Abstract
Motor neuron diseases (MNDs) are a class of progressive neurological diseases that damage the motor neurons. An accurate diagnosis is important for the treatment of patients with MNDs because there is no standard cure for the MNDs. However, the rates of false positive and false negative diagnoses are still very high in this class of diseases. In the case of Amyotrophic Lateral Sclerosis (ALS), current estimates indicate 10% of diagnoses are false-positives, while 44% appear to be false negatives. In this study, we developed a new methodology to profile specific medical information from patient medical records for predicting the progression of motor neuron diseases. We implemented a system using Hbase and the Random forest classifier of Apache Mahout to profile medical records provided by the Pooled Resource Open-Access ALS Clinical Trials Database (PRO-ACT) site, and we achieved 66% accuracy in the prediction of ALS progress.
Keywords: ALS, Cloud computing, Hbase, Mahout, Randomforest, Medical decision support sytem, Big data
I. Introduction
Motor neuron diseases (MNDs) are a class of progressive neurological diseases that damage the motor neurons, which control essential voluntary muscle activity such as speaking, walking, breathing, swallowing, as well as general movement of the body [1]. Based on the nature of inheritance, manifestation, and site of motor neuron degeneration, MNDs are classified into five groups - Amyotrophic lateral sclerosis (ALS), primary lateral sclerosis (PLS), progressive muscular atrophy (PMA), progressive bulbar palsy (PBP), and pseudobulbar [1]. Symptoms can occur at any age from children to adults, but the causes of most MNDs are not known except through inheritance [2].
Since there is no standard cure for the MNDs, multiple treatments such as physical and occupational therapy, rehabilitation, and balanced diet are used to help patient maintain their quality of life. However, this means that a precise patient diagnosis is very important to decide the most appropriate treatment. Although many diagnostic techniques including MRI and genetic testing are suggested in the literature [3], the rate of false positive and false negative diagnoses are still very high in this class of diseases. In ALS, current estimates indicate 10% of diagnoses are false-positives, while 44% appear to be false negatives [3]. In addition, reaching a final diagnosis can be delayed because the symptoms of MNDs are similar with those of other diseases such as cancer, depression, and anemia [3]. Therefore, a rapid and accurate diagnosis is a critical need for the treatment of MND patients.
As computational science is incorporated into medical and biological science, huge amounts of medical and experimental data are accumulated rapidly in public repositories. Many research groups have studied the efficacy of applying computational modalities to develop personalized treatments using these large datasets. G-DOC is a recent example of a clinical database that combines medical profiling data from patients with breast and colorectal cancers [4], while CARE makes use of collaborative filtering techniques on “big data” to predict personalized disease risks [5]. Although these previous studies open new doors for personalized treatments, they are still for very specific medical cases and the accuracy of prediction or diagnosis are suboptimal. To increase the accuracy of diagnosis for chronic disease, we need to develop new methods to profile huge amounts of data from heterogeneous domains such as genomics, laboratory, and electrical patient records.
In this study, we developed a new methodology to profile specific medical information from patient medical records for predicting the progression of motor neuron diseases. We implemented a system using Hbase and the Random forest classifier of Apache Mahout to profile 1883793 records of clinical data and increased the accuracy of prediction using an open cloud computing environment. In addition, we implemented a web-based interface for an end-user to access the Hbase and easily control Mahout parameters. Training and testing data were generated from features of medical records provided by the Pooled Resource Open-Access ALS Clinical Trials Database (PRO-ACT) site. The accuracy of prediction based on testing after training the random forest classifier with the training dataset, is 66% using just 3 profiling features. If we increase the number of profiling features, we expect that accuracy will be further improved. In this paper, we first summarize the backgrounds of our research, and describe our method and datasets. Finally, we discuss our results and future research directions.
II. Background
MNDs damage motor neurons that control voluntary muscle activity including speaking, walking, breathing, and swallowing. Normally, messages from upper motor neurons in a brain are transmitted to particular muscles through lower motor neurons in the brain stem and spinal cord [1]. Upper motor neurons send directions that trigger the lower motor neurons to make movements. Lower motor neurons control muscles in the arms, legs, chest, face, throat, and tongue [3]. If there are problems in signal transmission between the upper motor neurons and the lower motor neurons, the limb muscles develop stiffness, and movements become slow and laborious. When the signals are disrupted between lowest motor neurons and the muscle, the muscles gradually weaken and may begin wasting away as well as developing uncontrollable twitching [3].
