Abstract
Background:
Aging is one of the most important risk factors for cancer. It appears that aberrant epigenetic changes might be a common driver of aging and cancer. Among them are changes in DNA methylation and DNA hydroxymethylation. The 5′ carbon of cytosines in CpG dinucleotides of DNA can be either methylated or hydroxymethylated. Like 5′-methylcytosine, changes in 5′-hydroxymethylcytosine may occur due to aging, potentially leading to downstream changes in transcription and cancer development.
Methods:
We set up a method to measure 5′-methyl-2′-deoxycytidine and 5′-hydroxymethyl-2′-deoxycytidine in DNA using liquid chromatography/mass spectrometry (LC/MS-MS) and used this method to measure the percentage of total cytosine that was either methylated or hydroxymethylated in the liver tissues of young and old C57Bl/6 male mice. The DNA was enzymatically hydrolyzed by sequential digestion with nuclease P1, phosphodiesterase I and alkaline phosphatase. The isotopomers [15N3]-2′-deoxycytidine and (methyl-d3, ring-6-d1)-5-methyl-2′-deoxycytidine were added to the DNA hydrolysates as internal standards. DNA methylation and hydroxymethylation were calculated as a percentage of total deoxycytidine in genomic DNA.
Results:
Within day variations for DNA methylation and hydroxymethylation were 3.45% and 8.40%, while day to day variations were 6.14% and 17.68%, respectively. Using this method it was determined that hepatic DNA of old mice had increased levels of hydroxymethylation relative to young (0.32 ± 0.02% vs. 0.24 ± 0.01%, P = 0.02), with no significant changes in 5′-methylcytosine.
Conclusions:
DNA hydroxymethylation measured by LC/MS-MS method can be a novel biomarker of aging. It will be useful to investigate the potential role of DNA hydroxymethylation in the development and prevention of age-associated cancer.
Keywords: DNA hydroxymethylation, DNA methylation, Aging, Liver, Mouse
INTRODUCTION
The environment can influence gene transcription through a series of covalent modifications added to the DNA and DNA supporting structures referred to as epigenetic marks. DNA methylation is the most widely researched epigenetic mark, and is characterized by the addition of a methyl group to the 5′ carbon of the cytosine nucleotide, creating 5′-methylcytosine. This addition of methyl groups to cytosine bases throughout the genome can affectively increase or decrease the transcription of certain genes.1 The location and density of 5′-methylcytosine relative to the gene seems to have great importance, as this may determine whether that gene is actively transcribed or silenced.2,3 Recently the hydroxymethylation of cytosine nucleotides at the same 5′ carbon site has been discovered.4 It has been suggested that this hydroxymethylation is a transient status of the active demethylation process of DNA methylation, resulting in unmodified cytosine.5,6 DNA methylation is converted to hydroxymethylation through an oxidative reaction carried out by the ten-eleven translocation, or TET family of enzymes.4,7 This hydroxymethylated cytosine is further oxidized to 5′-formylcytosine and 5′-carboxylcytosine until ultimately being replaced by an unmodified cytosine by a thymine-DNA glycosylase.6,8
Like DNA methylation, the genomic location of 5′-hydroxymethylcytosine may be a factor in transcriptional regulation, possibly due to its decreased binding affinity with methylbinding proteins.9,10 In contrast to methylation, acquisition of hydroxymethylation in a regulatory region of a gene might enhance expression of that gene.7,11 The abundance of hydroxymethylated DNA is fairly low and varies across tissue types, with the highest amount located in the brain.6,12 Outside of the central nervous system, the distribution of hydroxymethylation is widely variable and tends to decrease in cell cultures.13 Loss of global hydroxymethylation is associated with cancer such as melanoma14 and can be a diagnostic and prognostic marker. Mutations in TET2 are commonly observed in hematopoietic malignancies.15
The aging process has previously been shown to alter DNA methylation in the mammalian genome.16,17 DNA hydroxymethylation is no exception, particularly in regards to changes of 5′-hydroxymethylcytosine in the aging brain.18–20 Age associated changes in global hydroxymethylation in other tissues are not yet known, but are equally as intriguing. Because the liver has many metabolic regulatory functions, epigenetic alterations that may change the function of the liver could have meaningful downstream effects. In this research we attempt to determine the effect aging has on hepatic DNA hydroxymethylation at the global level. We also measure the expression of the three Tet genes in both age groups in an effort to associate abundance of global 5′-hydroxymethylcytosine to expression of the Tet enzymes.
