Abstract
Resting-state functional connectivity (FC) fMRI (rs-fcMRI) offers an appealing approach to mapping the brain’s intrinsic functional organization. Blood oxygen level dependent (BOLD) and arterial spin labeling (ASL) are the two main rs-fcMRI approaches to assess alterations in brain networks associated with individual differences, behavior and psychopathology. While the BOLD signal is stronger with a higher temporal resolution, ASL provides quantitative, direct measures of the physiology and metabolism of specific networks. This study systematically investigated the similarity and reliability of resting brain networks (RBNs) in BOLD and ASL. A 2 × 2 × 2 factorial design was employed where each subject underwent repeated BOLD and ASL rs-fcMRI scans on two occasions on two MRI scanners respectively. Both independent and joint FC analyses revealed common RBNs in ASL and BOLD rs-fcMRI with a moderate to high level of spatial overlap, verified by Dice Similarity Coefficients. Test–retest analyses indicated more reliable spatial network patterns in BOLD (average modal Intraclass Correlation Coefficients: 0.905 ± 0.033 between-sessions; 0.885 ± 0.052 between-scanners) than ASL (0.545 ± 0.048; 0.575 ± 0.059). Nevertheless, ASL provided highly reproducible (0.955 ± 0.021; 0.970 ± 0.011) network-specific CBF measurements. Moreover, we observed positive correlations between regional CBF and FC in core areas of all RBNs indicating a relationship between network connectivity and its baseline metabolism. Taken together, the combination of ASL and BOLD rs-fcMRI provides a powerful tool for characterizing the spatiotemporal and quantitative properties of RBNs. These findings pave the way for future BOLD and ASL rs-fcMRI studies in clinical populations that are carried out across time and scanners.
Keywords: Arterial spin labeling, Default mode network, Functional connectivity, Resting-state, Test–retest repeatability
Introduction
Since the seminal work by Biswal et al. in 1995 (Biswal et al., 1995), the study of resting brain networks (RBN) based on functional connectivity (FC) in resting state fMRI (rs-fcMRI) has experienced an upsurge from basic to clinical neuroscience. The majority of rs-fcMRI studies have used blood oxygen level dependent (BOLD) contrast due to its technical simplicity, high sensitivity and temporal resolution. Recently, a growing number of rs-fcMRI studies have employed arterial spin labeled (ASL) perfusion MRI (Chuang et al., 2008; Dai et al., 2013; Jann et al., 2013; Liang et al., 2011, 2012; Zou et al., 2009), which measures cerebral blood flow (CBF) using magnetically labeled arterial blood water as an endogenous tracer (Detre et al., 1992). Compared to BOLD, perfusion-based FC analysis provides more direct and quantitative measures of the physiology and metabolism of specific networks (Buxton et al., 2004). The inherently quantitative nature of ASL allows for the assignment of biologically meaningful values to the networks, thus may complement BOLD by providing a more interpretable biomarker.
To date, however, the application of perfusion-based rs-fcMRI in clinical neuroscience has been hampered by the relatively low sensitivity and temporal resolution of ASL compared to BOLD. The recent development of pseudo-continuous ASL (pCASL) with background suppressed (BS) 3D acquisitions (e.g. GRASE — a hybrid of spin and gradient echo and Stack-of-Spirals) has dramatically improved the sensitivity and temporal SNR of perfusion imaging series (Alsop et al., 2014; Fernandez-Seara et al., 2008), allowing the detection of CBF based RBNs while minimizing potential BOLD contaminations (Du et al., 2012; Liang et al., 2012). Another appealing feature of perfusion based rs-fMRI using pCASL with 3D BS GRASE or Stack-of-Spirals is the improved visualization of RBNs involving brain regions affected by susceptibility artifacts at the tissue–air interfaces (Fernandez-Seara et al., 2005).
Given the complementary nature of BOLD and perfusion rs-fcMRI —higher sampling rate/temporal resolution in BOLD and absolute CBF quantification in ASL, the combination of the two contrasts may offer a powerful tool for rs-fcMRI studies to fully characterize the spatiotemporal and quantitative properties of RBNs. The primary purpose of this study was to present a framework for independent and joint FC analyses of BOLD and perfusion based rs-fcMRI data to identify common and modality specific RBNs, using rigorous statistical approaches. For future applications of BOLD and perfusion-based functional connectivity analyses in clinical studies, it is critical to establish the reliability of RBNs across time (Meindl et al., 2010; Shehzad et al., 2009; Zuo et al., 2010), scanner platforms (Van Dijk et al., 2010) and modalities as well as their dependencies on imaging parameters (Birn et al., 2013; Patriat et al., 2013; Van Dijk et al., 2010)). For this purpose, a 2 × 2 × 2 factorial design was employed in the present study using repeated BOLD and ASL rs-fcMRI scans on two occasions on two MRI scanners respectively. We hypothesized that BOLD and ASL rs-fcMRI should show common RBNs that are reproducible across time and scanners. The overall FC in BOLD RBNs is stronger than that of ASL RBNs, yet ASL networks show higher FC in specific brain regions (e.g. orbitofrontal cortex). Finally, network specific quantitative CBF measured by ASL may indicate the baseline metabolic activity and may be associated with (or underlie) the strength of functional connectivity of the corresponding network (Aslan et al., 2011; Liang et al., 2013; Tomasi et al., 2013).
Materials and methods
Participants and data acquisition
Ten healthy volunteers (6f/4 m; Age [mean ± std] = 22 ± 3 years) underwent repeated MRI scans on two 3T Siemens TIM Trio MR systems using the standard 12-channel head coils and identical pulse sequences. A 2 × 2 × 2 factorial design was employed, i.e., 2 repeated scans on 2 scanners using 2 modalities (ASL and BOLD). On the first day they participated in two sessions approximately one hour apart on one of the two scanners, and on the second day (2.1 ± 1.3 days apart) the protocol was repeated on the other scanner (scanner order was counterbalanced across participants). Each session included resting state (rs-) BOLD imaging with 2D EPI readout and the following parameters: Volumes = 240, Matrix = 64 × 64, Slice Thickness = 4 mm with 1 mm gap, 30 slices, Repetition Time/Echo Time (TR/TE) = 2000/30 ms, Flip Angle = 77°, Pixel Bandwidth = 2298 Hz, Field of View = 220 mm; resting state pseudo-continuous ASL (rs-pCASL) with single-shot 3D background suppressed GRASE readout and the following parameters: Volumes = 120 (60 label/control pairs), Matrix = 64 × 64, Slice Thickness = 5 mm, 26 slices, Repetition Time/Echo Time/Label Time/Post Label Delay (TR/TE/τ/PLD) = 4000/22/1200/1000 ms, Pixel Bandwidth = 2003 Hz, Field of View = 220 mm, labeling offset = 9 cm, and 2 global inversion pulses were applied during the PLD for background suppression (the first BS pulse was applied immediately after the labeling pulses, the second BS pulse 700 ms after the labeling pulses. Overall these two BS pulses achieved ~85% suppression for gray and white matter signals. The residual image intensity was necessary for motion correction as well as to avoid zero crossing signals for subtraction between label and control images) (Kilroy et al., 2013; St Lawrence et al., 2012). For CBF quantification an additional volume (M0, equilibrium magnetization image) with the same parameters as described for pCASL but with a long PLD of 4000 ms and without BS was acquired. Finally, a T1 weighted, high resolution, anatomical scan was performed (MP-RAGE, 192 sagittal slices with 1 mm isotropic voxels, TR/TE/TI = 2170/4.33/1100 ms). Resting state was defined as lying still with eyes open while fixating a white cross at the center of a dark screen.
Data processing
Analysis of the MRI data was performed using SPM8 (http://www.fil.ion.ucl.ac.uk/spm/) and in-house Matlab (The MathWorks, Natick, USA) routines. Statistical analyses were performed using Matlab Statistics Toolbox, R Programming Project (http://www.r-project.org/) and IBM SPSS Statistics (Version 19).
CBF quantification
Raw pCASL GRASE images were motion corrected separately for control and label images (Wang et al., 2008) before perfusion-weighted time series were created by sinc-subtraction of label and control images (ΔM). Notably, sinc-subtraction has been demonstrated to efficiently minimize spurious BOLD contaminations within the ASL signal (Aguirre et al., 2002; Chuang et al., 2008; Liu and Wong, 2005). The computation of the quantitative CBF signal was based on a single compartment kinetic model (Chen et al., 2011; Wang et al., 2003, 2005). After quantification, the CBF images were co-registered to the individuals’ anatomical scans, normalized into MNI standard space (thereby resampled into 2 × 2 × 2 mm voxel resolution) and spatially smoothed with an 8 mm full-width at half maximum (FWHM) Gaussian Kernel.
BOLD images were first slice-time and motion corrected followed by coregistration, normalization and spatial smoothing identical to the CBF images. Anatomical images were normalized into standard MNI space and segmented into gray matter, white matter and cerebrospinal fluid (GM/WM/CSF) using the algorithms provided by SPM8. The individual GM images were averaged and thresholded at 0.3, providing a binary mask representing gray matter voxels only (GM mask).
Network decomposition using ICA
Networks were identified by means of the Group ICA of fMRI Tool-box (GIFT) using a concatenated group level ICA approach (Calhoun et al., 2001, 2004).
First, separate group ICAs for BOLD and ASL data were performed. The individual time series were zero-meaned and the GM mask was applied for the ICA infomax algorithm (Bell and Sejnowski, 1995). The ICA model order was estimated using the AIC/MDL criterion, which yielded an optimal number of components for each individual dataset with a median of 36 for BOLD and 20 components for ASL datasets. Group- and subject-specific maps (as back — reconstructed by the GICA procedure) were stored as z-maps. RBNs were identified by means of conjoint template correlation (GIFT) and visual inspection of the component maps according to the spatial distribution given in the literature (Beckmann et al., 2005; Franco et al., 2009).
