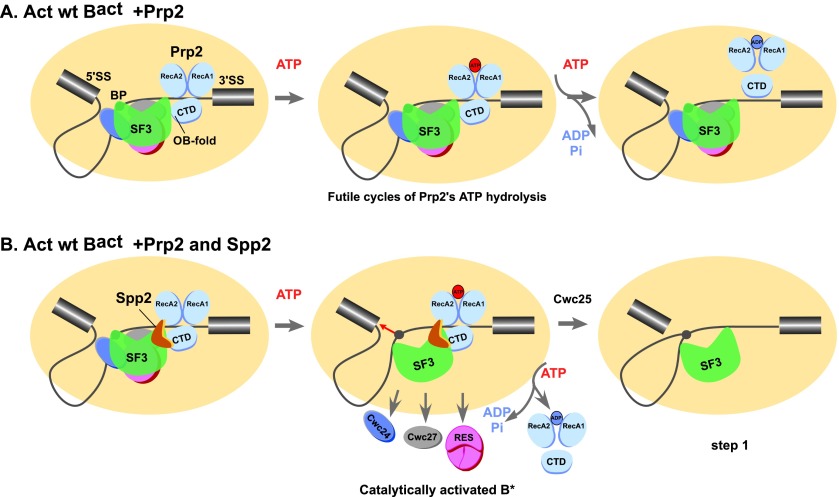
Structural rearrangement of the activated spliceosome (Bact) to yield a catalytically active complex (B*) is mediated by the DEAH-box NTPase Prp2 in cooperation with the G-patch protein Spp2. Warkocki et al. demonstrate that Spp2 is not required to recruit Prp2 to its bona fide binding site in the Bact spliceosome. However, transformation of the Bact to the B* spliceosome occurs only when Spp2 is present and is accompanied by dissociation of Prp2 and a reduction in its NTPase activity.
Keywords: spliceosome activation, DEAH-box helicase, Prp2, G-patch protein, Spp2, ATP hydrolysis
Abstract
Structural rearrangement of the activated spliceosome (Bact) to yield a catalytically active complex (B*) is mediated by the DEAH-box NTPase Prp2 in cooperation with the G-patch protein Spp2. However, how the energy of ATP hydrolysis by Prp2 is coupled to mechanical work and what role Spp2 plays in this process are unclear. Using a purified splicing system, we demonstrate that Spp2 is not required to recruit Prp2 to its bona fide binding site in the Bact spliceosome. In the absence of Spp2, the Bact spliceosome efficiently triggers Prp2’s NTPase activity, but NTP hydrolysis is not coupled to ribonucleoprotein (RNP) rearrangements leading to catalytic activation of the spliceosome. Transformation of the Bact to the B* spliceosome occurs only when Spp2 is present and is accompanied by dissociation of Prp2 and a reduction in its NTPase activity. In the absence of spliceosomes, Spp2 enhances Prp2’s RNA-dependent ATPase activity without affecting its RNA affinity. Our data suggest that Spp2 plays a major role in coupling Prp2’s ATPase activity to remodeling of the spliceosome into a catalytically active machine.
Pre-mRNA splicing proceeds by way of two phosphoester transfer reactions and is catalyzed by the spliceosome, which consists of the U1, U2, U4/U6, and U5 small nuclear ribonucleoproteins (snRNPs) and numerous non-snRNP proteins (Wahl et al. 2009). Spliceosome assembly occurs de novo on each pre-mRNA and follows an intricate pathway involving major structural rearrangements during each round of splicing. The various remodeling steps are driven in yeast by eight conserved DExD/H-box ATPases/RNA helicases. An interesting feature of the spliceosome is that it initially assembles into a multimegadalton ensemble—termed complex B—that contains all of the snRNPs but does not yet have an active site.
Activation of the spliceosome is then initiated by the combined action of the Prp28 and Brr2 RNA helicases, yielding the Bact complex. In this process, U1 and U4 snRNPs are displaced from the spliceosome, and new base-pair interactions between the U6 and U2 snRNAs and between U6 and the 5′ splice site (5′SS) are formed. The resulting RNA structure plays a central role in catalyzing both steps of pre-mRNA splicing (Staley and Guthrie 1998; Fica et al. 2013). During activation, 20 new proteins, including those of the NTC (nineteen complex), are stably integrated into the Bact complex and stabilize the newly formed RNA–RNA interaction network (Chan et al. 2003; Chan and Cheng 2005; Fabrizio et al. 2009). The final catalytic activation of the spliceosome requires an additional ATP-dependent remodeling step, yielding complex B*. This step is catalyzed by the DEAH-box ATPase Prp2 (Kim and Lin 1996).
Prp2 is structurally related to three other spliceosomal DEAH-box ATPases: Prp16, Prp22, and Prp43, which are involved, respectively, in the second catalytic step, the dissociation of the spliced mRNA from the post-catalytic spliceosome, and the final dismantling of the intron lariat spliceosome into its snRNP constituents. These four helicases possess a highly conserved core structure that includes two RecA-like domains, with a β hairpin in the RecA2 domain, a winged helix (WH), and a ratchet domain (He et al. 2010; Walbott et al. 2010; Cordin and Beggs 2013). The presence of the latter domains is typical for processive helicases such as Hel308 (Buttner et al. 2007). In addition, the spliceosomal DEAH-box helicases share a C-terminal domain that contains an oligonucleotide/oligosaccharide (OB)-fold that, in Prp43, is situated at the entry of its RNA-binding channel (Walbott et al. 2010). While Prp2, Prp16, and Prp22 have so far been shown to function exclusively during pre-mRNA splicing, Prp43 also plays a role in ribosome biogenesis (Combs et al. 2006; Leeds et al. 2006).
Purified Prp2 is an RNA-dependent ATPase that is capable of hydrolyzing any rNTP in the presence of ssRNAs but not DNA (Kim et al. 1992). However, no RNA helicase activity has yet been demonstrated for the isolated Prp2 in vitro (Kim et al. 1992). While the exact target of Prp2 in the spliceosome is not known, the impact of its action on the structure of the Bact spliceosome is substantial. Not only are the two proteins Cwc24 and Cwc27 discarded from the spliceosome, but the binding of other proteins—in particular the U2 snRNP SF3a and SF3b proteins—is weakened (Ohrt et al. 2012). As the U2 SF3 proteins are known to interact with pre-mRNA sequences near the branch site (BS) (Gozani et al. 1996; McPheeters and Muhlenkamp 2003), it has been hypothesized that their remodeling by Prp2 makes the BS adenosine accessible for nucleophilic attack at the 5′SS (Warkocki et al. 2009; Lardelli et al. 2010). Prp2 activity also creates a high-affinity binding site for Yju2 and the step 1 factor Cwc25, the binding of which may lead to a closed conformation of the step 1 catalytic center and thus facilitate efficient step 1 catalysis, leading to formation of the spliceosomal complex C (Konarska and Query 2005; Warkocki et al. 2009; Ohrt et al. 2012; Krishnan et al. 2013). Finally, Prp2’s activity may also directly or indirectly destabilize RNA elements that comprise the catalytic core, such as the U2/U6 helix I, to promote a fully competent catalytic conformation of the step 1 catalytic center (Wlodaver and Staley 2014).
While the structural changes that occur in the spliceosome during its Prp2-mediated catalytic activation have been investigated in some detail, the mechanism by which Prp2 promotes the observed structural rearrangement of the spliceosome is only poorly understood. Like all other spliceosomal DEAH-box ATPases, Prp2 interacts only transiently with the spliceosome (King and Beggs 1990), dissociating after hydrolyzing ATP (Roy et al. 1995; Kim and Lin 1996). Thus, the strength of interaction between Prp2 and its binding site in the spliceosome is significantly weakened during catalytic activation. It is not clear, however, whether this occurs at the B* complex stage or concomitant with step 1 catalysis.
One important requirement for efficient catalytic activation by Prp2 is a minimum length of intron nucleotides downstream from the BS adenosine of the pre-mRNA (hereafter also termed the 3′ tail). Truncated pre-mRNAs retaining only a 5- to 6-nucleotide (nt)-long 3′ tail allow efficient activation of the spliceosome (i.e., Bact complex formation) but do not allow catalytic activation or the subsequent catalysis of step 1 of splicing (Cheng 1994; Fabrizio et al. 2009). Catalytic activation occurs efficiently only when the 3′ tail has a length of ∼25 nt or more (Rymond and Rosbash 1985; Cheng 1994). Prp2 also cross-links in the Bact spliceosome to the 3′ tail of the pre-mRNA, primarily between nucleotide 28 (downstream from the BS) and the 3′ splice site (3′SS), suggesting that the interaction of Prp2 with this region of the intron plays a key role in the catalytic activation process (Teigelkamp et al. 1994; Liu and Cheng 2012).
DExD/H-box ATPases often have protein cofactors that modulate and coordinate their function or recruit them to their substrates (Silverman et al. 2004; Cordin et al. 2012; Cordin and Beggs 2013). A protein that is critically required for the Prp2-mediated catalytic activation of the spliceosome is its cofactor, Spp2. Spp2 was originally identified as a multicopy suppressor of a temperature-sensitive Prp2 mutant strain (Last et al. 1987). Prp2 interacts physically with Spp2 in a two-hybrid assay, although its binding may not be very strong (Roy et al. 1995; Silverman et al. 2004). Spp2 contains a glycine-rich “G-patch” domain (Aravind and Koonin 1999), and the interaction of Spp2’s G-patch with a region (termed the OB-fold) (He et al. 2010; Walbott et al. 2010) in the C-terminal domain of Prp2 appears to be critical for Prp2’s function in the spliceosome (Silverman et al. 2004). Interestingly, Prp43 also requires a G-patch cofactor, Ntr1 (also named Spp382), to facilitate the disassembly of the intron–lariat spliceosome (Tsai et al. 2005; Pandit et al. 2006; Tanaka et al. 2007), while the G-patch protein Pfa1 cooperates with Prp43 in the pre-rRNA processing pathways (Lebaron et al. 2009).