To reach our research goals we set up a mass spectrometrymethod to precisely measure global DNA hydroxymethylation using only 1 μg of genomic DNA. We have improved a previous liquid chromatography/electrospray ionization-mass spectrometry method that we invented to measure genomic DNA methylation, simultaneously measuring both modifications using the same internal standard.21 Similar to other accounts of mass spectrometry to measure hydroxymethylation, the method reported here is both sensitive and accurate.22,23
MATERIALS AND METHODS
1. Apparatus
All experiments were performed on an Agilent 1100 high-performance liquid chromatography (HPLC) (Palo Alto, CA, USA) with an Applied Biosystems 3200 Q Trap mass spectrometermass spectrometer system (Concord, ON, Canada) equipped with a turbospray ionization source. The liquid chromatograph used was an Agilent 1100 Series with a quaternary pump, a vacuum degasser and an autosampler. A SuplexpKb 100 analytical column (25 cm × 2.1 mm) protected by a 5-μm SuplexpKb 100 precolumn (2 cm × 2.1 mm) (Supelco, Bellefonte, PA, USA) was used. The liquid chromatography/mass spectrometry (LC/MS-MS) was controlled through the Analyst 1.4 software (Applied Biosystems) run on a Dell Precision T3400 computer under the Microsoft Windows XP operating system.
2. Reagents
The mobile phase for all reactions consisted of 7 mmol/L ammonium acetate pH 6.7 and HPLC-grade methanol 5% (v/v), and was prepared with HPLC-grade water (Fisher Scientific, Pittsburgh, PA, USA). Before use, the mobile phase was filtered through a 0.2-μm nylon membrane (Millipore, Billerica, MA, USA).
The stable isotope-labeled compounds [15N3]2′-deoxycytidine and the custom-made (methyl-d3, ring-6-d1)-5′-methyl-2′- deoxycytidine (both from Cambridge Isotopes Laboratories, Inc., Andover, MA, USA) were used as internal standards for 2′-deoxycytidine, 5′-methyl-2′-deoxycytidine and 5′-hydroxymethyl-2′-deoxycytidine residues. Purified 5′-hydroxymethyl-2′-deoxycytidine (Berry & Associates, Dexter, MI, USA) was used as an external standard, as well as to develop a standard curve, determine the limits of detection and quantification and to determine the percent recovery.
3. DNA extraction and hydrolysis
Extraction of DNA was conducted through the standard phenol/chloroform/isoamyl alcohol [25:24:1 (v/v/v)] method with precipitation with 100% ethanol and 3 mol/L sodium acetate, pH 5.2. Following extraction DNA was re-dissolved in TE buffer. DNA quantification and 260/280 ratios were measured on a Nanodrop 1000 (Thermo Scientific, Wilmington, DE, USA). All DNA samples used had a 260/280 ratio equal to or greater than 1.80. DNA hydrolysis was performed as previously described.21 In brief, 1 μg of genomic DNA was denatured by heating the sample at 100°C for 3 minutes and subsequently chilled on ice. Next, 0.1 mol/L ammonium acetate (pH 5.3) and 2 units of nuclease P1 (Sigma, St. Louis, MO, USA) were then added to each sample. The mixture was incubated at 45°C for 2 hours. Subsequently, 1 μL of 1 mol/L ammonium bicarbonate (pH 7.8) (Sigma) and 0.002 units of venom phosphodiesterase I (Sigma) were added to each sample. All samples were incubated for an additional 2 hours at 37°C, then 0.5 units of alkaline phosphatase (Sigma) was added to the mixture and another 1 hour of incubation at 37°C took place. The stable isotopes [15N3]-2′-deoxycytidine and (methyl-d3, ring-6-d1)-5′-methyl-2′-deoxycytidine were then added to the samples to reach a final concentration of 1.00 and 0.25 ng/μL, respectively. The total volume for each sample was 35 μL at the end of hydrolysis.
