Abstract
Mutations in TNFRSF1A encoding TNF receptor 1 (TNFR1) cause the autosomal dominant TNF receptor-associated periodic syndrome (TRAPS): a systemic autoinflammatory disorder. Misfolding, intracellular aggregation, and ligand-independent signaling by mutant TNFR1 are central to disease pathophysiology. Our aim was to understand the extent of signaling pathway perturbation in TRAPS. A prototypic mutant TNFR1 (C33Y), and wild-type TNFR1 (WT), were expressed at near physiological levels in an SK-Hep-1 cell model. TNFR1-associated signaling pathway intermediates were examined in this model, and in PBMCs from C33Y TRAPS patients and healthy controls. In C33Y-TNFR1-expressing SK-Hep-1 cells and TRAPS patients’ PBMCs, a subtle, constitutive upregulation of a wide spectrum of signaling intermediates and their phosphorylated forms was observed; these were associated with a proinflammatory/antiapoptotic phenotype. In TRAPS patients’ PBMCs, this upregulation of proinflammatory signaling pathways was observed irrespective of concurrent treatment with glucocorticoids, anakinra or etanercept, and the absence of overt clinical symptoms at the time that the blood samples were taken. This study reveals the pleiotropic effect of a TRAPS-associated mutant form of TNFR1 on inflammatory signaling pathways (a proinflammatory signalome), which is consistent with the variable and limited efficacy of cytokine-blocking therapies in TRAPS. It highlights new potential target pathways for therapeutic intervention.
Keywords: Autoinflammation, Protein microarray, Signalome, TNF receptor 1, TRAPS
Introduction
TNF receptor-associated periodic syndrome (TRAPS) is an autoinflammatory disease that is associated with autosomal dominant mutations in the gene encoding the TNF receptor 1 (TNFR1) located on chromosome 12p13 [1]. More than 91 different mutations in TNFR1 have been identified (http://mf.igh.cnrs.fr/infevers/) and over 60% of these mutations are associated with TRAPS [2].
The link between the presence of TNFR1 mutations and the inflammatory conditions associated with TRAPS is still the subject of intense investigations. Defective receptor shedding was originally thought to reduce neutralization of TNF-α by soluble TNFR1, thus resulting in the continuous inflammatory stimulus that is notable in TRAPS [1]. However, defective shedding is variable between different TNFR1 mutants and different cell types and thus cannot explain all the clinical features of TRAPS [3]. Mechanisms associated with intracellular retention of the mutant TNFR1 and sequelae of this are now widely believed to be major factors in the pathogenesis of TRAPS, including ligand-independent signaling, misfolding and an unfolded protein response, induction of reactive oxygen species (ROS) and impaired autophagy [4–13].
Recently, we demonstrated that there is upregulated expression of multiple proinflammatory genes in transfected cells expressing mutant TNFR1 relative to those expressing normal (WT) TNFR1 [8]. Our transcriptome studies demonstrated chronic alteration in gene expression, but did not elucidate posttranslational modifications such as phosphorylation or ubiquitination of proteins, which are intimately associated with the acute control of proinflammatory signaling. Therefore, studying the activity of multiple signaling pathways that constitute a proinflammatory signalome is essential for a better understanding of the pathogenesis of TRAPS.
TNFR1-TNF-α-mediated signaling is complex; a well established consequence is the activation of the canonical pathway of NF-κB leading ultimately to IκB degradation and the activation of the NF-κB p50/p65 heterodimer. Subsequent formation of the proapoptosis, death-inducing signaling complex (complex II or DISC) leads to caspase 8 activation [14,15]. However, normal physiological signal induction through TNFR1 has also been demonstrated to activate the p38, Erk, and JNK mitogen activated protein kinases (MAPK) [16], the Jak/STAT and PI3K/Akt pathways. This has been shown to occur through an increasing number of intracellular associations, including sphingomyelinases [17], SHP-1, c-Src, and Jak2 [18,19]. The fate of the cell is, therefore, determined by a complex balance between these activated pathways [20].
In TRAPS, both activation and inhibition of NF-κB have been reported [7,12,21,22] and differential activation of NF-κB subunit members may be a feature of different TNFR1 mutations. Furthermore, TRAPS-associated mutations lead to activation of JNKs and P38 MAPK that may result from spontaneous signaling by intracellular misfolded TNFR1 mutants or indirectly through other mediators such as ROS that inactivate MAPK phosphatases resulting in sustained MAPK signaling [11,13,23]. Resistance to apoptosis is also detected in neutrophils and PBMCs isolated from TRAPS patients [24,25].
For many years, clinical management of TRAPS has involved treatment with corticosteroids and, more recently, the use of biological agents that block the action of proinflammatory cytokines—particularly the TNF-binding agent etanercept and the interleukin (IL)-1 receptor antagonist anakinra. A recent comprehensive review of the treatment of autoinflammatory diseases demonstrates that, for TRAPS, corticosteroids, etanercept, and anakinra show broadly similar levels of efficacy, with anakinra being most successful at inducing a complete clinical response (although response duration was not defined) [26]. However, for many TRAPS patients, none of these therapies bring about permanent remission, or they induce only a partial response, and 5–15% of patients fail to respond to corticosteroids, etanercept or anakinra [26]. We previously reported that levels of IL-6 are frequently raised in TRAPS patients with a C33Y TNFR1 mutations [27]; one of these patients, in whom either etanercept or anakinra had limited efficacy, was treated with the IL-6 receptor blocking antibody tocilizumab [28]. This patient initially responded well to the treatment, although blood levels of proinflammatory cytokines (TNF-α, IL-1α, IL-6, and IL-8) were not reduced [28]. Thus cytokine blockade in TRAPS has variable, and often transient, efficacy and does not target the central pathogenetic processes that drive the inflammatory response.
We applied proteomic techniques to elucidate the variety and complexity of intracellular signaling pathways affected by a TRAPS-associated TNFR1 mutation. This study thus provides the most comprehensive picture to date of the proinflammatory signalome that is constitutively activated by mutant forms of TNFR1, even in TRAPS patients being treated with glucocorticoids and/or cytokine-blocking biologics.
Results
Optimization of reverse phase protein microarrays
Reverse phase protein microarrays (RPPAs) use femtomolar quantities of protein from clinical samples or cell lines; thus, optimization is crucial to the reproducibility of the technique. To achieve this goal SK-Hep-1 cells transfected with WT or mutant TNFR1 were treated with different stimulants (IL-1 (10 μg/mL), IL-6 (10 μg/mL), TNF-α (50 ng/mL) or LPS (20 μg/mL)) and were pooled for use as a positive control lysate in the following set of experiments.
Printing and blocking buffer optimization
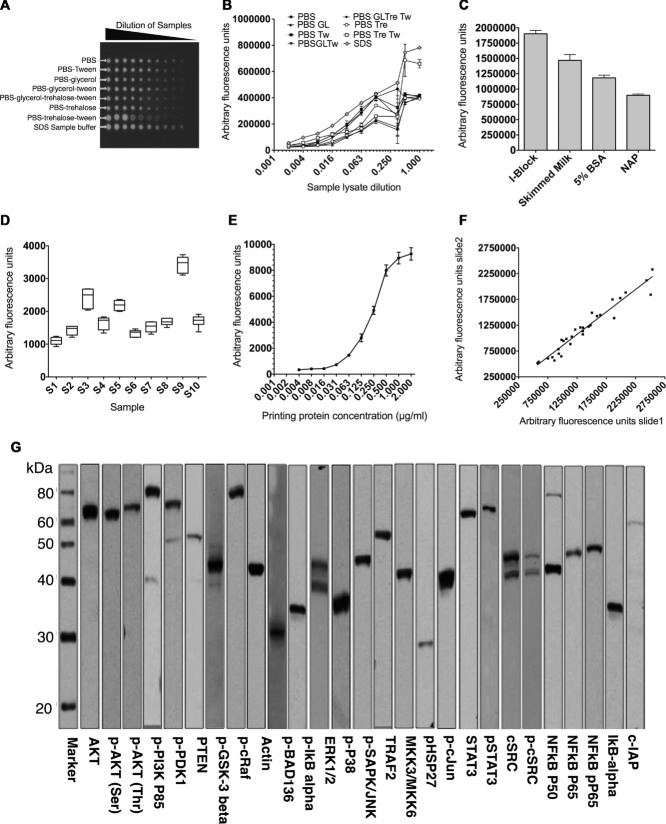
To identify the optimal printing buffer, the positive control lysate was serially diluted ten times in eight different buffers, namely, PBS, PBS-0.01% Tween, PBS-5% glycerol, PBS + glycerol + Tween, PBS + trehalose (50 mM) + glycerol + Tween, PBS + trehalose, PBS + trehalose + Tween or SDS sample buffer. Samples were then printed onto 16-pad nitrocellulose slides. To identify optimal blocking buffer, slides were blocked with four commonly used buffers, namely, I-Block, 5% skimmed milk, 5% BSA or non-animal protein (NAP) and then probed for β-actin. Optimal features, with regular circular shapes and high signal intensities, were achieved using SDS sample buffer (Fig. 1A and B). The strongest signals, after background subtraction, were achieved using I-Block (Fig. 1C). As a consequence, for all further experiments, SDS sample buffer was used as the printing buffer and I-Block as the blocking buffer.
