Abstract
Platinum-based chemotherapeutics exert their therapeutic efficacy via the formation of DNA adducts which interfere with DNA replication, transcription and cell division and ultimately induce cell death. Repair and tolerance of these Pt-DNA lesions by nucleotide excision repair (NER) and homologous recombination (HR) can substantially reduce the effectiveness of therapy. Inhibition of these repair pathways, therefore, holds the potential to sensitize cancer cells to Pt treatment and increase clinical efficacy. Replication Protein A (RPA) plays essential roles in both NER and HR, along with its role in DNA replication and DNA damage checkpoint activation. Each of these functions is, in part, mediated by RPA binding to single-stranded DNA (ssDNA). Here we report the synthesis and characterization of novel derivatives of RPA small molecule inhibitors and their activity in models of epithelial ovarian cancer (EOC) and non-small cell lung cancer (NSCLC). We have synthesized analogs of our previously reported RPA inhibitor TDRL-505 and determined the structure activity relationships. These data led us to the identification of TDRL-551, which exhibited a greater than 2-fold increase in in vitro activity. TDRL-551 showed synergy with Pt in tissue culture models of EOC and in vivo efficacy, as a single agent and in combination with platinum, in a NSCLC xenograft model. These data demonstrate the utility of RPA inhibition in EOC and NSCLC and the potential in developing novel anticancer therapeutics that target RPA-DNA interactions.
1. Introduction
Platinum (Pt)-based combination chemotherapy has been the front-line treatment for a variety of malignancies including testicular, lung, and ovarian cancer [1]. However, resistance to Pt-based regimens remains a major limitation in the successful treatment for many of these cancers including epithelial ovarian cancer (EOC) and non-small cell lung cancer (NSCLC) [2;3]. More than 80% of EOC patients relapse with Pt-resistant disease, where second line therapies are largely ineffective. Thus, ovarian cancer has been clinically designated as the most deadly gynecological cancer owing to extremely poor prognosis and overall low survival rates [4]. The clinical efficacy of cisplatin is a function of its ability to cross-link DNA thereby blocking DNA replication, transcription and cell division. Ultimately Pt- treatment induces apoptosis [5;6], however, the balance between DNA damage and DNA repair dictates the extent of tumor death. While Pt-resistance is multifactorial, increased DNA repair is a major contributor [7]. Hence, exploiting DNA repair as a target to sensitize cells to Pt-based chemotherapy holds immense potential for increasing the survival rates in cancer therapy.
Repair and tolerance of cisplatin-DNA adducts occur primarily via nucleotide excision repair (NER) and homologous recombination (HR) [4;8;9]. Approximately 95% of Pt-DNA lesions formed by cisplatin are intrastrand crosslinks with the remaining ~5% being interstrand crosslinks and a small number of mono-lesions [10]. There is evidence for and against each lesion type being the cytotoxic lesion caused by cisplatin. Interstrand lesions are less abundant and repaired more efficiently than intrastrand lesions [11;12], and involve the HR pathway in conjunction with the FANC protein complex (a group of proteins associated with Fanconi anemia)[13]. Interstand adducts are more cytotoxic with estimates to as few as 20 interstrand crosslinks causing cell death if left unrepaired [14]. While more abundant and repaired slower [15;16], intrastrand lesions are better tolerated via HR and bypass polymerases [17]. Repair of intrastrand crosslinks occurs via the NER pathway [4] Therefore, while the exact lesion responsible for clinical efficacy remains to be determined, what is clear is that both NER and HR have differential and contributory roles in the cellular sensitivity to cisplatin.
Replication protein A (RPA) is the major human ssDNA binding protein and is required for both NER and HR [18]. The RPA heterotrimer consists of 70 kDa, 32 kDa and 14 kDa subunits with the 70-kDa subunit containing the two major high affinity DNA binding domains (DBD) DBD A and B, as well as DBD C and F. DBD D and E are in the 32-kDa and 14-kDa subunit, respectively. Binding to short stretches of ssDNA (~ 8–10 nucleotides) is primarily mediated by DBD A and B, while intermediate length ssDNA (~ 12–23 nucleotides) also involves DBD C. Longer length ssDNA (~ 28–30 nucleotides) engages DBD D in addition to DBDs A, B and C [19–21]. RPA plays essential and non-redundant roles in both NER and HR, apart from its role in replication and DNA damage checkpoint activation [18]. Each of these roles requires binding of RPA to ssDNA, making RPA-DNA interaction a promising target for anti-cancer therapeutic activity in combination with cisplatin.
Structural analysis of RPA reveals unique protein-DNA interactions that would facilitate the design of potent and selective small molecule inhibitors (SMIs) [22]. It has been also shown that genetic mutants of RPA display defects in DNA repair without impacting DNA replication and vice versa [18;23;24]. This separation of function can be exploited by using chemical probes that exclusively interfere with the DNA repair pathway and that, in conjunction with DNA-damaging agents, would offer a new possibility for cancer treatment. Our group has previously reported both reversible and irreversible chemical inhibitors of RPA [25–28]. The reversible inhibitor TDRL-505 exhibits synergistic effects with DNA damaging agents in a lung cancer cell model. This small molecule hinders the binding of DBD A and B of RPA to ssDNA, which according to in silico docking analysis occurs as a consequence of its interaction with DBD B and the DBD A-B interdomain [27]. In the present study we screened a series of analogs TDRL-505 in vitro and evaluated their activity in an EOC cell culture model. Structure activity relationship (SAR) data led us to an enhanced lead compound, TDRL-551. Herein we report the in vitro, cellular and in vivo activity of the RPA inhibitor TDRL-551 in models of lung and ovarian cancer.
2. Materials and Methods
2.1 Protein Purification
Full length, heterotrimeric human RPA (fl-RPA) was expressed in E. coli and purified by a three column procedure as previously described [29]. The DBD-A/B construct was expressed as a SUMO-His6-RPA181–432 fusion protein. E. coli BL21 (DE3) cells in log growth were induced for 3 hours with 0.5mM IPTG at 37°C. The cells were lysed in buffer containing 50mM Tris pH 7.5, 300 mM NaCl, 10% sucrose, 10mM imidazole, 25 μg/ml lysozyme, 1 μg/ml leupeptin, 1 μg/ml pepstatin and 0.5 mM PMSF. The lysate was loaded onto a Ni-NTA column washed and then incubated overnight with wash buffer containing 3 μg/ml ULP1 protease to cleave the SUMO tag. The cleaved His6-RPA181–432 was eluted from the Ni-NTA column with elution buffer containing 350mM imidazole. The His6-RPA was then further purified on a size exclusion column (SEC) to remove the cleaved SUMO tag fragment. The SEC pool was then concentrated and stored at -80°C.