The function of Spp2 in the catalytic activation of the spliceosome is not known. Spp2 is detected in spliceosomes lacking Prp2, as assayed by immunoprecipitation experiments, suggesting that Spp2 might help recruit Prp2 to the spliceosome (Roy et al. 1995; Silverman et al. 2004). As most studies of the catalytic activation of spliceosomes have been conducted using yeast whole-cell extracts, which pose certain limitations, it cannot be excluded that the Bact spliceosome has a specific binding site for Prp2 alone and that the cooperation between the two factors serves another purpose during catalytic activation. For example, it is still unknown what is required to stimulate Prp2’s ATPase activity in the spliceosome and how Prp2-catalyzed ATP hydrolysis is mechanically transmitted to the conformational changes that lead to the transformation of the Bact to the catalytically activated B* complex.
We addressed these questions here using a purified splicing system. Collectively, our data indicate that Spp2 is not required for the recruitment of Prp2 to its bona fide binding site at the Bact spliceosome but that it plays a major role in coupling spliceosome-dependent Prp2 ATPase activity to the remodeling of Bact into B* complexes.
Results
Prp2 binds to the activated spliceosome in the absence of Spp2
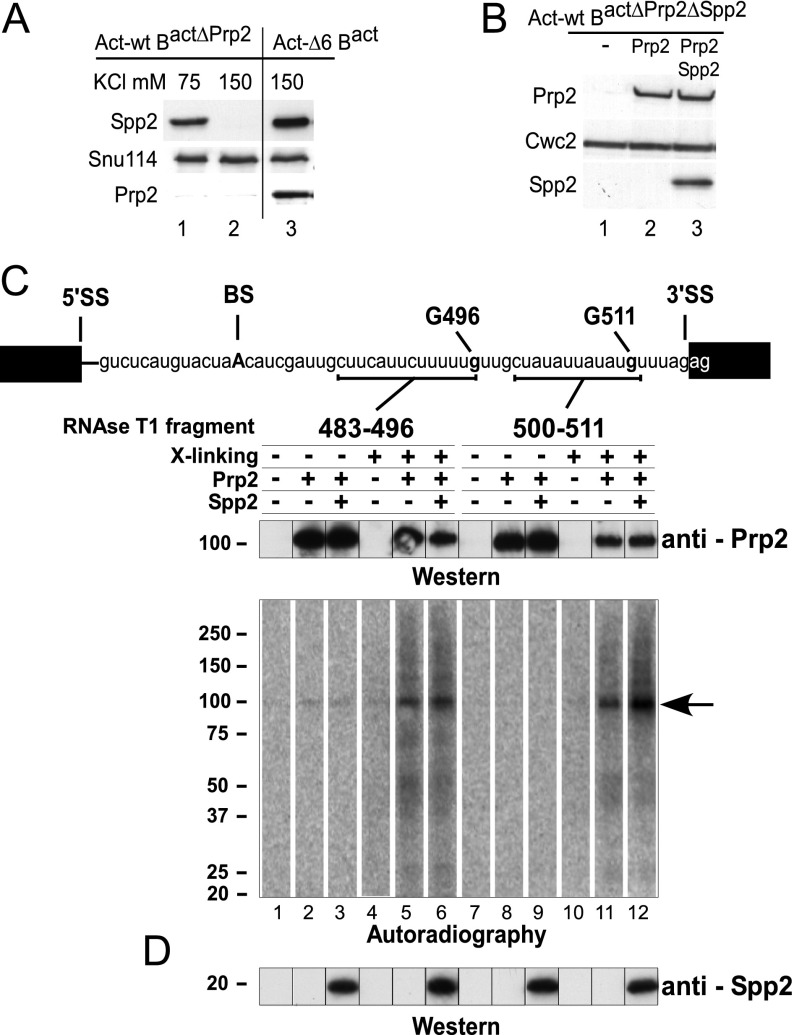
We first addressed whether Spp2 is required to recruit Prp2 to the Bact spliceosome using our purified splicing system (Warkocki et al. 2009). Bact ΔPrp2 spliceosomes were prepared by incubating heat-inactivated splicing extract from a temperature-sensitive prp2-1 yeast strain with an actin wild-type pre-mRNA that contained MS2-binding sites at its 5′ end (Warkocki et al. 2009). The Bact ΔPrp2 spliceosomes were purified by a combination of glycerol gradient centrifugation and MS2–MBP affinity chromatography with amylose beads. When the salt concentration during both purification steps was 75 mM, the purified spliceosomes contained endogenous Spp2 but no Prp2, as revealed by immunoblotting (Fig. 1A, lane 1). Thus, at near-physiological salt concentrations, Spp2 binds independently of Prp2 to activated spliceosomes, an observation consistent with previous findings (Roy et al. 1995). However, the interaction of Spp2 with the Bact ΔPrp2 spliceosomes was abolished when the purification was performed at 150 mM KCl (Fig. 1A, lane 2), and, in this way, we could isolate Bact complexes that lacked both Spp2 and Prp2. Purified Bact ΔPrp2 ΔSpp2 spliceosomes bound to the amylose matrix were then incubated with a twofold molar excess of recombinant Prp2 in the absence or presence of recombinant Spp2. The spliceosomes were washed with a buffer containing 150 mM KCl to remove the excess of the recombinant proteins, eluted, and fractionated by glycerol gradient centrifugation at 150 mM KCl. Immunoblotting revealed that Prp2 is present in the purified spliceosomes irrespective of the presence or absence of Spp2 (Fig. 1B, lanes 2,3). These data indicate that Spp2 is not required for the recruitment of Prp2 to the Bact spliceosome and thus that these two factors can bind independently of each other. While the binding of Spp2 to the spliceosome in the absence of Prp2 is abolished at 150 mM salt (Fig. 1A), it is stabilized in the presence of Prp2 (Fig. 1B, lane 3). This is consistent with the idea that the two proteins interact directly with each other in the spliceosome, as suggested by previous yeast two-hybrid analyses (Silverman et al. 2004).
Figure 1.
Prp2 and Spp2 bind to the spliceosome independently. (A) Western blot analysis of Spp2 association with the spliceosome. Spliceosomes were assembled on wild-type (lanes 1,2) or truncated (ActΔ6) actin pre-mRNAs (which leads to stalling of the spliceosome assembly at the Bact stage) (lane 3) in heat-treated prp2-1 extract (lanes 1,2) or wild-type extract (lane 3). Spliceosomes were affinity-purified in parallel in the presence of 75 mM (lane 1) or 150 mM (lanes 2,3) KCl. Immunoblotting was performed with rabbit polyclonal antibodies against GST-Spp2, Prp2, and Snu114 as indicated. (B) Prp2 and Spp2 association with the spliceosome at 150 mM KCl. The Bact ΔPrp2 ΔSpp2 spliceosomes were incubated with buffer (lane 1), Prp2 (lane 2), or Prp2 and Spp2 (lane 3). Unbound proteins were removed by washing with buffer containing 150 mM KCl, and spliceosomes were fractionated by glycerol gradient centrifugation at 150 mM KCl. Immunoblotting was performed with rabbit polyclonal antibodies against Prp2, Spp2, and Cwc2. (C) Cross-linking of Prp2 to pre-mRNA in purified spliceosomes. Two distinct site-specifically labeled pre-mRNAs were created, each carrying a single 32P-labeled phosphate 5′ at nucleotide G 496 or G 511. The theoretical RNA fragments remaining after digestion with RNase T1 are indicated by a bar below the sequence. Bact ΔPrp2 ΔSpp2 spliceosomes were assembled on site-specifically modified actin pre-mRNAs and purified at 150 mM KCl. After incubating with Prp2 or Prp2 plus Spp2, the complexes were UV cross-linked and then digested with RNase T1. Proteins cross-linked to 32P-labeled RNA were precipitated with antibodies against Prp2 and analyzed by SDS PAGE. Cross-linked 32P-labeled proteins were visualized by Western blot analysis (top panel) and autoradiography (bottom panel) as described above. The arrow indicates the 32P-labeled RNA cross-linked to Prp2. (D) Bact spliceosomes before cross-linking were probed with antibodies against Spp2.
Next, we investigated whether Spp2 is required to direct Prp2 to its bona fide binding site on the spliceosome. While little is known about potential protein interaction partners of Prp2 within the spliceosome, it was previously shown that in the presence of Spp2, Prp2 interacts directly with the intron downstream from the BS (Teigelkamp et al. 1994; Liu and Cheng 2012). We therefore investigated Prp2–pre-mRNA interactions in the presence or absence of Spp2 by performing UV cross-linking with purified Bact spliceosomes assembled on actin pre-mRNA that carried a single 32P-labeled phosphate 5′ of either G 496 or G 511 in the 3′ tail (i.e., 23 or 38 nt downstream from the BS adenosine, respectively) (Fig. 1C). Bact spliceosomes were then digested with RNase T1, and proteins cross-linked to 32P-labeled RNA were precipitated with antibodies against Prp2 and analyzed by SDS-PAGE. Prp2 was cross-linked to RNA preceding both nucleotides G 496 and G 511 at the 3′ end of the intron (Fig. 1C). The extent of Prp2 cross-linking to the region upstream of G 496 was nearly identical in the absence or presence of Spp2, while its interaction with the region upstream of G 511 was stimulated in the presence of Spp2 (Fig. 1C; cf. lanes 5 and 6 and lanes 11 and 12, respectively). Thus, in the absence of Spp2, Prp2 is recruited to a binding site on the Bact spliceosome that is very similar if not identical to its binding site in the presence of Spp2.