Patients with MNDs can range in age from childhood to adult. While the symptoms appears at birth or before the child starts to walk in children, they can sometimes present in adults 40 years of age or later and are more commonly seen in men than women [3]. Among different MNDs, amyotrophic lateral sclerosis (ALS) is the most common MND in adults, and affects both upper and lower motor neurons [3]. A rapid and accurate diagnosis is very important to the patient because ALS is fatal and there is no cure or treatment. Symptoms include weakness, muscle wasting, fasciculations, and hyperreflexia [3], and it takes on average, 12 months from onset of symptoms to an established diagnosis [3]. However, many factors can delay reaching a proper diagnosis; 10% of potential cases are false-positives, and up to 40% may be false-negatives because the symptoms of ALS may mimic other fatal diseases such as cancer, depression, and heart failure [3]. Therefore, we need to improve the accuracy of ALS diagnosis.
Recently many groups have initiated studies and suggested methods to manage and analyze “Big Data” in other fields such as economics and advertising. Among these methods, cloud computing is one of popular solutions for the “big data” issues. The basic idea of cloud computing is to divide a large task into small subtasks, and allocate them to a number of parallel systems for execution. To implement this, cloud computing needs a flexible and scalable infrastructure and an algorithm to manage distributed processing and storage capacity dynamically.
Among the many available open-source infrastructures, Hadoop is one of most popular cloud infrastructures based on GFS (Google File system) implemented by Google [6, 7]. It manages resources in a distributed computing environment using YARN [6, 7], and it can manage a large scale of data processing through the MapReduce algorithm [8, 9]. Hadoop is also a fundamental platform for other cloud packages such as Apache Pig, Hive, and HBase [8].
HBase is an open-source and non-relational database working on top of Hadoop in a distributed computing system [10]. It is a column-oriented database based on the description of the “Big Table” algorithm developed by Google [11]. A database in HBase consists of a set of tables, and each table contains row, column family, column qualifier, cell, and version. Data are stored in tables following its row, and rows are identified by rowkeys. Data within a row is grouped by column family, and data within a column family is addressed by a column qualifier. A combination of rowkey, column family, and column qualifier uniquely identifies a cell, which contains specific data [10]. In addition, data in a cell can have versions, and versions are differentiated by their timestamp [10].
Mahout is a set of open-source libraries to implement distributed or scalable machine learning algorithms for collaborative filtering, clustering and classification on the Hadoop platform [12]. In addition, Mahout provides a variety of functional libraries for mathematical and statistical analyses. Mahout's core algorithms are implemented using the MapReduce paradigm [12].
One of technical challenges in this study was to integrate these three platforms, Hadoop, Hbase, and Mahout to permit easy management of data and to rapidly enable the prediction of ALS progress with minimal project delay. To achieve these purposes, we implemented a web-based interface to transfer data among the three platforms using a Java servlet. In this manner, we developed a medical decision support system, which a physician can access profiling medical data anytime and anywhere.
III. Methods
A. Datasets
Data used in this article were obtained from the Pooled Resource Open-Access ALS Clinical Trials (PRO-ACT) Database. In 2011, Prize4Life, in collaboration with the Northeast ALS Consortium, and with funding from the ALS Therapy Alliance, formed the Pooled Resource Open-Access ALS Clinical Trials (PRO-ACT) Consortium. [14].
The database consists of 11 datasets-ALSFRS, Death Report, Demographics, Family History, Forced Vital Capacity, Laboratory Data, Riluzole use, Slow Vital Capacity, Subject ALS History, Treatment Group, and Vital Signs. Using a subset of the PRO-ACT database, there was a. challenge competition to predict the progression of ALS. In the challenge, the subset including 1822 out of 8635 patients are used after removing less standardized, misspellings and lack of standard units for lab data [15]. All types of dataset are also combined into the same file as the subset [15]. Following the same formats described in PRO-ACT site for the competition, we generated training and testing datasets including 918 and 279 patient records [14].
