Abstract
Follow-up and Extension of a Prior Genome-wide Association Study of Posttraumatic Stress Disorder: Gene × Environment Associations and Structural Magnetic Resonance Imaging in a Highly Traumatized African-American Civilian Population
To the Editor
A genome-wide significant single nucleotide polymorphism (SNP) was recently reported (p = 3.97 × 10−8) on chromosome 7p12, rs406001, in a genome-wide association study (GWAS) of posttraumatic stress disorder (PTSD) (1). Although this SNP was found significant in a European American (EA) population (n = 1578), it was not significant in their African-American (AA) cohort (n = 2766) or in additional replication samples (approximately 2000 EAs). We sought to replicate the top associations from this prior work in our large GWAS for PTSD and to extend these findings with structural magnetic resonance imaging (MRI) to examine potential intermediate neural phenotypes of this polymorphism.
Our sample consisted of study participants from Grady Memorial Hospital (Atlanta, Georgia) as part of the Grady Trauma Project. As previously shown (2), these participants were adult, primarily female, highly traumatized, impoverished, primarily AA, with high rates of PTSD. We assessed current PTSD symptoms in over 3000 AA subjects with GWAS data with the PTSD symptom scale (mPSS) (3). Other phenotype measurements included in the data collection were Childhood Trauma Questionnaire (4) as a measure of childhood trauma; and Beck Depression Inventory (5) as a measure of depressive symptoms.
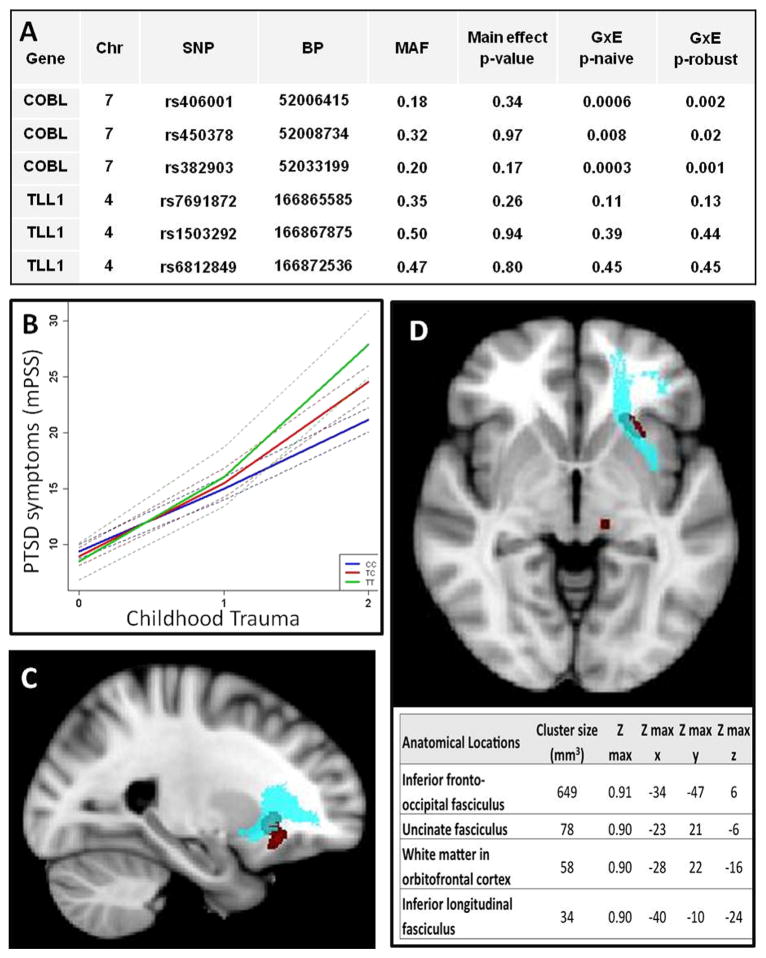
The DNA was extracted from saliva in Oragene collection vials (DNA Genotek, Ontario, Canada) with the DNAdvance kit (Beckman Coulter Genomics, Danvers, Massachusetts). The top SNPs from Xie et al. (1) were genotyped on the Illumina Omni1-Quad microarray, and statistical genetic analyses were performed in PLINK (6) and R statistical software (http://www.r-project.org/). The genotyping call rate for the SNPs was >.99, and the distribution did not depart from Hardy-Weinberg equilibrium (p > .1). To reduce potential bias by population stratification, we restricted the analysis to 3215 AA subjects who clustered together on principal component analysis. With linear regression, we found that the top SNPs reported in Xie et al. (1) did not replicate as main effects in associations with PTSD symptoms in our study (Figure 1A); however, we found a significant genotype × environment interaction (G × E) with childhood trauma in their top SNP rs406001 (n = 3076, t = 14.98, p = .0006) (Figure 1B). In fact, all three tested SNPs near the gene COBL had significant G × E interactions with childhood trauma in our AA population.
Figure 1.