2.2 Electrophoretic Mobility Shift Assays (EMSA)
EMSA reactions (20 μL) were performed with 50 nM fl-RPA and 2.5 nM 5’[32P]-labeled 34-base DNA in buffer containing 20mM HEPES (pH 7.0), 1mM DTT, 0.001% NP-40, 100 mM NaCl, 5 mM MgCl2 and 50 μg/ml bovine serum albumin (BSA). Chemical compounds, either purchased from ChemDiv or synthesized in our laboratory, were suspended in DMSO and titrated as detailed in each figure. The DMSO concentration in the reaction mixture was kept constant at or below5%. RPA was incubated with inhibitor or DMSO control in reaction buffer for 30 minutes before the addition of DNA. Reactions were incubated for 5 minutes at room temperature and products separated via 6% native polyacrylamide gel electrophoresis. The bound and unbound fractions were then quantified by phosphor-imager analysis using ImageQuant software (Molecular Dynamics, CA) and IC50 values calculated by non-linear regression using SigmPlot (Sysat). For EMSA reactions with RPA DBD-A/B, 150 nM DBD-A/B was used and electrophoresis was performed at 4° C. All other conditions were identical to those described for the full length RPA.
2.3 Chemical Synthesis
2.3.1 General
All solvents and chemicals were used as purchased from commercial suppliers. 1H NMR spectra were obtained on a Bruker Avance III 500 MHz NMR spectrometer. Chemical shifts are expressed in parts per million (ppm, δ), relative to tetramethylsilane (TMS) as internal reference. Signals are described as s (singlet), d (doublet), dd (doublet of doublets), dt (doubles of triplets), t (triplet), q (quartet), or p (pentet).
2.3.2 2-chloro-7-ethoxy-3-(3-(4-iodophenyl)-4,5-dihydro-1H-pyrazol-5-yl)quinoline (7a)
NaOH (0.83 mL, 2.5 M in water, 2.07 mmol) was added dropwise to a solution of 4-iodoacetophenone (0.36 g, 1.47 mmol) and 2-chloro-7-ethoxyquinoline-3-carbaldehyde (0.35 g, 1.47 mmol) in EtOH (12 mL). After stirring for a 45 min at 40 °C, the reaction mixture was quenched with HCl (1.38 mL, 3 M). The crude mixture containing the resulting enone was then filtered, thoroughly washed with EtOH, and used in the next step without further purification. Hydrazine monohydrate (0.36 mL, 7.33 mmol) was added dropwise to a suspension of the enone obtained in the previous step in EtOH (30 mL). The mixture was refluxed for 1.5 h with stirring, after which it was allowed to cool to room temperature. The obtained solid was filtered and washed with EtOH. Further purification by trituration with EtOH furnished the title compound as an off-white solid (0.57 g, 81% over 2 steps). 1H NMR (500 MHz, DMSO-d6) δ 1.41 (t, J=7.0 Hz, 3H), 2.89 (dd, J=16.5, 10.0 Hz, 1H), 3.67 (dd, J=16.5, 11.0 Hz, 1H), 4.20 (q, J=7.0 Hz, 2H), 5.19 (dt, J=10.5, 3.5 Hz, 1H), 7.27 (dd, J=9.0, 2.5 Hz, 1H), 7.34 (d, J=2.5 Hz, 1H), 7.44 (d, J=8.5 Hz, 2H), 7.74 (d, J=8.5 Hz, 2H), 7.84 (d, J=3.5 Hz, 1H), 7.97 (d, J=9.0 Hz, 1H), 8.42 (s, 1H).
2.3.3 4-(5-(2-chloro-7-ethoxyquinolin-3-yl)-3-(4-iodophenyl)-4,5-dihydro-1H-pyrazol-1-yl)-4-oxobutanoic acid (9a or TDRL-551)
A round-bottom flask coupled with a reflux condenser and containing a dry mixture of 2-chloro-7-ethoxy-3-(3-(4-iodophenyl)-4,5-dihydro-1H-pyrazol-5-yl)quinoline (7a) (0.6 g, 1.25 mmol) and glutaric anhydride (0.14 g, 1.25 mmol) was immersed into a preheated oil bath (65 °C). CHCl3 (24 mL) was then added through the condenser in one portion. The resulting solution was refluxed for 1.5 h with stirring, after which it was allowed to cool to room temperature. The obtained solid was filtered and washed with ethyl acetate. Further purification by trituration with ethyl acetate yielded acid 9a as an off-white solid (0.53 g, 72%). 1H NMR (500 MHz, DMSO-d6) δ 41.40 (t, J=7.0 Hz, 3H), 1.83 (p, J=7.5 Hz, 2H), 2.30 (t, J=7.5 Hz, 2H), 2.82 (dt, J=15.0, 7.5 Hz, 1H), 2.91 (dt, J=15.0, 7.5 Hz, 1H), 3.28 (dd, J=18.0, 5.5 Hz, 1H), 3.97 (dd, J=18.0, 12.0 Hz, 1H), 4.19 (q, J=7.0 Hz, 2H), 5.83 (dd, J=12.0, 5.5 Hz, 1H), 7.26 (dd, J=9.0, 2.5 Hz, 1H), 7.35 (d, J=2.5 Hz, 1H), 7.57 (d, J=8.5 Hz, 2H), 7.84 (d, J=8.5 Hz, 2H), 7.93 (d, J=9.0 Hz, 1H), 7.99 (s, 1H), 12.09 (s, 1H).
2.4 Cell Culture
A2780 cells and A2780/R cells were purchased from Sigma. All other cell lines were from ATCC and routinely tested for mycoplasma contamination. Cells were maintained in RPMI media supplemented with 10% FBS (Atlanta Biological), penicillin and streptomycin. Cultures were incubated at 37 °C in 5% CO2 and sub-cultured 2–3 times per week.
2.4.1 Clonogenic survival assays
Cells were plated in a 6 well (100,000 cells/well) or 24 well (25,000 cells/well) plate, incubated for at least 18 hours and then treated with Pt/etoposide and/or RPA inhibitors. After 48 hours of treatment, the cells were re-plated in 10 cm dishes (500–1000 cells/dish) and incubated for 8–10 days to allow colony formation. Plates were then washed with PBS, fixed with glutaraldehyde (Fisher Scientific) and stained with crystal violet (Acros Organics). The colonies were then counted using an Acolyte Synbiosis colony counter, viability determined versus vehicle controls which were plotted versus drug concentration.