Since different ICA model orders can lead to unequal decompositions thus splitting components into subnetworks or merging of networks (Abou-Elseoud et al., 2010; Calhoun et al., 2009), matching and ordering of components across ICA runs are not a straightforward procedure from a mathematical point of view. To address this issue on drawing inferences about, and comparisons between subject cohorts, group approaches and joint ICA approaches have been suggested (Calhoun et al., 2001, 2009), which perform the unmixing on the temporally concatenated dataset. These approaches assure that subject or modality specific components are based on the same group component hence enabling comparative statistics for BOLD and ASL derived networks without bias of separate decompositions. Accordingly we employed a joint-ICA where the BOLD and CBF datasets were assigned to 8 (i.e., 2 × 2 × 2) sessions per subject and temporally concatenated. The same ICA parameters were used for this approach with subsequent back-reconstruction for sessions and modalities. The MDL criterion for the joint dataset yielded a median of 16 components for the aggregated group dataset.
We computed t-maps to display the separate BOLD and ASL RBNs in joint and separate ICA. Specifically in the joint ICA the group component is the same for ASL and BOLD as it is computed across all datasets. The back-regressed individual subject and session IC-maps were then used to generate modality specific ASL and BOLD RBNs using one-sample t-tests against zero (significance set at p < 0.001). While the t-maps were used for displaying the networks, the z-scores representing the ICA group components were used to generate RBN-masks that were used in the analyses below.
Statistics
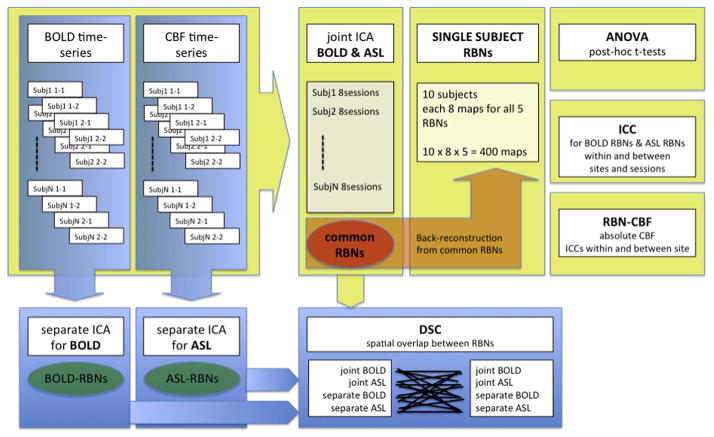
Spatial similarity and overlap of BOLD and ASL based RBNs were assessed by Dice Similarity Coefficients (DSC), while test–retest repeatability of ASL and BOLD based RBNs was estimated using Intraclass Correlation Coefficients (ICCs). Statistical analyses were performed on the network as well as on a voxel-wise level. Repeated-measures ANOVAs and post-hoc t-tests were computed to identify differences between modalities, scanners and sessions, as well as possible interaction effects. A schematic overview of analyses is given in Fig. 1.
Fig. 1.
Schematic workflow of ICA of BOLD and ASL data. ICA decompositions were performed separately for BOLD and ASL datasets as well as in a combined joint-ICA to compute Resting Brain Networks (RBNs). Dice Similarity Coefficients (DSC) were calculated to compare spatial overlap of resulting RBNs across different ICA runs. Test–retest reliability of RBNs across session and scanner was estimated by means of Intra-class correlation coefficients (ICC). An ANOVA with post-hoc t-test was employed to statistically compare RBNs from BOLD and ASL. Finally, baseline activity of RBNs (RBNCBF) was calculated and associations between RBNCBF and functional connectivity strength were tested by Pearson correlation coefficients.
Statistical maps were corrected for multiple comparisons (type I errors) using AlphaSim (Ward, 2000). This procedure estimates the distribution of random cluster sizes given a statistical map and threshold taking into consideration the spatial smoothness of the data (Bennett et al., 2009; Forman et al., 1995). Accordingly, it provides the minimal cluster size required for clusters to be at a level above random clustering at a chosen correction level. We performed 1000 iterations and selected the correction level at alpha < 0.05.
Dice Similarity Coefficients (DSC) of RBNs
To quantify the degree of similarity and spatial overlap of RBNs, we computed the Dice’s Similarity Coefficient (DSC) (Dice, 1945; Zhu et al., 2013) according to the formula:
| (1) |
where, A and B represent sets of voxels within two given RBNs (thresholded at z > 2) and the parallel brackets denote the number of voxels in the set within the brackets.
Network based CBF
Mean network perfusion was defined as the spatially and temporally averaged CBF values across all voxels within the group RBN map where z > 2, here dubbed as RBNCBF. The resulting four RBNCBF metrics (one for each of the four ASL scans) per subject were subjected to a 2 × 2 repeated measures ANOVA with within-subject factors of site (1; 2) and session (1; 2) to test for the consistency of CBF quantification for each network respectively.
Network Amplitudes of Low Frequency Fluctuations (ALFF)
In addition to RBN-CBF we also computed the Amplitudes of Low Frequency Fluctuations (ALFF) (Zang et al., 2007), which provides a measure of regional spontaneous activity fluctuations. Using dynamic CBF to compute ALFFs instead of relative BOLD signal fluctuations, the ALFFs have a direct physiological meaning and a quantitative unit (i.e. ml/100 g/min). Accordingly we computed the CBF-ALFF for each session in each subject respectively, and extracted the corresponding mean ALFF in each network. ALFF differences across networks and subjects were then tested using an ANOVA. Additionally, we normalized the ALFF values with regard to subjects’ specific RBN-CBF (adjusted for global GM-CBF) providing a %ALFF for each subject with respect to their baseline RBN-CBF values (Chuang et al., 2008). These %ALFF measurements were also subjected to an ANOVA.
Intraclass Correlation Coefficient (ICCs)
Voxel wise maps for FC and regional CBF
The ICC estimation was based on a repeated-measure mixed effects ANOVA model with absolute agreement of values (A-k) (Landis and Koch, 1977; McGraw and Wong, 1996; Shrout and Fleiss, 1979). The ICC measures the proportion of total variance that is accounted for by the variation between subjects against the variance associated with either between-site or within-site effects. Hence, ICC over 0.5 indicates that scanner or session variation is lower than between-subjects variance. ICC was calculated by
| (2) |
where MSw and MSb are the within- and between-subject errors respectively, MSe is the mean residual error and n is the sample size.
Voxel-wise maps of ICC for either FC or CBF were computed for within-site (averaging z-/CBF-maps across sites) and between-site (averaging z-/CBF-maps across sessions) variance. Further, to facilitate interpretation, we calculated a single ICC value for each network (using only voxels within a given RBN where z > 2) by taking the mode of the ICC value distribution histogram between 0 and 1 (Zuo et al., 2010). The mode of a histogram distribution represents the most prevalent value in the distribution. Besides the modal-ICC we also computed the percentage of voxels with ICC > 0.6 within each RBN as compared the total number of voxels within this RBN (threshold z > 2).
ANOVA
Voxel-wise maps for FC
As separate ICAs can result in unequal decompositions of networks as outlined in Materials and methods, the statistical comparison of RBNs was performed using single-subject maps resulting from the joint ICA. This assured that different model orders, component unmixing, network splitting/merging or component matching across ICA runs does not bias possible network differences.
We performed voxel-wise 2 × 2 × 2 repeated measures ANOVAs on the single subject z-maps with within-subject factors of modality (BOLD; CBF), site (1,2) and session (1,2) for each RBN. Statistical thresholds for main and interaction effects were set at p < 0.01 (F (1,9) = 10.56). Maps with significant effects were further subjected to two-sample two-sided post-hoc t-tests (significance level p < 0.001).
Correlation between RBNCBF and network z-scores
To assess the potential relationships between individual network connectivity and baseline network perfusion, we computed the voxel-wise Pearson correlation between the RBN z-scores and the respective regional (voxel-level) CBF from the same scan session. In addition, we compared the network connectivities between the two modalities by correlating the average z-scores of all joint BOLD-RBNs (10 subjects across 4 sessions) to those of the corresponding ASL-RBNs. Using ICA as compared to Seed Based Approaches (SBA) where a direct correlation between any two specified brain areas is computed and considered as FC strength, the FC strength used here (ICA z-scores) represents the degree to which a given voxel is integrated within a given network component, i.e. its relative connectivity strength to all other voxels in the specific network. To provide an estimate of how prevalent an CBF-FC relation is within each RBN, we also computed the percentage of voxels with significant correlations above r > 0.4 within each RBN as compared to the total number of voxels within this RBN (threshold z > 2).
Results
Common RBNs in BOLD and ASL rs-fMRI
The three different ICA decompositions (i.e., ASL-only, BOLD-only and joint ASL/BOLD) revealed five common RBNs: the Default Mode Network (DMN, correlation of ICA group component to template networks for: BOLD-only ICA R = 0.37, ASL only ICA R = 0.37, joint ICA R = 0.28), the two lateralized Executive Control Networks (ECNs) (RECN; RBOLD = 0.36, RASL = 0.22, Rjoint = 0.28/LECN; RBOLD = 0.33, RASL = 0.24, Rjoint = 0.24) the Occipital Visual Network (OVN; RBOLD = 0.34, RASL = 0.28, Rjoint = 0.35) and the Auditory Network (AUN; RBOLD = 0.32, Rjoint = 0.18). The spatial pattern of the 5 networks is depicted in Fig. 2 for separate ICAs and Fig. 3 for joint ICA (multi-slice views of RBNs displayed in Fig. S1) respectively. The results consistently showed similar RBNs in each modality and assessment of spatial similarity using DSC demonstrated a moderate to high level of overlap for all RBNs across ICA runs (Table 1, Fig. S6).