Main effects, gene × environment (G × E) interactions, and neuroimaging phenotypes associated with top posttraumatic stress disorder (PTSD) hits from Xie et al. (1). (A) Association tests (linear regression) of main effects with PTSD symptoms (mPSS) and G × E with childhood trauma. (B) Regression lines with 95% confidence intervals (dotted lines) showing predicted PTSD symptom severity on the basis of childhood trauma load. These lines illustrate that there is an interaction between childhood trauma load and T carriers of SNP rs406001. Voxel-wide t test results indicating (in red) lower fractional anisotropy in TC compared with CC genotype (for SNP rs406001) in sagittal (C) and axial (D) sections as well as locations in accompanying table. Blue voxels depict the uncinate fasciculus (defined by the Johns Hopkins White Matter Atlas). BP, base pair; Chr, chromosome; MAF, minor allele frequency; Max, maximum; p-naïve, standard regression model p value; p-robust, p value adjusted with robust standard errors with R package geepack (11); SNP, single nucleotide polymorphism.
To examine the potential effects of this genotype on brain structure, we used MRI data from a subset of subjects on whom we had collected imaging data. Diffusion-weighted images (isotropically distributed along 60 directions; 39 × 2.5-mm-thick axial slices, matrix = 128 × 128, field of view = 220 × 220 mm, voxel size = 1.72 × 1.72 × 2.5 mm) were acquired from AA women between 18 and 62 years of age (n = 66, mean age = 38.5, SD = 12.7) with parameters previously described (7). All image processing and analysis were conducted with FMRIB Software Library (FSL version 4.1; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki/) (8), similar to a previous study (7). Tract-based spatial statistics (version 1.2) were used to examine associations between rs406001 genotype, childhood trauma, and fractional anisotropy (FA) across the entire brain. There were no significant main effects of childhood trauma exposure or G × E interactions with regard to FA values (p < .1, family-wise error corrected threshold). However, we observed a main effect of genotype. Compared with the CC genotype, carriers of the risk (T) allele demonstrated significantly lower FA in the following areas: left inferior-fronto occipital fasciculus; left inferior longitudinal fasciculus; white matter in the left frontal orbital cortex; and the left uncinate fasciculus (p < .1, family-wise errorcorrected) (Figure 1C,D). Univariate analysis of covariance results revealed that, after statistically controlling for age, current PTSD (mPSS), and depression symptoms (Beck Depression Inventory), the relationships between rs406001 genotype and FA within these clusters remained statistically significant (all p values < .01).
In summary, we found a significant effect of rs406001 (and other SNPs in linkage disequilibrium) interacting with childhood trauma exposure in a large AA cohort. These data suggest that examining trauma exposure in combination with genetic risk might allow more robust integration across population cohorts, because the original study found no main effect in AA subjects.
The rs406001 SNP is intergenic with no known function; however, the closest gene is COBL, which might be related to actin polymerization and neuronal development and function. Our brain imaging findings indicate that risk allele carriers demonstrate alterations in white matter integrity in brain regions associated with emotion processing; in particular, the uncinate fasciculus serves as a primary connection between the amygdala and ventral aspects of the prefrontal cortex, which is thought to play a role in extinction of learned fear (9,10). Thus, it is possible that risk allele carriers are more vulnerable to the development of anxious psychopathology via decrements in these white matter pathways. These data serve as a partial replication and extension of the first large GWAS for PTSD and suggest a putative white matter intermediate phenotype that might underlie PTSD risk.
Acknowledgments
This work was primarily supported by the National Institutes of Mental Health (MH071537 and MH096764 to KJR). Support was also received from Emory and Grady Memorial Hospital General Clinical Research Center, National Institutes of Health National Centers for Research Resources (M01RR00039), and Howard Hughes Medical Institute (KJR). We would like to thank staff and participants from the Grady Trauma Project for their time and effort in supporting this research.
Footnotes
The authors report no biomedical financial interests or potential conflicts of interest.
References
- 1.Xie P, Kranzler HR, Yang C, Zhao H, Farrer LA, Gelernter J. Genome-wide association study identifies new susceptibility loci for posttraumatic stress disorder. Biol Psychiatry. 2013;74:656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillespie CF, Bradley B, Mercer K, Smith AK, Conneely K, Gapen M, et al. Trauma exposure and stress-related disorders in inner city primary care patients. Gen Hosp Psychiatry. 2009;31:505–514. doi: 10.1016/j.genhosppsych.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falsetti SA, Resnick HS, Resick PA, Kilpatrick D. The Modified PTSD Symptom Scale: A brief self-report measure of posttraumatic stress disorder. The Behavior Therapist. 1993;16:161–162. [Google Scholar]
- 4.Bernstein DP, Fink L. Childhood Trauma Questionnaire Manual. San Antonio, TX: Psychological Corporation; 1998. [Google Scholar]
- 5.Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. 1961;4:561–571. doi: 10.1001/archpsyc.1961.01710120031004. [DOI] [PubMed] [Google Scholar]
- 6.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fani N, King TZ, Jovanovic T, Glover EM, Bradley B, Choi K, et al. White matter integrity in highly traumatized adults with and without post-traumatic stress disorder. Neuropsychopharmacology. 2012;37:2740–2746. doi: 10.1038/npp.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23(suppl 1):S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 9.Phelps EA, Delgado MR, Nearing KI, LeDoux JE. Extinction learning in humans: Role of the amygdala and vmPFC. Neuron. 2004;43:897–905. doi: 10.1016/j.neuron.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 10.Maren S, Phan KL, Liberzon I. The contextual brain: implications for fear conditioning, extinction and psychopathology. Nat Rev Neurosci. 2013;14:417–428. doi: 10.1038/nrn3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halekoh U, Højsgaard S, Yan J. The R package geepack for generalized estimating equations. J Stat Softw. 2006;15:1–11. [Google Scholar]