Figure 1.
Validation of reverse phase protein microarray. (A) Positive control lysate was prepared from SK-Hep-1 cells transfected with WT or mutant TNFR1 stimulated with IL-1 (10 μg/mL), IL-6 (10 μg/mL), and TNF-α (50 ng/mL) or LPS (20 μg/mL); cells were then lysed and the lysates pooled. The lysates were serially diluted in eight different printing buffers, arrayed on 16- pad nitrocellulose-coated slides and probed for β-actin by RPPA using infrared reporter-conjugate. (B) Multiple printing buffers were used to print the positive lysates on nitrocellulose slides and feature signal intensities were assessed by probing the slide for β-actin using RPPA. Data are shown as mean of three replicates and the experiment was repeated three times with similar results. All fluorescent signals are reported as arbitrary fluorescence units (AFU). (C) Fluorescent signal intensities obtained for RPPA analysis using a range of blocking buffers. Data are shown as mean = SD of three samples in one experiment representative of three independent experiments. (D) Intraslide reproducibility of RPPA: ten different lysates (S1-S10) were printed (replicate n = 6) on 16-pad slides. A box and whisker plot is shown, with median represented by a black line within the box representing the interquartile range, using Tukeys estimation for whisker length. The coefficient of variation (%) is indicated for each sample. (E) Feature-associated β-actin signal associated with protein concentration of source lysate, up to 2 μg/mL. Positive control lysates were serially diluted and spotted on nitrocellulose slides. The slides were probed by RPPA for β-actin, p-AKT Threonine, p-AKT serine, and p-PDK1. Data are shown as mean ± SD of 18 samples from one experiment representative of three independent experiments. (F) Interslide reproducibility between signals from the same lysates (n = 30) printed on two different nitrocellulose slides and probed for RPPA (R2 = 0.942). (G) Primary antibodies were validated using western blotting and strip images are shown for selected antibodies in this study. Images are representative of two independent experiments.
Reproducibility and validity of RPPA
To test the reproducibility of microarray, both intraslide and interslide variability was assessed. For intraslide validations, ten different positive control lysates were serially diluted in 1× SDS sample buffer and printed onto 16-pad nitrocellulose slides in six replicates each; slides were then processed for the detection of β-actin. The coefficient of variation (CV%) within the replicates was less than 12% for all the analyzed samples (Fig. 1D). Protein concentration in the printing buffer and protein binding to the array demonstrated a characteristic saturation curve, with protein concentrations of over 1 μg/mL achieving >90% maximum signal intensity (Fig. 1E).
Interslide reproducibility was examined by printing the lysates used above onto two different 16-pad nitrocellulose slides in six replicates. The printed slides were then probed for β-actin, p-AKT-threonine, p-AKT-serine, and p-PDK1. The results indicated high correlation coefficient between signals of corresponding samples printed on the two different slides (r2 = 0.9401) (Fig. 1F).
Validation of primary antibodies for microarray experiments
A key challenge in RPPA is the availability of high specificity antibodies that reveal only bands of the expected sizes. Positive control lysate were run in SDS-PAGE, and transferred to nitrocellulose membranes and assembled within a strip western device (Miniblotter 20SL, Immunetics). Diluted antibodies (120 μL per strip) were incubated with the individual wells (strips), followed by secondary antiimmunoglobulin-reporter conjugate and substrate. Only antibodies that stained for specific bands of the correct molecular weights were included in this study (Fig. 1G). The optimal dilution of each antibody was confirmed by microarray using 64-pad nitrocellulose slides and infrared signal detection (Table 1).
Table 1.
List of primary antibodies used in TNFR1 signaling study
| NF-κB Pathway | PI3K/AKT Pathway | MAPK Pathway | Src/JAK/STAT3 | ||||
|---|---|---|---|---|---|---|---|
| Antibody | Dilution | Antibody | Dilution | Antibody | Dilution | Antibody | Dilution |
| NF-κB p65 | 1:250 | AKT | 1:100 | P38 MAP K | 1:000 | c-SRC | 1:000 |
| pNF-κB p65 | 1:100 | P-AKT (T308) | 1:50 | pP38 MAP | 1:000 | pC-SRC | 1:1000 |
| p105/p50 NF-κB | 1:1000 | P-AKT (Ser.) | 1:25 | pHSP27 | 1:50 | Jak2 | 1:000 |
| IκB-α | 1:250 | p-eNos | 1:500 | ERK1/2 | 1:2000 | pJak2 | 1:1000 |
| P-IκB-α | 1:500 | pC-raf | 1:1000 | pERK1/2 | 1:2000 | Stat3 | 1:1000 |
| IKK-α | 1:250 | GSK-3β | 1:500 | pSAPK/JNK | 1:1000 | pStat3 | 1:500 |
| IKK-β | 1:250 | pGSK-3β(Ser9) | 1:500 | pMEK3/MEK6 | 1:100 | ||
| A20 | 1:500 | PDK1 | 1:1000 | pMAPKAPK 2 | 1:1000 | ||
| c-IAP2 | 1:500 | pPDK1 | 1:1000 | pSek1/MKK4 | 1:250 | c-FLIP | 1:500 |
| pTAKI | 1:100 | PTEN | 1:1000 | pC-Jun (Ser) | 1:100 | P-ATF2 | 1:250 |
| pPTEN | 1:1000 | TRAF2 | 1:500 | P-ELK-1 | 1:500 | ||
| PI3K 110 | 1:250 | Actin (Mouse) | 1:1000 | ||||
| pPI3K P85 | 1:250 | NU P98 | 1:1000 | ||||
| pBad 136 | 1:000 | HSP 90 | 1:1000 | ||||
All antibodies were rabbit antibodies and were purchased from Cell Signaling Technology. For western blotting, the antibodies were diluted 1:1000 in antibody diluent (Dako). For lysate microarray, they were diluted as shown in the table in the same dilution buffer.
Constitutive activation of inflammatory signaling pathways in SK-Hep-1 cells expressing mutant TNFR1
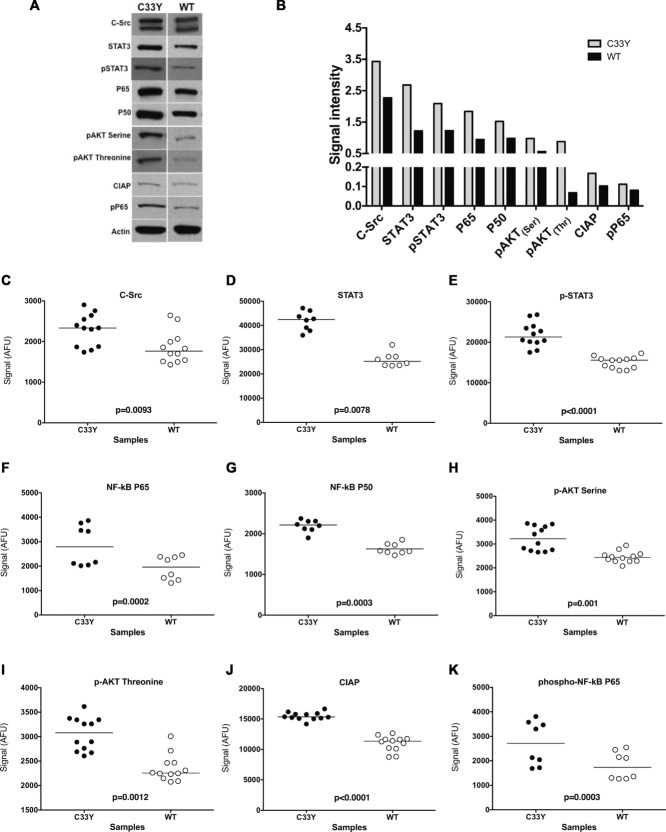
Intracellular signaling pathway intermediates that govern inflammation (MAPK, PI3K/AKT, c-Src/Jak2/STAT3, NF-κB) and are activated by TNFR1 were examined in cell lysates from SK-Hep-1 cells transfected with C33Y TNFR1 or WT TNFR1 constructs. Separation of protein from the lysates was performed by SDS-PAGE followed by electrotransfer to nitrocellulose membranes (Invitrogen, UK). Immunoblotting was conducted using the primary antibodies for the inflammatory target proteins such as c-Src, STAT3, p-STAT3, p65, p50 p-AKT threonine, p-AKT serine, CIAP, and p-P65. In addition, β-actin was used as an internal control for protein loading normalization for all samples. The western blotting results revealed a generalized increase in expression of the tested inflammatory markers in cells transfected with mutant TNFR1 (C33Y) compared with cells transfected with WT TNFR1 (Fig. 2A and B). However, due to the limitations in scalability and quantification of western blotting, the optimized RPPA was applied to profile these signaling pathways within TNFR1 expressing cells. The same lysates used in western blotting above were also tested by RPPA using the same primary antibodies. There was a strong correlation between the results obtained from western blotting and those from microarray, as significant elevations of the tested inflammatory molecules were found in cells expressing mutant TNFR1 (C33Y) compared to WT TNFR1 (Fig. 2C–K). These preliminary results allowed us to apply RPPA, with its high throughput capability, to evaluate more signaling molecules in response to different stimuli under a variety of conditions.