The Bact spliceosome strongly stimulates Prp2’s NTPase activity in the absence of Spp2
Our finding that Spp2 is not required to recruit Prp2 to the spliceosome raised the question of whether the Bact complex harbors an intrinsic ability to trigger the ATPase activity of Prp2. The inherent ATPase activity of Prp2—or, indeed, that of any other spliceosomal helicase—has, to date, never been investigated directly within spliceosomes. Therefore, we first developed an assay to measure Prp2’s ATPase activity in the context of the spliceosome. Bact spliceosomes contain stoichiometric quantities of the helicase Brr2 (which hydrolyzes solely ATP) (Santos et al. 2012) and the obligate GTPase Snu114 (Fabrizio et al. 1997). In contrast, Prp2 hydrolyzes all rNTPs and can use UTP for efficient catalytic activation of the spliceosome (Kim et al. 1992; Ohrt et al. 2012). Thus, to selectively measure Prp2’s NTPase activity in the presence of purified Bact spliceosomes (which, aside from these three enzymes, do not contain any other abundant NTPases), we used UTP as the sole energy source.
We first determined the UTP concentration that is needed for efficient step 1 catalysis by our purified Bact spliceosomes upon addition of recombinant Prp2, Spp2, and Cwc25 (Supplemental Fig. S1A). The catalysis of step 1 reached a high-efficiency plateau at 100–150 µM UTP (Supplemental Fig. S1A). A similar result was obtained with CTP, while the required concentrations of ATP and GTP were lower, suggesting stronger binding (lower Km) of the purine triphosphates to Prp2. At their saturating concentrations, all rNTPs allowed efficient (>50%) step 1 catalysis. Analytical glycerol gradient centrifugation showed that the Prp2/Spp2-induced transformation of the 45S Bact complex into the 40S B* complex with UTP as an energy source was nearly quantitative (Supplemental Fig. S1B). Furthermore, these results confirmed that the ATPase activity of Brr2 and the GTPase activity of Snu114 are not required for the final catalytic activation of the spliceosome by Prp2 and Spp2 once Bact has formed.
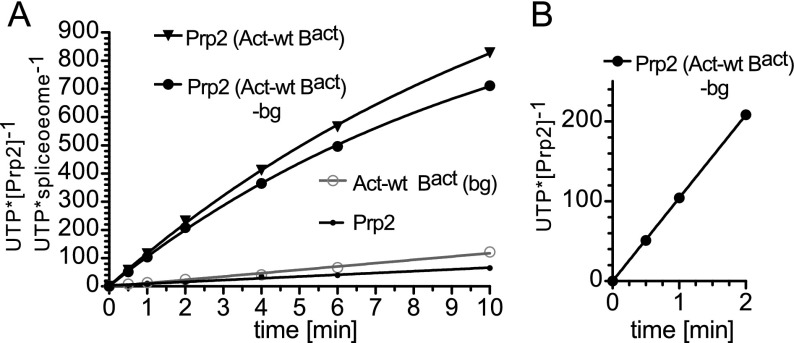
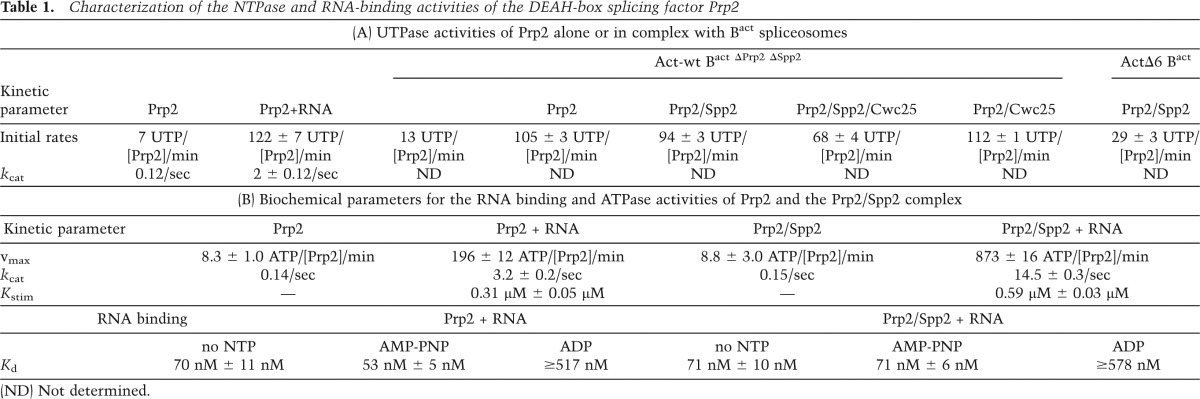
To monitor UTP hydrolysis by Prp2 in the presence of stoichiometric concentrations of the spliceosome, we concentrated the purified Bact complexes by ultrafiltration to ∼50 nM. Purified Bact ΔPrp2 ΔSpp2 spliceosomes (Act-wt Bact) (Fig. 2A) were then supplemented with either buffer or Prp2, the latter in a 1:1 molar ratio with the spliceosomes (both 50 nM). Under the latter conditions, the binding of Prp2 to the spliceosome was nearly quantitative, as determined by Western blot analysis (data not shown). UTP (containing α32P-UTP tracer) was subsequently added, and hydrolysis was monitored by thin-layer chromatography (TLC) analysis and quantified with a PhosphorImager. The number of UTP molecules hydrolyzed by a single Prp2 molecule at each time point was calculated (Supplemental Table S1) and is plotted in Figure 2A. The Act-wt Bact spliceosomes alone hydrolyzed UTP at a rate of ∼13 UTP/min (Supplemental Table S1, column 5). This value remained nearly constant within the experimental time frame of 10 min [Act-wt Bact (bg) background]. For Prp2 alone, the corresponding rate was 7 UTP/min (Prp2) (see also Table 1A for a summary). However, when both Prp2 and Act-wt Bact spliceosomes were present, the rate of UTP hydrolysis increased dramatically [Fig. 2, Prp2 (Act-wt Bact); Supplemental Table S1]. To estimate the number of UTP molecules hydrolyzed exclusively by Prp2 in the presence of the spliceosome, we subtracted the amount of UTP hydrolysis by Act-wt Bact alone (i.e., the background) from that of Bact bound by Prp2, thus obtaining the amount of UTP hydrolyzed by Prp2 when incorporated in the Bact complex [Prp2 (Act-wt Bact) (−bg)] (Supplemental Table S1, column 6). We then focused on the first 2 min of the time course (Fig. 2B), during which the amount of UTP hydrolyzed was linear with respect to time, and determined an initial rate of 105 UTP/min ± 3 UTP/min per Prp2/Bact spliceosome (Table 1A; Supplemental Table S1). This is comparable with the UTPase activity of Prp2 stimulated by saturating quantities (i.e., 2 µM) of an RNA oligo (U)30 (122 ± 7 UTP/min) (Table 1A). Thus, the spliceosome increased the rate of Prp2’s UTP hydrolysis in the absence of Spp2 by ∼15-fold and is therefore a potent stimulator of Prp2’s NTPase activity.
Figure 2.
The spliceosome is a potent stimulator of Prp2’s NTPase activity. (A) UTP hydrolysis was investigated by TLC and quantified by a PhosphorImager. UTP hydrolysis by the purified Bact ΔPrp2 ΔSpp2 spliceosomes in either the absence [Act-wt Bact (bg)] or presence (Prp2 Act-wt Bact) of Prp2 or by Prp2 alone (Prp2) was determined. The amount of UTP hydrolyzed by Prp2 in spliceosomes (“UTP•spliceosome−1”) and hydrolyzed by Prp2 in the absence of the spliceosome (Prp2) (“UTP•[Prp2]−1”) is plotted as a function of time. Prp2 Act-wt Bact (−bg) was generated by subtracting the spliceosome alone [Act-wt Bact (bg)] values from those obtained with Prp2 Act-wt Bact. (B) Initial rate of UTP hydrolysis by Prp2 in spliceosomes within the first 2 min of the time course. Data points were analyzed by linear regression to derive the initial rate of Prp2-catalyzed UTP hydrolysis in the context of the spliceosome.
Table 1.
Characterization of the NTPase and RNA-binding activities of the DEAH-box splicing factor Prp2
Prp2’s spliceosome-stimulated UTPase activity is unproductive in the absence of Spp2
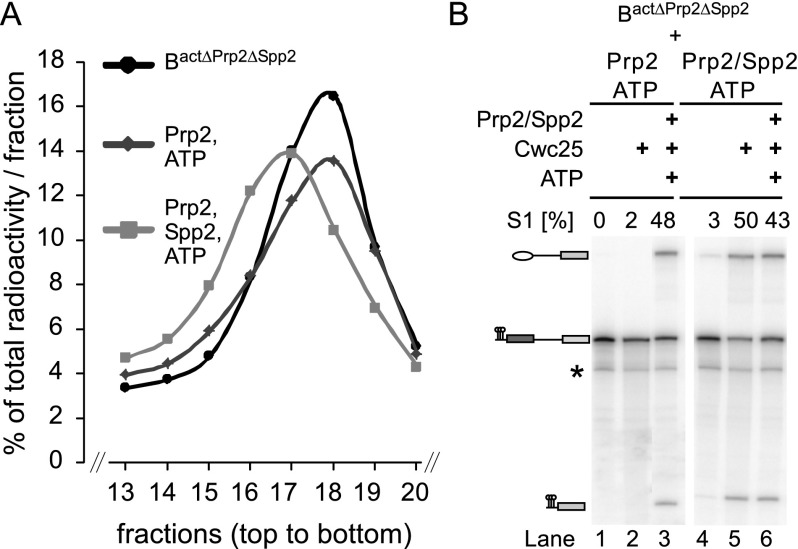
Our unexpected finding that isolated Bact spliceosomes bind Prp2 independently of Spp2 and strongly stimulate Prp2’s UTPase activity in the absence of Spp2 raised the question of whether catalytic activation of Bact complexes might already occur to some extent when the Bact complex is incubated solely with Prp2 and ATP. The transformation from Bact to B* involves a change in the sedimentation behavior of these complexes from 45S (Bact) to 40S (B*) during glycerol gradient centrifugation (Kim and Lin 1996; Warkocki et al. 2009). As shown in Figure 3A, incubation of Bact ΔPrp2 ΔSpp2 spliceosomes with Prp2 and ATP did not change the sedimentation profile of Bact complexes as compared with complexes incubated with buffer only. Moreover, these complexes could not catalyze splicing efficiently if supplemented additionally with the step 1 splicing factor Cwc25 (Fig. 3B, lane 2). In contrast, incubation of 45S Bact ΔPrp2 ΔSpp2 with both Spp2 and Prp2 in the presence of ATP led to its almost quantitative transformation into the 40S B* complex (Fig. 3A), which carried out step 1 catalysis after complementation with Cwc25 only (Fig. 3B, lane 5). These results show that Spp2 is required for the Prp2 NTP-dependent remodeling of the Bact spliceosome and that without it, this structural rearrangement does not take place.