B. System Architecture
The purpose of this project is to implement a medical decision support system for the fast and accurate prediction of the rapidity of ALS progression. To achieve this purpose, we designed and implemented a cloud computing system to manage and analyze thousands of diagnosis data using Hbase on Hadoop. For final decision support, we incorporated the random forest module from Mahout into the system. To visualize the final results, we implemented a web-based interface. Therefore, the decision supporting system consists of these three modules: data management, generation of feature matrix, and prediction of ALS progress (Figure 1).
Fig 1. The data flow of a disease progress decision support system.
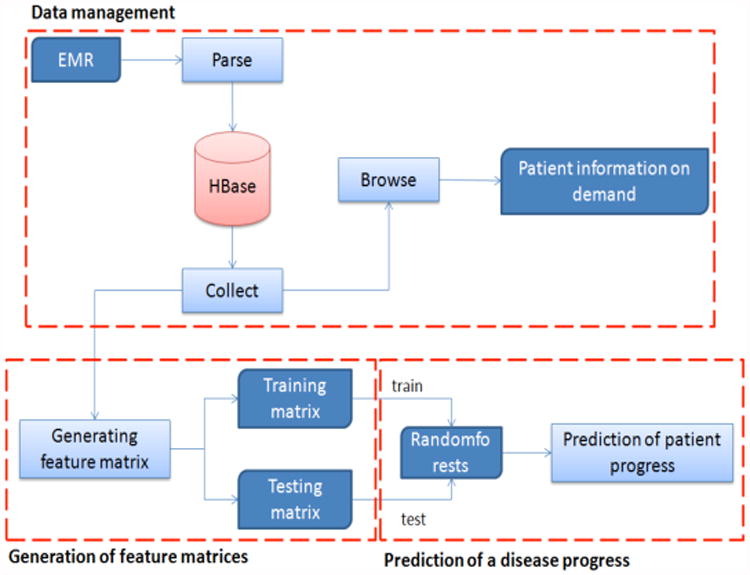
In the data management module, Hbase, one of the most popular NOSQL databases, was used to manage millions of records fast and efficiently. While SQL databases need time to parse SQL commands and generate indexes when databases exceed millions of rows of data, NOSQL databases are able to manage millions of data directly. After collecting key fields in medical records from PRO-ACT sites, we divided the fields into groups of indexes and values. Using the index groups as the column indexes, we first produced a big patient table. The data management can also extract a personal medical record using a patient id, and collect data in specific fields for building a feature matrix.
The second module needed generates a feature matrix from the ALS medical records stored in Hbase to use for prediction. Since the purpose of this study is to predict the speed at which patients lose neuromuscular function, some care is needed to select fields that contribute to the prediction. Generally, ALS medical records can be divided into dynamic and static data. Dynamic data contain ALSFRS (ALS Functional Rating Scale) question scores, alternative ALS measures, vital sign, and lab tests. Static data include demographics, ALS history, family history, and so on. To predict ALS progression, we selected dynamic data elements as key features because dynamic data show the temporal variation of patient. Among many methods available to compute features from time-series data, we calculated the slopes of the data using a least square method. The least square method is one of the most accurate methods to compute a line of best fit, the best approximation of the given set of data using equation (1).
| (1) |
In this study, ALSFRS question score is a key feature among available dynamic data because this score quantifies the physical muscle capability of patients. However, ALSERS score is also an incomplete feature because the score can depend on the subjective decisions of the examining physician. Thus, we try to enrich this score by including objective features such as vital signs and lab tests to complement potential limitations of the ALSERS score. After calculating the slope of ALS scores for 3 months and the slopes of other features for 12 months, we combine them to build a training feature matrix from a training dataset. Since we use a supervised learning algorithm for the classification, we tagged a target class to features of each patient by taking a threshold from the slope of ALS score for 12 months. In the same way, we generated a testing matrix from testing dataset.
For the third module, we divided patients into fast and slow-progress groups. Among the algorithms of classification in Mahout, we used a random forest classification. Random Forest is an ensemble machine learning algorithm consisting of many decision trees [13]. If the Random Forest is composed of N decision trees, we define the trees as
| (2) |
where V is a u-dimensional vector.
Then, the ensemble classifier generates N outputs from N trees. Among these outputs, one final prediction is selected by the most dominant tree after they vote for the most popular tree.