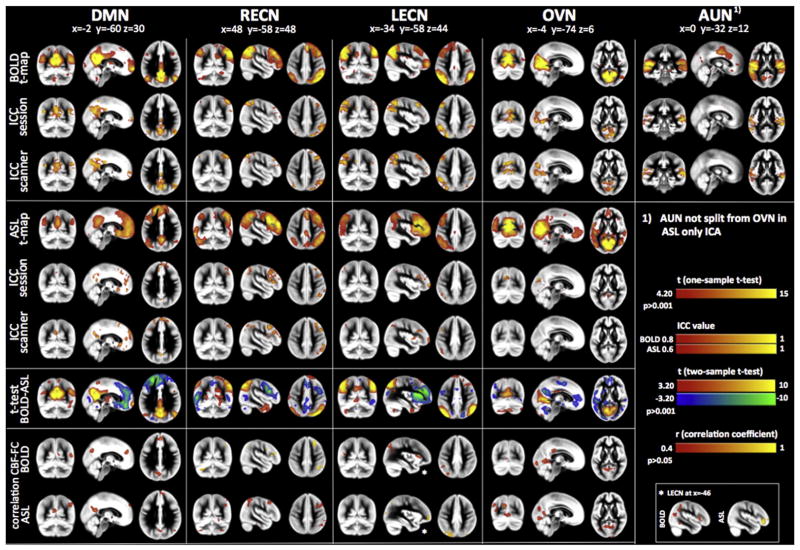
Fig. 2.
Results from separate ICAs for BOLD and ASL. Modality specific group RBNs were computed as one-sample t-test across the specific single subject maps. Left column displays BOLD RBN results while right column those for ASL. Five common RBNs were analyzed: Default Mode Network (DMN), left and right Executive Control Networks (L/R-ECNs), Occipital Visual Network (OVN) and Auditory Network (AUN). Test–retest reliability between sessions and scanners are displayed as Intraclass Correlation Coefficient (ICC) maps for BOLD and ASL networks (ICC maps were masked by ICA group RBN maps thresholded at z > 2). Differences between the two modalities (BOLD vs. ASL) were assessed by means of two-sample two-sided t-tests (significance threshold was set at p < 0.001). The bottom row displays voxel-wise correlation maps for CBF and Functional Connectivity strength (z-scores) for the five distinct networks (only significant (p < 0.05) correlations above r > 0.4 are displayed. Correlation maps have been masked by ICA group RBN maps thresholded at z > 2). Multi-slice views of all analyses can be found in Supplemental Figs. S1–S5.
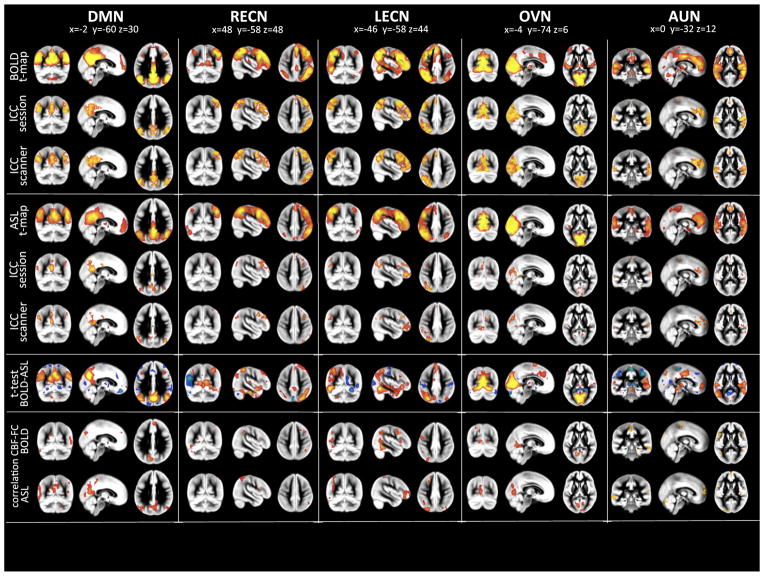
Fig. 3.
Results from joint-ICAs for BOLD and ASL (Display, organization and color-scaling are analog to Fig. 2). Modality specific group RBNs were computed as one-sample t-test across the specific single subject maps. Left column displays BOLD RBN results while right column those for pCASL. Five common RBNs were analyzed: Default Mode Network (DMN), left and right Executive Control Networks (L/R-ECNs), Occipital Visual Network (OVN) and Auditory Network (AUN). Test–retest reliability for session and scanner are displayed as Intraclass Correlation Coefficient (ICC) maps for BOLD and ASL networks (ICC maps were masked by ICA group RBN maps thresholded at z > 2). Differences between the two modalities (BOLD vs. ASL) were assessed by means of two-sample two-sided t-tests (significance threshold was set at p < 0.001). The bottom row displays voxel-wise correlation maps for CBF and Functional Connectivity strength (z-scores) for the five distinct networks (only significant (p < 0.05) correlations above r > 0.4 are displayed. Correlation maps have been masked by ICA group RBN maps thresholded at z > 2). Multi-slice views of all analyses can be found in Supplemental Figs. S1–S5.
Table 1.
Dice Similarity Coefficients (DSCs) between RBNs resulting from different ICA runs (joint-ICA group component GC, joint-ICA ASL networks, joint-ICA BOLD networks and RBNs from separate BOLD (BOLD-only) or ASL (ASL-only) ICAs. Highlighted are the DSCs between RBNs from ASL and BOLD based on the joint-ICA approach indicating high concordance of network patterns.
| Joint ASL | Joint BOLD | Only ASL | Only BOLD | |
|---|---|---|---|---|
| DMN | ||||
| Joint GC | 0.81 | 0.89 | 0.29 | 0.61 |
| Joint ASL | 0.71 | 0.35 | 0.57 | |
| Joint BOLD | 0.27 | 0.61 | ||
| Only ASL | 0.35 | |||
| RECN | ||||
| Joint GC | 0.76 | 0.82 | 0.67 | 0.62 |
| Joint ASL | 0.59 | 0.67 | 0.54 | |
| Joint BOLD | 0.60 | 0.64 | ||
| Only ASL | 0.56 | |||
| LECN | ||||
| Joint GC | 0.80 | 0.85 | 0.61 | 0.52 |
| Joint ASL | 0.66 | 0.61 | 0.45 | |
| Joint BOLD | 0.58 | 0.55 | ||
| Only ASL | 0.41 | |||
| OVN | ||||
| Joint GC | 0.77 | 0.94 | 0.46 | 0.69 |
| Joint ASL | 0.71 | 0.47 | 0.59 | |
| Joint bold | 0.44 | 0.69 | ||
| Only ASL | 0.50 | |||
| AUN | ||||
| Joint GC | 0.79 | 0.86 | – | 0.57 |
| Joint ASL | 0.66 | – | 0.47 | |
| Joint BOLD | – | 0.60 | ||
| Only ASL | – | |||
Between BOLD-only and ASL-only RBNs, the left and right ECN (DSC = 0.42/0.56) and the DMN (0.35) showed a moderate to high level of overlap, whereas the OVN and AUN appeared as a single component in ASL-only ICA and a direct quantitative comparison was thus inappropriate (Fig. 3). Comparing the RBNs detected by separate ICA to the joint-ICA networks revealed a high level of agreement between BOLD-only and joint RBNs (average DSC across networks [mean ± SD] = 0.58 ± 0.05), while for ASL-only ICA the two ECNs reached the same high degree of similarity (R/L-ECN = 0.65/0.60) but the DMN (0.30) and OVN (0.45) had reduced spatial overlap with the corresponding networks in joint ICA. Finally, RBNs from joint-ICA for BOLD and ASL showed the highest agreement to the common group component (DSC range 0.76–0.94) as well as between each other (DSC range 0.59–0.71). Although this high agreement is somewhat expected as the RBNs were derived from the same concatenated ICA decomposition, it is required to compare networks across modalities while ruling out effects of different ICA model orders and decompositions.
Repeatability of RBNs across time and scanner
Test–retest repeatability of ASL and BOLD based RBNs was estimated using ICCs on a voxel-wise basis. The spatial distribution of ICC values overlapped with the pattern of z-maps for the respective networks (Figs. 2 & 3 and S2 & S3). Higher ICCs were found in areas with high FC. Separate ICA for BOLD and ASL demonstrated good to excellent ICCs in BOLD (average modal ICCs across RBNs: between sessions 0.800 ± 0.094; between scanners 0.735 ± 0.074), while ASL yielded lower but still fairly reliable ICC values (0.619 ± 0.080/0.506 ± 0.109) (Table 2). For joint-ICA based RBNs reliability was higher for BOLD RBNs showing average modal ICCs of 0.905 ± 0.033 between sessions and 0.885 ± 0.052 between scanners, and modal ICCs for ASL RBNs were similar to ASL-only for reliability between sessions (0.545 ± 0.048) and between scanners (0.575 ± 0.059) (Table 2). Voxel-wise display of ICC maps showed reliable test–retest repeatability between sessions and scanners (ICC > 0.6, threshold in Figs. 2 & 3) for all networks within their core areas (Table 3a).
Table 2.