Figure 3.
Spp2 is required for Prp2-catalyzed NTP-dependent remodeling of the Bact spliceosome. (A) Glycerol gradient sedimentation profiles of Act-wt Bact ΔPrp2 ΔSpp2 spliceosomes incubated with buffer alone (black) or in the presence of Prp2 and ATP in either the absence (dark gray) or presence (light gray) of Spp2. Radioactivity contained in each fraction was determined by Cherenkov counting and calculated as the percentage of total radioactivity in one gradient. The percentage of total radioactivity present in each gradient fraction is plotted. Ten percent to 30% (v/v) glycerol gradients containing 75 mM KCl were loaded with 400-µL samples and centrifuged at 60,000 rpm for 2 h in a TH660 rotor (Sorvall). (B) The Bact ΔPrp2 ΔSpp2 spliceosomes complemented with Prp2/ATP or Prp2/Spp2/ATP were recovered from the peak fractions of the glycerol gradients shown in A and then incubated for 1 h at 23°C under reconstitution conditions with buffer (lanes 1,4), Cwc25 (lanes 2,5), or Prp2, Spp2, Cwc25, and ATP (lanes 3,6, positive controls). Thus, in lanes 4–6, spliceosomes that had been catalytically activated during the preincubation step were used. The formation of step 1 splicing products was monitored by 8% denaturing RNA PAGE and quantified by a PhosphorImager. The percentage of step 1 (S1) products (compared with the total RNA signal in a lane) is indicated above each lane. RNA species are indicated at the left (from the top): lariat–intron–3′ exon, pre-mRNA, uncharacterized RNA species, and 5′ exon.
Spp2 couples NTP hydrolysis by Prp2 to catalytic activation of the spliceosome
Our data suggest that in the absence of Spp2, Prp2 in the Bact complex hydrolyzes high amounts of UTP that are not coupled to any notable structural rearrangement of the spliceosome. We thus assayed whether Prp2’s UTPase activity in the Bact ΔPrp2 ΔSpp2 spliceosome is altered after complementation with Spp2 (which transforms Bact to B*) and after addition of both Spp2 and Cwc25 (which leads to step 1 catalysis).
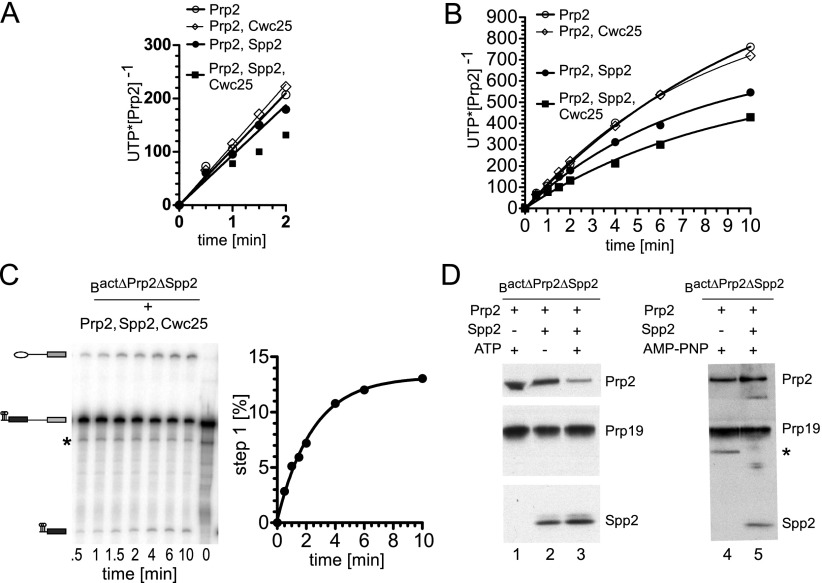
Figure 4A shows that the initial rates of UTP hydrolysis by Prp2/spliceosomes in the absence or presence of Spp2 were similar (see also Supplemental Table S2). However, the rate of UTP hydrolysis in the presence of Spp2 was slower at later time points so that, after 10 min, each Prp2/spliceosome had hydrolyzed ∼500 UTP molecules, as compared with ∼800 UTPs hydrolyzed by each Prp2/spliceosome in the absence of Spp2 (Fig. 4B; Supplemental Table S2). Additionally, when Cwc25 was present, the rate of UTP hydrolysis decreased even more so that, after 10 min, each Prp2/spliceosome had hydrolyzed ∼400 UTP molecules (Fig. 4B; Supplemental Table S2). Importantly, Cwc25 had no effect on the rate of UTP hydrolysis by Prp2/spliceosomes if Spp2 was omitted; i.e., under conditions that do not allow catalytic activation of the spliceosome and subsequent step 1 catalysis (Fig. 4A,B; Supplemental Table S2).
Figure 4.
UTP hydrolysis by Prp2 in the spliceosome is reduced after B* formation. (A) UTP hydrolysis was monitored by TLC and quantified by a PhosphorImager. Bact ΔPrp2 ΔSpp2 spliceosomes were incubated with Prp2 at an ∼1:1 ratio in the absence or presence of a 3–5 molar excess (140–150 nM) of Spp2, Spp2 and Cwc25, or Cwc25 as indicated. The reactions were started by addition of UTP. Values were obtained after subtraction of the background hydrolysis by spliceosomes without added Prp2 and represent mean values from two experiments (Supplemental Table S2). Data points for all but the “Prp2, Spp2, and Cwc25” condition were analyzed by linear regression to obtain initial rates of UTP hydrolysis. (B) As in A, except the entire 10-min time course is shown. Data points are fitted with single exponential functions. (C) Time course of step 1 of the splicing reaction catalyzed by Bact ΔPrp2 ΔSpp2 spliceosomes supplemented with Prp2, Spp2, and Cwc25. Splicing was analyzed by denaturing PAGE and quantified by a PhosphorImager. Experimental points were fitted with a single exponential curve. (D) Western blot analysis of Prp2 and Prp2/Spp2 association with purified Bact and B* spliceosomes. Bact ΔPrp2 ΔSpp2 spliceosomes were affinity-purified, bound to the amylose matrix, and incubated with Prp2 alone or Prp2 and Spp2 ± ATP (lanes 1–3) or AMP-PNP (lanes 4,5). Unbound proteins were removed by washing, and spliceosomes subsequently eluted from the matrix were fractionated by glycerol gradient centrifugation at 75 mM (lanes 1–3) or 150 mM (lanes 4,5) KCl. Probing was performed with rabbit polyclonal antibodies against Prp2, Spp2, and Prp19.
To test whether the substantial decrease in the rate of UTP hydrolysis by Prp2/spliceosomes in the presence of Spp2 and Cwc25 coincided with step 1 catalysis, we followed the progress of step 1 under our test conditions (Fig. 4C). After 10 min, ∼15% of the pre-mRNA was converted into step 1 products, indicating in turn that formation of B* was also successful. Thus, Spp2 helps to couple UTP hydrolysis by Prp2 to the formation of B* spliceosomes, and this leads to a reduction in the amount of UTP hydrolyzed by Prp2 over time.
The observed reduction in the amount of UTP hydrolysis that accompanies the Prp2/Spp2-mediated Bact-to-B* transformation could be due to displacement of Prp2 from the rearranged spliceosome. Indeed, immunoprecipitation analyses in splicing extracts previously showed that catalytically active spliceosomes have a greatly reduced affinity for Prp2 (Plumpton et al. 1994; Edwalds-Gilbert et al. 2000). However, these studies could not distinguish precisely at which step Prp2’s affinity is decreased (i.e., already when B* is generated or concomitant with step 1 catalysis). To investigate whether the B* spliceosome has a lower affinity for Prp2 compared with Bact, the binding of Prp2 to Bact and B* spliceosomes was compared. Affinity-purified Bact ΔPrp2 ΔSpp2 spliceosomes bound to the amylose matrix were incubated with Prp2 alone or Prp2 and Spp2 with or without ATP. The matrix-bound spliceosomes were then washed to remove the excess of the recombinant proteins, eluted, and fractionated by glycerol gradient centrifugation. Immunoblotting revealed that both Prp2 and Spp2 were present in the Bact spliceosome (Fig. 4D, lane 2). However, the majority of Prp2 dissociated from the B* spliceosome, while Spp2 remained bound (Fig. 4D, lane 3). This indicates that the affinity of Prp2 decreases significantly during the transformation of Bact to the B* complex. In contrast, in the absence of Spp2, Prp2 remained bound to the Bact complex in the presence of both ATP (Fig. 4D, lane 1) and AMP-PNP (Fig. 4D, lane 4). Prp2 did not dissociate from the spliceosome even in the presence of Spp2 when AMP-PNP was added to the reaction (Fig. 4D, lane 5), which blocks B* formation (Warkocki et al. 2009; Ohrt et al. 2012), confirming that the decrease in affinity of Prp2 is dependent on B* formation.