2.4.2 Assessment of Synergy via Combination Index
In the combination index studies, the A2780 cells were treated with RPA inhibitor and Pt/etoposide alone as well as the combination of both – the inhibitor and the DNA damaging chemotherapeutic agent. The range of treatment was dependent on the IC50 of each inhibitor/drug. If the IC50 was X, then the cells were treated at a range of X/4 to 3X concentration in a colony formation assay. The kill curves from both the single agent treatments as well as the combination treatment were used in a Chou-Talalay based method to determine the combination index (CI) at different fractions of cells affected [30;31]. A CI > 1 indicates antagonism between the two agents, while a CI < 1 indicates synergy. A CI of 1 demonstrates an additive effect.
2.5 Compound-DNA binding assay
A competitive DNA intercalation assay was performed using SYBR-Green (Sigma) and salmon sperm DNA (Fisher). Reactions were carried out in 25 mM MOPS (pH 6.5) containing 30 μM sonicated salmon sperm DNA, SYBR-Green and varying concentrations of RPA inhibitors. Reactions were performed in a black 96-well plate in a final volume of 110 μl. Doxorubicin, a known non-covalent DNA binding chemotherapeutic, was used as a positive control. Fluorescence was measured using a BioTek® Synergy™ H1 hybrid multi-mode microplate reader with an excitation wavelength of 485 nm, emission wavelength of 528 nm and a read height of 7 mm. Data were collected using BioTek® Gen5™ reader software. Reactions were incubated a maximum of 5 minutes before measurements were collected.
2.6 In vivo Analysis of TDRL-551
Non-obese diabetic/severe combined immunodeficient mice (NOD/SCID) were obtained from The Jackson Laboratory (http://www.jax.org). All animal studies were conducted under the guidelines of the NIH and were approved by the Institutional Animal Care and Use Committee of Indiana University School of Medicine. Animals were maintained under pathogen-free conditions and a 12-hour light-dark cycle. The safety and tolerability of TDRL-551 was assessed in naïve NOD/SCID mice. Mice were treated IP with increasing concentrations of TDRL-551 in a formulation consisting of 20% DMSO, 10% Tween 80, 70% PBS. Based on preliminary PK analysis and a half-life of ~7 hours (data not shown) we administered 3 doses per week for two weeks and measured body weight every other day.
To assess anti-cancer efficacy the hind flanks of sixty 8–10 week old mice were implanted with 2 × 106 H460 NSCLC cells in matrigel. Tumor volumes were monitored by caliper measurement [tumor volumes = length × (perpendicular width)2 × 0.5]. Mice with tumors ranging between 32–152.5mm3 8 days following implantation were randomized into 4 treatment arms. Carboplatin was dissolved in water and administered via intraperitoneal injection at 50 mg/kg on days 8, 14, and 20 following implantation. TDRL-551 was suspended in 20% DMSO, 10% Tween 80, 70% PBS and administered via intraperitoneal injection at 200 mg/kg biweekly on days 8, 10, 14, 17, and 20. Vehicle controls were administered to arms not receiving indicated treatments. Tumor volumes were monitored biweekly as indicated and the results are presented as the average tumor volume ± standard error of the mean for each group (n=14 per group).
3. Results
3.1. Screening TDRL-505 analogs
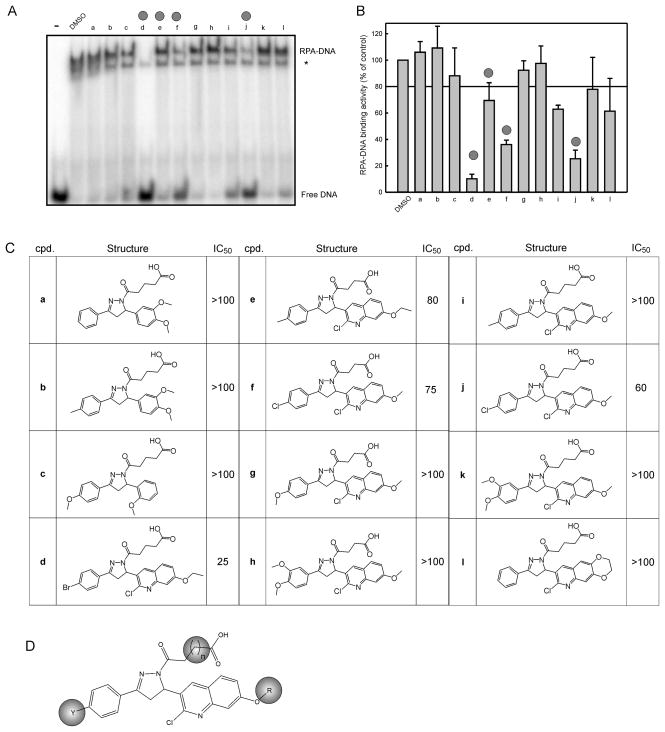
Toward improving the potency and physiochemical properties of TDRL-505, we screened 26 analogous compounds to assess their ability to inhibit RPA-DNA binding activity. These 26 compounds shared the same 2-pyrazoline core structure as TDRL-505, but differed in either the type/length of the side chain attached to N1, the substitution of the phenyl group at C3, or the type of aromatic ring at C5. Data from a representative of 12 compounds are shown in Figure 1A along with quantification of the data in Figure 1B. Each of the analogs that displayed inhibitory activity towards RPA was titrated over a range of concentrations to determine IC50 values (Figure 1C). These data were used to determine SAR. We identified three important aspects of the molecules: the length of the carboxylic acid chain, the halogen on the phenyl ring, and the alkyl ether in the quinoline ring (Figure 1D). Consequently, we pursued an organic synthesis scheme to prepare additional TDRL-505 analogs and further interrogate the structure activity relationships.
Figure 1. EMSA screening of TDRL-505 analogs.
A) 12 TDRL-505 analogs were screened using the EMSA assay for RPA-DNA inhibition activity at 100 μM. The free DNA and RPA-DNA complexes are indicated. The asterisk indicated the position of the E. coli SSB-DNA complex. B) Quantification of duplicate determinations were averaged and plotted as a percent of control with bars representing the range of values C) Structures of compounds with their corresponding IC50 values (μM) calculated from EMSA reactions as described in Methods. Compounds were titrated at a range of 1–125 μM. D) SAR identified areas of interest and they are highlighted by the colored ovals.
