Abstract
The Cpx envelope stress response mediates a complex adaptation to conditions that cause protein misfolding in the periplasm. A recent microarray study demonstrated that Cpx response activation led to changes in the expression of genes known, or predicted, to be involved in cell wall remodeling. We sought to characterize the changes that the cell wall undergoes during activation of the Cpx pathway in Escherichia coli. Luminescent reporters of gene expression confirmed that LdtD, a putative l,d-transpeptidase; YgaU, a protein of unknown function; and Slt, a lytic transglycosylase, are upregulated in response to Cpx-inducing conditions. Phosphorylated CpxR binds to the upstream regions of these genes, which contain putative CpxR binding sites, suggesting that regulation is direct. We show that the activation of the Cpx response causes an increase in the abundance of diaminopimelic acid (DAP)-DAP cross-links that involves LdtD and YgaU. Altogether, our data indicate that changes in peptidoglycan structure are part of the Cpx-mediated adaptation to envelope stress and indicate a role for the uncharacterized gene ygaU in regulating cross-linking.
INTRODUCTION
The envelope of Gram-negative bacteria is a complex multilayer structure that protects bacteria from changing conditions in the environment. This structure consists of the outer membrane, the inner membrane, and the periplasmic space. The periplasm contains the cell wall, which provides protection against osmotic stresses and maintains the cell shape. It also provides an oxidizing environment where periplasmic proteins that are translocated from the cytoplasm can be stabilized via disulfide bond formation (1). The periplasm is more sensitive to the conditions outside the cell than the cytoplasm due to the porous nature of the outer membrane (2). Consequently, bacteria have various stress responses whose function consists of sensing and responding to stresses in the periplasm. Among these, the sigma E and Cpx envelope stress responses are thought to respond to conditions that lead to misfolded outer membrane and periplasmic proteins, respectively (3–5, 66). Four additional pathways that respond to envelope stress have been described: the Bae (BaeSR), phage shock (Psp), Rcs, and vesicle release responses. The first three of these pathways have been linked to the efflux of toxic compounds, disruptions of proton motive force, and cell wall perturbations, respectively (6–8).
Regulation of the Cpx pathway is carried out by the products of the cpxRA operon that encodes the response regulator CpxR and the histidine kinase CpxA. The Cpx pathway regulates numerous genes for periplasmic protein folding and degrading factors to alleviate envelope stress, including the protease/chaperone DegP, the disulfide oxidase DsbA, and the peptidyl-prolyl isomerase PpiA (9, 10). A recent microarray analysis (11) identified new members of the Cpx regulon by analyzing changes in gene expression after overexpression of NlpE (12), a well-known inducing cue of the Cpx pathway. Some of the novel genes identified included the known or predicted cell wall modification genes ygaU, ldtD, and slt. All of these genes were upregulated when the Cpx pathway was activated. LdtD is a 67-kDa l,d-transpeptidase that synthesizes DAP (diaminopimelic acid)-DAP cross-links by the removal of the fourth d-alanine residue of an acyl donor tetrapeptide stem and the attachment of the remaining meso-DAP residue to the meso-DAP residue of a second acyl acceptor peptide (13). YgaU is a 16-kDa conserved hypothetical protein with a LysM domain, found in enzymes that interact with and degrade the cell wall (14, 15), and a BON motif that has a membrane binding domain consisting of a conserved glycine residue and several hydrophobic regions (16). Slt is a 65-kDa lytic transglycosylase that cleaves the β-1,4 glycosidic bond between N-acetylmuramic acid and N-acetylglucosamine (NAG) residues and concomitantly catalyzes an intramolecular glycosyl transfer reaction forming a 1,6-anhydro-N-acetylmuramic acid (17). In this study, we confirmed that the activation of the Cpx pathway causes changes in the regulation of ldtD, ygaU, and slt, and we present evidence suggesting that this regulation could be direct. Furthermore, we showed that the cell wall undergoes changes in response to the activation of the Cpx pathway. These changes are partly dependent on LdtD and YgaU. Our data suggest a previously undescribed regulatory role in peptidoglycan (PG) modification for YgaU.