Taken together, our data suggest that following NTP-dependent RNP remodeling by Prp2 and Spp2, yielding the B* complex, Prp2 is ejected from its NTPase-stimulating binding site on the spliceosome. This in turn leads to the significant decrease in UTP hydrolyzed by Prp2/spliceosomes in the presence of Spp2 (Fig. 4B). The observation that the number of UTP molecules hydrolyzed by Prp2 was reduced by only ∼40%–50% in the presence of Spp2 is likely due to a poor transformation of the Bact to B* complexes under the conditions used for this experiment, which in turn resulted in a low yield of step 1 catalysis (i.e., ∼15%) (Fig. 4C). Thus, very likely, the Bact complexes remaining in the reaction mixture continue to stimulate Prp2’s UTPase activity.
Spp2 acts as a Prp2 cofactor, enhancing Prp2’s RNA-stimulated ATPase activity
The effects of Spp2 on important kinetic parameters of Prp2’s NTPase activity (e.g., the turnover number [kcat]) could not be reliably measured in the context of the spliceosome, as the presence of Spp2 leads to a mixed population of spliceosomal complexes with different Prp2 NTPase activities. Thus, to assay the effect of Spp2 on Prp2’s enzymatic properties and thereby potentially gain insight into the mechanism by which Spp2 might couple Prp2’s NTPase activity to catalytic activation, we carried out RNA binding and ATPase assays solely with purified Prp2 and Spp2. To investigate whether Spp2 affects Prp2’s ability to bind RNA, we performed fluorescence anisotropy experiments using a 24-nt fluorescein-labeled RNA oligonucleotide. While Spp2 alone did not bind RNA under our experimental conditions (i.e., 8 µM Spp2) (data not shown), Prp2 bound the 24-nt ssRNA with a dissociation constant (Kd) of 70 nM ± 11 nM (Table 1B), which was only slightly lower in the presence of AMP-PNP (Kd of 53 nM ± 5 nM). However, incubation with ADP increased the Kd by an order of magnitude (Kd ≥ 517 nM), indicating that Prp2 binding to RNA is destabilized after ATP hydrolysis (Table 1B). Addition of Spp2 to the reaction mixtures did not significantly change the strength of ssRNA binding to Prp2 irrespective of whether AMP-PNP or ADP was present (Table 1B).
We next assayed whether Spp2 influences Prp2’s intrinsic or RNA-stimulated ATPase activity. In the absence of RNA, Prp2 has a low intrinsic ATPase activity with a kcat of 0.14/sec−1 (Table 1B). In the presence of increasing concentrations of a (U)30 RNA, Prp2’s ATPase activity was significantly increased, reaching a maximum hydrolysis rate of 196 ± 12 ATP molecules hydrolyzed by a single Prp2 protein within a minute and a kcat (Prp2 and RNA) of 3.2 ± 0.2/sec (Fig. 5A; Table 1B). A similar dependence on RNA concentration was observed for UTP hydrolysis (Fig. 5B; Table 1A), although the kcat for Prp2’s RNA-stimulated UTPase activity was 30% lower (2 ± 0.12/sec) than its ATPase activity (Table 1B). Addition of an excess of Spp2 (up to 50-fold molar excess; 5 µM) over Prp2 alone did not stimulate Prp2’s intrinsic ATPase activity in the absence of RNA (Fig. 5C). However, at saturating (U)30 RNA concentrations, the ATPase activity of Prp2 was significantly enhanced in the presence of increasing concentrations of Spp2, reaching a maximum rate of 873 ± 16 ATP molecules hydrolyzed per Prp2 molecule per minute at 5–8 µM Spp2 (Fig. 5A,D). Thus, with a kcat of 14.5 ± 0.3/sec, Spp2 accelerates Prp2’s RNA-stimulated ATPase activity by a factor of ∼4.5 (Table 1B). As the concentration of RNA that results in a half-maximal stimulation of Prp2’s RNA-dependent ATPase activity (kstim) is only slightly increased in the presence of Spp2 (Table 1B), Spp2 primarily enhances the RNA-stimulated NTP kcat of Prp2 without significantly affecting Prp2’s RNA-binding affinity.
Figure 5.
Prp2 NTPase activity is stimulated by RNA and Spp2. (A) Prp2 (15–30 nM) was incubated with increasing concentrations of (U)30 RNA oligo in the absence or presence of 5 µM Spp2. Reactions were initiated by the addition of 1 mM ATP/MgCl2. ATP molecules hydrolyzed by a single Prp2 protein within 1 min are plotted (error bars indicate the standard errors of the means of three independent experiments). (B) The UTPase activity of Prp2 alone was measured and plotted as described in A. (C) The ATPase activity of Prp2 was monitored without RNA and ±Spp2 at 0.2, 1, or 5 µM for 10 min at 23°C. (D) Prp2’s relative RNA-stimulated ATPase activity was investigated as in C in the presence of 2 µM (U)30 RNA oligo. The reactions were incubated for 2–4 min at 23°C. The Prp2 ATPase rates were normalized by setting the value obtained without Spp2 to 1.
It was previously shown that isolated recombinant Prp2 does not exhibit RNA helicase activity (Kim et al. 1992). In view of our finding that Spp2 enhances Prp2’s ATPase activity, we investigated whether Prp2 can unwind dsRNA in the presence of Spp2. As shown in Supplemental Figure S2, Prp2 is not able to unwind a U4/U6 RNA duplex with both single-stranded 3′ and 5′ overhangs either alone (left panel) or in the presence of saturating amounts of Spp2 (middle panel), whereas the spliceosomal DEAH-box RNA helicase Prp22 completely unwinds this U4/U6 RNA duplex (right panel). Thus, in this reductionist system, Spp2 does not enable Prp2 to unwind dsRNA, and it will be interesting to see whether Prp2 remodels the spliceosome without changing base-pairing interactions within the spliceosomal RNA–RNA network.
A long 3′ tail is required for efficient and productive Prp2 spliceosome-stimulated UTPase activity
Efficient catalytic activation and step 1 catalysis by the spliceosome requires ∼25 nt downstream from the pre-mRNA’s BS. Truncated pre-mRNAs retaining only a 5- to 6-nt-long 3′ tail allow efficient activation of the spliceosome (i.e., Bact complex formation) but do not allow catalytic activation or the subsequent catalysis of step 1 of splicing (Cheng 1994; Fabrizio et al. 2009). We therefore investigated the role of this region of the pre-mRNA in the binding of Prp2 and Spp2 and in the spliceosome-mediated activation of Prp2’s NTPase activity. We thus assembled Bact spliceosomes on a pre-mRNA retaining only 6 nt downstream from the BS (i.e., ActΔ6); due to the strong reduction in stable Bact formation, pre-mRNAs lacking all nucleotides downstream from the BS were not tested. Immunoblotting revealed that purified ActΔ6 Bact spliceosomes assembled in wild-type extracts contained Prp2 and Spp2 at levels similar to Bact complexes formed on wild-type Act pre-mRNA (Fig. 1A, lane 3), an observation consistent with previous results obtained by mass spectrometry (Fabrizio et al. 2009) or pull-down experiments (Liu and Cheng 2012). Moreover, recombinant Prp2 and Spp2 bind to purified ActΔ6 Bact ΔPrp2 ΔSpp2 (Supplemental Fig. S3A; Supplemental Table S3) as efficiently as they do to spliceosomes containing Act-wt pre-mRNA (Fig. 1B, lane 3). We conclude that the presence of a long RNA stretch downstream from the BS does not per se affect Prp2 or Spp2 binding to the spliceosome.
These results imply that the lack of catalytic activation with ActΔ6 pre-mRNA is not due to the absence of Prp2 and/or Spp2 binding. We thus next checked whether Prp2’s UTPase activity is compromised. To test this, we assembled Bact ΔPrp2 ΔSpp2 spliceosomes on the ActΔ6 pre-mRNA and performed the UTPase assay as described above in the presence of stoichiometric amounts of Prp2 (50 nM) and an excess of Spp2 (150 nM). Supplemental Figure S4 shows that ActΔ6 Bact spliceosomes stimulate Prp2’s UTPase activity only moderately, with an initial rate that is about two-thirds lower (29 ± 3 UTP/min per Prp2/Spp2/spliceosome) than those observed with Act-wt Bact spliceosomes (94 ± 3 UTP/min per Prp2/Spp2/spliceosome) (Table 1A), and, in addition, it does not level off with time (Supplemental Fig. S4). Importantly, this stimulation of Prp2’s UTPase activity by ActΔ6 Bact spliceosomes is unproductive; i.e., it does not lead to significant transformation of Bact to B* complexes, as evidenced by their sedimentation behavior on glycerol gradients (Supplemental Fig. S3B). Thus, UTP hydrolysis by Prp2 in ActΔ6 Bact spliceosomes does not appear to be coupled to RNP remodeling even in the presence of Spp2. In summary, our data indicate that efficient and productive Prp2 spliceosome-stimulated UTPase activity requires a long 3′ tail of the pre-mRNA intron.
Discussion
Prp2-mediated catalytic activation of the spliceosome is only poorly understood. Here we showed that Prp2 and Spp2 can bind independently to the spliceosome and that in the absence of Spp2, the activated (Bact) spliceosome efficiently triggers Prp2’s NTPase activity. However, these cycles of NTP hydrolysis appear to be futile in that they do not lead to catalytic activation of the Bact spliceosome. Transformation of the Bact to the B* spliceosome occurs only when Spp2 is present and is accompanied by dissociation of Prp2 and a reduction in its NTPase activity. Finally, we showed that in the absence of spliceosomes, Spp2 does not alter Prp2’s affinity for RNA or its intrinsic ATPase activity but greatly enhances its RNA-dependent ATPase activity, probably by modulating Prp2’s ATPase center. Collectively, our data suggest that Spp2 plays a major role in coupling Prp2’s ATPase activity to catalytic activation of the spliceosome.
