DK 10017-01; D.M.M., and DK-56112, J.A.H.), the Pennington Biomedical Research Foundation (M.W.H.) and the American Diabetes Association (M.W.H, D.M.M, and G.L.D).
Footnotes
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, three tables, and one figure and can be found with this article online at http://www.cellmetabolism.org/cgi/content/full/2/4/251/DC1/.
References
- An J, Muoio DM, Shiota M, Fujimoto Y, Cline GW, Shulman GI, Koves TR, Stevens R, Millington D, Newgard CB. Hepatic expression of malonyl-CoA decarboxylase reverses muscle, liver and whole-animal insulin resistance. Nat Med. 2004;10:268–274. doi: 10.1038/nm995. [DOI] [PubMed] [Google Scholar]
- Barakat HA, Dohm GL, Loesche P, Tapscott EB, Smith C. Lipid content and fatty acid composition of heart and muscle of the BIO 82.62 cardiomyopathic hamster. Lipids. 1976;11:747–751. doi: 10.1007/BF02533049. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dryer WJ. A rapid method of total lipid extraction and purification. Can J Biochem. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Caro JF, Dohm LG, Pories WJ, Sinha MK. Cellular alterations in liver, skeletal muscle, and adipose tissue responsible for insulin resistance in obesity and type II diabetes. Diabetes Metab Rev. 1989;5:665–689. doi: 10.1002/dmr.5610050804. [DOI] [PubMed] [Google Scholar]
- Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol. 2002;545:27–41. doi: 10.1113/jphysiol.2002.021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Miyazaki M, Socci ND, Hagge-Greenberg A, Liedtke W, Soukas AA, Sharma R, Hudgins LC, Ntambi JM, Friedman JM. Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science. 2002;297:240–243. doi: 10.1126/science.1071527. [DOI] [PubMed] [Google Scholar]
- Cortright RN, Azevedo JL, Jr, Zhou Q, Sinha M, Pories WJ, Itani SI, Dohm GL. Protein kinase C modulates insulin action in human skeletal muscle. Am J Physiol Endocrinol Metab. 2000;278:E553–E562. doi: 10.1152/ajpendo.2000.278.3.E553. [DOI] [PubMed] [Google Scholar]
- Dobrzyn A, Dobrzyn P, Lee SH, Miyazaki M, Cohen P, Asilmaz E, Hardie DG, Friedman JM, Ntambi JM. Stearoyl-CoA de-saturase 1 deficiency reduces ceramide synthesis by down-regulating serine palmitoyltransferase and increasing {beta}-oxidation in skeletal muscle. Am J Physiol Endocrinol Metab. 2004a;288:E599–E607. doi: 10.1152/ajpendo.00439.2004. [DOI] [PubMed] [Google Scholar]
- Dobrzyn P, Dobrzyn A, Miyazaki M, Cohen P, Asilmaz E, Hardie DG, Friedman JM, Ntambi JM. Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proc Natl Acad Sci U S A. 2004b;101:6409–6414. doi: 10.1073/pnas.0401627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm GL, Tapscott EB, Pories WJ, Dabbs DJ, Flickinger EG, Meelheim D, Fushiki T, Atkinson SM, Elton CW, Caro JF. An in vitro human muscle preparation suitable for metabolic studies. Decreased insulin stimulation of glucose transport in muscle from morbidly obese and diabetic subjects. J Clin Invest. 1988;82:486–494. doi: 10.1172/JCI113622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- Enoch HG, Catala A, Strittmatter P. Mechanism of rat liver microsomal stearyl-CoA desaturase. Studies of the substrate specificity, enzyme-substrate interactions, and the function of lipid. J Biol Chem. 1976;251:5095–5103. [PubMed] [Google Scholar]
- Gaster M, Rustan AC, Aas V, Beck-Nielsen H. Reduced lipid oxidation in skeletal muscle from type 2 diabetic subjects may be of genetic origin: evidence from cultured myotubes. Diabetes. 2004;53:542–548. doi: 10.2337/diabetes.53.3.542. [DOI] [PubMed] [Google Scholar]
- Gavrilova O, Marcus-Samuels B, Graham D, Kim JK, Shulman GI, Castle AL, Vinson C, Eckhaus M, Reitman ML. Surgical implantation of adipose tissue reverses diabetes in lipoatrophic mice. J Clin Invest. 2000;105:271–278. doi: 10.1172/JCI7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpaster BH, Theriault R, Watkins SC, Kelley DE. Intramuscular lipid content is increased in obesity and decreased by weight loss. Metabolism. 2000;49:467–472. doi: 10.1016/s0026-0495(00)80010-4. [DOI] [PubMed] [Google Scholar]