4. Liquid chromatography/mass spectrometry procedure
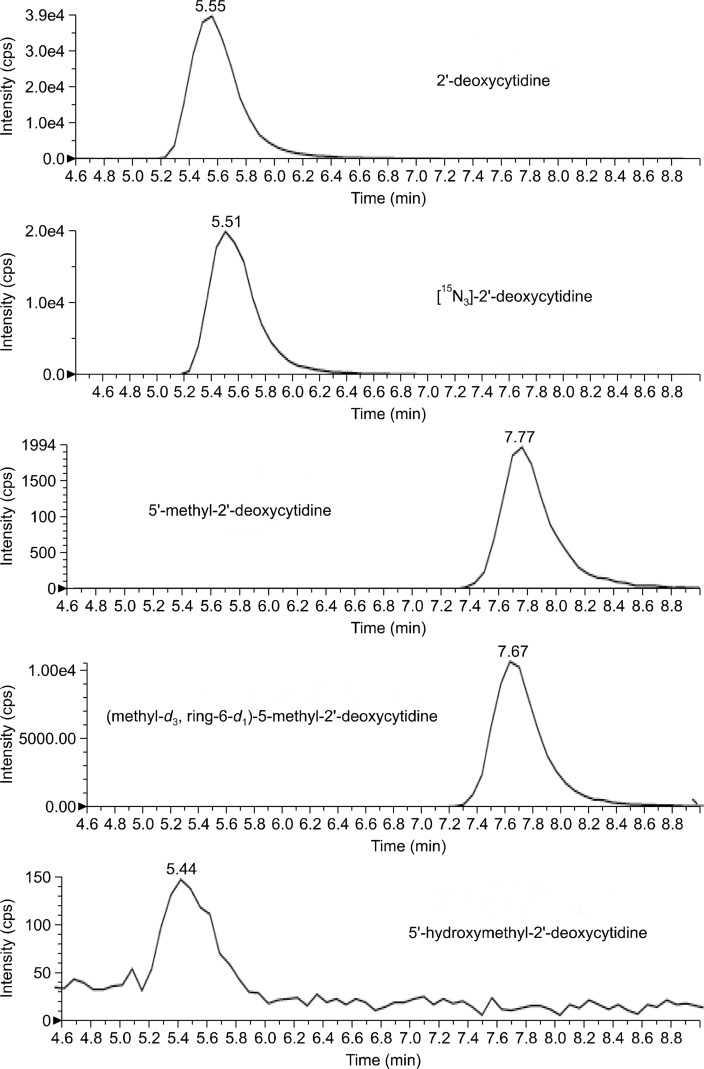
The analytical column was equilibrated with the mobile phase (ammonium acetate, methanol and water) at a flow rate of 300 μL/min. A volume of 10 μL of hydrolyzed DNA was injection into the column that was kept at 25°C to separate the DNA bases by isocratic elution. The total run time for each was 14.0 minutes. The turbospray source was heated to 400°C, and included nitrogen gas at a pressure of 60 psi. After optimizing the turbo ion spray voltage of the mass spectrometer for several runs, it was decided to keep voltage constant at 5,000 V for the best sensitivity. The mass spectrometer was operated in positive ion mode, and the declustering potential was set to 22.0 V. The mass spectrometer was set to collect data in multiple reaction monitoring mode, using the following mass transitions after ion fragmentation: 2′-deoxycytidine at m/z 228.1 → 112.1, 5′-methyl- 2′-deoxycytidine at m/z 242.1 → 126.1, and 5′-hydroxymethyl- 2′-deoxycytidine at m/z 258.1 → 142.1, as previously reported (Fig. 1).22
Figure 1.
Typical liquid chromatography/mass spectrometry (LC/MS-MS) chromatogram of DNA digests with peaks of each deoxycytidine and internal standard. cps, counts per second.
5. Calculation of % 5′-methylcytosine and % 5′-hydroxymethylcytosine
Using the known masses of isotope-labeled internal standards added to each sample the absolute amount of cytosine, 5′-methylcytosine and 5′-hydroxymethylcytosine per 1 μg of DNA can be calculated using the area of the peaks:
Furthermore, this amount can be expressed as a percentage of total cytosine by using the following equation:
By expression the amount of 5′-methylcytosine and 5′-hydroxymethylcytosine as a percentage of total cytosine any variation in the initial amount of genomic DNA is accounted for.