Prp2 binds to the Bact spliceosome independently of Spp2
Here we showed that Prp2 can bind to the spliceosome even in the absence of Spp2. This is consistent with recent work on the human PRP2 ortholog, which was shown to bind to spliceosomes in the absence of hSPP2 (Zang et al. 2014). Likewise, consistent with previous studies (Teigelkamp et al. 1994; Liu and Cheng 2012), Prp2 could be cross-linked to the 3′ tail of the pre-mRNA intron upon UV irradiation of Bact spliceosomes lacking Spp2. Therefore, we conclude that Prp2 binds to a similar if not identical site on the Bact spliceosome independent of the presence of Spp2. Nonetheless, Prp2 appears to reinforce Spp2’s binding to the Bact spliceosome, as evidenced by our finding that Spp2 is bound in a more salt-resistant manner when Prp2 is also present (Fig. 1B). This effect is probably due to an interaction between spliceosome-bound Prp2 and Spp2. Indeed, earlier studies showed that an interaction between the G-patch region of Spp2 and the OB-fold of the C-terminal region of Prp2 is important for spliceosome function in vivo (Silverman et al. 2004; He et al. 2010; Walbott et al. 2010). As ySpp2 did not show any RNA-binding activity when tested in a binary system (data not shown), Spp2’s binding site in the spliceosome is probably largely determined by protein–protein interactions. Interestingly, hSPP2 (GPKOW) was shown to bind RNA, presumably due to the presence of additional domains in GPKOW that are absent from ySpp2 (Zang et al. 2014).
The Bact spliceosome triggers Prp2’s NTPase activity even in the absence of Spp2
The ability to isolate biochemically defined Act-wt Bact complexes that contained Prp2 either alone or together with its cofactor, Spp2, allowed us for the first time to measure directly Prp2’s NTPase activity within the spliceosome. Our NTP hydrolysis measurements were performed with [α32P]-UTP instead of ATP in order to exclude any effects of the U5 snRNP-associated Brr2 ATPase and Snu114 GTPase. Interestingly, the Act-wt Bact-Prp2 complex hydrolyzed a high amount of UTP (105 ± 4 UTP molecules per Prp2/Bact complex per minute), which was almost constant over the first 10 min (Fig. 2). As the intrinsic UTPase activity of Prp2 in the absence of the spliceosome was only 7 UTP/min, the Bact spliceosome stimulates the UTPase activity by a factor of ∼15.
The observed stimulation of Prp2’s NTPase activity when bound to Bact complexes lacking Spp2 is likely due to its interaction with intron nucleotides downstream from the BS (i.e., the region that we denote as the 3′ tail) as opposed to nonspecific interactions with other spliceosomal RNAs. First, as discussed above, our data suggest that Prp2 binds to its bona fide spliceosomal binding site also in the absence of Spp2, contacting nucleotides in the intron’s 3′ tail (Fig. 1C), and, consistent with previous data (Kim et al. 1992), ssRNA greatly enhanced Prp2’s ATPase activity in a binary reaction (Fig. 5; Table 1B). When we measured the UTPase activity of Prp2, stimulated solely by an RNA oligo (U)30 (Fig. 5B), 1–2 µM RNA (U)30 oligo was needed to induce efficient Prp2 UTPase activity; at 50 nM RNA (U)30 oligo, only ∼14 ± 2 UTP molecules were hydrolyzed per Prp2 per minute (Fig. 5B). In contrast, we observed a powerful stimulation of Prp2’s UTPase activity by the ΔSpp2 spliceosome already at a concentration of 50 nM (in which the pre-mRNA or other spliceosomal RNAs are also at 50 nM), which was comparable with the UTPase activity of Prp2 stimulated by saturating quantities (2 µM) of an RNA (U)30 oligo as revealed by similar initial rates: 105 ± 3 UTP/min per Prp2/Bact spliceosome versus 122 ± 7 UTP/min per Prp2/RNA (U)30 oligo (Table 1A). Thus, we conclude that the Bact spliceosome provides a favorable microenvironment for Prp2 binding by placing it in close proximity to the pre-mRNA so that the high local concentration of the long 3′ intron tail efficiently triggers Prp2’s NTPase activity, and Spp2 does not appear to play a major role in this. However, Prp2 hydrolyzes NTP in a “futile cycle” most likely because, in the absence of Spp2, the energy produced by NTP hydrolysis cannot be coupled to mechanical work (i.e., transformation of the Bact to the B* complex), and Prp2 remains trapped in its high-affinity binding site in the Bact spliceosome. Indeed, we show that in the absence of Spp2, Prp2 remains bound to the Bact spliceosome even in the presence of ATP (Fig. 4D).
Spp2 is required for coupling of Prp2’s spliceosome-triggered UTP hydrolysis to catalytic activation
When we measured UTP hydrolysis by the Bact spliceosome in the presence of Spp2, the situation differed markedly. While the initial rate of UTP hydrolysis by Prp2 was nearly similar to that observed in the absence of Spp2, the amount of UTP hydrolyzed decreased significantly with increasing incubation time in the presence of Spp2 (500 UTP molecules hydrolyzed per Prp2/Spp2/spliceosome, in contrast to 800 UTP molecules per Prp2/spliceosome after 10 min) (Fig. 4B). This reduction in UTP hydrolysis correlated with the successful catalytic activation of the spliceosome, as evidenced by the ability of these spliceosomes to catalyze the first step of splicing when Cwc25 was additionally added (Fig. 4C). As under our experimental conditions, the Bact-to-B* transition was not quantitative, Bact complexes remaining in the reaction mixture would continue to stimulate the spliceosome/Prp2-catalyzed UTP hydrolysis, explaining why the number of hydrolyzed UTP molecules did not drop below 40%–50% of the initial value (Fig. 4B). For this reason, it was also not possible to calculate the number of hydrolyzed UTP molecules required to transform one molecule of the Bact spliceosome into a catalytically activated complex.
Taken together, our data suggest that following the Spp2-assisted NTP-dependent RNP remodeling by Prp2, yielding the B* complex, Prp2 makes no further contribution to the splicing process and is ejected from its NTPase-stimulating binding site on the spliceosome. Release of Prp2, as opposed to direct inhibition of its NTPase activity by Spp2 (or some other spliceosomal protein), in turn leads to the significant decrease in UTP molecules hydrolyzed by Prp2/spliceosomes in the presence of Spp2. This idea is supported by our finding that only the Bact spliceosome has a high-affinity binding site for Prp2, which is lost in the B* spliceosome (Fig. 4D). This in turn implies that the productive Prp2/Spp2-mediated remodeling of the spliceosome not only leads to its catalytic activation but, at the same time, abolishes the high-affinity binding of Prp2. At first sight, it is surprising that Spp2 significantly enhances Prp2’s RNA-dependent NTPase activity when it is not part of the spliceosome, while, in the spliceosome, the initial rates of UTP hydrolysis by Prp2 are not affected by Spp2 (cf. Figs. 4A and 5A; see also the Discussion below). A trivial explanation for this could be the limitation of the assay system (see the Materials and Methods) that we used here to measure spliceosome-dependent Prp2 UTPase activity, which exhibited a very low transformation of Bact to B*. An alternative explanation could be the following: Assuming that, in the presence of Spp2, Prp2 would remodel the Bact spliceosome in a hit-and-run manner, Prp2 would leave the spliceosome following the first UTP hydrolysis-mediated power stroke; thus, we would not necessarily expect to observe an increased initial rate of UTP hydrolysis but a reduction over time, consistent with the result shown in Figure 4B. Clearly, additional experiments are needed to clarify this point.
Potential mechanisms by which Spp2 couples Prp2 ATP hydrolysis to catalytic activation
Our results clearly indicate that Spp2 is required to couple Prp2’s spliceosome-triggered NTP hydrolysis to mechanical work; i.e., remodeling of the spliceosome’s structure. What could be the mechanism of this coupling process? An initial clue to this question was revealed by our investigation of the modulation of Prp2’s enzymatic properties by Spp2 in the absence of spliceosomes. First, Spp2 significantly accelerated the kcat of the RNA-stimulated ATPase activity of Prp2 by a factor of ∼4.5 (Table 1B), thereby establishing a central role for Spp2 as a Prp2 cofactor. As Spp2 did not increase the affinity of Prp2 for a ssRNA substrate in the presence of ADP or a nonhydrolyzable ATP analog, these findings indicate that Spp2 may enhance the RNA-stimulated NTPase activity of Prp2 by modulating/changing the structure of Prp2’s NTP-binding center.
A high rate (i.e., high kcat) of Prp2-catalyzed NTP hydrolysis could be relevant for coupling of NTP hydrolysis with the remodeling of the spliceosome; that is, catalytic activation of the spliceosome by Prp2 and Spp2 involves the destabilization of several spliceosomal proteins, including some (e.g., U2-Hsh155 and RES-Snu17) that interact with the 3′ tail of the pre-mRNA intron (McPheeters and Muhlenkamp 2003; Wysoczanski et al. 2014). At the same time, a 25- to 30-nt-long 3′ tail is also necessary for efficient catalytic activation, and Prp2 contacts the distal part of the 3′ tail (Fig. 1C; Teigelkamp et al. 1994; Liu and Cheng 2012). Thus, it is possible that Prp2 translocates along the 3′ tail (Liu and Cheng 2012), thereby destabilizing RNA-bound proteins. This would likely require that Prp2 acts processively and that the latter does not occur in the absence of Spp2 despite the fact that ATP is continuously hydrolyzed. Indeed, faster turnover of ATP by Prp2 (in the presence of Spp2) may lead to higher rate of mechanical translocation along the RNA, which in turn may be required for the efficient remodeling activity of Prp2. The enhancement of Prp2’s NTPase activity that we observed in the presence of Spp2 may therefore be essential for coordinated RNA binding, ATP hydrolysis, and translocation steps along the RNA (i.e., productive RNPase activity). In the absence of Spp2, these steps would not be coupled, and Prp2 would engage in futile cycles of RNA binding, ATP hydrolysis, release from the 3′ tail RNA, and rebinding without completely dissociating from the spliceosome due to its interactions with one or more proteins (see Fig. 6).