3.2. Chemistry
The synthetic approach developed for the preparation of TDRL-505 analogs is depicted in Figure 2 and involved 5 steps. Quinolines carbaldehydes 4 were prepared by acylation of alkoxyanilines 2 with acetic anhydride 1, followed by Vilsmeier-Haack formylation [Herbert]. Aldol condensation/dehydration with a corresponding methyl ketone 5 and sodium hydroxide yielded enones 6, which, upon treatment with hydrazine, afforded 2-pyrazolines 7. Lastly, acylation at N1 of the pyrazoline core with a cyclic anhydride 8 furnished oxoacids 9. The list of all the synthesized TDRL-505 analogs is shown in Table1.
Figure 2. Synthesis of TDRL-505 analogs.
Reagents and conditions: (a) DIEA, DMAP, DCM, rt, 2 h, 90–100%. (b) (i) DMF, POCl3, 0 °C, 20 min, (ii) Amide, 110 °C, 2.5 h, 44–64%. (c) NaOH 10%, EtOH, 45 °C, 45 min. (d) H2N-NH2.H2O, EtOH, reflux, 1.5 h, 73–81% (over 2 steps). (e) CHCl3, reflux 1.5 h, 40–72%.
Table 1. In vitro and cellular IC50 values of TDRL-505 and its analogs.
The in vitro IC50 values are based on EMSA reactions as described in Methods. RPA was incubated with the above compounds at concentration range of 1–125 μM. The cellular IC50 values are calculated from clonogenic survival assays as described in Methods. The cells were treated with the above compounds at a concentration range of 1–200 μM. Only four compounds were tested for cellular activity, the rest are indicated with ‘na’ meaning data not available since the experiments were not performed.
| ENTRY | COMPOUND | Y | R | n | In vitro IC50 (μM) | Cellular IC50 (μM) |
|---|---|---|---|---|---|---|
| 1 | TDRL-505 | p-Br | Et | 1 | 38 | >50 |
| 2 | TDRL-533 | p-Br | Me | 1 | >50 | >50 |
| 3 | TDRL-540 | p-F | Me | 1 | >100 | na |
| 4 | TDRL-543 | p-Br | Et | 2 | 25 | 50 |
| 5 | TDRL-539 | p-F | Me | 2 | >100 | na |
| 6 | TDRL-534 | p-Br | Me | 2 | 35 | na |
| 7 | TDRL-556 | p-Br | iPr | 2 | 43 | na |
| 8 | TDRL-551 | p-I | Et | 2 | 18 | 25 |
| 9 | TDRL-557 | p-F3CO | Et | 2 | 30 | na |
| 10 | TDRL-652 | m-I | Et | 2 | 15 | na |
| 11 | TDRL-617 | See below | >100 | na | ||
| 12 | TDRL-634 | See below | >100 | na | ||
|
| ||||||
|
| ||||||
3.3. In vitro inhibition of RPA’s DNA binding activity
The in vitro inhibition of RPA’s DNA binding activity was determined by titrating all synthesized TDRL-505 analogs over a range of concentrations from 0–125 μM in an EMSA based assay (Table 1). We observed a slight increased potency by addition of a methylene group to the oxocarboxylic acid moiety (entries 1 and 4, and 2 and 6). Additionally, we found a correlation between the identity of the halogen atom on the phenyl ring and the effectiveness of the compound. Iodine imparted the best inhibitory activity, followed by bromine, chlorine (data not shown) and fluorine, in that order (entries 2 and 3, 4 and 8, and 5 and 6). The pattern of halo-substitution on the phenyl ring was also evaluated. Since the meta-iodo isomer did not exhibit any effect over its para analog (entries 8 and 10), we pursued the latter due to its simpler purification process. Lastly, a fluorinated substituent, the trifluoromethoxy group, did not alter the potency of the compound when compared to the parental bromo substitution (entries 4 and 9). Another part of the molecule that was subject of analysis was the alkyl ether moiety on the quinoline ring. The replacement of the ethyl group by either a methyl or isopropyl counterpart resulted in a slight decrease in inhibitory activity (entries 1 and 2, and 4, 6 and 7). Of all analogs tested, TDRL-551 (entry 8) exhibited the highest in vitro, as well as cellular, activity.
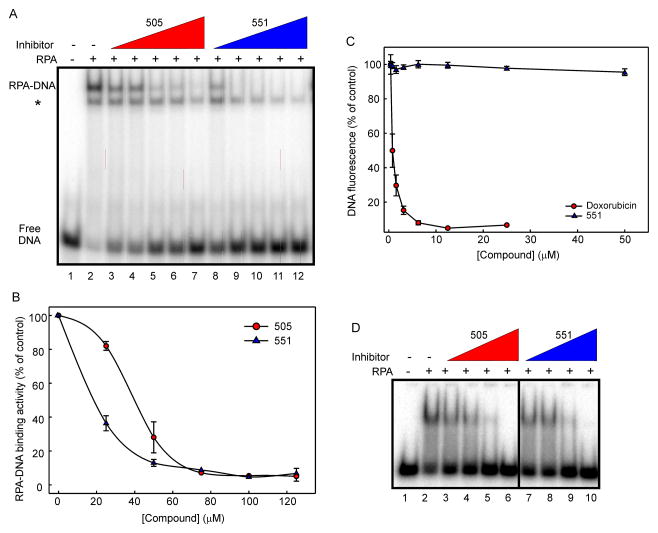
To further interrogate the most potent compound TDRL-551, we directly compared its activity to that of the parent compound, TDRL-505. The data presented in Figure 3A compare the EMSA based in vitro inhibitory activity of TDRL-551 with that of TDRL-505. The IC50 values, calculated form the plotted graphs (Figure 3B), were found to be 18 and 38 μM, respectively, making TDRL-551 greater than twice as potent than its predecessor. The two potential mechanisms for inhibition are either a direct interaction with the protein or an interaction with the DNA that renders it unable to bind to the protein. Our previous in silico docking analyses suggested that the 505 class of compounds inhibits DNA binding activity via a direct interaction with the protein RPA [27]. To confirm the mechanism of inhibition for 551 was via an interaction with RPA and not DNA we assessed the ability of TDRL-551 to bind to DNA using a fluorescence displacement assay. The results presented in Figure 3C demonstrate that no DNA binding activity was observed for 551 and confirm that the compound inhibits the protein-DNA interaction by binding directly to the RPA protein and not via binding to the DNA. To further delve into the mechanism of binding, we tested whether TDRL-551 could inhibit RPA DBD-A/B binding (the major high affinity DNA binding core) to DNA. Both TDRL-551 and TDRL-505 inhibit RPA DBD-A/B-DNA interaction and hence employ a similar inhibition mechanism (Figure 3D).