Figure 2.
Comparison of western blot and RPPA detection of intracellular signaling intermediates and phosphorylation states in cultured SK-Hep-1 cells. (A) SK-Hep-1 cells stably transfected with either normal TNFR1 (WT; right column) or TNFR1-C33Y variant (C33Y; left column) at 80% confluency were lysed and assessed for expression of signaling intermediates by SDS-PAGE and western blotting. Representative results from three independent experiments are shown. Detection of β-actin was used as an internal control and as a normalizer for protein loading for all samples used. (B) Densitometric quantification of samples shown in panel (A). Data are shown as mean + SD of three samples pooled from three independent experiments. (C–K) Target protein abundance was assessed by RPPA using dual color fluorometric detection for the specific target protein and β-actin, respectively. Scatter plots are shown, representing mean signal after background subtraction and normalization to β-actin signal from each individual lysate spot of the technical replicates. The bar indicates the median signal, n = 8 biological replicates, or greater, for each plot. The experiment was repeated three times with similar results. Significance values were derived using the Wilcoxon Test for repeated measures. All fluorescent signals are reported as arbitrary fluorescence units (AFU), after normalization to β-actin signal.
Stimulation with TNF-α does not significantly change status of inflammatory signaling pathways
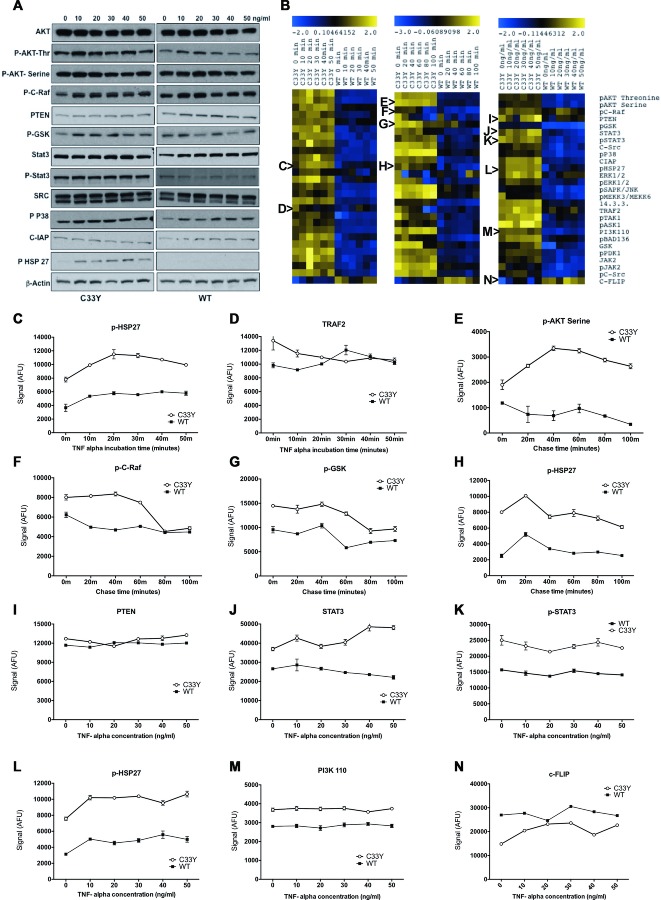
Both C33Y TNFR1- and WT TNFR1-transfected SK-Hep-1 cells were treated with TNF-α under three different conditions: time course of response to constant exposure to 10 ng/mL TNF-α; pulse-chase response to 2 min exposure to 10 ng/mL TNF-α; dose-response of exposure to various concentrations of TNF-α for 30 min. Cells were lysed and aliquots of the lysates were analyzed by western blotting (Fig. 3A); the remaining volumes of the lysates were transferred to 384-well plates for array printing. Arrays were probed with antibodies for the specific targets in addition to probing for β-actin for standardization of the protein loading. RPPAs were analyzed with an infrared scanner and normalized signal intensities were calculated using RPP analyzer software. The results for each condition are shown in Figure 3B as heat maps of the log2 relative expression levels detected for each sample; they are also shown graphically for selected exemplar molecules in Figure 3C–N as the mean ± SD of three different biological replicates; statistical data (two-way ANOVA) for Figure 3C–N are shown in Supporting Information Table 2. Continuous stimulation with 10 ng/mL TNF-α induced a slight, parallel increase in levels of p-HSP27 in both C33Y and WT cells (Fig. 3C), but had opposite effects on TRAF2 expression by the two cell lines, causing decreased expression in C33Y cells and increased expression in WT cells, thereby resulting in convergence of TRAF2 levels (Fig. 3D). Pulse-chase with 10 ng/mL TNF-α had various effects on different signaling molecules: it initially enhanced p-AKT-serine expression in C33Y cells, whilst suppressing it in WT cells (Fig. 3E); it induced suppression of p-C-Raf and p-GSK expression at later time points, with a greater effect on C33Y than WT cells (Fig. 3F and G); and it induced a short-term rise in p-HSP27 levels in both C33Y and WT cells (Fig. 3H). The dose-response experiments indicated that any effects of TNF-α on signaling molecule expression were apparent with 10 ng/mL TNF-α, and higher doses did not generally show further effects (Fig. 3I–N).
Figure 3.
Activation of inflammatory signaling intermediates by the TNFR1 C33Y mutation. (A) SK-Hep-1 transfectants were stimulated with different concentrations of TNF-α (10, 20, 30, 40, and 50 ng/mL) for 30 min. Cell lysates from these cells were tested by western blotting for detection of various signaling molecules. β-actin was used as an internal control for protein loading normalization and data shown are representative of three independent experiments. (B) Blue (low) to yellow (high) heat maps representing the relative abundance of signaling pathways intermediates using RPPA in SK-Hep-1 transfectants stimulated with TNF-α under three different conditions: Time, 10 ng/mL for 0, 10, 20, 30, 40, and 50 min; Pulse, 10 ng/mL for 2 min then lyse the cells after 0, 20, 40, 60, 80, and 100 min; Concentration, different concentrations of TNF-α 0, 10, 20, 30, 40, and 50 ng/mL for 30 min, respectively. Each column of the heat map represents an SK-Hep-1 cell culture. Rows represent the signaling molecules studied. Signal values represented on the color scale for the heat maps are log2 transformed from the AFU. Letters at the side of the heat maps refer to the relevant graphs, depicting the mean ± SD signal intensities of that particular heat map row as follows: (C) p-HSP27 (time), (D) TRAF2 (time), (E) pAKT serine (pulse), (F) p-C-Raf (pulse), (G) pGSK (pulse), (H) p-HSP27 (pulse), (I) PTEN (concentration), (J) STAT3 (concentration), (K) phospho-STAT3 (concentration), (L) phospho-HSP27 (concentration), (M) Phospho-inositide 3 kinase P110 (concentration) and (N) cFLIP (concentration). All fluorescent signals are reported as arbitrary fluorescence units (AFU), with β-actin signal normalisation. Data in (C–N) are shown as mean ± SD of three samples pooled and are representative of three independent experiments. Statistical analysis of RPPA data is presented in Supporting Information Table 2.