Figure 6.
Schematic representation of the major role played by Spp2 in coupling Prp2’s ATPase activity to remodeling of the spliceosome into a catalytically active machine. Prp2’s structure is depicted schematically as proposed for the structurally related DEAH-box helicase Prp43, with the canonical helicase core comprising the RecA1 and RecA2 domains. The conserved C-terminal domain (CTD) is also shown (He et al. 2010; Walbott et al. 2010; Cordin et al. 2012). (A) In the absence of Spp2, Prp2 binds to the 3′ intron tail of the wild-type pre-mRNA in the Bact spliceosome. In the presence of ATP, the interaction of Prp2 with the Bact spliceosome leads to a stimulation of Prp2’s ATPase activity that does not result in catalytic activation of the spliceosome. (B) Instead, when Spp2 also binds, its interaction with the OB-fold domain of Prp2 may induce structural changes in the RecA domains, leading to a productive conformation of Prp2 and thereby influencing the rate of ATP hydrolysis and likely translocation along the ssRNA intron (Cordin et al. 2012; Liu and Cheng 2012). This results in remodeling of target protein-binding sites and catalytic activation of the spliceosome (B*) (Warkocki et al. 2009; Ohrt et al. 2012). Following catalytic activation, Prp2 is released from its binding site, the BS adenosine becomes accessible for nucleophilic attack at the 5′SS, and Cwc25 promotes efficient step 1 catalysis.
In this context, it is interesting to note that Bact spliceosomes, which are assembled on a pre-mRNA containing only a 6-nt-long 3′ tail (ActΔ6 Bact), stimulated Prp2’s UTPase activity considerably less efficiently (by a factor of 3) than Act-wt Bact spliceosomes (Table 1A). Importantly, under these conditions, NTP hydrolysis did not lead to significant catalytic activation despite the presence of Spp2 (Supplemental Fig. S3B). Indeed, Cwc24, Cwc27, and the RES complex proteins are equally represented in the absence or presence of ATP (Supplemental Table S3), while they are displaced from a catalytically activated Act-wt B* complex (Warkocki et al. 2009; Ohrt et al. 2012). It is possible that Prp2 may still transiently interact with the 6-nt-long RNA tail, leading to the observed moderate Prp2-mediated UTPase activity. However, Prp2 will not be able to efficiently translocate along the short 3′ tail RNA. This in turn would suggest that Spp2 may couple NTP hydrolysis to spliceosome activation only when Prp2 translocates along a long 3′ tail RNA (see also above).
Spp2’s function as a Prp2 cofactor could extend beyond that of its influence on Prp2’s ATPase center; that is, Prp2 and Spp2 can bind independently to the Bact spliceosome (Fig. 1), and a long 3′ tail is not essential for this binding (Fabrizio et al. 2009), indicating that both proteins likely are involved in multiple protein–protein interactions in the Bact spliceosome. It is thus conceivable that the power stroke, fueled by ATP hydrolysis and accompanying structural changes within the motor module of Prp2, is transmitted through the various anchor points that Prp2 and Spp2 share with other components of the spliceosome. This could also explain why Prp2/Spp2-mediated ATP hydrolysis has such a dramatic impact on the structure of the spliceosome. This model would predict that in the absence of a protein that may be important for this relay system, Prp2 might hydrolyze ATP in a futile cycle even in the presence of Spp2. Indeed, Cwc22 may be such a candidate, as spliceosomes lacking Cwc22 can still bind Prp2 and hydrolyze ATP without coupling it to catalytic activation (Yeh et al. 2011). It will be very interesting in the future to learn more about the interaction partners of Prp2 and Spp2 in the spliceosome and analyze their mutual functional interplay in more detail.
Interestingly, significant amounts of Spp2 remain bound to purified spliceosomes after catalytic activation, while Prp2 is largely displaced (Fig. 4D). It was recently shown in a yeast two-hybrid screen that hPRP2 interacts with not only hSPP2/GPKOW but, surprisingly, also hPRP16 (Hegele et al. 2012). It will therefore be interesting to investigate whether, in yeast, Spp2 cooperates with Prp16 in remodeling the spliceosome prior to the second catalytic step of splicing.
Functional similarities between the G-patch proteins Spp2 and Ntr1/Pfa1
Interestingly, the G-patch protein Spp2 appears to modulate the enzymatic properties of the Prp2 ATPase much the same way as the G-patch proteins Ntr1 and Pfa1 influence the Prp43 ATPase. First, all three proteins enhance the RNA-stimulated ATPase activity of their respective enzymes (Fig. 5; Tanaka et al. 2007; Lebaron et al. 2009). Second, both Prp2 and Prp43 bind ssRNA less tightly in the presence of ADP compared with AMP-PNP (He et al. 2010), and RNA binding by these enzymes is not significantly influenced by their cognate G-patch cofactors (Christian et al. 2014; this study). Third, not only do Prp2 and Prp43 share the same domain organization—including a ratchet domain, a β-hairpin in the RecA2 domain (typical of processive helicases), and a C-terminal OB-fold domain (He et al. 2010; Walbott et al. 2010; Cordin and Beggs 2013)—but the regulation of the ATPase activity of Prp2 and Prp43 may be mediated by structurally equivalent interactions between their OB-fold domains and the G-patch domains of Spp2 and Ntr1 or Pfa1, respectively (Tanaka et al. 2007; Lebaron et al. 2009; He et al. 2010; Walbott et al. 2010; Christian et al. 2014). Finally, contacts of residues in the G-patch domain of Ntr1 with residues of the β-hairpin loop of Prp43 are essential for the function of Prp43 in the spliceosome (Tanaka et al. 2007; He et al. 2010; Walbott et al. 2010; Christian et al. 2014). As these residues are evolutionarily conserved and present also in Spp2 and Prp2, this indicates that interactions of corresponding residues in Spp2 with equivalent regions of the β-hairpin loop of Prp2 are also crucial for the function of the DEAH-box ATPase Prp2 in the spliceosome. The only difference is that Prp2 does not exhibit dsRNA unwinding activity in vitro either alone or in the presence of Spp2 (Supplemental Fig. S2; Kim et al. 1992), while Ntr1 (or Pfa1) stimulates the low intrinsic dsRNA unwinding activity of Prp43 (Tanaka et al. 2007).
However, given the striking overall similarities in their biochemical and structural properties, it is tempting to speculate that Spp2 and Ntr1 (or Pfa1) help couple the energy of ATP hydrolysis by their cognate DEAH-box ATPases to mechanical work via a common overall mechanism. Indeed, it is currently not clear whether Prp43 acts in the context of the intron–lariat spliceosome as a bona fide RNA unwindase. A major target for Prp43 is probably the interaction of the pre-mRNA BS with U2 RNA, which is still stabilized by U2 proteins in the purified intron–lariat spliceosome (Fourmann et al. 2013). Thus, it is also possible that Prp43 may have to strip the U2 proteins from the BS and that Ntr1 may employ a mechanism similar to the one proposed here for Spp2 to activate Prp43 and couple its NTPase activity to RNP remodeling in the spliceosome.
In this study, we show for the first time that it is possible to reliably measure the NTPase activity of Prp2 and its modulation by Spp2 in the context of purified spliceosomes. For most of the spliceosomal ATPase/helicases, including Prp2, evidence has been provided that they also function by proofreading certain steps of spliceosome assembly (Burgess and Guthrie 1993; Semlow and Staley 2012; Koodathingal and Staley 2013; Wlodaver and Staley 2014). The exact mechanisms by which this is achieved by the various helicases are not always clear; in particular whether they indeed act as molecular timers. The system described here should pave the way to address these questions more directly.
Materials and methods
Analysis of Prp2/pre-mRNA interactions in purified Bact spliceosomes by UV cross-linking
Approximately 0.1–2 pmol of purified Bact ΔPrp2 ΔSpp2 spliceosomes was incubated with Prp2 or Prp2 plus Spp2; the complexes were then pipetted in a thin layer onto precooled 10-well multitest slides and then irradiated for 30 sec with UV light at 254 nm on ice, essentially as described previously (Urlaub et al. 2002). SDS and EDTA were added to the irradiated and nonirradiated control samples to a final concentration of 0.1% and 10 mM, respectively, and spliceosomes were incubated for 10 min at 70°C. The reaction was then allowed to cool to 37°C, and 1 μL of 1000 U/μL RNase T1 (Ambion) was added and incubated for 30 min at 37°C followed by 30 min at 55°C. After addition of 0.3 M NaOAc, 1 μL of Glycoblue, and 4 vol of ethanol, the proteins cross-linked to 32P-labeled RNA were precipitated overnight at −20°C. After centrifugation at 13000 rpm for 30 min, the pellet was washed once with 70% ethanol, resuspended in NET-150 buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 0.1% NP-40), and subjected to immunoprecipitation.
Immunoprecipitation of Prp2/RNA cross-links
Protein A Sepharose resin (GE Healthcare) was prebound with anti-Prp2 antibody in NET-150 buffer and incubated at 4°C with UV cross-linked or non-cross-linked spliceosomal proteins prepared as above. The resin was then washed three times with NET-150 buffer, and protein loading buffer was added. The samples were denatured for 10 min at 70°C and loaded on SDS-PAGE. The gel was transferred to a nitrocellulose membrane, which was analyzed by autoradiography. Probing of the membrane was performed with rabbit polyclonal antibodies against Prp2.