Figure 3. In vitro analysis of TDRL-551 inhibitory activity.
A) RPA was incubated with compounds TDRL-505 and TDRL-551 ranging from 1–125 μM. DNA binding was analyzed by EMSA as described in Methods. The position of free DNA and the DNA–RPA complex is denoted in the figure. The asterisk indicates the position of the E. coli SSB-DNA complex. B) Quantification of the data presented in panel A. Data represent the average and SD of three independent determinations and the data were fitted using nonlinear regression analysis (Sigmaplot) to obtain IC50 values. C) Fluorescent displacement assays were performed as described and the indicated concentration of 551 or doxorubicin was titrated into reactions containing DNA and SybrGreen. The fluorescence was measured and data represent the average and SD of three independent determinations. D) RPA-A/B Box was incubated with compounds TDRL-505 and TDRL-551 ranging from 25–100 μM. DNA binding was analyzed by EMSA as described in Methods. The position of free DNA and the DNA–RPA A/B complex is denoted in the figure.
3.4. Cellular activity of TDRL-551
3.4.1. Single agent anti-cancer activity of RPA inhibitors in EOC cell line
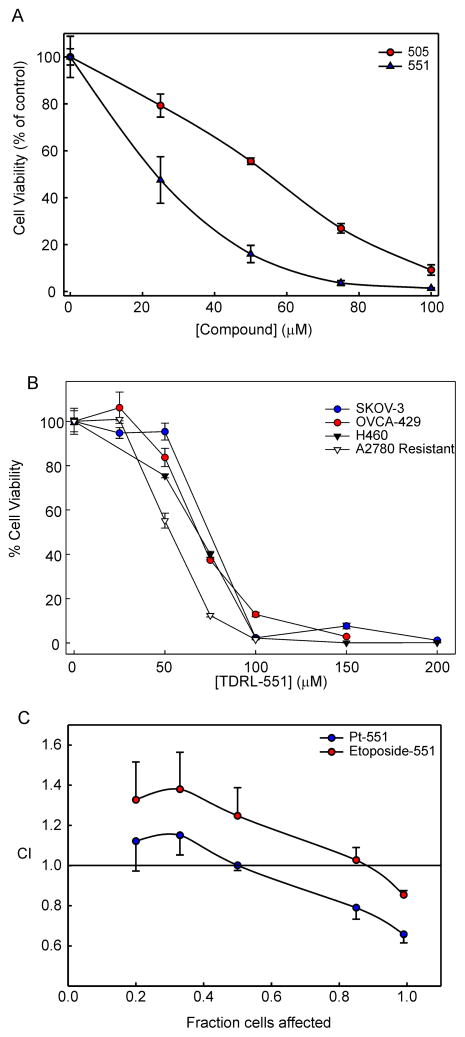
Considering the essential role of RPA in S-phase DNA replication [32] and our previous data with both reversible and irreversible RPA inhibitors [25;27], we evaluated the TDRL-505 analogs for single agent anti-cancer activity in the A2780 EOC cell line using clonogenic survival assays (Table 1). Consistent with our in vitro EMSA based studies, TDRL-551 showed the best single agent activity in these cells. Also in line with the in vitro results was the relative cellular inhibitory activity of all tested compounds. Figure 4A shows the data obtained from clonogenic survival assays, comparing the single agent activity of our original lead TDRL-505 and our optimized lead, TDRL-551. TDRL-505’s IC50 was found to be 55 μM, while TDRL-551 was twice as potent with an IC50 of 25μM. Interestingly, the degree of improvement in potency remained consistent between the in vitro and cellular assays. To ensure that the activity was not cell line specific, we also assessed the single agent activity of TDRL-551 in three other EOC cell lines, SKOV3 and OVCA429 (ATCC) both of which were isolated from patients with recurrent ovarian cancer following platinum therapy and the cisplatin resistant A2780 derivative [33]. We also assessed activity in the H460 NSCLC cell line. In each case, TDRL-551 displayed single agent activity similar to that observed in the parental A2780 EOC cells (Figure 4B) demonstrating that, as would be expected for an RPA inhibitor, the mode of activity is not restricted to a single cell line or cancer type.
Figure 4. Cellular activity of TDRL-551.
A) A2780 cells were treated with RPA inhibitor TDRL-505 or TDRL-551 for 48 hours and viability was assessed in a colony formation assay as described in Methods. The colonies were counted and normalized to the untreated controls to determine cellular viability. The data represent the average and SEM from three independent determinations and the data were fit using non- linear regression analysis (Sigmaplot) to calculate cellular IC50s. B) Analysis of TDRL551 single agent activity in H460 NSCLC, SKOV3, A2780/R and OVCA429 EOC cells as described in panel A. C) The CI of TDRL-551 with Pt and Etoposide were determined through a Chou-Talalay based approach as described in the Methods. The data represent the average and SEM from three independent determinations
3.4.2. Synergy with DNA damaging chemotherapeutic agents in EOC
Since repair and tolerance of Pt-DNA lesions predominantly occur via NER and HR, cellular inhibition of RPA should have a dramatic effect on the sensitization of cancer cells to Pt. In order to determine whether inhibition of RPA with TDRL-551 synergizes with Pt in EOC cells, we performed combination treatment studies with TDRL-551 and cisplatin. We used the platinum sensitive A2780 cell line as our cell culture model for EOC. Figure 4C shows an average of three biological replicate experiments for combination studies of TDRL-551 with Pt in A2780 cell line with appropriate single agent controls. Our data show a synergistic effect indicated by a CI < 1 at 0.5 or higher fraction of cells affected. The data obtained are consistent with our hypothesis that RPA inhibition makes cancer cells more sensitive towards Pt and hence acts synergistically with cisplatin treatment. Since RPA also plays a crucial role in replication, we also tested TDRL-551 in combination with etoposide, a topoisomerase II inhibitor. TDRL-551 was mildly synergistic with etoposide at the highest fraction of cells affected (> 0.8) (Figure 4C).
3.5. RPA Inhibitor TDRL-551 displays single agent anti-cancer activity and sensitizes NSCLC tumors to platinum based treatment in vivo.