6. Gene expression
Changes in expression of the Tet genes were determined by quantitative real-time polymerase chain reaction (PCR). An RNA extraction was performed using Trizol reagent and cDNA was synthesized using a standard reverse transcription kit (Invitrogen, Carlsbad, CA, USA). Quantitative real-time PCR was conducted on the LightCycler 480 real-time PCR machine (Roche, Indianapolis, IN, USA). Gene expression was determined as ΔCt, following normalization to the housekeeping gene GAPDH (ΔCt = CtGeneX –CTGAPDH). Expression was normalized to the young control group and is displayed as fold difference.
7. Animals and diets
This study was reviewed and approved by the Institutional Animal Care and Use Committee of the United States Department of Agriculture (USDA) Human Nutrition Research Center on Aging at Tufts University. Ten weanling (“Young”) and ten 18 month (“Old”) C57BL/6 male mice were fed an amino-acid defined diet described by Walzem and Clifford24 for 20 weeks. Animals were individually housed and group pair-fed to limit variability in food intake. At the end of the study the young mice were 6 months of age and the old mice were 23 months of age. All tissues in this study were obtained at necropsy, flash frozen in liquid nitrogen and stored at −80°C until the time of DNA and RNA extraction.
8. Statistics
To determine differences between the young and old age groups a Student’s t-test was performed. If variables were skewed a logarithmic transformation was first conducted, resulting in normal distributions of all variables to be used in statistical comparisons. Differences were considered statistically significant if the P-value < 0.05. Values in the text are means ± standard error of the mean unless otherwise noted. Statistical procedures were carried out on Statistical Analysis System software version 9.3 (SAS Institute Inc., Cary, NC, USA).
RESULTS
1. Mass spectrometry quantification of 5′-hydroxymethylcytosine
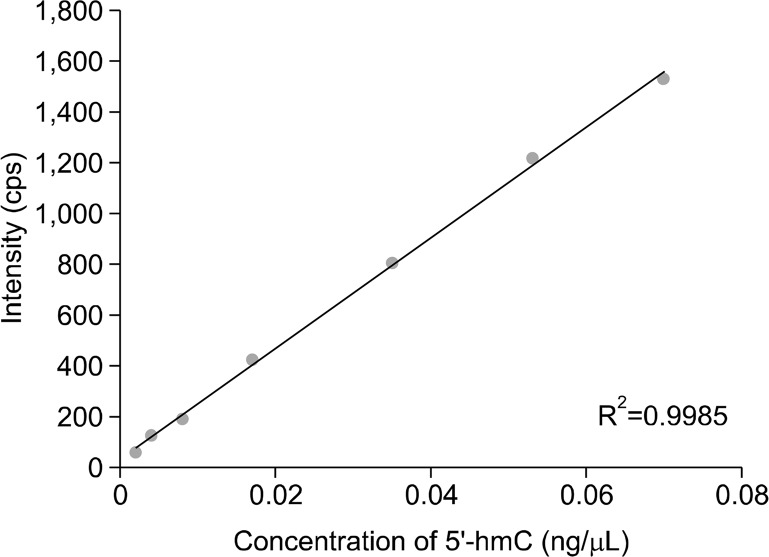
Purified 5′-hydroxymethylcytosine standard was dissolved in water and used to create a standard curve of known concentrations. Calibration curves of 5′-hydroxymethylcytosine by the mass spectrometry were linear (Fig. 2). The limit of detection of 5′-hydroxymethylcytosine, defined as a signal-to-noise ratio of at least 3, was determined to be 2 pg/μL, and the limit of quantification, defined as a signal-to-noise ratio of at least 10, was determined to be 4 pg/μL.
Figure 2.
Standard curve of known concentrations of dissolved 5′-hydroxymethylcytosine (5′-hmC). cps, counts per second.
To determine the retention time of 5′-hydroxymethylcytosine relative to the other forms of cytosine, a hydrolyzed DNA sample was spiked with 0.035 ng of 5′-hydroxymethylcytosine before being injected into the LC/MS-MS. As expected, the retention time was determined to be similar to that of unmodified cytosine (Fig. 1). To ensure that sensitivity for 5′-hydroxymethylcytosine was similar when detected by the mass spectrometry alone as when detected along with cytosine, the percent recovery was calculated by adding a known amount of 5′-hydroxymethylcytosine to hydrolyzed DNA samples. After multiple trials, the average recovery was 82.05%.