Binding of Prp2 and Spp2 to the spliceosome
Purified Bact ΔPrp2 ΔSpp2 spliceosomes bound to the amylose affinity resin were incubated with a 1.5–2 molar excess of Prp2 and/or Spp2 in GK75 buffer for 10 min at 23°C. Unbound proteins were removed by washing with buffer containing 150 mM KCl. The spliceosomes were eluted and subjected to glycerol gradient centrifugation in buffer containing 75 or 150 mM KCl. Proteins recovered from the peak fractions were analyzed by Western blotting with rabbit polyclonal antibodies against Prp2, Spp2 (kind gifts from Ren-Jang Lin), Snu114, Cwc2, and Prp19 (kind gift from Kum-Loong Boon).
Analysis of Prp2’s UTPase activity in the context of the spliceosome
To monitor UTP hydrolysis by Prp2 in the presence of stoichiometric concentrations of purified Bact ΔPrp2 or Bact ΔPrp2 ΔSpp2 spliceosomes, we concentrated purified Bact complexes by ultrafiltration to ∼30–60 nM (Amicon 0.5-mL 100-kDa cutoff filter, Millipore). Nine microliters of concentrated spliceosomes were combined on ice with GK75 buffer (containing 0.15 U/µL RNAsin) mixed with 1.8 µL of protein mix containing Prp2, Spp2, and/or Cwc25 to a final concentration of 30–60 nM Prp2 and excess Spp2 or Cwc25, as indicated, or with GK75 buffer alone and incubated for 2–5 min at 23°C. The reactions were started by addition of 1.2 µL of 1 mM UTP with α32P UTP tracer (∼5000 cpm/pmol). Aliquots of 1 µL were taken at the indicated time points and stopped by mixing with 12 µL of ice-cold 50 mM EDTA and 10 mM Tris-HCl (pH 8.0). Subsequently, 1.2 µL of the stopped aliquots were spotted on a CEL 300 PEI TLC plate (Macherey Nagel) and resolved in 0.25 M KH2PO4 (pH 3.5) for 2 h (Cashel et al. 1969). Spots were visualized and quantitated using a PhosphorImager (GE Healthcare).
RNA-binding analysis of Prp2 in the absence of spliceosomes
In vitro RNA-binding studies were performed by fluorescence anisotropy. Briefly, a FAM 5′ end-labeled 24-nt ssRNA oligo (5′-GGCCGCGAGAAAAAAAAAAAAAAA-3′) (5 nM) was incubated with increasing concentrations of Prp2 (2 nM to 2 µM) in the absence or presence of 8 µM Spp2 in binding buffer (20 mM Hepes-KOH at pH 7.9, 75 mM KCl, 1.5 mM MgCl2, 5% [v/v] glycerol, 100 ng/µL BSA, 1 mM DTT) followed by addition of 1 mM AMP-PNP or ADP, when indicated, at room temperature. Fluorescence anisotropy and the apparent equilibrium Kds were measured as previously described (Santos et al. 2012).
Analysis of Prp2’s ATPase activity in the absence of spliceosomes
In vitro ATPase assays were performed in binding buffer using 15–30 nM Prp2 and the indicated concentrations of a (U)30 RNA oligonucleotide in the absence or presence of the indicated concentrations of Spp2 (Fig. 5C,D). Reactions were preincubated for 5 min at 23°C, initiated by addition of 1 mM ATP/MgCl2, and incubated for an additional 5–10 min. ATP hydrolysis was monitored using a colorimetric assay (PiColorLock, Innova Biosciences) as previously described (Santos et al. 2012), and the ATP kcats were calculated as the number of ATP molecules hydrolyzed per Prp2 molecule per second at the saturated concentrations of ATP and RNA substrates.
Supplementary Material
Acknowledgments
We thank T. Conrad and W. Behrens-Kranz for excellent technical assistance, R.-J. Lin and K.-L. Boon for providing antibodies, K. Hartmuth for helpful discussions, and C. Will for critical reading of the manuscript. Z.W. was supported by a grant (“Fuga”) from the National Science Centre (PL) and a scholarship (“START”) of the Foundation for Polish Science. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 860) to R.L.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.253070.114.
References
- Aravind L, Koonin EV. 1999. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem Sci 24: 342–344. [DOI] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C. 1993. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci 18: 381–384. [DOI] [PubMed] [Google Scholar]
- Buttner K, Nehring S, Hopfner KP. 2007. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol 14: 647–652. [DOI] [PubMed] [Google Scholar]
- Cashel M, Lazzarini RA, Kalbacher B. 1969. An improved method for thin-layer chromatography of nucleotide mixtures containing 32P-labelled orthophosphate. J Chromatogr 40: 103–109. [DOI] [PubMed] [Google Scholar]
- Chan SP, Cheng SC. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199. [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. [DOI] [PubMed] [Google Scholar]
- Cheng SC. 1994. Formation of the yeast splicing complex A1 and association of the splicing factor PRP19 with the pre-mRNA are independent of the 3′ region of the intron. Nucleic Acids Res 22: 1548–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian H, Hofele RV, Urlaub H, Ficner R. 2014. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res 42: 1162–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs DJ, Nagel RJ, Ares M Jr, Stevens SW. 2006. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol 26: 523–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Beggs JD. 2013. RNA helicases in splicing. RNA Biol 10: 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Hahn D, Beggs JD. 2012. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol 24: 431–438. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Kim DH, Kim SH, Tseng YH, Yu Y, Lin RJ. 2000. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. RNA 6: 1106–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Laggerbauer B, Lauber J, Lane WS, Lührmann R. 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J 16: 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608. [DOI] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann JB, Schmitzová J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R. 2013. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 27: 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozani O, Feld R, Reed R. 1996. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev 10: 233–243. [DOI] [PubMed] [Google Scholar]
- He Y, Andersen GR, Nielsen KH. 2010. Structural basis for the function of DEAH helicases. EMBO Rep 11: 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. 2012. Dynamic protein–protein interaction wiring of the human spliceosome. Mol Cell 45: 567–580. [DOI] [PubMed] [Google Scholar]
- Kim SH, Lin RJ. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol 16: 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Smith J, Claude A, Lin RJ. 1992. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J 11: 2319–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DS, Beggs JD. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res 18: 6559–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Query CC. 2005. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev 19: 2255–2260. [DOI] [PubMed] [Google Scholar]
- Koodathingal P, Staley JP. 2013. Splicing fidelity: DEAD/H-box ATPases as molecular clocks. RNA Biol 10: 1073–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R, Blanco MR, Kahlscheuer ML, Abelson J, Guthrie C, Walter NG. 2013. Biased Brownian ratcheting leads to pre-mRNA remodeling and capture prior to first-step splicing. Nat Struct Mol Biol 20: 1450–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR III, Stevens SW. 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last RL, Maddock JR, Woolford JL Jr. 1987. Evidence for related functions of the RNA genes of Saccharomyces cerevisiae. Genetics 117: 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Papin C, Capeyrou R, Chen YL, Froment C, Monsarrat B, Caizergues-Ferrer M, Grigoriev M, Henry Y. 2009. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J 28: 3808–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. 2006. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol 26: 513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Cheng SC. 2012. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol 32: 5056–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Muhlenkamp P. 2003. Spatial organization of protein-RNA interactions in the branch site–3′ splice site region during pre-mRNA splicing in yeast. Mol Cell Biol 23: 4174–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T, Prior M, Dannenberg J, Odenwälder P, Dybkov O, Rasche N, Schmitzová J, Gregor I, Fabrizio P, Enderlein J, et al. 2012. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 18: 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S, Lynn B, Rymond BC. 2006. Inhibition of a spliceosome turnover pathway suppresses splicing defects. Proc Natl Acad Sci 103: 13700–13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumpton M, McGarvey M, Beggs JD. 1994. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J 13: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL Jr. 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1: 375–390. [PMC free article] [PubMed] [Google Scholar]
- Rymond BC, Rosbash M. 1985. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature 317: 735–737. [DOI] [PubMed] [Google Scholar]
- Santos KF, Jovin SM, Weber G, Pena V, Lührmann R, Wahl MC. 2012. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci 109: 17418–17423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semlow DR, Staley JP. 2012. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci 37: 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. 2004. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 24: 10101–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Aronova A, Schwer B. 2007. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes & Dev 21: 2312–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S, McGarvey M, Plumpton M, Beggs JD. 1994. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J 13: 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub H, Hartmuth K, Lührmann R. 2002. A two-tracked approach to analyze RNA-protein cross-linking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods 26: 170–181. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N. 2010. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J 29: 2194–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243. [DOI] [PubMed] [Google Scholar]
- Wlodaver AM, Staley JP. 2014. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA 20: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysoczanski P, Schneider C, Xiang S, Munari F, Trowitzsch S, Wahl MC, Lührmann R, Becker S, Zweckstetter M. 2014. Cooperative structure of the heterotrimeric pre-mRNA retention and splicing complex. Nat Struct Mol Biol 21: 911–918. [DOI] [PubMed] [Google Scholar]
- Yeh TC, Liu HL, Chung CS, Wu NY, Liu YC, Cheng SC. 2011. Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol Cell Biol 31: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang S, Lin TY, Chen X, Gencheva M, Newo AN, Yang L, Rossi D, Hu J, Lin SB, Huang A, et al. 2014. GPKOW is essential for pre-mRNA splicing in vitro and suppresses splicing defect caused by dominant-negative DHX16 mutation in vivo. Biosci Rep doi: 10.1042/BSR20140142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.