Within and between-scanner modal-ICC and standard deviations for BOLD and ASL resting brain networks (RBNs) for joint as well as separate ICA runs and average values over the four chopped-BOLD runs (for chopped-BOLD please see Supplemental Material). Lower right lists the RBNs respective ICC values for CBF as well as for the global GM.
| Modal | std | ||||
|---|---|---|---|---|---|
| Within | Between | Within | Between | ||
| BOLD joint | DMN | 0.925 | 0.900 | 0.174 | 0.173 |
| LECN | 0.900 | 0.925 | 0.183 | 0.179 | |
| RECN | 0.875 | 0.875 | 0.194 | 0.194 | |
| OVN | 0.950 | 0.925 | 0.110 | 0.144 | |
| AUN | 0.875 | 0.800 | 0.179 | 0.210 | |
| Modal | std | ||||
| Within | Between | Within | Between | ||
|
| |||||
| BOLD separate | DMN | 0.825 | 0.800 | 0.209 | 0.219 |
| LECN | 0.925 | 0.825 | 0.229 | 0.228 | |
| RECN | 0.675 | 0.700 | 0.229 | 0.223 | |
| OVN | 0.750 | 0.700 | 0.215 | 0.210 | |
| AUN | 0.825 | 0.650 | 0.229 | 0.229 | |
| Modal* | std* | ||||
| Within | Between | Within | Between | ||
|
| |||||
| BOLD chopped | DMN | 0.681 | 0.700 | 0.214 | 0.215 |
| LECN | 0.744 | 0.725 | 0.209 | 0.812 | |
| RECN | 0.731 | 0.706 | 0.219 | 0.226 | |
| OVN | 0.794 | 0.713 | 0.226 | 0.221 | |
| AUN | 0.669 | 0.656 | 1.056 | 3.059 | |
| Modal | std | ||||
| Within | Between | Within | Between | ||
|
| |||||
| ASL joint | DMN | 0.550 | 0.625 | 0.240 | 0.232 |
| LECN | 0.625 | 0.650 | 0.223 | 0.226 | |
| RECN | 0.525 | 0.525 | 0.221 | 0.223 | |
| OVN | 0.500 | 0.525 | 0.218 | 0.215 | |
| AUN | 0.525 | 0.550 | 0.230 | 0.231 | |
| Modal | std | ||||
| Within | Between | Within | Between | ||
|
| |||||
| ASL separate | DMN | 0.675 | 0.425 | 2.871 | 0.342 |
| LECN | 0.550 | 0.400 | 0.225 | 1.826 | |
| RECN | 0.700 | 0.600 | 0.217 | 0.226 | |
| OVN/AUN | 0.550 | 0.600 | 0.233 | 3.124 | |
| Modal | std | ||||
| Within | Between | Within | Between | ||
|
| |||||
| CBF | DMN | 0.975 | 0.950 | 0.057 | 0.041 |
| LECN | 0.950 | 0.975 | 0.051 | 0.053 | |
| RECN | 0.950 | 0.975 | 0.043 | 0.057 | |
| OVN | 0.975 | 0.975 | 0.024 | 0.040 | |
| AUN | 0.925 | 0.975 | 0.074 | 0.049 | |
| Global GM | 0.916 | 0.915 | 0.065 | 0.060 | |
avarage over 4 runs
Table 3.
Percentage of voxels within each of the five RBNs showing ICC > 0.6 or a significant CBF-FC correlation in either BOLD or ASL.
| DMN | RECN | LECN | OVN | AUN | |
|---|---|---|---|---|---|
| A) Percentage of voxels within each RBN exhibiting ICC > 0.6
| |||||
| ASL intra | 31.28% | 22.28% | 25.32% | 23.57% | 19.46% |
| ASL inter | 25.74% | 19.27% | 21.71% | 22.19% | 19.94% |
| BOLD intra | 80.46% | 74.64% | 81.09% | 96.52% | 80.56% |
| BOLD inter | 82.40% | 75.77% | 82.79% | 90.58% | 63.44% |
| B) Percentage of voxels within each RBN displaying significant CFB-FC correlation
| |||||
| BOLD | 3.54% | 3.35% | 8.62% | 18.66% | 2.97% |
| ASL | 28.64% | 2.11% | 15.09% | 16.33% | 9.77% |
Analysis of quantitative CBF within RBNs
Quantitative CBF values were obtained within masks of the gray matter (GMCBF = 62.1 ± 13.1 ml/100 g/min) and the identified RBNs (RBNCBF) by joint ICA respectively (DMN 71.1 ± 3.0 ml/100 g/min; LECN 62.2 ± 2.83 ml/100 g/min; RECN 61.7 ± 3.6 ml/100 g/min; OVN 61.7 ± 4.4 ml/100 g/min; AUN 71.6 ± 5.9 ml/100 g/min; see Fig. 4).
Fig. 4.
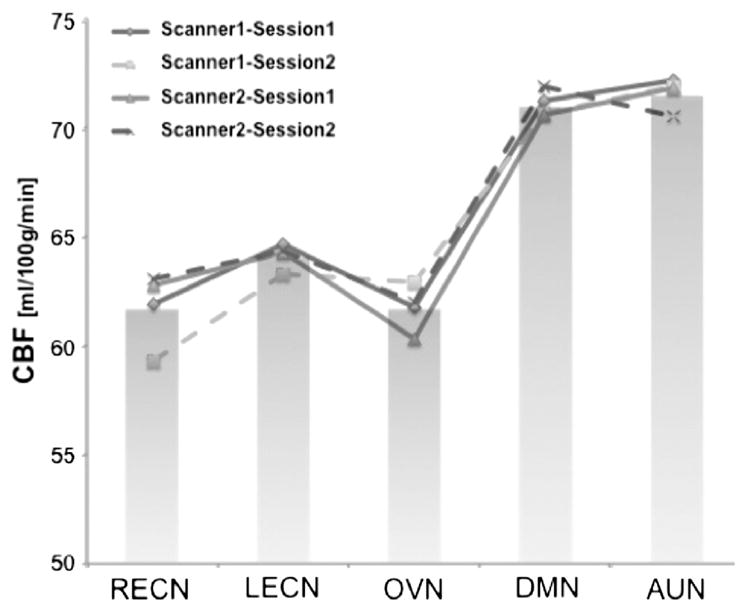
Mean CBF values for RBNs (bar-plot). Line-plot represents mean values of separate ASL sessions. Default Mode Network (DMN), left and right Executive Control Networks (L/R-ECNs), Occipital Visual Network (OVN) and Auditory Network (AUN).
The 2 × 2 repeated measures ANOVAs of global mean GMCBF as well as GM-adjusted RBNCBF values for the DMN, the L-ECN and the AUN did not show a statistically significant effect of scanner or session. The R-ECN showed an interaction effect of scanner * session (Fscanner = 9.13, p = 0.01/Fsession = 2.33, p = 0.16/Fscanner * session = 10.88, p = 0.01) while the OVN showed an effect of session only (Fscanner = 2.04, p = 0.19/Fsession = 5.29, p = 0.05/Fscanner * session = 0.18, p = 0.68). Furthermore, a main effect of network on RBNCBF values was observed (F(4,36) = 14.97, p < 0.001) indicating that mean baseline perfusion varies significantly across brain networks, with the DMN and the AUN exhibiting higher CBF than the other 3 RBNs.
Repeatability of RBNCBF values across time and scanner was further evaluated by ICC and showed highly reproducible global mean GMCBF (modal ICC = 0.915 between sessions and 0.914 between scanners, voxel-wise maps of CBF ICCs can be found in Fig. S7). Average modal ICC values across RBNCBF were 0.955 ± 0.021 between sessions and 0.970 ± 0.011 between scanners (see Table 2 bottom right).
Analysis of CBF based ALFF
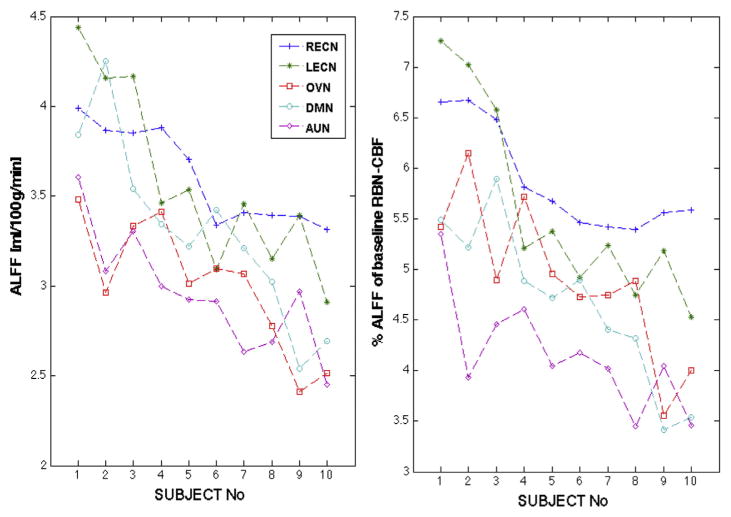
The ALFF analysis (Fig. 5A) revealed significant differences across networks (ANOVA F-statistic F = 5.69, p < 0.001) and post-hoc t-tests (Tukey–Kramer) revealed that R/L-ECNs were significantly different from OVN and AUN at p < 0.05 level, whereas DMN did not show differences to any other network. Furthermore, RBN-ALFF was significantly different across subjects (F = 4.37, p < 0.0005). Similarly, the normalized ALFF (%ALFF signal of respective RBN-CBF; Fig. 5B) showed also a significant effect of RBN (F(4) = 8.9 p < 0.00003) with significant differences between RECN and DMN, OVN and AUN, as well as LECN and AUN, where the two ECNs showed higher % fluctuations than other networks. Furthermore, the %ALFF with regard to RBN-CBF was different across subjects (F(9) = 2.92, p < 0.01).
Fig. 5.