After training a Random Forest in Mahout using a training matrix, we measure the performance of the classification using a testing matrix, calculating sensitivity, specificity, and accuracy with equation (3), (4), and (5).
| (3) |
| (5) |
| (5) |
C. Implementation
A system was implemented in Java on a dual core (INTEL) machine running a Linux operating system. After installing Hadoop in pseudo-distributed mode, we installed the Hbase, Hive, and Mahout packages. We also developed a web-based user interface using a Java servlet in an Apache Tomcat application server to transfer data between Hbase and Mahout.
IV. Result
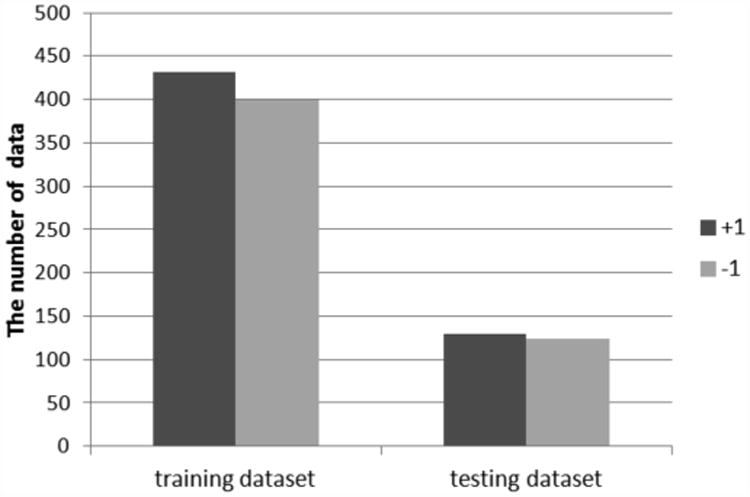
To discover essential features for the prediction, we selected several features such as ALSFRS, weight in Vital Signs, and Forced Vital Capacity. Then we computed their slopes for 3 months and 12 months from the first diagnosis using a least square method. Since the random forest approach is a supervised artificial intelligence algorithm, we first assigned target classes to each row in the feature matrix. In fact, if the slope of ALSFRS for 12 months is lower than threshold 0.6, +1(slow) is assigned, otherwise, -1(fast) is assigned. Figure 2 shows the distributions of classes in training and testing datasets.
Fig 2. The distributions of classes in training and testing datasets.
Although ALSFRS plays an important role in the diagnosis of ALS, because ALSFRS can depend on subjective decisions made by physicians, it has limitations. To overcome its limitations, we added other features such as weight and FVC (Forced Vital Capacity) in the feature matrix, and measured the variations of sensitivity, specificity, and accuracy of the prediction (the number of tree=100). Table I shows the improvement accuracy in proportion of the number of features used.
Table I. The variations of sensitivity, specificity, and accuracy of the prediction.
| Feature | Specificity | Sensitivity | Accuracy |
|---|---|---|---|
| The slope of ALSFRS | 47.58% | 57.36% | 52.57% |
| The slope of ALSFRS + The slope of weight | 62.09% | 65.89% | 64.03% |
| The slope of ALSFRS + The slope of weight + The slope of FVC | 66.12% | 65.89% | 66.01% |
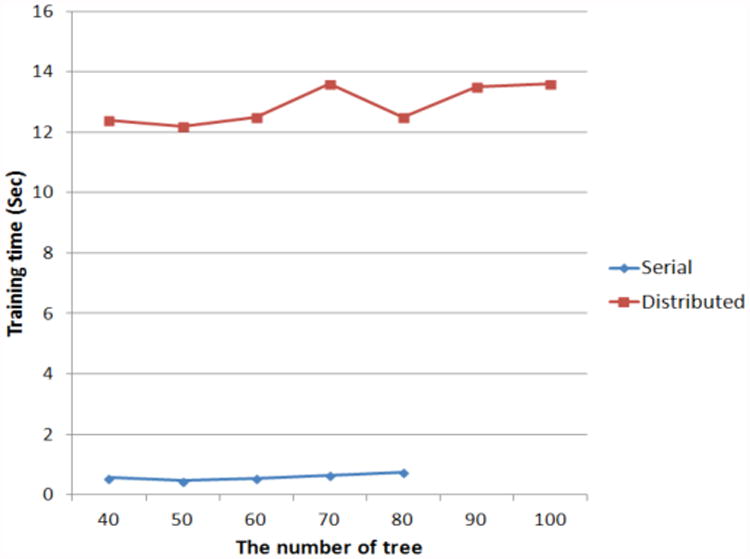
We also compared the prediction performance of distributed and serial random forest approaches in the Mahout and Weka packages. For the comparison, we changed the number of nodes for the random forest from 40 to 100, and measured the speed of training, sensitivity, specificity, and accuracy of the prediction.