Overall, the most significant factor causing elevation of the inflammatory signaling intermediates (including PI3K, MAPK, STAT3, Jak2/c-Src) and activation of transcription factors (such as ATF, Jun, and ELK) was expression by the SK-Hep-1 cells of the mutant C33Y TNFR1 compared with WT TNFR1 expression, regardless of stimulation by TNF-α (Fig. 3). An exception to this pattern was c-FLIP, which showed significantly higher levels in WT than C33Y TNFR1-expressing cells (bottom row of the heat maps in Figure 3B and N).
Increased NF-κB subunit activity in nuclear lysates of SK-Hep-1 cells carrying mutant TNFR1
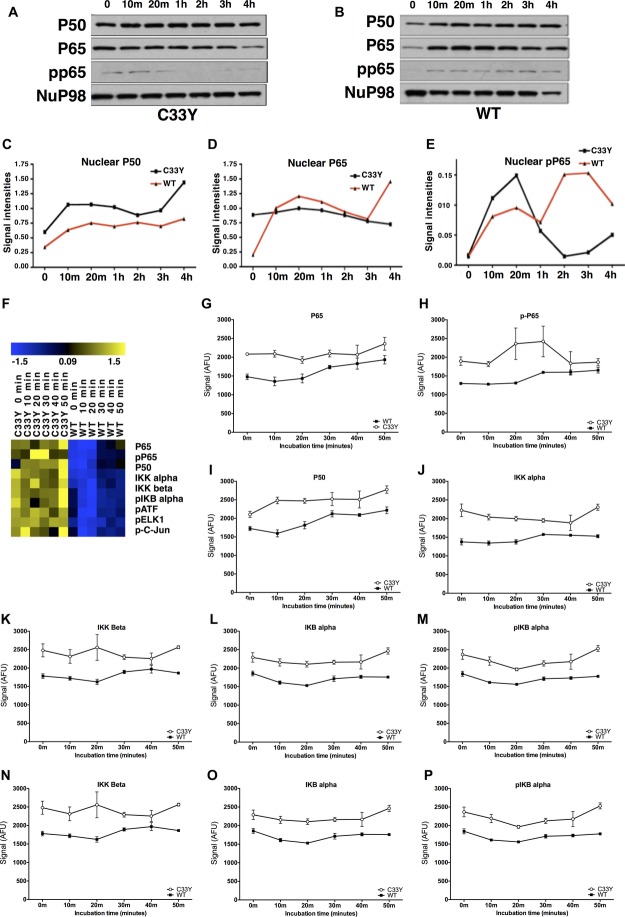
Lysates of nuclear subcellular fractions were prepared from the SK-Hep-1 cells stably transfected with C33Y mutant TNFR1 or with WT TNFR1, either without stimulation, or following stimulation with 50 ng/mL TNF-α for various lengths of time. Western blot analysis of these lysates to detect nuclear localization of NF-κB subunits is shown in Figure 4A–E. Expression of NuP98 was used as an internal control for normalization of nuclear protein loading. Densitometric quantification of the western blots in Figure 4A and B is shown in Figure 4C–E. These data show that nuclear localization of NF-κB P50 and P65 is constitutively higher in C33Y SK-Hep-1 transfectants than in WT. The nuclear levels of P50 increase in response to TNF-α in both C33Y and WT cells (Fig. 4C), whereas nuclear P65 is increased in WT cells only to levels similar to those seen in the nuclei of C33Y cells (Fig. 4D). Nuclear levels of pP65 are much lower, but initially increase in both C33Y and WT transfectants in response to TNF-α; they then fall again in C33Y cells after 1 hour of stimulation, but continue to rise in WT cells (Fig. 4E).
Figure 4.
NF-κB activation in TNFR1-C33Y-expressing SK-Hep-1 cells. (A) TNFR1-C33Y-transfected or (B) TNFR1-WT-transfected, SK-Hep-1 transfectants were stimulated with 50 ng/mL TNF-α for different times (0, 10 min, 20 min, 1, 2, 3, and 4 hours), lysed and assessed for NF-κB pathway intermediates by chemiluminescent imaging using SDS-PAGE and western blot. Representative results of three experiments are shown. NuP98 was used as internal control normalizer of protein loading for nuclear lysate samples. Densitometric quantification of (C) nuclear P50, (D) nuclear P65, and (E) nuclear phosphoP65 were obtained using the RPPA method. Data are shown as mean ± SD of three samples pooled from three independent experiments. (F) Heat maps representing NF-κB signaling intermediates determined by RPPA in SK-Hep-1 transfectants stimulated with 10 ng/mL TNF-α for various times (0, 10, 20, 30, 40, and 50 min). Each column represents separate SK-Hep-1 cell nuclear lysates organized by the transfected variant, and then by time of stimulation; each row represents a separate signaling molecule. The blue to yellow heat map indicates relative abundance from lower and higher levels, respectively. Signal values represented on the color scale for the heat maps are log2 transformed from the AFU obtained. (G–P) Graphical representation of the signals visualized on the heat map (F): (G) P65, (H) p-P65, (I) P50, (J) IKKα, (K) IKKβ, (L) IKBα, (M) p-IKBα, (N) p-ATF, (O) p-ELK1, and (P) p-C-Jun. All fluorescent signals are reported as arbitrary fluorescence units (AFU), with β-actin signal normalization. Data are shown as mean ± SD of three samples pooled from three independent experiments. Statistical analysis of RPPA data is presented in Supporting Information Table 3.
Nuclear fractions of C33Y- and WT TNFR1-transfected SK-Hep-1 cells were also analyzed by RPPA for expression of the NF-kB subunits P65, pP65, and P50; for members of the IKK regulatory system; and for the transcription factors p-ATF, p-ELK1, and p-C-Jun (the findings are shown as a heat map in Figure 4F and graphically in Figure 4G–P; statistical analysis by two-way ANOVA shown in Supporting Information Table 3). All of these proteins were constitutively upregulated in the nuclei of C33Y cells compared with WT cells. Incubation with TNF-α slightly increased nuclear expression of NF-κB P65 and P50 (Fig. 4G and I, respectively; Supporting Information Table 3), and induced slight, but statistically significant, changes in levels of IKB alpha and pIKB alpha (Fig. 4L and M, respectively; Supporting Information Table 3). By contrast, nuclear levels of NF-κB pP65 (Fig. 4H), IKK alpha/beta (Fig. 4J and K), or the phosphorylated transcription factors (Fig. 4N–P) were not affected by incubation with TNF-α (Supporting Information Table 3). Thus, the most significant effect on nuclear localization of transcription factors and regulatory proteins was that of the C33Y TNFR1 mutation per se.
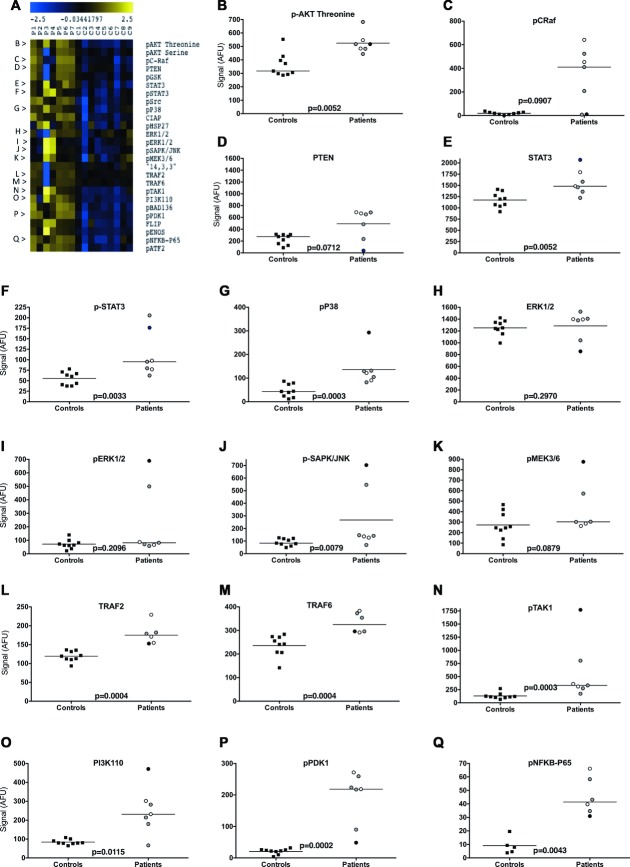
PBMC of C33Y-variant TRAPS patients confirm the subtle activation of multiple inflammatory pathways
In order to corroborate the above findings, PBMCs from TRAPS patients carrying C33Y mutation and PBMCs from sex- and age-matched healthy controls were separated and lysed, and the same inflammatory signaling intermediates were investigated. The RPPA results reveal that the majority of the tested proteins and their phosphorylated forms were constitutively upregulated in TRAPS patients compared with healthy controls with high levels of statistical significance (p values range from 0.012 to 0.0002) — these findings are shown as a heat map in Figure 5A, and graphically in Figure 5B–Q. The only tested proteins not to differ significantly between the TRAPS patients and controls were pC-Raf, ERK1/2, pERK1/2, pMEK3/6, pHSP-27, and PTEN; indeed, even for these proteins, two or more patients showed elevated levels, which is consistent with the behavior of the SK-Hep-1 cells expressing C33Y-TNFR1 (Fig. 3). Thus, the results were mainly consistent with the results obtained with transfected SK-Hep-1 cell lines, thus supporting the use of the SK-Hep-1 cell line as a model for studying TRAPS.