- Goodyear LJ, Giorgino F, Sherman LA, Carey J, Smith RJ, Dohm GL. Insulin receptor phosphorylation, insulin receptor substrate-1 phosphorylation, and phosphatidylinositol 3-kinase activity are decreased in intact skeletal muscle strips from obese subjects. J Clin Invest. 1995;95:2195–2204. doi: 10.1172/JCI117909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RR, Abrams L, Nikoulina S, Ciaraldi TP. Insulin action and glucose metabolism in nondiabetic control and NIDDM subjects. Comparison using human skeletal muscle cell cultures. Diabetes. 1995;44:936–946. doi: 10.2337/diab.44.8.936. [DOI] [PubMed] [Google Scholar]
- Henry RR, Ciaraldi TP, Abrams-Carter L, Mudaliar S, Park KS, Nikoulina SE. Insulin-dependent Diabetes Mellitus Subjects. Biochemical and Molecular Mechanisms. J Clin Invest. 1996;98:1231–1236. doi: 10.1172/JCI118906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EP, Awad T, Palma J, Webster T, Hubbell E, Warrington JA, Spira A, Wright G, Buckley J, Triche T, et al. Guidelines: Expression profiling—best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5:229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- Houdali B, Wahl H, Kresi M, Nguyen V, Haap M, Machicao F, Ammon H, Renn W, Schleicher E, Haring H. Glucose oversupply increases Delta9-desaturase expression and its metabolites in rat skeletal muscle. Diabetologia. 2003;46:203–212. doi: 10.1007/s00125-002-1015-2. [DOI] [PubMed] [Google Scholar]
- Hulver MW, Berggren JR, Cortright RN, Dudek RW, Thompson RP, Pories WJ, MacDonald KG, Cline GW, Shulman GI, Dohm GL, Houmard JA. Skeletal muscle lipid metabolism with obesity. Am J Physiol Endocrinol Metab. 2003;284:E741–E747. doi: 10.1152/ajpendo.00514.2002. [DOI] [PubMed] [Google Scholar]
- Hulver MW, Zheng D, Tanner CJ, Houmard JA, Kraus WE, Slentz CA, Sinha MK, Pories WJ, MacDonald KG, Dohm GL. Adiponectin is not altered with exercise training despite enhanced insulin action. Am J Physiol Endocrinol Metab. 2002;283:E861–E865. doi: 10.1152/ajpendo.00150.2002. [DOI] [PubMed] [Google Scholar]
- Itani SI, Zhou Q, Pories WJ, MacDonald KG, Dohm GL. Involvement of protein kinase C in human skeletal muscle insulin resistance and obesity. Diabetes. 2000;49:1353–1358. doi: 10.2337/diabetes.49.8.1353. [DOI] [PubMed] [Google Scholar]
- Kim JK, Gavrilova O, Chen Y, Reitman ML, Shulman GI. Mechanism of insulin resistance in A-ZIP/F-1 fatless mice. J Biol Chem. 2000;275:8456–8460. doi: 10.1074/jbc.275.12.8456. [DOI] [PubMed] [Google Scholar]
- Kruszynska YT, Mukherjee R, Jow L, Dana S, Paterniti JR, Jr, Olefsky JM. Skeletal muscle peroxisome proliferator-activated receptor-gamma expression in obesity and non-insulin-dependent diabetes mellitus. J Clin Invest. 1998;101:543–548. doi: 10.1172/JCI1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenfant P, Joanisse DR, Theriault R, Goodpaster BH, Kelley DE, Simoneau JA. Fat content in individual muscle fibers of lean and obese subjects. Int J Obes Relat Metab Disord. 2001;25:1316–1321. doi: 10.1038/sj.ijo.0801733. [DOI] [PubMed] [Google Scholar]
- Millington DS, Kodo N, Norwood DL, Roe CR. Tandem mass spectrometry: a new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J Inherit Metab Dis. 1990;13:321–324. doi: 10.1007/BF01799385. [DOI] [PubMed] [Google Scholar]
- Mills S, Foster D, McGarry J. Interaction of malonyl-CoA and related compounds with mitochondria from different rat tissues. Relationship between ligand binding and inhibition of carnitine palmitoyltransferase I. Biochem J. 1983;214:83–91. doi: 10.1042/bj2140083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Kim YC, Gray-Keller MP, Attie AD, Ntambi JM. The biosynthesis of hepatic cholesterol esters and triglycerides is impaired in mice with a disruption of the gene for stearoyl-CoA desaturase 1. J Biol Chem. 2000;275:30132–30138. doi: 10.1074/jbc.M005488200. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Kim YC, Ntambi JM. A lipogenic diet in mice with a disruption of the stearoyl-CoA desaturase 1 gene reveals a stringent requirement of endogenous monounsaturated fatty acids for triglyceride synthesis. J Lipid Res. 2001;42:1018–1024. [PubMed] [Google Scholar]
- Muoio DM, MacLean PS, Lang DB, Li S, Houmard JA, Way JM, Winegar DA, Corton JC, Dohm GL, Kraus WE. Fatty acid homeostasis and induction of lipid regulatory genes in skeletal muscles of peroxisome proliferator-activated receptor (PPAR) alpha knock-out mice. Evidence for compensatory regulation by PPAR delta. J Biol Chem. 2002a;277:26089–26097. doi: 10.1074/jbc.M203997200. [DOI] [PubMed] [Google Scholar]