A) Average CBF-ALFF for each network and subject. B) %ALFFs with respect to RBN-CBF (adjusted for global GM-CBF). Subjects were ordered according to their overall mean ALFF. ANOVA revealed significant ALFF differences across RBNs and subjects for both absolute and normalized ALFF.
Differences in RBNs between BOLD and ASL
Analyses of the variations of the FC strength (or z values) of the detected joint-ICA based RBNs (between modalities, scanners and sessions, as well as associated interaction effects) were performed on a voxel-wise level, using repeated-measures ANOVAs and post-hoc t-tests. These analyses indicated a significant main effect of modality (BOLD vs. ASL) in all networks. The majority of the differences between BOLD and ASL RBNs were observed within the brain areas constituting the corresponding networks. In general, BOLD networks showed a stronger overall level of FC, with the exception of higher FC in several specific regions of CBF networks (Figs. 3 & S4):
The DMN showed higher FC for BOLD in the posterior areas (Precuneus and bilateral angular gyrus) but higher FC in orbital-medial frontal cortex in ASL.
The two ECNs displayed higher FC in BOLD within network areas on the respective hemisphere (inferior and superior frontal gyri as well as temporal gyrus), but increased FC on the contralateral homotopic areas in ASL.
The AUN showed a difference in areas of the DMN (Precuneus and medial frontal gyrus) where ASL showed higher FC.
The OVN showed significantly stronger FC for BOLD in the primary visual cortex.
The coordinates and cluster sizes of detected significant FC differences are reported in Table 4.
Table 4.
Summary table of two-sample t-test between single subject RBN maps from BOLD and ASL respectively. Anatomical label, peak voxels of clusters, cluster sizes and t-value at peak voxels are listed.
| DMN | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster |
t-Value at peak coordinate |
Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster |
t-Value at peak coordinate |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||||||||||
| ASL > BOLD | −54 | 10 | 34 | Left | Inferior frontal gyrus | 501 | −6.26 | BOLD > ASL | −56 | −10 | −12 | Left | Middle temporal gyrus | 1372 | 6.46 |
| −52 | −40 | 54 | Left | Inferior parietal lobe | 1221 | −5.56 | −44 | −70 | 30 | Left | Angular Gyrus | 2241 | 9.52 | ||
| −46 | 40 | 24 | Left | Middle frontal gyrus | 312 | −5.43 | −32 | 52 | −2 | Left | Middle frontal gyrus | 319 | 6.81 | ||
| −32 | −2 | 64 | Left | Precentral gyrus | 343 | −5.46 | −20 | −88 | −8 | Left | Lingual gyrus | 606 | 7.57 | ||
| −20 | −88 | 24 | Left | Cuneus | 633 | −5.95 | 48 | −64 | 28 | Right | Angular gyrus | 1872 | 10.01 | ||
| −4 | 44 | −20 | Left | Orbito-frontal gyrus | 692 | −7.68 | 44 | −2 | 18 | Right | Insula | 377 | 5.60 | ||
| 60 | −42 | 48 | Right | Inferior parietal lobe | 752 | −5.67 | 38 | 22 | 24 | Right | Inferior frontal gyrus | 272 | 6.11 | ||
| 50 | 20 | −6 | Right | Inferior frontal gyrus | 287 | −5.31 | 36 | −24 | −10 | Right | Hippocampal gyrus | 419 | 5.94 | ||
| 40 | 44 | 32 | Right | Middle frontal gyrus | 427 | −5.80 | 26 | −80 | −6 | Right | Fusiform gyrus | 463 | 6.44 | ||
| 30 | 0 | 66 | Right | Superior frontal gyrus | 441 | −4.79 | 26 | −36 | 58 | Right | Postcentral gyrus | 335 | 8.14 | ||
| 28 | −80 | 22 | Right | Cuneus | 329 | −5.61 | 4 | −70 | 34 | Right | Precuneus | 5129 | 18.86 | ||
| 18 | −64 | −12 | Right | Lingual gyrus | 723 | −5.59 | |||||||||
| 6 | 26 | 34 | Right | Anterior cingulate gyrus | 220 | −4.34 | |||||||||
| LECN | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||||||||||
| ASL > BOLD | −60 | −6 | 34 | Left | Postcentral Gyrus | 559 | −4.89 | BOLD > ASL | −54 | 16 | 28 | Left | Inferior frontal gyrus | 205 | 5.99 |
| −42 | −84 | 4 | Left | Occipital gyrus | 649 | −5.69 | −30 | −66 | 50 | Left | Superior parietal lobe | 12497 | 12.24 | ||
| 22 | 14 | −34 | Right | Temporal pole | 264 | −6.54 | −20 | 0 | 56 | Left | Superior frontal gyrus | 2355 | 6.50 | ||
| 10 | −66 | 22 | Right | Precuneus | 3326 | −7.15 | 42 | 32 | 16 | Right | Inferior frontal gyrus | 207 | 4.34 | ||
| 6 | 62 | −4 | Right | Orbito-frontal gyrus | 321 | −4.72 | 38 | −68 | 44 | Right | Angular gyrus | 228 | 4.96 | ||
| 28 | 8 | −6 | Right | Putamen | 260 | 5.05 | |||||||||
| 20 | 4 | 56 | Right | Superior frontal gyrus | 288 | 5.17 | |||||||||
| RECN | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||||||||||
| ASL > BOLD | −20 | 0 | −28 | Left | Parahippocampal gyrus | 2272 | −7.54 | BOLD > ASL | −42 | 6 | 26 | Left | Inferior frontal gyrus | 329 | 5.01 |
| −2 | −88 | 20 | Left | Cuneus | 5871 | −9.37 | −36 | −92 | −12 | Left | Inferior occipital gyrus | 472 | 6.01 | ||
| −2 | 42 | −26 | Left | Rectal gyrus | 449 | 7.21 | |||||||||
| 52 | −20 | −26 | Right | Inferior temporal gyrus | 6564 | 9.15 | |||||||||
| 46 | 6 | −18 | Right | Temporal pole | 1888 | 7.92 | |||||||||
| 30 | 6 | 64 | Right | Middle frontal gyrus | 437 | 6.90 | |||||||||
| 24 | −74 | 50 | Right | Superior parietal lobe | 2708 | 9.70 | |||||||||
| 20 | −20 | 70 | Right | Precentral gyrus | 236 | 5.37 | |||||||||
| 8 | −6 | 12 | Right | Thalamus | 339 | 6.52 | |||||||||
| 4 | −36 | 44 | Right | Cingulate gyrus | 292 | 5.11 | |||||||||
| AUN | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||||||||||
| ASL > BOLD | −60 | −6 | 34 | Left | Postcentral gyrus | 559 | −4.89 | BOLD > ASL | −54 | 16 | 28 | Left | Inferior frontal gyrus | 205 | 5.99 |
| −42 | −84 | 4 | Left | Middle occipital gyrus | 649 | −5.69 | −30 | −66 | 50 | Left | Superior parietal lobe | 12497 | 12.24 | ||
| 22 | 14 | −34 | Right | Temporal pole | 264 | −6.54 | −20 | 0 | 56 | Left | Superior frontal gyrus | 2355 | 6.50 | ||
| 10 | −66 | 22 | Right | Precuneus | 3326 | −7.15 | 42 | 32 | 16 | Right | Inferior frontal gyrus | 207 | 4.34 | ||
| 6 | 62 | −4 | Right | Orbito-medial frontal gyrus | 321 | −4.72 | 38 | −68 | 44 | Right | Angular gyrus | 228 | 4.96 | ||
| 28 | 8 | −6 | Right | Putamen | 260 | 5.05 | |||||||||
| 20 | 4 | 56 | Right | Superior frontal gyrus | 288 | 5.17 | |||||||||
| OVN | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | Peak MNI coordinate
|
Hemisphere | Anatomical label | #Voxels in cluster | t-Value at peak coordinate | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| x | y | z | x | y | z | ||||||||||
| ASL > BOLD | −52 | −22 | 42 | Left | Supramarginal gyrus | 437 | −5.74 | BOLD > ASL | −6 | 32 | 40 | Left | Medial frontal gyrus | 680 | 5.01 |
| −22 | −40 | 6 | Left | Hippocampal gyrus | 454 | −8.04 | 0 | −80 | 2 | Left | Lingual gyrus | 11213 | 17.46 | ||
Relationship between regional CBF and FC (z-scores)
Correlation between the network average z-scores for BOLD and ASL RBNs revealed a significant correlation (r = 0.2; p < 0.005). The voxel-wise correlations between RBN z-scores and the corresponding regional CBF of the same sessions resulted in significant correlations within the network specific nodes of each RBN from separate as well as joint ICA for both ASL and BOLD modalities (r > 0.4; p < 0.05) (Figs. 2, 3 & S5). It is worth noting that the correlation between regional CBF and FC was more pronounced in ASL than in BOLD RBNs, and specifically the correlation was more prevalent within the DMN, OVN and LECN (Table 3b).
Discussion
ASL perfusion MRI has received considerable attention in clinical neuroscience due to its quantitative and non-invasive nature. Absolute CBF values obtained using ASL in the whole brain and specific brain regions have been shown to be reproducible across time scales of minutes, hours to days (Chen et al., 2011; Jain et al., 2012; Jann et al., 2013; Wu et al., 2011). There is a good correlation between ASL CBF and the gold standard of 15O-PET in both resting state and activation studies (Feng et al., 2004; Kilroy et al., 2013; Ye et al., 2000). Besides providing a robust mean CBF averaged across a few minute scan, it has been shown to be feasible to perform dynamic FC analysis of the ASL perfusion image series (Chuang et al., 2008; Dai et al., 2013; Jann et al., 2013; Liang et al., 2012; Zou et al., 2009), similar to BOLD FC analysis. To date, however, very few studies have systematically addressed the similarity of RBNs detected using BOLD and ASL contrasts, as well as their reliability across sessions and scanners. The present study attempted to fill in this gap using the state-of-the-art pCASL with single-shot 3D BS GRASE pulse sequence as well as rigorous statistical approaches to evaluate the similarity and repeatability of RBNs in BOLD and ASL rs-fcMRI.