(Some parts of this paper have been reported online as part of the M.Sc. thesis of M. Bernal-Cabas [67]).
MATERIALS AND METHODS
Growth conditions.
Escherichia coli MC4100 was used as the parental strain in this study. Strains were grown in LB (Luria-Bertani) medium at 30°C, unless specified otherwise, with shaking and the proper antibiotics. The antibiotics used in this study were amikacin (3 μg/ml), chloramphenicol (25 μg/ml), kanamycin (50 μg/ml), and tetracycline (10 μg/ml) (Sigma-Aldrich).
Plasmids and strains.
Plasmids and strains used for this study are listed in Table 1. The promoter regions of various genes were cloned into lux vector pJW15 (18) to assess transcriptional activity, as previously described (4, 19). EcoRI- or XhoI-tagged primers were used to amplify the promoter of each gene via PCR (Table 2). Each amplified promoter region contained the putative CpxR binding site(s), the promoter, and the translational start site. To verify that the promoter of each gene was cloned properly into lux vector pJW15, the multiple-cloning site was amplified by using PCR (Table 2). The PCR products were purified by using the QIAquick PCR purification kit (Qiagen), and the DYEnamic ET terminator cycle sequencing kit was used for sequencing (Amersham) (Molecular Biology Service Unit, Department of Biological Sciences, University of Alberta). Knockout mutants were constructed via P1 phage transduction of the desired alleles obtained from the KEIO library (20). Transduction was verified by colony PCR (Table 2). In some cases, kanamycin resistance cassettes were removed from mutant strains using the Flp recombinase encoded on plasmid pFLP2, as described in references 20 and 63. Plasmids pCA-24N, pCA-nlpE, and pCA-ygaU were obtained from the ASKA library (21).
TABLE 1.
List of strains and plasmids used in this study
| Plasmid or E. coli strain | Relevant genotype or description | Reference(s) |
|---|---|---|
| Plasmids | ||
| pJW15 | pJW15 with p15 ori | 4 |
| pCA-ygaU | pCA-24N that overexpresses ygaU | 21 |
| pCA-24N | Cloning vector with IPTG-inducible expression | 21 |
| pFLP2 | Plasmid expressing Flp recombinase | 63 |
| pJW15-ygaU | pJW15 with ygaU promoter | This study |
| pJW15-ldtD | pJW15 with ldtD promoter | This study |
| pJW15-slt | pJW15 with slt promoter | This study |
| Strains | ||
| MC4100 | F− araD139Δ(argF-lac)U169 rpsL150 (Strr) relA1 flbB5301 deoC1 ptsF25 rbsR | 64 |
| TR10 | MC4100 cpxA24 | 22, 29 |
| TR11 | MC4100 cpxA102 | 19 |
| TR15 | MC4100 cpxA711 | 22 |
| TR50 | MC4100 λRS88[cpxP-lacZ] | 22 |
| TR51 | MC4100 cpxR::spc | 29 |
| TR70 | TR50 cpxR1::spc | 65 |
| pJW15 | MC4100(pJW15) | 4 |
| MBC1 | MC4100(ygaU-pJW15) | This study |
| MBC2 | TR10(ygaU-pJW15) | This study |
| MBC3 | TR51(ygaU-pJW15) | This study |
| MBC4 | MC4100(pCA-NlpE, ygaU-pJW15) | This study |
| MBC5 | MC4100(ygaU-pJW15, pCA-24N) | This study |
| MBC6 | MC4100(ldtD-pJW15) | This study |
| MBC7 | TR10(ldtD-pJW15) | This study |
| MBC8 | TR51(ldtD-pJW15) | This study |
| MBC9 | MC4100(pCA-NlpE, ldtD-pJW15) | This study |
| MBC10 | MC4100 (ldtD-pJW15, pCA-24N) | This study |
| MBC11 | MC4100(slt-pJW15) | This study |
| MBC12 | TR10(slt-pJW15) | This study |
| MBC13 | TR51(slt-pJW15) | This study |
| MBC14 | MC4100(pCA-NlpE, slt-pJW15) | This study |
| MBC15 | MC4100(slt-pJW15, pCA-24N) | This study |
| MBC16 | MC4100 ygaU::kan | This study |
| MBC20 | MC4100 ygaU::kan(pCA-ygaU) | This study |
| MBC21 | TR10 ygaU::kan(pCA-ygaU) | This study |
| MBC23 | TR50 ΔygaU | This study |
| MBC24 | TR50 ΔldtD | This study |
| MBC25 | TR50 ΔldtD ygaU | This study |
TABLE 2.