- Muoio DM, Way JM, Tanner CJ, Winegar DA, Kliewer SA, Houmard JA, Kraus WE, Dohm GL. Peroxisome proliferator-activated receptor-alpha regulates fatty acid utilization in primary human skeletal muscle cells. Diabetes. 2002b;51:901–909. doi: 10.2337/diabetes.51.4.901. [DOI] [PubMed] [Google Scholar]
- Nikoulina SE, Ciaraldi TP, Mudaliar S, Carter L, Johnson K, Henry RR. Inhibition of glycogen synthase kinase 3 improves insulin action and glucose metabolism in human skeletal muscle. Diabetes. 2002;51:2190–2198. doi: 10.2337/diabetes.51.7.2190. [DOI] [PubMed] [Google Scholar]
- Ntambi J. The regulation of stearoyl-CoA desaturase (SCD) Prog Lipid Res. 1995;34:139–150. doi: 10.1016/0163-7827(94)00010-j. [DOI] [PubMed] [Google Scholar]
- Ntambi JM, Buhrow SA, Kaestner KH, Christy RJ, Sibley E, Kelly TJ, Jr, Lane MD. Differentiation-induced gene expression in 3T3–L1 preadipocytes. Characterization of a differentially expressed gene encoding stearoyl-CoA desaturase. J Biol Chem. 1988;263:17291–17300. [PubMed] [Google Scholar]
- Ntambi J, Miyazaki M. Regulation of stearoyl-CoA desaturases and role in metabolism. Prog Lipid Res. 2004;43:91–104. doi: 10.1016/s0163-7827(03)00039-0. [DOI] [PubMed] [Google Scholar]
- Ntambi JM, Miyazaki M, Stoehr JP, Lan H, Kendziorski CM, Yandell BS, Song Y, Cohen P, Friedman JM, Attie AD. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc Natl Acad Sci USA. 2002;99:11482–11486. doi: 10.1073/pnas.132384699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Ciaraldi T, Abrams-Carter L, Mudaliar S, Nikoulina S, Henry R. PPAR-gamma gene expression is elevated in skeletal muscle of obese and type II diabetic subjects. Diabetes. 1997;46:1230–1234. doi: 10.2337/diab.46.7.1230. [DOI] [PubMed] [Google Scholar]
- Rahman SM, Dobrzyn A, Dobrzyn P, Lee SH, Miyazaki M, Ntambi JM. Stearoyl-CoA desaturase 1 deficiency elevates insulin-signaling components and down-regulates protein-tyrosine phosphatase 1B in muscle. Proc Natl Acad Sci USA. 2003;100:11110–11115. doi: 10.1073/pnas.1934571100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Peiffer C, Oakes ND, Browne CL, Kraegen EW, Biden TJ. Reversal of chronic alterations of skeletal muscle protein kinase C from fat-fed rats by BRL-49653. Am J Physiol. 1997;273:E915–E921. doi: 10.1152/ajpendo.1997.273.5.E915. [DOI] [PubMed] [Google Scholar]
- Singh Ahuja H, Liu S, Crombie DL, Boehm M, Leibowitz MD, Heyman RA, Depre C, Nagy L, Tontonoz P, Davies PJ. Differential effects of rexinoids and thiazolidinediones on metabolic gene expression in diabetic rodents. Mol Pharmacol. 2001;59:765–773. doi: 10.1124/mol.59.4.765. [DOI] [PubMed] [Google Scholar]
- Storlien LH, Pan DA, Kriketos AD, O’Connor J, Caterson ID, Cooney GJ, Jenkins AB, Baur LA. Skeletal muscle membrane lipids and insulin resistance. Lipids. 1996;31(Suppl):S261–S265. doi: 10.1007/BF02637087. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–68. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Way JM, Harrington WW, Brown KK, Gottschalk WK, Sundseth SS, Mansfield TA, Ramachandran RK, Willson TM, Kliewer SA. Comprehensive messenger ribonucleic acid profiling reveals that peroxisome proliferator-activated receptor {{gamma}} activation has coordinate effects on gene expression in multiple insulin-sensitive tissues. Endocrinology. 2001;142:1269–1277. doi: 10.1210/endo.142.3.8037. [DOI] [PubMed] [Google Scholar]
- Ye JM, Doyle PJ, Iglesias MA, Watson DG, Cooney GJ, Kraegen EW. Peroxisome proliferator–activated receptor (PPAR)-{alpha} activation lowers muscle lipids and improves insulin sensitivity in high fat–fed rats: comparison with PPAR-{gamma} activation. Diabetes. 2001;50:411–417. doi: 10.2337/diabetes.50.2.411. [DOI] [PubMed] [Google Scholar]
- Zierath JR, Ryder JW, Doebber T, Woods J, Wu M, Ventre J, Li Z, McCrary C, Berger J, Zhang B, Moller DE. Role of skeletal muscle in thiazolidinedione insulin sensitizer (PPAR{gamma} agonist) action. Endocrinology. 1998;139:5034–5041. doi: 10.1210/endo.139.12.6364. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.