Shown in Figures 3 and 4, we cannot precisely measure the training speeds and the accuracies when going beyond 80 trees using the serial random forest algorithm because the training time increases very rapidly. In instances when starting with fewer than 80 trees, the accuracies of the distributed approach are better than those of the serial. Although the speed of the serial method is faster than that of the distributed method when starting with fewer than 80 trees, the variation of the training speed in the distributed method is more stable than that in the serial one. Based on all results, if the number of features increases, the speed of the serial random forest is expected to decrease. In addition, we may implement a system to train a classifier and predict the results without as long a time delay because the training time of the random forest in a cloud environment takes tens of seconds.
Fig 3. The comparison of the accuracies in proportion to the number of trees between serial and distributed random forest.
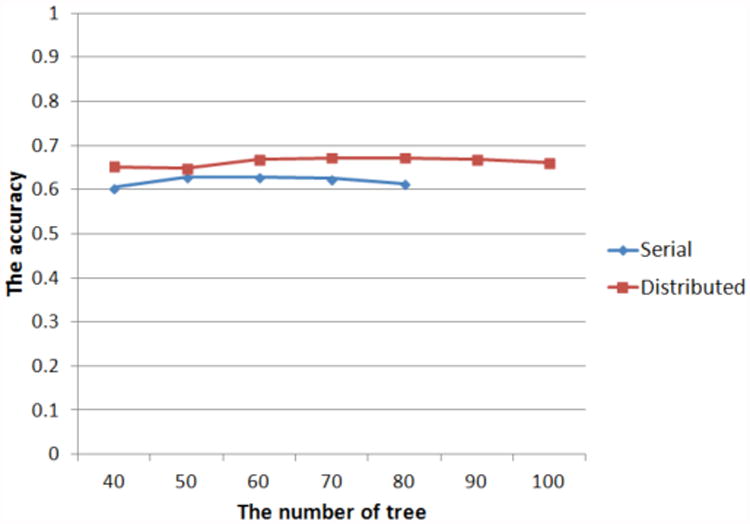
Fig 4. The comparison of the training speed in proportion to the number of trees between serial and distributed random forest.
V. Discussion and Future Directions
The advent of information technology has changed the environment of medical care, and many research groups are discovering new cures and diagnosis methods for fatal diseases by leveraging the overwhelming amount of heterogeneous data that has accumulated. One of the primary obstacles to achieving these discoveries is how to efficiently manage big heterogeneous datasets. In this study, we implemented a system to diagnose ALS disease as one approach to solve this issue.
ALS is a fatal disease, which damages motor neurons needed to control voluntary muscle activity, and patients die within 2 years after a definitive diagnosis. Since there is no effective cure for the disease, an accurate diagnosis is important to treat ALS patients. However, 10% of the diagnoses are false-positives, and 40% may be false-negatives because the symptoms of ALS mimic other fatal diseases. In addition, the treatments need to be adjusted depending on the rate of ALS progression in patients because some ALS patients can survive over decades.
To solve these problems, we suggest a new decision supporting framework in a cloud-computing environment utilizing “big data” in this study. We integrated Mahout with Hbase and added our analysis modules to the system using a Java servlet to analyze ten thousands of medical records from the PRO-ACT database to predict the rate of ALS progression. We tried to discover the key features of the diagnosis, and determined that the variations of weight and FVC (Forced Vital Capacity) are complementary features to ALSFRS. Finally, we achieved 66% accuracy in the prediction of ALS progress using a random forest algorithm with the slopes of ALSFRS, weight, and FVC for 3 months in this study. In addition, we showed it was possible to implement a real-time decision supporting using a supervised classifier with big data because of the short training time needed for the Mahout based random forest algorithm.
Even our results were overwhelmingly positive, there still remain several issues and tasks to address. The first issue is to find the correlations between each field in medical data and how those relate to the overall accuracy of the diagnosis. In this study, only three fields, ALSFRS, weight, and FVC were used. Therefore, a more detailed analysis of the correlations of other parameters and the inclusion of other key features, are expected to improve the performance of the diagnosis.