Figure 5.
Signaling pathway snapshot from TRAPS patient PBMCs. (A) PBMCs from TRAPS patients (P1–7) and controls (C1–9) were lysed and assessed for intracellular signaling molecules using RPPA. Untreated PMBCs were lysed after separation from blood. Heat map showing various inflammatory signaling pathway intermediates is shown. Rows represent the various signaling molecules studied. Blue and yellow denote markers that are present at lower and higher levels, respectively. (B–Q) Scatter plots representing measurement of selected target proteins from the heat map data, including (B) p-AKT threonine, (C) pC-Raf, (D) PTEN, (E) STAT-3, (F) pSTAT3, (G) pP38, (H) ERK1/2, (I) pERK1/2, (J) pSAPK/JNK, (K) pMEK3/6, (L)TRAF2, (M) TRAF6, (N) pTAK1, (O) PI3K110, (P) pPDK1, and (Q) pNFκB-p65. Data are shown as individual sample data (mean of four technical replicates per sample) of nine patient samples or seven control samples. The black bar indicates the median signal for all samples in each group. Signal represents arbitrary fluorescence units; AFU, after background subtraction and normalization to β-actin. Patient treatment is represented by shading of the symbols: anakinra, gray; etanercept, white; steroid, black. Significance values were derived using the Wilcoxon Test for repeated measures.
It is important to note that the raised levels of signaling intermediates in the TRAPS patients’ PBMCs occurred despite all seven patients being free of overt clinical symptoms, and being on treatment with potent antiinflammatory therapies at the time that blood was taken (Fig. 5); four were being treated with anakinra (IL-1 receptor antagonist—symbols shaded gray), two with etanercept (a TNF neutralizing agent — symbols shaded white) and one with steroids (symbol shaded black). Thus, blocking the action of proinflammatory cytokines associated with TRAPS (e.g. IL-1, TNF) can improve clinical signs and symptoms, but not the underlying activation of intracellular inflammatory signaling pathways.
Discussion
This study details, for the first time, the pleiotropic effect of a TRAPS-associated mutant form of TNFR1 on multiple inflammatory signaling pathways — a proinflammatory signalome. It is therefore the most comprehensive report to date evidencing mutant forms of TNFR1 as promoters of a proinflammatory state and may help to identify potential targets for future specific therapeutic options for a variety of inflammatory disorders.
The examination of multiple TNFR1 signaling pathways by reverse-phase protein microarray analysis methods demonstrates not only that such large scale signaling studies can be performed with relative ease, but that our model system, SK-Hep-1 cells transfected with C33Y or WT TNFR1 constructs, and C33Y TRAPS patients’ PBMCs behave consistently with each other and show pleiotropic alterations due to the presence of the C33Y mutated form of TNFR1. Overall, there is a generalized increase in relative protein levels for a host of proinflammatory and antiapoptotic signaling intermediates (nonactivated and activated) in both transfected cells and patients’ PBMCs carrying the C33Y TNFR1 mutation. The C33Y-associated upregulation of signaling intermediates is apparent with or without stimulation by TNF-α. This is fully consistent with the ligand-independent signaling hypothesis, with constitutive signaling by mutant TNFR1 driving the cells into a sensitized, proinflammatory state.
The expression of C33Y-mutant TNFR1 influences NF-κB related signaling, but also activation of PI3-kinase/AKT, MEK/ERK, JNK, and P38 MAPK pathways and STAT3. Our results are consistent with increased MAPK and JNK activation reported previously [11]. However, our further observation of increased levels of the inactive forms suggests that effects of ROS in limiting phosphatase activity related to MAPKs is not the sole reason for the increase in active forms seen, but that there is a shift in the homeostatic balance within these pathways in cells expressing the C33Y variant of TNFR1. It will be interesting and important to extend these observations to cells expressing other TRAPS-associated TNFR1 mutations, as we are currently doing for the R92Q variant of TNFR1 (Abduljabbar et al., in preparation).
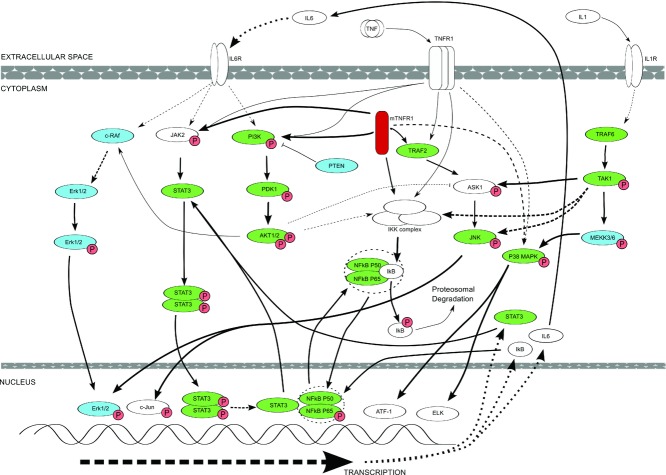
Figure 6 summarizes the pathway associations of many of the signaling intermediates and transcription factors investigated in the present study in both the C33Y and WT TNFR1-transfected SK-Hep-1 cell lines and in PBMCs from C33Y TNFR1 TRAPS patients and healthy controls. Intracellular mutant TNFR1 is highlighted in red; signaling/transcription proteins that are significantly upregulated in the TRAPS patients’ PBMCs are shown in green and those that do not differ significantly between patients and controls are shown in blue. The diagram also shows how the relevant signaling pathways may be triggered physiologically by exogenous TNF binding to WT TNFR1 at the cell surface, and/or pathologically by constitutive activation of mutant, misfolded TNFR1 in the cytoplasm. Furthermore, some of the pathways may also be triggered via the interaction of other exogenous proinflammatory cytokines with their cell surface receptors — for example, IL-6 (activating STAT3 and AKT1/2) and IL-1 (activating TRAF6) (Fig. 6). Thus, the pathways shown could be activated directly by ligand-independent activity of cytoplasmic mutant TNFR1 and/or indirectly by exogenous proinflammatory cytokines (TNF, IL-6, IL-1). The latter could be generated as a consequence of the constitutive proinflammatory effects of mutant TNFR1 itself and/or due to exogenous stimuli such as infecting microbes. It is relevant, in this context, that signaling pathways more distal to those triggered directly by TNFR1 do not show significant differences between the PBMCs of the TRAPS patients and controls — i.e. c-Raf and Erk1/2 associated with IL-6 signaling, and pMEKK3/6 in the IL-1 dependent TRAF6 pathway (Fig. 5 and 6). Furthermore, the negative regulator PTEN did not differ significantly between patients and controls.
Figure 6.
Diagrammatic representation of key alterations in signaling molecule expression and phosphorylation states as determined by examination of both SK-Hep-1 cell line models and TRAPS patient PBMCs expressing the C33Y TNFR1 variant, compared with cells expressing the WT TNFR1, or normal control sample PBMCs. The diagram does not show every target examined during this research project, but only indicates known signaling pathway interactions between different molecules. Dashed lines represent interactions involving more than one intermediate. Thick lines indicate where there is likely to be increased interaction and activation of downstream molecules or events. Signaling intermediates highlighted in green are those that are significantly upregulated in C33Y TRAPS patients’ PBMCs compared with those of healthy control individuals. The signaling intermediates highlighted in blue show no significant difference in levels between patient and control PBMCs.