The internal standard (methyl-d3, ring-6-d1)-5-methyl-2′-deoxycytidine was used to calculate the absolute mass of both 5′-methylcytosine and 5′-hydroxymethylcytosine. The coefficient of determination, or R2 between expected and calculated concentration of 5′-hydroxymethylcytosine using this internal standard in a series of standard solutions was 0.996.
The variability between runs performed on the same day and between runs performed on different days was calculated as coefficient of variance (Table). In both cases, the variation was lower for calculating the mass and percent of total cytosine that was methylated than calculating the mass and percentage of cytosine that was hydroxymethylated. Furthermore, intra-day variation for all calculations was much lower than the variability in runs that occurred on different days. Therefore, all samples from our animal experiments were run on the same day.
Table.
Variability in 5′-methylcytosine (5′-mC) and 5′-hydroxymethylcytosine (5′-hmC) as measured by liquid chromatography/mass spectrometry (LC/MS-MS)
| Intra-day variationac (n=16) | % CV Intra-daya (n = 16) | Inter-day variationbc (n = 48) | % CV Inter-dayb (n = 48) | |
|---|---|---|---|---|
| % 5′-mC of total cytosine | 4.54 ± 0.14% | 3.45% | 5.22 ± 0.11% | 6.14% |
| % 5′-hmC of total cytosine | 0.23 ± 0.01% | 8.40% | 0.10 ± 0.02% | 17.68% |
| Calculated mass of 5′-mC (ng) | 3.34 ± 0.25 | 2.66% | 6.37 ± 1.13 | 5.84% |
| Calculated mass of 5′-hmC (ng) | 0.17 ± 0.01 | 7.92% | 0.07 ± 0.01 | 17.43% |
DNA samples were extracted from liver tissue of C57Bl/6 mice.
DNA samples were extracted from colonic scrapings.
Expressed as mean ± standard error of the mean. CV, coefficient of variance.
2. Animal study
Over the 20 week feeding period four of the ten mice in the old age group died, while none of the young mice died. Deaths were attributed to liver cancer or unknown causes.25 The animals in the young age group gained weight consistently throughout the feeding period, whereas the old mice had little variation in weight. There was no difference in animal weight at the end of the feeding period.
3. Measuring DNA methylation and hydroxymethylation in hepatic tissue
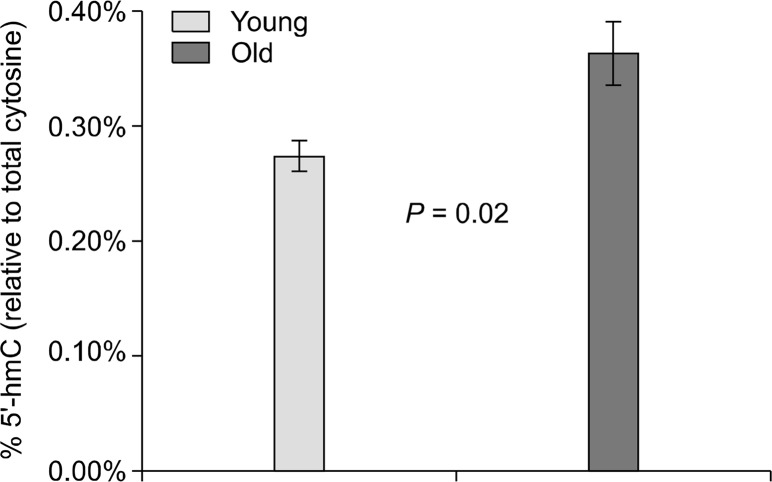
There was no change in % 5′-methylcytosine when comparing our young and old age groups and values were nearly identical (4.56 ± 0.20% vs. 4.57 ± 0.21%, P = not significant). However, there was a statistically significant increase in % 5′-hydroxymethylcytosine in the livers from elder mice relative to the young (0.24 ± 0.01% vs. 0.32 ± 0.02%, P = 0.02) (Fig. 3).
Figure 3.