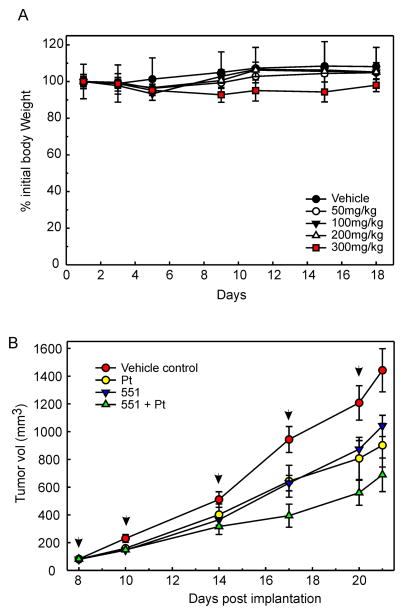
To determine the effect of RPA inhibition via TDRL-551 treatment in vivo we first assessed tolerability and experiments demonstrated a good safety profile with no weight loss observed with intraperitoneal administration up to 200 mg/kg (Figure 5A). A slight decrease in body weight was observed at 300 mg/kg, but still did not reach greater than 10% loss of weight. Co-treatment with carboplatin was also assessed and again, no adverse effects or loss of weight was observed up to 200 mg/kg (data not shown). We then determined anti-cancer activity in H460 NSCLC xenografts. Initial pharmacokinetic analysis revealed that ability to achieve a plasma concentration of >20 μM with a half-life of over 7 hours (data not shown). Tumor cells were therefore implanted in NOD/SCID mice that were randomized and treated with vehicle, TDRL-551; carboplatin; or the combination of TDRL-551 and carboplatin (Figure 5B). Carboplatin is often used in the treatment of NSCLC and forms DNA adducts chemically indistinguishable from those forms with cisplatin. As a result of the similarity in the DNA adducts formed between carboplatin and cisplatin, the repair pathways that impact sensitivity are identical. Tumor volumes were monitored for 2 weeks following initiation of treatment regimens and averages for each treatment arm are reported. Each of the treatment arms is clearly distinct from the untreated control group. Carboplatin treatment and TDRL-551 displayed similar growth inhibition of tumors. This demonstrates single agent anti-cancer activity of TDRL-551 in vivo that is consistent with the cellular assays reported above. Interestingly, mice receiving carboplatin and TDRL-551 demonstrated the slowest tumor growth, consistent with TDRL-551 sensitizing cells to platinum. These data provide strong evidence that TDRL-551 can be used to sensitize NSCLC tumors to Pt-based therapy.
Figure 5. In vivo analysis of TDRL-551.
A) Acute toxicity and tolerability of TDRL-551 was assessed via body weight determinations following triweekly dosing at the indicated drug concentrations. Mice were treated on days 1, 3, 5, 8, 10, and 12 IP as described in Methods. Data are reported as the percent of body weight on day 1 and represent the mean ± standard error of the mean (n=3). B) In vivo anti-cancer activity was assessed in human H460 NSCLC tumor xenografts in NOD/SCID mice. Mice were implanted on day 1, tumor measured by calipers and individual mice randomly assigned to one of 4 treatment arms. Carboplatin was administered once per week on days 8, 14 and 20. TDRL 551 was administered biweekly on days 8, 10, 14, 17, and 20. Tumor volumes were monitored by caliper measurement [tumor volume = length (perpendicular width)2 × 0.5] biweekly as indicated. Average tumor volume ± standard error of the mean for each group is reported in mm3 (n=14).
4. Discussion
The research presented in this article describes the synthesis, structure activity relationships and in vitro and cellular activity of novel reversible RPA inhibitors in EOC and NSCLC. We have demonstrated both single agent activity and synergy in combination with DNA damaging chemotherapeutic agents; cisplatin and etoposide. In vivo data demonstrate no overt toxicity and good clinical efficacy in combination with carboplatin in a NSCLC xenograft model. This represents the first in vivo deployment of a small molecule inhibitor targeting the RPA-DNA interaction. The SARs defined the necessary substituents for activity while maintaining excellent bioavailability. These data demonstrate that to achieve in vivo activity a balance between potency and bioavailability can lean towards lower affinity as long as PK parameters allow clinically effective concentrations to be maintained. This balance is especially important in targeting RPA an essential protein with homozygous mutations being embryonically lethal in mice, while heterozygous mutants having an early predisposition to cancer [34]. No loss of function mutation for RPA has been reported in humans and genetic knockdown of RPA affects cellular viability [32]. Consequently, targeting RPA could have potential negative effects on rapidly dividing healthy cells, such as gut epithelial, hematopoietic, or hair follicle, and it could lead to unwanted side effects. For this reason, exploiting the separation of function phenomena in RPA in a manner amenable for therapeutic intervention is crucial. Our laboratory has previously published minimal cytotoxic effects on peripheral blood lymphocytes with the TDRL-505 inhibitor [27]. Moreover, our preliminary mouse toxicity studies indicate no significant overall change in body weight for doses up to 200 mg/kg, but show anti-tumor activity at the same dosage in a lung cancer xenograft model. We hypothesize that since cancer cells are undergoing an abnormal unregulated rate of proliferation, they are in a state of replicative stress and their dependence on RPA can be used to obtain a therapeutic window without harming the normal cells. This can also be understood by analogy to an oncogene addiction model, in which cancer cells have a higher dependence on the oncogene compared to normal cells and hence can be selectively targeted. Finally, RPA’s overexpression has been correlated with multiple cancers like breast, lung and colon, and it has also been associated with metastasis [35–38]. Thus, clinical reports of altered RPA expression in a variety of cancers make RPA a promising novel therapeutic target.
It is also important to elucidate whether our inhibitors exclusively impair the repair function of RPA without compromising its role in replication. Our previously published data with inhibitor TDRL-505 demonstrate a G1 cell cycle arrest, however the cells that are already in S-phase progress through the replication phase [27]. This indicates that our inhibitors are either blocking the initial phase of replication initiation or early origin firing and inhibiting the transition from G1 to S phase, or causing an alteration in the DNA damage checkpoint signaling. In either case, the 505 class of inhibitors do not block progression through S-phase once the G1-S transition has occurred. We also have demonstrated that 505 and 551 have a similar mechanism and target the DBD-A/B domain of RPA. Interestingly, we did not observe a significant difference in the inhibition potency of TDRL-505 and TDRL-551 for RPA DBD-A/B. We speculate the improved potency of TDRL-551 for full length RPA could be due to its binding at other sites in RPA and thus the overall potency of the molecule could be a result of multiple binding sites.