Spatial similarity and repeatability of RBNs
The five RBNs analyzed in this study represented the DMN, left and right ECNs, OVN and AUN. Their spatial pattern was consistent with commonly reported networks in literature (Beckmann et al., 2005). Specifically, these five networks were identified objectively by spatial correlation of the ICA results to reference networks (Shirer et al., 2012). Regarding the spatial similarity between BOLD and ASL RBNs, we found a moderate to high level of concordance between ASL and BOLD RBNs using both independent and joint ICA. DSC values indicated substantial overlap between RBNs of BOLD-only and joint-ICA. For ASL-only ICA, however, while ECNs presented similar spatial patterns to the remaining ICA runs, DMN, OVN, and AUN revealed differences. Specifically, the DMN showed more widespread involvement of anterior areas whereas AUN and OVN did not separate in ASL-only ICA but were represented as a single component. This represents a well known problem in ICA where different model orders can lead to splitting networks into subnetworks or merging them into one component (Abou-Elseoud et al., 2010; Calhoun et al., 2001, 2009; Kiviniemi et al., 2009). These differences in ICA decompositions resulting in heterogeneous RBNs pose a problem when matching RBNs from different ICA runs and consequentially can bias further statistical analyses. Hence, to avoid such a bias for statistical comparison of ASL and BOLD RBNs, single subject maps for both modalities have to be computed within the same framework. This can be achieved using identical seeds (Viviani et al., 2011) or by integrating both modalities into a common ICA. The joint group ICA computed all networks simultaneously across modalities, which substantially improved their spatial concordance (i.e. DSCs). Thus, while separate and joint analyses demonstrated that ASL-based FC analysis is feasible and yields group RBNs consistent with known BOLD-RBNs, only joint-ICA provided an unbiased decomposition necessary for performing a proper statistical comparison. The voxel-wise ANOVA comparing the RBNs of individual subjects computed by the joint-ICA revealed a significant main effect of modality in all studied networks that was attributed to generally lower FC (z-scores) in ASL as revealed by the post-hoc t-test, while scanner and session effects were negligible. Lower FC in ASL has been previously observed in seed-based network analyses (Viviani et al., 2011) and thought to be related to lower SNR or fewer volumes, as discussed below.
The test–retest repeatability of BOLD-RBNs was found to be high, both between sessions and scanners (short (hours) and long (days) term reliability; Figs. 3 & 4 and Table 1). This finding was consistent with earlier studies showing that RBNs can be reliably identified across time (Shehzad et al., 2009) and resting conditions (i.e., eyes open, closed or fixating on a cross) (Patriat et al., 2013) using BOLD rs-fcMRI. Moreover, BOLD-RBNs showed higher reliability than components related to physiological noise and imaging artifacts (Zuo et al., 2010). These findings further established/corroborated the reliability of ICA-based FC analyses in BOLD rs-fcMRI data. In the present study, RBNs from ASL-only ICA exhibited relatively smaller yet still adequate test–retest repeatability across sessions and scanners. Joint-ICA showed generally slightly increased modal ICCs for both BOLD and ASL, but markedly reduced the standard deviation of ICC values within a network. These findings herald the potential of performing rs-fcMRI studies in individual subjects using BOLD and ASL contrasts, although to a lesser degree with the latter. Furthermore, since ASL showed stronger FC in medial prefrontal regions of the DMN compared to BOLD due to reduced sensitivity to susceptibility artifacts (Fernandez-Seara et al., 2005; Liang et al., 2012), it may be particularly valuable in studying psychiatric disorders involving the orbitofrontal cortex.
To make a note of caution, functional connectivity analyses present a coarse measure of brain organization while the true organization of brain networks is still unknown. Moreover, since BOLD and ASL measure different contrasts of neurovascular coupling but are physiologically related and reflect hemodynamic fluctuations, it is reasonable to assume that they share common RBNs. It is likely that both resting state BOLD and ASL data will be acquired in future neuroimaging studies. The proposed joint ICA may offer an appealing approach to identify more reliable findings in terms of network connectivity, since the findings need to be replicated in both BOLD and ASL data, compared to performing ICA on each modality separately.
Reliable network-specific CBF quantification
While the spatial pattern of RBNs appears more reliable using BOLD, ASL provides network specific CBF measurements, a physiologically meaningful parameter inaccessible by BOLD. We found that both global and network CBF can be reliably assessed between sessions and scanners (modal ICCs above 0.9). This high modal ICC might be due to the fact that global CBF differences contribute to a large inter-individual variance compared to intra-subject variance thus increasing voxel-level ICCs. This finding has implications for rs-fcMRI studies in clinical populations. Specifically, the capability for reliable CBF quantification becomes crucial in patient studies where pathophysiology (Alsop et al., 2010; Detre et al., 2012; Detre et al., 2009; Grieder et al., 2013) or medication (Chen et al., 2011; Wang et al., 2011) is likely to alter global or regional CBF. In support for this notion, the major RBNs exhibit consistent, systematic differences in baseline CBF levels, regardless whether correction for global CBF is performed or not.
The DMN and the AUN showed the highest levels of CBF among all networks, which were also higher than the global CBF. This finding is consistent with earlier work measuring CBF in brain networks and suggests that the DMN may retain a higher level of metabolic activity during baseline (Li et al., 2012b), which is suppressed during task performance (Rao et al., 2007; Zhu et al., 2013). The DMN initially was described as a network that exhibits higher metabolic activity (in FDG-PET) during rest and shows deactivation during task execution (Raichle et al., 2001). For the AUN it is argued that it is not completely at rest since there is continuous sound as a consequence of scanning. CBF is proposed as a close marker for metabolic activity as it is normally coupled with glucose uptake of neuronal populations during activity (Fox et al., 1988) and further highly correlates with baseline GABA concentrations and thus indicates changes in excitatory (glutamatergic) and inhibitory (GABA-ergic) neurotransmitters fundamental to the regulation of neuronal firing rates (Donahue et al., 2010) (for review see (Raichle, 1998)). To sum, these findings suggest that network specific CBF may represent the metabolic activity of the associated network that is inaccessible by BOLD rs-fcMRI. Since the repeatability of both BOLD RBNs and network specific ASL CBF were high, the combination of ASL and BOLD into a joint FC analysis may provide a powerful tool in future rs-fcMRI studies carried out across time and scanners.
Besides baseline metabolism of brain areas within a network assessed by CBF, the amplitudes of fluctuations have been argued to present another characteristic of networks and brain areas. ALFF was introduced as a measure related to the amount of spontaneous neuronal activity within brain areas (Zang et al., 2007). However, while conventionally computed on relative BOLD signal fluctuations these amplitudes have no unit, performing the computation on CBF fluctuations however, it becomes possible to attribute a quantitative physiological unit [ml/100 g/min] to these fluctuations. Our results show that there are systematic differences between subjects as well as across networks in the amplitudes of their fluctuations. Notably, the %ALFF normalized to RBN-CBF (Chuang et al., 2008) showed a similar pattern as the absolute ALFF, suggesting that ALFFs are independent of baseline CBF and might provide an additional physiological marker to characterize temporal fluctuations of RBNs in future studies.
Relation between network CBF and network functional connectivity
The correlation analysis between regional CBF and ASL-FC strength (z-scores) revealed a positive relation between these two physiological measures in certain areas specific for the distinct networks. These observations are in line with recent reports showing evidence for specific coupling between regional CBF and functional connectivity from separate ASL and BOLD scans in certain nodes of a network (Li et al., 2012c; Liang et al., 2013). Accordingly, these findings of FC–CBF relationships indicate that CBF and FC provide complementary yet related information on the brain’s baseline functional organization. Recent reports suggest that rCBF in specific network nodes present the cost to maintain proper network integrity (Liang et al., 2013; Tomasi et al., 2013) and that in disease both markers show alterations (Kindler et al., 2013). In line with this notion, a study in Alzheimer’s disease patients showed an association of CBF and FC in nodes of the DMN that were correlated with cognitive performance. Furthermore, medical treatment enhanced both measures along with reductions in disease severity (Li et al., 2012a). Hence including global and regional CBF into FC analysis might contribute to the understanding of inter-individual variability, specifically in clinical populations where alterations in both characteristics have been observed and might influence each other as well as BOLD responses during task performance (Liu et al., 2012).