Percentage of total cytosine that was hydroxymethylated in each age group. Old mice had significantly higher levels of 5′-hydroxymethylcytosine (5′-hmC). Error bars represent standard error of the mean.
4. Expression of the Tet genes in hepatic tissue
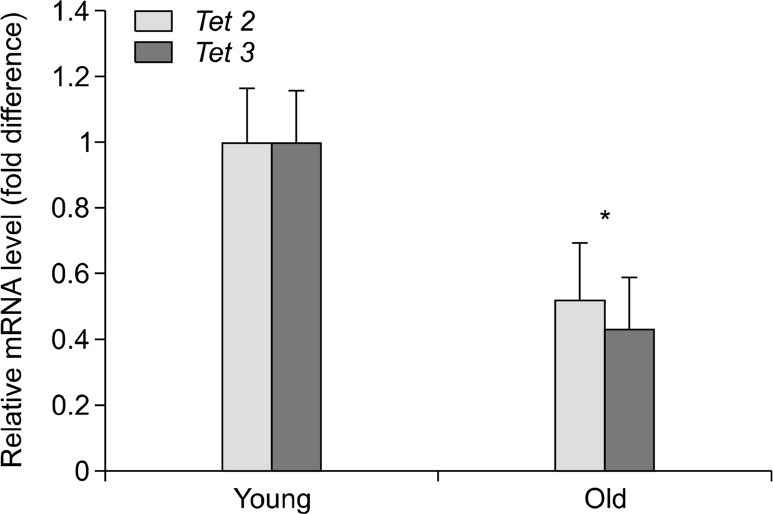
The mRNA expression of all three of the known Tet enzymes was measured to determine age-associated differences. As expected, little Tet1 mRNA was expressed in all samples (data not shown).26 There were, however, discernable differences in the amount of Tet3 mRNA between the young and the old age groups (1.0 ± 0.17 vs. 0.52 ± 0.17, P = 0.03) (Fig. 4). Likewise, the expression of Tet2 was also reduced in the old age group relative to the young, although this difference did not reach statistical significance (1.0 ± 0.16 vs. 0.43 ± 0.16, P = 0.07).
Figure 4.
Expression of Tet2 and Tet3 mRNA normalized to the young group. *P < 0.05. Error bars represent standard error of the mean.
DISCUSSION
Through the adaption of a previous LC/MS-MS method to measure DNA methylation we can now simultaneously measure DNA methylation and hydroxymethylation, an epigenetic mark that is gaining relevance in regards to biological mechanisms. The procedure described here makes use of an internal standard for 5′-methylcytosine to also measure the relative abundance of 5′-hydroxymethylcytosine. We used 8.75 ng of (methyl-d3, ring-6-d1)-5-methyl-2′-deoxycytidine in each sample, an amount that was low enough to accurately calculate the small amount of 5′-hydroxymethylcytosine in the DNA. We measured the linearity of the method through the correlation of a set of 5′-hydroxymethylcytosine standards with known concentrations to their respective calculated concentrations (R2 = 0.9996). This is a time saving and cost effective alternative to synthesizing an isotopelabeled internal standard specific to 5′-hydroxymethylcytosine.
Depending on the retention time of the analytes, LC/MS-MS can be a high-throughput technique. In this protocol each sample takes 14 minutes to run through the instrument, allowing for up to 102 samples to be analyzed in a day. The variability of measured 5′-methylcytosine and 5′-hydroxymethylcytosine was larger when samples were run in the LC/MS-MS on separate days instead of on the same day. This may be due to the cooling and subsequent re-heating of the electrospray ion source or variability in the mobile phase that was made fresh daily. To reduce this variability it was determined that all samples from the same subject be analyzed in the same day or that the date of the run be a factor in the model for statistical analysis.