The model helps to explain why upregulation of multiple signaling pathways was observed in TRAPS patients’ PBMCs in the present study in spite of concurrent treatment with corticosteroids, anakinra or etanercept and being free of overt clinical symptoms. It may also help to explain why various cytokine/receptor neutralizing biologics (e.g. etanercept, anakinra, tocilizumab) may have variable effects in TRAPS patients or may be effective for limited periods in particular patients [26,29]. These biologics inhibit the binding of the exogenous proinflammatory cytokines to their cell surface receptors, thereby downregulating ligand-dependent activation of particular signaling pathways. However, in some patients, these may not be the key pathways driving the inflammatory response and/or cells may adapt to the blockade by diverting activity into other proinflammatory pathways. More importantly, none of these neutralizing biologics will prevent the constitutive activity of the cytoplasmic mutant TNFR1. This explains why, in the present study, upregulation of signaling pathways was observed in TRAPS patients on biologic therapies and is consistent with the observations that TRAPS patients may show a clinical response to anticytokine or steroid treatments, but still have symptoms of on-going subclinical systemic inflammation [28]. Flares of clinical inflammation may then be due to synergy between the constitutive signaling activity of mutant TNFR1 and enhanced signaling by proinflammatory cytokines produced in response to exogenous stimuli (e.g. infection) [11]. The importance of identifying new therapeutic modalities that directly target the signaling pathways constitutively activated by mutant TNFR1 is thus apparent. It would also be desirable to extend the current studies to TRAPS patients with different levels of disease activity, and before and after treatment with particular therapeutic agents to see how this affects the signaling pathways studied.
Activation of STAT3 is particularly interesting because this can also be stimulated by IL-6 via Jak2 or c-Src. We have previously shown that, amongst inflammatory cytokines, IL-6 in particular is significantly elevated in C33Y TRAPS patients [27], and that the anti-IL6 receptor therapeutic antibody tocilizumab can have benefits in the clinical management of TRAPS [28]. Thus, enhanced Jak/STAT signaling may be a consequence of the combined effects of enhanced ligand-independent signaling by mutant TNFR1 and an autocrine feedback loop resulting from IL-6 production. The previously described ‘‘IL-6 amplifier’’ positive feedback loop of inflammatory activity may be particularly relevant in this context [30,31]; this was first elucidated in the form of synergy between IL-17A and IL-6, inducing increased IL-6 production by fibroblasts through NF-κB and STAT3 signaling. However, other cytokines and TLR-agonists may also synergize with IL-6 to enhance further production of this important inflammatory cytokine — including stimulation via TNFR1 [31]. We therefore hypothesize that, in TRAPS, ligand-independent signaling by mutant TNFR1 stimulates production of IL-6, and the two act synergistically to drive the ‘‘IL-6 amplifier’’ cycle in tissue cells; this may help to explain some of the tissue-localized inflammatory features of attacks in TRAPS, including monocytic fasciitis and the associated migratory erythematous skin rashes [32,33]. Furthermore, since IL-1 can also drive IL-6 production by various cell types [34–36], this hypothesis is also consistent with the therapeutic effect in TRAPS of the IL-1 antagonist anakinra [28]. In addition, a recent case report demonstrated the acute hyperresponsiveness of a TRAPS patient's PBMCs to inflammasome stimulation, resulting in significant IL1-β production [37], once again supporting both the case for hyperresponsiveness to innate inflammatory signals other than TNF-α and possible mechanisms by which IL-1 antagonists act in TRAPS patients.
STAT3 activation also promotes its own gene expression, and STAT3 has been shown to interact with NF-κB in both its phosphorylated (pSTAT3) and unphosphorylated (U-STAT3) forms. U-STAT3 competes with IκB to bind nuclear NF-κB and can therefore act as an activator (as well as an inhibitor) of NF-κB driven transcription, depending upon the promoter targeted and the balance of pSTAT3/U-STAT3. Increased abundance of U-STAT3 is reported initially to sustain NF-κB stimulated IL-6/IL-8/CCL5 expression for a prolonged period [38,39]; this is consistent with the high levels of IL-8, as well as IL-6, that we have observed in C33Y TRAPS patients [8,27]. The increases in both U-STAT3 and pSTAT3, irrespective of the mechanism for their upregulation, may therefore have significant consequences for the longevity of the inflammatory response in TRAPS patients.
Our present findings indicate that a complex network of signaling pathways are perturbed in both cultured cells and patients’ PBMCs that express a prototypic TRAPS-related TNFR1 mutation (C33Y). Both the levels of the total protein and activated forms of many TNFR1-related signaling molecules are elevated, suggesting a shift in the homeostasis within cells to accommodate the ongoing TNFR1-mediated signaling, resulting in cells which are potentially more hyperacutely sensitive to other stimuli, yet not necessarily to TNF-α itself. This complex signalome links NF-κB, p38, Jnk and Erk, MAPK, Akt, Jak2, and STAT3 signaling pathways, and is constitutively driven by the mutant TNFR1, irrespective of the proinflammatory cytokines secreted as a consequence of these intracellular events. This helps to explain the diverse pathophysiology of TRAPS, the variable and limited efficacy of cytokine-blocking agents in the management of TRAPS, and offers new targets and outcome measures for future therapeutic interventions. These insights also have much wider implications for other inflammatory and autoinflammatory diseases, where cytokine neutralizing therapies are highly useful, but may have limited degree or duration of efficacy. Delineation of signalomes in other autoinflammatory diseases will also help to clarify whether the signalome defined in this study is TRAPS-specific, or shares features with other autoinflammatory diseases.
Materials and methods
Cell culture, stimulation, and lysis
The human liver adenocarcinoma SK-Hep-1 endothelial-like cell line (which naturally expresses low levels of TNFR1) was stably transfected with WT or C33Y mutant TNFRSF1A gene constructs as previously described [5]. Cells were grown in 6-well plates at a density of 2 × 105 cells per well in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 units/mL of penicillin, 10 mg/mL of streptomycin, and 5 μg/mL of blasticidin (Invitrogen Life Technologies, USA) in a 37°C incubator with 5% CO2 until 80% confluent. To produce lysates, cells were serum starved for 24 hours before stimulation with TNF-α (R&D System) as follows:
TNF-α time course: TNF-α (10 ng/mL) was used to stimulate the cells over a time course (0, 10, 20, 30, 40, and 50 min)
TNF-α pulse-chase experiment: cells were stimulated with TNF-α (10 ng/mL) for 2 min and then immediately changed to fresh medium and cells were lysed at different times (0, 20, 40, 60, 80, and 100 min); and
TNF-α concentration experiment: different concentrations of TNF-α (0, 10, 20, 30, 40, and 50 ng/mL) were used for 30 min.
The cells were then washed with ice-cold phosphate buffered saline (PBS), and lysed in 200 μL of RIPA buffer (Pierce, Thermo Fisher Scientific) containing protease and phosphatase inhibitors (Pierce, Thermo Fisher Scientific). Protein concentrations were determined by BCA protein assay (Pierce, Thermo Fisher Scientific). Lysates were mixed with 4× SDS (0.25 M Tris HCl pH 6.8, 30% Glycerol, 8% SDS, and 10% 2-mercaptoethanol) at a ratio of 3:1 and were boiled for 5 min at 95°C to obtain complete protein denaturation prior to use in microarray or western blotting.
Nuclear /cytoplasmic fractionation
NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Pierce, Thermo Fisher Scientific) were used to separate the cytoplasm from the nucleus to study the NF-κB signaling pathway. According to the manufacturer's recommendations, cells were washed twice with ice-cold PBS and the cell pellet was resuspended in 200 μL of cytoplasmic extraction reagent (cer1) mix containing protease and phosphatase inhibitor cocktail, vortexed for 15 s and incubated on ice for 10 min. Eleven microliters of ice-cold Cytosol Extraction Buffer-B was added, vortexed for 5 s and incubated on ice for 1 min. After vortexing the samples were centrifuged for 5 min at 16 000 × g. Supernatants (cytoplasmic extract) were transferred immediately to clean prechilled tubes and placed on ice. Pellets (containing the nuclei) were washed twice with ice-cold PBS to decrease contamination with cytoplasmic components, followed by lysis with 100 μL of ice-cold Ner1 buffer mix containing 1× phosphatase and protease inhibitors and were vortexed for 15 s. Samples were then incubated on ice and the last step was repeated every 10 min for 40 min. Samples were centrifuged at 16 000 × g for 10 min. Immediately, supernatants (nuclear extract) were transferred to clean prechilled tubes. Protein concentrations were measured by BCA protein assay.