Study limitations and further development of ASL rs-fcMRI
Besides considerable overlap between ASL and BOLD RBNs, there are significant differences in their spatial patterns in specific areas of the RBNs. These differences primarily arise from the imaging modality rather than effects of different scanners or sessions. There are several potential causes underlying the overall lower connectivity strength in ASL and its lower reliability. First, although recordings had the same duration (i.e. 8 min) ASL had only one fourth of the image volumes of the BOLD scan. This is due to the requirement of ASL for labeling of the inflowing blood, a post-labeling delay to account for arterial transit times and the fact that always a control and a label image have to be acquired in order to quantify CBF. Secondly, ASL has lower SNR compared to BOLD (Aguirre et al., 2002) which hampers FC analysis using ICA or cross-correlation. To explore if the lower FC in ASL was due to reduced temporal resolution or SNR we explored the effects of both possible causes. We subdivided (chopped) the BOLD timeseries to match the temporal resolution of ASL by taking every fourth volume of the BOLD run (see Supplemental Material). Performing ICA and subsequent analyses on the chopped BOLD datasets, we found only minor reductions in modal ICCs (Table 2) and DSCs (Table S1). These results confirm earlier studies demonstrating that temporal resolution has minor effects on FC analysis and that rather the total scan time (which was the same for all datasets in the present study) is a critical factor affecting FC reliability (Birn et al., 2013; Van Dijk et al., 2010). The analysis of temporal SNR revealed that BOLD rs-fcMRI had overall higher temporal SNR than CBF (see Supplemental Material). However, the t-test also revealed medial and orbitofrontal areas with higher SNR in CBF, the same area that showed increased CBF functional connectivity (Fig. S8. (Liang et al., 2012)). This increased sensitivity in areas close to tissue–air boundaries are known to cause susceptibility artifacts in EPI image acquisition which are greatly alleviated in 3D GRASE readout (Vidorreta et al., 2012). While increased FC in areas of susceptibility can be explained by the better sensitivity of ASL in these areas, each RBN showed additional clusters of increased FC in ASL. These clusters are mostly outside the z-maps of the distinct RBNs and could suggest decreased sensitivity of ASL to separate RBNs. The reduced tSNR combined with shorter timeseries might cause fluctuations across voxels to share more mutual information and hence increased correlation with the group component template fluctuation. As a consequence, ASL-RBNs might show more widespread and less well segregated networks than BOLD since the information content of voxel timeseries is less separable than in BOLD. This limitation might be overcome in future studies by using longer CBF timeseries or increasing the model-order of the ICA for ASL.
Another issue that is known to influence FC is motion (Power et al., 2012; Van Dijk et al., 2012). Especially if systematic motion differences exist between two groups or, in the case of the present study, modalities this has to be considered. However, statistical analyses of translational and rotational motion yielded neither significant differences between modalities nor between sessions or scanners (see Supplemental Material). Hence, the differences in FC between BOLD and ASL RBNs might be mainly attributed to SNR. We did not report motor networks due to the presence of wrap around signals in the top slices of 3D GRASE images. While possessing great promise for rs-fcMRI studies, the spatial resolution and image coverage still need to be improved in pCASL with single-shot 3D BS GRASE for widespread applications in clinical neuroscience.
Conclusion
To conclude, the combination of quantitative information on network metabolism from ASL and spatial organization of functional networks from BOLD rs-fMRI provides a powerful tool for characterizing RBNs. While BOLD RBNs showed excellent test–retest reliability across sessions and scanners in their spatial pattern, ASL RBNs showed reduced yet still adequate repeatability. The highly reproducible network-specific ASL CBF measurements may complement BOLD rs-fMRI by providing quantitative CBF as an index of the metabolic activity of specific networks.
Moreover, we found that FC strength in RBNs is correlated with the baseline CBF in core areas of the corresponding networks. This suggests that joint FC and network CBF analyses using BOLD and ASL may fully characterize the spatiotemporal and quantitative properties of RBNs that are especially desirable for longitudinal rs-fcMRI studies, pharmacological MRI studies as well as for the comparison of RBNs across different subject groups.
Supplementary Material
Acknowledgments
Funding
This work was supported by the US National Institutes of Health grants U01-MH081902, P50-HD055784, R01-MH080892, R01-NS081077, and R01-EB014922 as well as partly by Garen and Shari Staglin and the International Mental Health Research Organization. KJ has a fellowship of the Swiss National Science Foundation & Swiss Foundation for Grants in Biology and Medicine grant 142743.
We would like to thank Jennifer Andreotti for the insightful discussions on the ICC and Lirong Yang for the help with SNR analysis.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neuroimage.2014.11.028.
References
- Abou-Elseoud A, Starck T, Remes J, Nikkinen J, Tervonen O, Kiviniemi V. The effect of model order selection in group PICA. Hum Brain Mapp. 2010;31:1207–1216. doi: 10.1002/hbm.20929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre GK, Detre JA, Zarahn E, Alsop DC. Experimental design and the relative sensitivity of BOLD and perfusion fMRI. NeuroImage. 2002;15:488–500. doi: 10.1006/nimg.2001.0990. [DOI] [PubMed] [Google Scholar]
- Alsop DC, Dai W, Grossman M, Detre JA. Arterial spin labeling blood flow MRI: its role in the early characterization of Alzheimer’s disease. J Alzheimers Dis. 2010;20:871–880. doi: 10.3233/JAD-2010-091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsop DC, Detre JA, Golay X, Gunther M, Hendrikse J, LH-G Recommended implementation of arterial spin labeling perfusion MRI for clinical applications: a consensus of the ISMRM perfusion study group and the European ASL in dementia consortium. Magn Reson Med. 2014 doi: 10.1002/mrm.25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslan S, Huang H, Uh J, Mishra V, Xiao G, van Osch MJ, Lu H. White matter cerebral blood flow is inversely correlated with structural and functional connectivity in the human brain. NeuroImage. 2011;56:1145–1153. doi: 10.1016/j.neuroimage.2011.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Bennett CM, Wolford GL, Miller MB. The principled control of false positives in neuroimaging. Soc Cogn Affect Neurosci. 2009;4:417–422. doi: 10.1093/scan/nsp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Molloy EK, Patriat R, Parker T, Meier TB, Kirk GR, Nair VA, Meyerand ME, Prabhakaran V. The effect of scan length on the reliability of resting-state fMRI connectivity estimates. NeuroImage. 2013;83C:550–558. doi: 10.1016/j.neuroimage.2013.05.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Buxton RB, Uludag K, Dubowitz DJ, Liu TT. Modeling the hemodynamic response to brain activation. NeuroImage. 2004;23(Suppl 1):S220–S233. doi: 10.1016/j.neuroimage.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pekar JJ. A method for comparing group fMRI data using independent component analysis: application to visual, motor and visuomotor tasks. Magn Reson Imaging. 2004;22:1181–1191. doi: 10.1016/j.mri.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Liu J, Adali T. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. NeuroImage. 2009;45:S163–S172. doi: 10.1016/j.neuroimage.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wan HI, O’Reardon JP, Wang DJ, Wang Z, Korczykowski M, Detre JA. Quantification of cerebral blood flow as biomarker of drug effect: arterial spin labeling phMRI after a single dose of oral citalopram. Clin Pharmacol Ther. 2011;89:251–258. doi: 10.1038/clpt.2010.296. [DOI] [PubMed] [Google Scholar]
- Chuang KH, van Gelderen P, Merkle H, Bodurka J, Ikonomidou VN, Koretsky AP, Duyn JH, Talagala SL. Mapping resting-state functional connectivity using perfusion MRI. NeuroImage. 2008;40:1595–1605. doi: 10.1016/j.neuroimage.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Shankaranarayanan A, Schlaug G, DCA . Quantitative Measurement of Signal Fluctuations in ASL from Resting State Functional Networks. ISMRM; Salt Lake City, UT, USA: 2013. [Google Scholar]
- Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn Reson Med. 1992;23:37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]
- Detre JA, Wang J, Wang Z, Rao H. Arterial spin-labeled perfusion MRI in basic and clinical neuroscience. Curr Opin Neurol. 2009;22:348–355. doi: 10.1097/WCO.0b013e32832d9505. [DOI] [PubMed] [Google Scholar]
- Detre JA, Rao H, Wang DJ, Chen YF, Wang Z. Applications of arterial spin labeled MRI in the brain. J Magn Reson Imaging. 2012;35:1026–1037. doi: 10.1002/jmri.23581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice LR. Measurement of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- Donahue MJ, Near J, Blicher JU, Jezzard P. Baseline GABA concentration and fMRI response. NeuroImage. 2010;53:392–398. doi: 10.1016/j.neuroimage.2010.07.017. [DOI] [PubMed] [Google Scholar]
- Du Y, Yan L, Wang DJ, Fan Y. Hum Brain Mapp. Beijing China: 2012. Resting-state brain networks in BOLD fMRI and perfusion fMRI. [Google Scholar]
- Feng CM, Narayana S, Lancaster JL, Jerabek PA, Arnow TL, Zhu F, Tan LH, Fox PT, Gao JH. CBF changes during brain activation: fMRI vs. PET NeuroImage. 2004;22:443–446. doi: 10.1016/j.neuroimage.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Fernandez-Seara MA, Wang Z, Wang J, Rao HY, Guenther M, Feinberg DA, Detre JA. Continuous arterial spin labeling perfusion measurements using single shot 3D GRASE at 3 T. Magn Reson Med. 2005;54:1241–1247. doi: 10.1002/mrm.20674. [DOI] [PubMed] [Google Scholar]