We did not see any differences in the amount of cytosine that was methylated between our two age groups. Some studies have indicated that aging does in fact alter global hepatic methylation levels, while others seem to show that this only occurs in cancerous tissue.27–29 It is possible that changes in DNA methylation with age are diet-dependent, and that the amino-acid defined diet fed here, or the diets fed to the mice before they reached our animal facility, prevented any age-associated changes in methylation at the global level. The Walzem and Clifford amino acid defined diet has a higher level of methionine (8.2 g/kg) compared with AIN 93M (3.3 g/kg) and AIN 76A (3.0 g/kg).24,30,31 Because methionine is the precursor to the donor of methyl groups to DNA methylation, S-adenosylmethionine, this high level of methionine may mask the hypomethylation tendencies often associated with aging.32,33
The percent of total cytosine that was hydroxymethylated in the livers of old mice was significantly higher than that of the younger mice. While this is intriguing, it is important to note that less than one percent of cytosine was shown to be hydroxymethylated in these samples, and that the location of this hydroxymethylation within the genome is unknown. This increase in hydroxymethylation is out of concordance with the decrease in expression of Tet2 and Tet3 mRNA that was exhibited in the older mice. It may be that the activity of the TET enzymes increases with age, possibly due to the physiologic process of aging itself or an increased availability of coenzymes such as iron.4 Because the turnover of 5′-methylcytosine to 5′-hydroxymethylcytosine is an oxidative reaction, one can speculate that the oxidative environment that is associated with aging may in fact increase the activity of the TET enzymes, especially at the elder age of these mice (24 months).34 This may produce the increase in 5′-hydroxymethylcytosine levels that were found in this study, regardless of mRNA expression of the Tet genes.
Besides active removal of 5′-hydroxymethylcytosine through the oxidation to 5′-formylcytosine and 5′-carboxylcytosine, 5′-hydroxymethylcytosine can also be lost passively through cell divisions.35 Rapidly dividing cells, such as embryonic stem cells, have an increased turnover of 5′-hydroxymethylcytosine to unmodified cytosine, possibly through the inability of the maintenance DNA methyltransferase DNMT1 to methylate a target cytosine that is hydroxymethylated on the parent strand of DNA.4,26,36 Conversely, cells that do not rapidly divide may have an increase in hydroxymethylation, such as is observed in the central nervous system.37 A decrease in cellular replication may be the cause of the increase in hydroxymethylation shown in the liver tissue from the old age group here.
In this study we have demonstrated the usefulness of a new LC/MS-MS method to measure DNA hydroxymethylation. Through this method we have determined changes in hydroxymethylation in hepatic DNA from young and old C57Bl/6 male mice, and those changes were associated with Tet enzyme mRNA expression. DNA hydroxymethylation measured by LC/MS-MS method can be a novel biomarker of aging and will be useful to investigate the mechanisms underlying the development of age-associated cancer.
Acknowledgments
This material is based upon work supported by the USDA, under agreement No. 51000-074-01S. Any opinions, findings, conclusion, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the USDA. This project has been supported in part by the National Institute of Health Grants R01 AG025834 (Sang-Woon Choi). This work was also supported by the Bio-Synergy Research Project (NRF-2012M3A9C4048735) of the Ministry of Science, ICT and Future Planning through the National Research Foundation (Sang-Woon Choi). All authors participate in the conception, design or conduction of the study as well as interpretation of data and drafting the manuscript.
Footnotes
CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
REFERENCES
- 1.Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 2.Boyes J, Bird A. Repression of genes by DNA methylation depends on CpG density and promoter strength: evidence for involvement of a methyl-CpG binding protein. EMBO J. 1992;11:327–33. doi: 10.1002/j.1460-2075.1992.tb05055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irvine RA, Lin IG, Hsieh CL. DNA methylation has a local effect on transcription and histone acetylation. Mol Cell Biol. 2002;22:6689–96. doi: 10.1128/MCB.22.19.6689-6696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–5. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat Rev Genet. 2011;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 6.Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PloS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Zhang X, Clark E, Mulcahey M, Huang S, Shi YG. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–3. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 8.He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–84. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–8. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sérandour AA, Avner S, Oger F, Bizot M, Percevault F, Lucchetti-Miganeh C, et al. Dynamic hydroxymethylation of deoxyribonucleic acid marks differentiation-associated enhancers. Nucleic Acids Res. 2012;40:8255–65. doi: 10.1093/nar/gks595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids. 2011;2011:870726. doi: 10.4061/2011/870726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nestor CE, Ottaviano R, Reddington J, Sproul D, Reinhardt D, Dunican D, et al. Tissue type is a major modifier of the 5-hydroxymethylcytosine content of human genes. Genome Res. 2012;22:467–77. doi: 10.1101/gr.126417.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JJ, Murphy GF, Lian CG. Melanoma epigenetics: novel mechanisms, markers, and medicines. Lab Invest. 2014;94:822–38. doi: 10.1038/labinvest.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroeze LI, Aslanyan MG, van Rooij A, Koorenhof-Scheele TN, Massop M, Carell T, et al. Characterization of acute myeloid leukemia based on levels of global hydroxymethylation. Blood. 2014;124:1110–8. doi: 10.1182/blood-2013-08-518514. [DOI] [PubMed] [Google Scholar]
- 16.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richardson B. Impact of aging on DNA methylation. Ageing Res Rev. 2003;2:245–61. doi: 10.1016/S1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 18.Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2010;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, Dzitoyeva S, Manev H. Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restor Neurol Neurosci. 2012;30:237–45. doi: 10.3233/RNN-2012-110223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Hove DL, Chouliaras L, Rutten BP. The role of 5-hydrox ymethylcytosine in aging and alzheimers disease: current status and prospects for future studies. Curr Alzheimer Res. 2012;9:545–9. doi: 10.2174/156720512800618008. [DOI] [PubMed] [Google Scholar]
- 21.Friso S, Choi SW, Dolnikowski GG, Selhub J. A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem. 2002;74:4526–31. doi: 10.1021/ac020050h. [DOI] [PubMed] [Google Scholar]
- 22.Le T, Kim KP, Fan G, Faull KF. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Anal Biochem. 2011;412:203–9. doi: 10.1016/j.ab.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Zhang L, Zhou K, Ye X, Zhang J, Xie A, et al. Simultaneous determination of global DNA methylation and hydroxymethylation levels by hydrophilic interaction liquid chromatography-tandem mass spectrometry. J Biomol Screen. 2012;17:877–84. doi: 10.1177/1087057112447946. [DOI] [PubMed] [Google Scholar]
- 24.Walzem RL, Clifford AJ. Folate deficiency in rats fed diets containing free amino acids or intact proteins. J Nutr. 1988;118:1089–96. doi: 10.1093/jn/118.9.1089. [DOI] [PubMed] [Google Scholar]
- 25.Kim KC, Jang H, Sauer J, Zimmerly EM, Liu Z, Chanson A, et al. Folate supplementation differently affects uracil content in DNA in the mouse colon and liver. Br J Nutr. 2011;105:688–93. doi: 10.1017/S0007114510004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singhal RP, Mays-Hoopes LL, Eichhorn GL. DNA methylation in aging of mice. Mech Ageing Dev. 1987;41:199–210. doi: 10.1016/0047-6374(87)90040-6. [DOI] [PubMed] [Google Scholar]
- 28.Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–51. [PubMed] [Google Scholar]
- 29.Pogribny IP, James SJ, Jernigan S, Pogribna M. Genomic hypomethylation is specific for preneoplastic liver in folate/methyl deficient rats and does not occur in non-target tissues. Mutat Res. 2004;548:53–9. doi: 10.1016/j.mrfmmm.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–51. doi: 10.1093/jn/123.11.1939. [DOI] [PubMed] [Google Scholar]
- 31.Medinsky MA, Popp JA, Hamm TE, Dent JG. Development of hepatic lesions in male Fischer-344 rats fed AIN-76A purified diet. Toxicol Appl Pharmacol. 1982;62:111–20. doi: 10.1016/0041-008X(82)90107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L, Wylie RC, Andrews LG, Tollefsbol TO. Aging, cancer and nutrition: the DNA methylation connection. Mech Ageing Dev. 2003;124:989–98. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006;136:1706S–10S. doi: 10.1093/jn/136.6.1706S. [DOI] [PubMed] [Google Scholar]
- 34.Chia N, Wang L, Lu X, Senut MC, Brenner C, Ruden DM. Hypothesis: environmental regulation of 5-hydroxymethylcy tosine by oxidative stress. Epigenetics. 2011;6:853–6. doi: 10.4161/epi.6.7.16461. [DOI] [PubMed] [Google Scholar]
- 35.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–50. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 37.Williams K, Christensen J, Helin K. DNA methylation: TET proteins-guardians of CpG islands? EMBO Rep. 2011;13:28–35. doi: 10.1038/embor.2011.233. [DOI] [PMC free article] [PubMed] [Google Scholar]