The second is to find an optimal classifier for our system to compare the performances of other classifiers such as Naive Bayesian and Logistic Regression. Although only several algorithms are parallelized in Mahout package, an optimal classifier for our features would also improve the performance.
The other task that remains is to improve the user interface from its current form, which only shows results of subsets of medical records from PRO-ACT. By attaching a user interface suitable for a data warehousing/business intelligence framework, we anticipate that our application will have a role as a decision support system enabling physicians to develop more appropriate personalized treatments for the patients with motor neuron diseases
Acknowledgments
This paper was supported by Dr. Tarek El-Ghazawi of the High-Performance Computing Laboratory (HPCL) at the George Washington University. Additional support was provided by Award Number UL1TR000075 from the NIH National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Advancing Translational Sciences or the National Institutes of Health. Data used in the preparation of this article were obtained from the Pooled Resource Open-Access ALS Clinical Trials (PRO-ACT) Database. In 2011, Prize4Life, in collaboration with the Northeast ALS Consortium, and with funding from the ALS Therapy Alliance, formed the Pooled Resource Open-Access ALS Clinical Trials (PRO-ACT) Consortium. The data available in the PRO-ACT Database has been volunteered by PRO-ACT Consortium members.
Contributor Information
Kyung Dae Ko, Email: kyung@gwu.edu, High-Performance Computing Laboratory (HPCL), The George Washington University, Ashburn, VA, United States.
Tarek El-Ghazawi, Email: tarek@gwu.edu, High-Performance Computing Laboratory (HPCL), The George Washington University, Ashburn, VA, United States.
Dongkyu Kim, Email: DKKim@childrensnational.org, Center for Translational Science, Children's National Medical Center, Washington DC, United States.
Hiroki Morizono, Email: HMorizono@childrensnational.org, Center for Genetic Medicine, Children's National Medical Center, Washington DC, United States.
References
- 1.Rosenfeld J, Swash M. What's in a name? Lumping or splitting ALS, PLS, PMA, and the other motor neuron diseases. Neurology. 2006 Mar 14;66(5):624–5. doi: 10.1212/01.wnl.0000205597.62054.db. [DOI] [PubMed] [Google Scholar]
- 2.Swash M. Motor neuron disease. Postgrad Med J. 1992 Jul;68(801):533–7. doi: 10.1136/pgmj.68.801.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristini J. Misdiagnosis and missed diagnoses in patients with ALS. JAAPA. 2006 Jul;19(7):29–35. [PubMed] [Google Scholar]
- 4.Madhavan S, Gusev Y, Harris M, et al. G-DOC: a systems medicine platform for personalized oncology. Neoplasia. 2011 Sep;13(9):771–83. doi: 10.1593/neo.11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chawla NV, Davis DA. Bringing Big Data to Personalized Healthcare: A Patient-Centered Framework. J Gen Intern Med. Jun 25; doi: 10.1007/s11606-013-2455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam C. Hadoop in action. Greenwich, Conn.: Manning Publications; 2010. [Google Scholar]
- 7.White T. Hadoop : the definitive guide. Beijing ; Sebastopol, CA: O'Reilly; 2009. [Google Scholar]
- 8.Taylor RC. An overview of the Hadoop/MapReduce/HBase framework and its current applications in bioinformatics. BMC Bioinformatics. 2010;11(Suppl 12):S1. doi: 10.1186/1471-2105-11-S12-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean J, Ghemawat S. MapReduce: A Flexible Data Processing Tool. Communications of the ACM. 2010;53(1):6. [Google Scholar]
- 10.Dimiduk N, Khurana A, Ryan MH. HBase in action. Shelter Island, NY: Manning; 2012. [Google Scholar]
- 11.Chang F, Dean J, Ghemawat S, et al. Seventh Symposium on Operating System Design and Implementation. Seattle, WA: Usenix Association; 2006. Bigtable: A distributed storage system for structured data. [Google Scholar]
- 12.Owen S. Mahout in action. Shelter Island, New York: Manning; 2012. [Google Scholar]
- 13.Svetnik V, Liaw A, Tong C, et al. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003 Nov-Dec;43(6):1947–58. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 14.Pooled Resource Open-Access ALS Clinical Trials Database (PRO-ACT) site, https://nctu.partners.org/ProACT.