Our current results demonstrating synergy of TDRL-551 with Pt in an EOC cell line, along with our published data showing synergy of TDRL-505 with Pt in a lung cancer cell line indicates that our RPA inhibitors are impairing the repair function of RPA. The major limitation for successful treatment of a variety of cancers, including EOC, has been the tolerance and repair of Pt-DNA adducts and has been specifically correlated to increased repair in a variety of ovarian cancer cell lines [12]. Hence, inhibiting DNA repair by targeting RPA could have a major significance for cancer therapy.
We also examined the ability of TDRL-551 to synergize with etoposide, a topoisomerase II inhibitor. Etoposide treatment leads to both single and double stranded DNA breaks as well as stalling and collapse of replication forks [39;40]. Inhibiting RPA’s replication function could further enhance the number of DNA breaks produced on etoposide treatment and improve the effectiveness of the treatment. Although TDRL-505 has been previously shown to be highly synergistic with etoposide in lung cancer cells through a flow cytometry based Annexin-PI staining assay [27], its optimized analog, TDRL-551, showed modest synergy with etoposide at the highest fractions of cells affected (> 0.8) in A2780 EOC cell line through colony formation assay. Compound TDRL-551 may be more specific in targeting the repair function of RPA than its predecessor TDRL-505 and hence doesn’t significantly impact replication and only mildly synergizes with etoposide. Alternatively, the differences may be a function of the cell lines used and cancer types being investigated. While cisplatin and etoposide are the standard drugs used in treatment against lung cancer, the therapy for ovarian cancer involves the combination of platinum and taxol. Etoposide is not a first line therapy for EOC and hence improving its effectiveness may be limited based on the cancer. Lastly, we cannot rule out that the differing synergy outcomes are a consequence of the type of assay performed in each case. Despite these caveats, the important finding is that RPA inhibition with TDRL-551 synergizes with cisplatin in EOC and may provide an avenue to increase sensitivity to platinum in the clinic.
RPA inhibitors can be used as DNA repair inhibitors to overcome resistance to platinum based chemotherapies. Inhibiting DNA repair with SMIs can be used in combination with Pt both at first line and second line stage of therapy. In first line therapy, Pt in combination with RPA inhibitors could lead to maximum effectiveness by killing the majority of cancer cells, which are now sensitized to Pt due to inhibition of DNA repair. This may certainly still lead to some surviving cancer cells that are resistant to the treatment due to other mechanisms, such as reduced uptake of platinum, increased drug efflux, or increased expression of proteins like glutathione that bind and inactivate Pt in the cells. However, since there will be less number of surviving cancer cells to relapse with platinum resistant forms, any increase in the effectiveness of the first line therapy would lead to an improved progression free survival (PFS), which can be clinically significant. As second line therapy, RPA inhibitors can be used in combination with Pt to re-sensitize the platinum resistant cancers, also leading to an increase in PFS. It is important to mention that platinum based chemotherapy is not the only scope for the utility of RPA inhibitors. Since RPA plays a variety of roles in different pathways, its other functions can also be targeted. For instance, RPA inhibitors can be used in combination with radiation therapy that induces double stranded breaks (DSB). Thus, inhibiting RPAs role in HR-dependent DSB repair would be expected to enhance the effectiveness of radiotherapy. In conclusion, RPA inhibitors can be used in a multitude of platforms with a special focus in the area of cancer therapy.
Acknowledgments
We thank the Indiana University in vivo Therapeutics Core facility for assistance with the in vivo studies, Jennifer Earley for the cloning, expression and purification of the RPA DBD-A/B construct and all the member of the lab for their input. This work was supported by NIH grants R01CA180710 and R41CA162648
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–48. doi: 10.1016/j.canlet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 2.Ozols RF, Bookman MA, Connolly DC, Daly MB, Godwin AK, Schilder RJ, Xu XX, Hamilton TC. Focus on epithelial ovarian cancer. Cancer Cell. 2004;5:19–24. doi: 10.1016/s1535-6108(04)00002-9. [DOI] [PubMed] [Google Scholar]
- 3.Einhorn LH. First-line chemotherapy for non-small-cell lung cancer: is there a superior regimen based on histology? J Clin Oncol. 2008;26:3485–6. doi: 10.1200/JCO.2008.17.2056. [DOI] [PubMed] [Google Scholar]
- 4.Saldivar JS, Wu X, Follen M, Gershenson D. Nucleotide excision repair pathway review I: implications in ovarian cancer and platinum sensitivity. Gynecol Oncol. 2007;107:S56–S71. doi: 10.1016/j.ygyno.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 5.Henkels KM, Turchi JJ. Cisplatin-induced apoptosis proceeds by caspase-3-dependent and -independent pathways in cisplatin-resistant and -sensitive human ovarian cancer cell lines. Cancer Res. 1999;59:3077–83. [PubMed] [Google Scholar]
- 6.Henkels KM, Turchi JJ. Induction of apoptosis in cisplatin-sensitive and -resistant human ovarian cancer cell lines. Cancer Res. 1997;57:4488–92. [PubMed] [Google Scholar]
- 7.Martin LP, Hamilton TC, Schilder RJ. Platinum resistance: the role of DNA repair pathways. Clin Cancer Res. 2008;14:1291–5. doi: 10.1158/1078-0432.CCR-07-2238. [DOI] [PubMed] [Google Scholar]
- 8.Al-Minawi AZ, Lee YF, Hakansson D, Johansson F, Lundin C, Saleh-Gohari N, Schultz N, Jenssen D, Bryant HE, Meuth M, Hinz JM, Helleday T. The ERCC1/XPF endonuclease is required for completion of homologous recombination at DNA replication forks stalled by inter-strand cross-links. Nucleic Acids Res. 2009;37:6400–13. doi: 10.1093/nar/gkp705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Enoiu M, Jiricny J, Scharer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012;40:8953–64. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decoville M, Schwartz A, Locker D, Leng M. Detection of minor adducts in cisplatin-modified DNA by transcription footprinting. FEBS Lett. 1993;323:55–8. doi: 10.1016/0014-5793(93)81447-8. [DOI] [PubMed] [Google Scholar]