- Fernandez-Seara MA, Edlow BL, Hoang A, Wang J, Feinberg DA, Detre JA. Minimizing acquisition time of arterial spin labeling at 3 T. Magn Reson Med. 2008;59:1467–1471. doi: 10.1002/mrm.21633. [DOI] [PubMed] [Google Scholar]
- Forman SD, Cohen JD, Fitzgerald M, Eddy WF, Mintun MA, Noll DC. Improved assessment of significant activation in functional magnetic resonance imaging (fMRI): use of a cluster-size threshold. Magn Reson Med. 1995;33:636–647. doi: 10.1002/mrm.1910330508. [DOI] [PubMed] [Google Scholar]
- Fox PT, Raichle ME, Mintun MA, Dence C. Nonoxidative glucose consumption during focal physiologic neural activity. Science. 1988;241:462–464. doi: 10.1126/science.3260686. [DOI] [PubMed] [Google Scholar]
- Franco AR, Pritchard A, Calhoun VD, Mayer AR. Interrater and intermethod reliability of default mode network selection. Hum Brain Mapp. 2009;30:2293–2303. doi: 10.1002/hbm.20668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieder M, Crinelli RM, Jann K, Federspiel A, Wirth M, Koenig T, Stein M, Wahlund LO, Dierks T. Correlation between topographic N400 anomalies and reduced cerebral blood flow in the anterior temporal lobes of patients with dementia. J Alzheimers Dis. 2013;36:711–731. doi: 10.3233/JAD-121690. [DOI] [PubMed] [Google Scholar]
- Jain V, Duda J, Avants B, Giannetta M, Xie SX, Roberts T, Detre JA, Hurt H, Wehrli FW, Wang DJ. Longitudinal reproducibility and accuracy of pseudo-continuous arterial spin-labeled perfusion MR imaging in typically developing children. Radiology. 2012;263:527–536. doi: 10.1148/radiol.12111509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jann K, Orosz A, Dierks T, Wang DJ, Wiest R, Federspiel A. Quantification of network perfusion in ASL cerebral blood flow data with seed based and ICA approaches. Brain Topogr. 2013 doi: 10.1007/s10548-013-0280-3. [DOI] [PubMed] [Google Scholar]
- Kilroy E, Apostolova L, Liu C, Yan L, Ringman J, Wang DJ. Reliability of two-dimensional and three-dimensional pseudo-continuous arterial spin labeling perfusion MRI in elderly populations: comparison with 15O-water positron emission tomography. J Magn Reson Imaging. 2013 doi: 10.1002/jmri.24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler J, Jann K, Homan P, Hauf M, Walther S, Strik W, Dierks T, Hubl D. Static and dynamic characteristics of cerebral blood flow during the resting state in schizophrenia. Schizophr Bull. 2013 doi: 10.1093/schbul/sbt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi V, Starck T, Remes J, Long X, Nikkinen J, Haapea M, Veijola J, Moilanen I, Isohanni M, Zang YF, Tervonen O. Functional segmentation of the brain cortex using high model order group PICA. Hum Brain Mapp. 2009;30:3865–3886. doi: 10.1002/hbm.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JR, Koch GG. Measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- Li W, Antuono PG, Xie C, Chen G, Jones JL, Ward BD, Franczak MB, Goveas JS, Li SJ. Changes in regional cerebral blood flow and functional connectivity in the cholinergic pathway associated with cognitive performance in subjects with mild Alzheimer’s disease after 12-week donepezil treatment. NeuroImage. 2012a;60:1083–1091. doi: 10.1016/j.neuroimage.2011.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Kadivar A, Pluta J, Dunlop J, Wang Z. Test–retest stability analysis of resting brain activity revealed by blood oxygen level-dependent functional MRI. J Magn Reson Imaging. 2012b;36:344–354. doi: 10.1002/jmri.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhu Y, Childress AR, Detre JA, Wang Z. Relations between BOLD fMRI-derived resting brain activity and cerebral blood flow. PLoS ONE. 2012c;7:e44556. doi: 10.1371/journal.pone.0044556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Tournier J, Masterton D, Connelly A, Calamante F. An Improved 3D GRASE PCASL Method for Whole-Brain Resting-State Functional Connectivity. ISMRM; Montreal, Canada: 2011. [Google Scholar]
- Liang XY, Tournier JD, Masterton R, Connelly A, Calamante F. A k-space sharing 3D GRASE pseudocontinuous ASL method for whole-brain resting-state functional connectivity. Int J Imaging Syst Technol. 2012;22:37–43. [Google Scholar]
- Liang X, Zou Q, He Y, Yang Y. Coupling of functional connectivity and regional cerebral blood flow reveals a physiological basis for network hubs of the human brain. Proc Natl Acad Sci U S A. 2013;110:1929–1934. doi: 10.1073/pnas.1214900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TT, Wong EC. A signal processing model for arterial spin labeling functional MRI. NeuroImage. 2005;24:207–215. doi: 10.1016/j.neuroimage.2004.09.047. [DOI] [PubMed] [Google Scholar]
- Liu J, Qiu M, Constable RT, Wexler BE. Does baseline cerebral blood flow affect task-related blood oxygenation level dependent response in schizophrenia? Schizophr Res. 2012;140:143–148. doi: 10.1016/j.schres.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw KO, Wong SP. Forming inferences about some intraclass correlation co-efficients. Psychol Methods. 1996;1:30–46. [Google Scholar]
- Meindl T, Teipel S, Elmouden R, Mueller S, Koch W, Dietrich O, Coates U, Reiser M, Glaser C. Test–retest reproducibility of the default-mode network in healthy individuals. Hum Brain Mapp. 2010;31:237–246. doi: 10.1002/hbm.20860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriat R, Molloy EK, Meier TB, Kirk GR, Nair VA, Meyerand ME, Prabhakaran V, Birn RM. The effect of resting condition on resting-state fMRI reliability and consistency: a comparison between resting with eyes open, closed, and fixated. NeuroImage. 2013;78:463–473. doi: 10.1016/j.neuroimage.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. Behind the scenes of functional brain imaging: a historical and physiological perspective. Proc Natl Acad Sci U S A. 1998;95:765–772. doi: 10.1073/pnas.95.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. A default mode of brain function. Proc Natl Acad Sci U S A. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Wang J, Tang K, Pan W, Detre JA. Imaging brain activity during natural vision using CASL perfusion fMRI. Hum Brain Mapp. 2007;28:593–601. doi: 10.1002/hbm.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, Petkova E, Castellanos FX, Milham MP. The resting brain: unconstrained yet reliable. Cereb Cortex. 2009;19:2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex. 2012;22:158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout PE, Fleiss JL. Intraclass correlations — uses in assessing rater reliability. Psychol Bull. 1979;86:420–428. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- St Lawrence KS, Owen D, Wang DJ. A two-stage approach for measuring vascular water exchange and arterial transit time by diffusion-weighted perfusion MRI. Magn Reson Med. 2012;67:1275–1284. doi: 10.1002/mrm.23104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Wang GJ, Volkow ND. Energetic cost of brain functional connectivity. Proc Natl Acad Sci U S A. 2013;110:13642–13647. doi: 10.1073/pnas.1303346110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. NeuroImage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidorreta M, Wang Z, Rodriguez I, Pastor MA, Detre JA, Fernandez-Seara MA. Comparison of 2D and 3D single-shot ASL perfusion fMRI sequences. NeuroImage. 2012;66C:662–671. doi: 10.1016/j.neuroimage.2012.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viviani R, Messina I, Walter M. Resting state functional connectivity in perfusion imaging: correlation maps with BOLD connectivity and resting state perfusion. PLoS ONE. 2011;6:e27050. doi: 10.1371/journal.pone.0027050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alsop DC, Song HK, Maldjian JA, Tang K, Salvucci AE, Detre JA. Arterial transit time imaging with flow encoding arterial spin tagging (FEAST) Magn Reson Med. 2003;50:599–607. doi: 10.1002/mrm.10559. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang Y, Wolf RL, Roc AC, Alsop DC, Detre JA. Amplitude-modulated continuous arterial spin-labeling 3.0-T perfusion MR imaging with a single coil: feasibility study. Radiology. 2005;235:218–228. doi: 10.1148/radiol.2351031663. [DOI] [PubMed] [Google Scholar]
- Wang Z, Aguirre GK, Rao H, Wang J, Fernandez-Seara MA, Childress AR, Detre JA. Empirical optimization of ASL data analysis using an ASL data processing tool-box: ASLtbx. Magn Reson Imaging. 2008;26:261–269. doi: 10.1016/j.mri.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DJ, Chen Y, Fernandez-Seara MA, Detre JA. Potentials and challenges for arterial spin labeling in pharmacological magnetic resonance imaging. J Pharmacol Exp Ther. 2011;337:359–366. doi: 10.1124/jpet.110.172577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward DB. AlphaSim. 2000. [Google Scholar]
- Wu WC, Jiang SF, Yang SC, Lien SH. Pseudocontinuous arterial spin labeling perfusion magnetic resonance imaging—a normative study of reproducibility in the human brain. NeuroImage. 2011;56:1244–1250. doi: 10.1016/j.neuroimage.2011.02.080. [DOI] [PubMed] [Google Scholar]
- Ye FQ, Berman KF, Ellmore T, Esposito G, van Horn JD, Yang Y, Duyn J, Smith AM, Frank JA, Weinberger DR, McLaughlin AC. H(2)(15)O PET validation of steady-state arterial spin tagging cerebral blood flow measurements in humans. Magn Reson Med. 2000;44:450–456. doi: 10.1002/1522-2594(200009)44:3<450::aid-mrm16>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Zang YF, He Y, Zhu CZ, Cao QJ, Sui MQ, Liang M, Tian LX, Jiang TZ, Wang YF. Altered baseline brain activity in children with ADHD revealed by resting-state functional MRI. Brain Dev. 2007;29:83–91. doi: 10.1016/j.braindev.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Zhu S, Fang Z, Hu S, Wang Z, Rao H. Resting state brain function analysis using concurrent BOLD in ASL perfusion fMRI. PLoS ONE. 2013;8:e65884. doi: 10.1371/journal.pone.0065884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q, Wu CW, Stein EA, Zang Y, Yang Y. Static and dynamic characteristics of cerebral blood flow during the resting state. NeuroImage. 2009;48:515–524. doi: 10.1016/j.neuroimage.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Kelly C, Adelstein JS, Klein DF, Castellanos FX, Milham MP. Reliable intrinsic connectivity networks: test–retest evaluation using ICA and dual regression approach. NeuroImage. 2010;49:2163–2177. doi: 10.1016/j.neuroimage.2009.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.