PBMC separation from TRAPS patient and control blood samples
Blood samples were obtained following informed consent from seven patients with the C33Y TNFR1 mutation and seven healthy age and sex-matched controls. The experiments conformed to the principles set out in the WMA Declaration of Helsinki and the NIH Belmont report. All the patients were free from overt symptoms of inflammation at the time that blood was taken. The mean age of the patients was 41.7 years (range 32–58) with a male:female ratio 5:2; four patients were being treated with anakinra, two with etanercept, and one with steroids. The seven healthy age and sex-matched controls were of mean age 44.7 years, range 29–65; male:female ratio 5:2. 20 mL blood was taken into a Falcon polypropylene tube (Becton Dickinson, Lincoln Park, NJ, USA) containing sterile citrate phosphate dextrose (ACD; Baxter Health Care, Norfolk, UK). Blood was then mixed 1:1 with PBS and 5 mL Histopaque was underlayered in 50 mL tubes followed by centrifugation at 900 × g for 20 min. The PBMCs were removed, washed twice in Hank's buffered saline (total volume 25 mL) and the cells were centrifuged at 400 × g for 10 min. The cell pellet was finally washed with PBS, centrifuged and lysed with lysis buffer (100 μL).
Immunoblotting
Proteins from the lysates were solubilized in 4× SDS loading buffer and were heated for 5 min at 100°C. Equal amounts of protein (50 μg) were separated on Novex 12% Tris glycine polyacrylamide gels (Invitrogen Life Technologies, USA) and transferred to nitrocellulose membranes. Membranes were blocked in PBS containing 0.1% Tween-20 and 5% nonfat dry milk powder, and incubated with primary antibodies (diluted 1:1000 in 5% BSA/PBST). All primary antibodies were purchased from Cell Signaling Technology. Table1 shows the list of the primary antibodies used in western blot and microarray. Blots were incubated overnight at 4°C then washed with PBST, and incubated with swine anti-rabbit immunoglobulin-HRP conjugate (DAKO) diluted 1:2000 in blocking buffer for 1 hour at room temperature. Following an additional PBST wash, membranes were incubated with Enhanced Chemiluminescence Detection reagent (ECL; Amersham, UK), exposed to film (Kodak, Sigma Aldrich, UK), and developed. The densities of resultant bands were measured with Un-SCAN-IT gel 5.3 software.
Reverse phase protein microarrays
Ten microlitres of cellular lysates were loaded into a 384-well plate and serially diluted 5 times with 1× SDS buffer. Samples were spotted onto nitrocellulose-coated glass slides (Grace Bio-labs) with a microarraying robot (MicroGrid 610, Digilab, Marlborough, MA, USA). Incubation of the slides took place overnight in blocking solution (0.2% I-block (Tropix, Bedford, MA, USA), 0.1% Tween-20 in PBS) at 4°C with constant rocking. After three washes of 5 min each, the slides were incubated with primary antibodies overnight at 4°C with shaking. In addition, β-actin antibody (mouse antibody) was included as a control for protein loading. After washing, slides were incubated with diluted infrared Licor secondary antibodies (680 CW anti-mouse Ig antibody for detection of β-actin and 800 CW anti-rabbit Ig antibody for detection of rabbit antibodies) for 30 min at room temperature in the dark with shaking. Slides were washed then dried by centrifugation at 500 × g for 5 min and scanned with a Licor Odyssey scanner (LI-COR, Biosciences) at 21 μm resolution at 700 nm (red) and 800 nm (green). The resultant TIFF images were processed with Genepix Pro-6 Microarray Image Analysis software (Molecular Devices Inc.) to obtain fluorescence data for each feature and generate standardized Genepix results (GPR) files. Protein signals were finally determined with background subtraction and normalization to the internal housekeeping targets using RPP analyzer, a module within the R statistical language [40].
Statistical analysis
Two-way ANOVA using GraphPad Prism was applied to compare the pattern of detection signaling molecules between C33Y and WT cells across the dose response to TNF-α, pulse chase experiments and time course experiments. Wilcoxon matched pairs test using GraphPad Prism was applied to compare the level of expression of signaling molecules in unstimulated C33Y and WT cells, or in TRAPS patients and healthy controls.
Acknowledgments
This project was supported by funds provided by The Jones 1986 Charitable Trust. We would like to thank Mr. Paul Radford, Mrs. Susan Bainbridge, and Mr. Colin Nicholson for their specialist technical support for elements of this project.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Supporting Information
Additional supporting information may be found in the online version of this article at the publisher's web-site
Supplementary
Supplementary
References
- 1.McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97:133–144. doi: 10.1016/s0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- 2.Sarrauste de Menthiere C, Terriere S, Pugnere D, Ruiz M, Demaille J, Touitou I. INFEVERS: the Registry for FMF and hereditary inflammatory disorders mutations. Nucleic Acids Res. 2003;31:282–285. doi: 10.1093/nar/gkg031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huggins ML, Radford PM, McIntosh RS, Bainbridge SE, Dickinson P, Draper-Morgan KA, Tighe PJ, et al. Shedding of mutant tumor necrosis factor receptor superfamily 1A associated with tumor necrosis factor receptor-associated periodic syndrome: differences between cell types. Arthritis Rheum. 2004;50:2651–2659. doi: 10.1002/art.20380. [DOI] [PubMed] [Google Scholar]
- 4.Todd I, Radford PM, Draper-Morgan KA, McIntosh R, Bainbridge S, Dickinson P, Jamhawi L, et al. Mutant forms of tumour necrosis factor receptor I that occur in TNF-receptor-associated periodic syndrome retain signalling functions but show abnormal behaviour. Immunology. 2004;113:65–79. doi: 10.1111/j.1365-2567.2004.01942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rebelo SL, Bainbridge SE, Amel-Kashipaz MR, Radford PM, Powell RJ, Todd I, Tighe PJ. Modeling of tumor necrosis factor receptor superfamily 1A mutants associated with tumor necrosis factor receptor-associated periodic syndrome indicates misfolding consistent with abnormal function. Arthritis Rheum. 2006;54:2674–2687. doi: 10.1002/art.21964. [DOI] [PubMed] [Google Scholar]
- 6.Todd I, Radford PM, Daffa N, Bainbridge SE, Powell RJ, Tighe PJ. Mutant tumor necrosis factor receptor associated with tumor necrosis factor receptor-associated periodic syndrome is altered antigenically and is retained within patients’ leukocytes. Arthritis Rheum. 2007;56:2765–2773. doi: 10.1002/art.22740. [DOI] [PubMed] [Google Scholar]
- 7.Yousaf N, Gould DJ, Aganna E, Hammond L, Mirakian RM, Turner MD, Hitman GA, et al. Tumor necrosis factor receptor I from patients with tumor necrosis factor receptor-associated periodic syndrome interacts with wild-type tumor necrosis factor receptor I and induces ligand-independent NF-kappaB activation. Arthritis Rheum. 2005;52:2906–2916. doi: 10.1002/art.21268. [DOI] [PubMed] [Google Scholar]
- 8.Rebelo SL, Amel-Kashipaz MR, Radford PM, Bainbridge SE, Fiets R, Fang J, McDermott EM, et al. Novel markers of inflammation identified in tumor necrosis factor receptor-associated periodic syndrome (TRAPS) by transcriptomic analysis of effects of TRAPS-associated tumor necrosis factor receptor type I mutations in an endothelial cell line. Arthritis Rheum. 2009;60:269–280. doi: 10.1002/art.24147. [DOI] [PubMed] [Google Scholar]
- 9.Dickie LJ, Aziz AM, Savic S, Lucherini OM, Cantarini L, Geiler J, Wong CH, et al. Involvement of X-box binding protein 1 and reactive oxygen species pathways in the pathogenesis of tumour necrosis factor receptor-associated periodic syndrome. Ann. Rheum. Dis. 2012;71:2035–2043. doi: 10.1136/annrheumdis-2011-201197. [DOI] [PubMed] [Google Scholar]
- 10.Bachetti T, Chiesa S, Castagnola P, Bani D, Di Zanni E, Omenetti A, D'Osualdo A, et al. Autophagy contributes to inflammation in patients with TNFR-associated periodic syndrome (TRAPS) Ann. Rheum. Dis. 2013;72:1044–1052. doi: 10.1136/annrheumdis-2012-201952. [DOI] [PubMed] [Google Scholar]