- 11.Johnson SW, Swiggard PA, Handel LM, Brennan JM, Godwin AK, Ozols RF, Hamilton TC. Relationship between platinum-DNA adduct formation and removal and cisplatin cytotoxicity in cisplatin-sensitive and -resistant human ovarian cancer cells. Cancer Res. 1994;54:5911–6. [PubMed] [Google Scholar]
- 12.Johnson SW, Perez RP, Godwin AK, Yeung AT, Handel LM, Ozols RF, Hamilton TC. Role of platinum-DNA adduct formation and removal in cisplatin resistance in human ovarian cancer cell lines. Biochem Pharmacol. 1994;47:689–97. doi: 10.1016/0006-2952(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106:277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Earley JN, Turchi JJ. Interrogation of nucleotide excision repair capacity: impact on platinum-based cancer therapy. Antioxid Redox Signal. 2011;14:2465–77. doi: 10.1089/ars.2010.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuertes MA, Castilla J, Alonso C, Perez JM. Cisplatin biochemical mechanism of action: From cytotoxicity to induction of cell death through interconnections between apoptotic and necrotic pathways. Current Medicinal Chemistry. 2003;10:257–66. doi: 10.2174/0929867033368484. [DOI] [PubMed] [Google Scholar]
- 17.Chou KM. DNA Polymerase Eta and Chemotherapeutic Agents. Antioxid Redox Signal. 2010;14:2521–9. doi: 10.1089/ars.2010.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haring SJ, Mason AC, Binz SK, Wold MS. Cellular functions of human RPA1. Multiple roles of domains in replication, repair, and checkpoints. J Biol Chem. 2008;283:19095–111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochkareva E, Korolev S, Lees-Miller SP, Bochkarev A. Structure of the RPA trimerization core and its role in the multistep DNA-binding mechanism of RPA. EMBO J. 2002;21:1855–63. doi: 10.1093/emboj/21.7.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bastin-Shanower SA, Brill SJ. Functional analysis of the four DNA binding domains of replication protein A - The role of RPA2 in ssDNA binding. J Biol Chem. 2001;276:36446–53. doi: 10.1074/jbc.M104386200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–47. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hass CS, Chen R, Wold MS. Detection of posttranslational modifications of replication protein A. Methods Mol Biol. 2012;922:193–204. doi: 10.1007/978-1-62703-032-8_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsey-Boltz LA, Reardon JT, Wold MS, Sancar A. In vitro analysis of the role of replication protein A (RPA) and RPA phosphorylation in ATR-mediated checkpoint signaling. J Biol Chem. 2012;287:36123–31. doi: 10.1074/jbc.M112.407825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neher TM, Bodenmiller D, Fitch RW, Jalal SI, Turchi JJ. Novel irreversible small molecule inhibitors of replication protein A display single-agent activity and synergize with cisplatin. Mol Cancer Ther. 2011;10:1796–806. doi: 10.1158/1535-7163.MCT-11-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neher TM, Shuck SC, Liu J, Zhang JT, Turchi JJ. Identification of novel small molecule inhibitors of the XPA protein using in silico based screening. ACS Chem Biol. 2010;5:953–65. doi: 10.1021/cb1000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shuck SC, Turchi JJ. Targeted inhibition of Replication Protein A reveals cytotoxic activity, synergy with chemotherapeutic DNA-damaging agents, and insight into cellular function. Cancer Res. 2010;70:3189–98. doi: 10.1158/0008-5472.CAN-09-3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anciano Granadillo VJ, Earley JN, Shuck SC, Georgiadis MM, Fitch RW, Turchi JJ. Targeting the OB-Folds of Replication Protein A with Small Molecules. J Nucleic Acids. 2010;2010:304035. doi: 10.4061/2010/304035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patrick SM, Turchi JJ. Replication Protein A (RPA) Binding to Duplex Cisplatin-damaged DNA Is Mediated through the Generation of Single-stranded DNA. J Biol Chem. 1999;274:14972–8. doi: 10.1074/jbc.274.21.14972. [DOI] [PubMed] [Google Scholar]
- 30.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 31.Chou TC, Talalay P. Quantitative-analysis of dose-effect relationships - the combined effects of multiple-drugs or enzyme-inhibitors. Advances in Enzyme Regulation. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 32.Chen R, Wold MS. Replication protein A: Single-stranded DNA's first responder: Dynamic DNA-interactions allow replication protein A to direct single-strand DNA intermediates into different pathways for synthesis or repair. Bioessays. 2014 doi: 10.1002/bies.201400107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton TC, Young RC, Ozols RF. Experimental model systems of ovarian cancer: applications to the design and evaluation of new treatment approaches. Semin Oncol. 1984;11:285–98. [Review] [87 refs] [PubMed] [Google Scholar]
- 34.Hass CS, Gakhar L, Wold MS. Functional characterization of a cancer causing mutation in human replication protein A. Mol Cancer Res. 2010;8:1017–26. doi: 10.1158/1541-7786.MCR-10-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomkiel JE, Alansari H, Tang N, Virgin JB, Yang X, VandeVord P, Karvonen RL, Granda JL, Kraut MJ, Ensley JF, Fernandez-Madrid F. Autoimmunity to the M(r) 32,000 subunit of replication protein A in breast cancer. Clin Cancer Res. 2002;8:752–8. [PMC free article] [PubMed] [Google Scholar]
- 36.Wong JMS, Ionescu D, Ingles CJ. Interaction between BRCA2 and replication protein A is compromised by a cancer-predisposing mutation in BRCA2. ONC. 2003;22:28–33. doi: 10.1038/sj.onc.1206071. [DOI] [PubMed] [Google Scholar]
- 37.Storr SJ, Chakrabarti J, Barnes A, Murray A, Chapman CJ, Robertson JF. Use of autoantibodies in breast cancer screening and diagnosis. Expert Rev Anticancer Ther. 2006;6:1215–23. doi: 10.1586/14737140.6.8.1215. [DOI] [PubMed] [Google Scholar]
- 38.Givalos N, Gakiopoulou H, Skliri M, Bousboukea K, Konstantinidou AE, Korkolopoulou P, Lelouda M, Kouraklis G, Patsouris E, Karatzas G. Replication protein A is an independent prognostic indicator with potential therapeutic implications in colon cancer. Mod Pathol. 2007;20:159–66. doi: 10.1038/modpathol.3800719. [DOI] [PubMed] [Google Scholar]
- 39.Hande KR. Etoposide: Four decades of development of a topoisomerase II inhibitor. European Journal of Cancer. 1998;34:1514–21. doi: 10.1016/s0959-8049(98)00228-7. [DOI] [PubMed] [Google Scholar]
- 40.Sinha BK, Haim N, Dusre L, Kerrigan D, Pommier Y. DNA strand breaks produced by etoposide (VP-16,213) in sensitive and resistant human breast tumor cells: implications for the mechanism of action. Cancer Res. 1988;48:5096–100. [PubMed] [Google Scholar]