- 11.Simon A, Park H, Maddipati R, Lobito AA, Bulua AC, Jackson AJ, Chae JJ, et al. Concerted action of wild-type and mutant TNF receptors enhances inflammation in TNF receptor 1-associated periodic fever syndrome. Proc. Natl. Acad. Sci. U.S.A. 2010;107:9801–9806. doi: 10.1073/pnas.0914118107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobito AA, Kimberley FC, Muppidi JR, Komarow H, Jackson AJ, Hull KM, Kastner DL, et al. Abnormal disulfide-linked oligomerization results in ER retention and altered signaling by TNFR1 mutants in TNFR1-associated periodic fever syndrome (TRAPS) Blood. 2006;108:1320–1327. doi: 10.1182/blood-2005-11-006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bulua AC, Simon A, Maddipati R, Pelletier M, Park H, Kim KY, Sack MN, et al. Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS) J. Exp. Med. 2011;208:519–533. doi: 10.1084/jem.20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schutze S, Tchikov V, Schneider-Brachert W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat. Rev. Mol. Cell. Biol. 2008;9:655–662. doi: 10.1038/nrm2430. [DOI] [PubMed] [Google Scholar]
- 15.Schneider-Brachert W, Tchikov V, Neumeyer J, Jakob M, Winoto-Morbach S, Held-Feindt J, Heinrich M, et al. Compartmentalization of TNF receptor 1 signaling: internalized TNF receptosomes as death signaling vesicles. Immunity. 2004;21:415–428. doi: 10.1016/j.immuni.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z, Connell MC, MacEwan DJ. TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation, mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial cells. Cell Signal. 2007;19:1238–1248. doi: 10.1016/j.cellsig.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 17.Adam-Klages S, Schwandner R, Adam D, Kreder D, Bernardo K, Kronke M. Distinct adapter proteins mediate acid versus neutral sphingomyelinase activation through the p55 receptor for tumor necrosis factor. J. Leukoc. Biol. 1998;63:678–682. doi: 10.1002/jlb.63.6.678. [DOI] [PubMed] [Google Scholar]
- 18.Pincheira R, Castro AF, Ozes ON, Idumalla PS, Donner DB. Type 1 TNF receptor forms a complex with and uses Jak2 and c-Src to selectively engage signaling pathways that regulate transcription factor activity. J. Immunol. 2008;181:1288–1298. doi: 10.4049/jimmunol.181.2.1288. [DOI] [PubMed] [Google Scholar]
- 19.Miscia S, Marchisio M, Grilli A, Di Valerio V, Centurione L, Sabatino G, Garaci F, et al. Tumor necrosis factor alpha (TNF-alpha) activates Jak1/Stat3-Stat5B signaling through TNFR-1 in human B cells. Cell Growth Diff.: Mol. Biol. J. American Assoc. Cancer Res. 2002;13:13–18. [PubMed] [Google Scholar]
- 20.MacEwan DJ. TNF receptor subtype signalling: differences and cellular consequences. Cell Signal. 2002;14:477–492. doi: 10.1016/s0898-6568(01)00262-5. [DOI] [PubMed] [Google Scholar]
- 21.Nedjai B, Hitman GA, Yousaf N, Chernajovsky Y, Stjernberg-Salmela S, Pettersson T, Ranki A, et al. Abnormal tumor necrosis factor receptor I cell surface expression and NF-kappaB activation in tumor necrosis factor receptor-associated periodic syndrome. Arthritis Rheum. 2008;58:273–283. doi: 10.1002/art.23123. [DOI] [PubMed] [Google Scholar]
- 22.Siebert S, Fielding CA, Williams BD, Brennan P. Mutation of the extracellular domain of tumour necrosis factor receptor 1 causes reduced NF-kappaB activation due to decreased surface expression. FEBS Lett. 2005;579:5193–5198. doi: 10.1016/j.febslet.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 23.Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 24.D'Osualdo A, Ferlito F, Prigione I, Obici L, Meini A, Zulian F, Pontillo A, et al. Neutrophils from patients with TNFRSF1A mutations display resistance to tumor necrosis factor-induced apoptosis: pathogenetic and clinical implications. Arthritis Rheum. 2006;54:998–1008. doi: 10.1002/art.21657. [DOI] [PubMed] [Google Scholar]
- 25.Nedjai B, Hitman GA, Quillinan N, Coughlan RJ, Church L, McDermott MF, Turner MD. Proinflammatory action of the antiinflammatory drug infliximab in tumor necrosis factor receptor-associated periodic syndrome. Arthritis Rheum. 2009;60:619–625. doi: 10.1002/art.24294. [DOI] [PubMed] [Google Scholar]
- 26.Ter Haar N, Lachmann H, Ozen S, Woo P, Uziel Y, Modesto C, Kone-Paut I, et al. Treatment of autoinflammatory diseases: results from the Eurofever Registry and a literature review. Ann. Rheum. Dis. 2013;72:678–685. doi: 10.1136/annrheumdis-2011-201268. [DOI] [PubMed] [Google Scholar]
- 27.Nowlan ML, Drewe E, Bulsara H, Esposito N, Robins RA, Tighe PJ, Powell RJ, et al. Systemic cytokine levels and the effects of etanercept in TNF receptor-associated periodic syndrome (TRAPS) involving a C33Y mutation in TNFRSF1A. Rheumatol. (Oxford) 2006;45:31–37. doi: 10.1093/rheumatology/kei090. [DOI] [PubMed] [Google Scholar]
- 28.Vaitla PM, Radford PM, Tighe PJ, Powell RJ, McDermott EM, Todd I, Drewe E. Role of interleukin-6 in a patient with tumor necrosis factor receptor-associated periodic syndrome: Assessment of outcomes following treatment with the anti-interleukin-6 receptor monoclonal antibody tocilizumab. Arthritis Rheum. 2011;63:1151–1155. doi: 10.1002/art.30215. [DOI] [PubMed] [Google Scholar]
- 29.Moll M, Kuemmerle-Deschner JB. Inflammasome and cytokine blocking strategies in autoinflammatory disorders. Clin. Immunol. 2013;147:242–275. doi: 10.1016/j.clim.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Murakami M, Hirano T. A four-step model for the IL-6 amplifier, a regulator of chronic inflammations in tissue-specific MHC class II-associated autoimmune diseases. Frontiers Immunol. 2011;2:22. doi: 10.3389/fimmu.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogura H, Murakami M, Okuyama Y, Tsuruoka M, Kitabayashi C, Kanamoto M, Nishihara M, et al. Interleukin-17 promotes autoimmunity by triggering a positive-feedback loop via interleukin-6 induction. Immunity. 2008;29:628–636. doi: 10.1016/j.immuni.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Quillinan N, Mohammad A, Mannion G, O'Keeffe D, Bergin D, Coughlan R, McDermott MF, et al. Imaging evidence for persistent subclinical fasciitis and arthritis in tumour necrosis factor receptor-associated periodic syndrome (TRAPS) between febrile attacks. Ann. Rheum. Dis. 2010;69:1408–1409. doi: 10.1136/ard.2009.118661. [DOI] [PubMed] [Google Scholar]
- 33.Hull KM, Wong K, Wood GM, Chu WS, Kastner DL. Monocytic fasciitis: a newly recognized clinical feature of tumor necrosis factor receptor dysfunction. Arthritis Rheum. 2002;46:2189–2194. doi: 10.1002/art.10448. [DOI] [PubMed] [Google Scholar]
- 34.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nature Rev. Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 35.Hu DN, Chen M, Zhang DY, Ye F, McCormick SA, Chan CC. Interleukin-1beta increases baseline expression and secretion of interleukin-6 by human uveal melanocytes in vitro via the p38 MAPK/NF-kappaB pathway. Invest. Ophthalmol. Vis. Sci. 2011;52:3767–3774. doi: 10.1167/iovs.10-6908. [DOI] [PubMed] [Google Scholar]
- 36.You M, Flick LM, Yu D, Feng GS. Modulation of the nuclear factor kappa B pathway by Shp-2 tyrosine phosphatase in mediating the induction of interleukin (IL)-6 by IL-1 or tumor necrosis factor. J. Exp. Med. 2001;193:101–110. doi: 10.1084/jem.193.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohmori S, Hino R, Nakamura M, Tokura Y. Heparin serves as a natural stimulant of the inflammasome and exacerbates the symptoms of tumor necrosis factor receptor-associated periodic syndrome (TRAPS) J. Dermatol. Sci. 2012;66:82–84. doi: 10.1016/j.jdermsci.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Cheon H, Yang J, Stark GR. The functions of signal transducers and activators of transcriptions 1 and 3 as cytokine-inducible proteins. J. Interferon Cytokine Res. 2011;31:33–40. doi: 10.1089/jir.2010.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR. Unphosphorylated STAT3 accumulates in response to IL-6 and activates transcription by binding to NFkappaB. Genes Develop. 2007;21:1396–1408. doi: 10.1101/gad.1553707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mannsperger HA, Gade S, Henjes F, Beissbarth T, Korf U. RPPanalyzer: Analysis of reverse-phase protein array data. Bioinformatics. 2010;26:2202–2203. doi: 10.1093/bioinformatics/btq347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary
Supplementary