Primer sequences used in this study
| Primer use and gene or site targeteda | Primer | Primer sequence |
|---|---|---|
| Construction of lux reporters | ||
| ldtD | pJW5-ldtD(R) | 5′-AAAGTAATTCTTTGCGTGCGGGGCTTT-3′ |
| ldtD | pJW5-ldtD(F) | 5′-AAACTCGAGTGGTACAAAGCTGGGAAGAT-3′ |
| ygaU | pJW5-ygaU(R) | 5′-AAAGAAATTCTACCGGTTTGTTCAGATGC-3′ |
| ygaU | pJW5-ygaU(F) | 5′-AAACTCGAGGGAACCGCTAAGCATGCACA-3′ |
| slt | pJW5-slt70(R) | 5′-AAAGAATTCTCAATGGACTCACGCACGGT-3′ |
| slt | pJW5-slt70(F) | 5′-AAAGGATCCTGCTCATCCAGTGAGTCGGC-3′ |
| Verification of the MCS in pJW15 | ||
| MCS-pJW15 | MCS-pJW15(R) | 5′-CACCAAAATTAATGGATTGCAC-3′ |
| MCS-pJW15 | MCS-pJW15(F) | 5′-GCTTCCCAACCTTACCAGAG-3′ |
| Amplification of gene promoters for EMSA | ||
| degP | PdegP-F | 5′-CGCTTATTCCACAAACTCTCG-3′ |
| degP | PdegP-R | 5′-CGGCTGAGACTTCTTCAGCAACGA-3′ |
| rpoD | PrpoD-F | 5′-CGAAGAACGCCTGGAGC-3′ |
| rpoD | PrpoD-R | 5′-TCACCTGAATGCCCATGTCG-3′ |
| ygaU | PygaU-F | 5′-AAATGCTGTGATGTCGCAGAGG-3′ |
| ygaU | PygaU-R | 5′-GCGTCCCAGAGTTTTTCTCC-3′ |
| ldtD | PldtD-F | 5′-AAATGGTACAAAGCTGGGAGAT-3′ |
| ldtD | PldtD-R | 5′-AAATTTGCGTGCGGGCTTTTTCT-3′ |
| slt | Pslt70-F | 5′-AAATCCTGCGGCAGATAACCAAT-3′ |
| slt | Pslt70-R | 5′-AAATCGGTGATCTGGCGGTATTC-3′ |
| KEIO or deletion verification | ||
| ldtD | KldtD-F | 5′-AAAGTGGCACATTACGGCG-3′ |
| ldtD | KldtD-R | 5′-AAAGAGAGTGTTGCAAACGCAGG-3′ |
| ygaU | KygaU-F | 5′-AAATCGGTAAAGGGTTCGGTTGG-3′ |
| ygaU | KygaU-R | 5′-AAATAAGCTTGAGGCCTGTGACG-3′ |
| slt | Kslt70-F | 5′-AAATCCTGCGGCAGATAACCAAT-3′ |
| slt | Kslt70-R | 5′-AAATCGGTGATCTGGCGGTATTC-3′ |
| cpxA* (cpxA24) | cpxA*-F | 5′-ATGACCGAGCTTCTGGATAGC-3′ |
| cpxA* (cpxA24) | cpxA*-R | 5′-CTTCGCCATCACGGCACGGGAG-3′ |
MCS, multiple-cloning site.
Luminescence assays.
Single colonies of bacterial strains were inoculated in 5 ml of LB medium with the indicated antibiotics and grown overnight at 30°C at 225 rpm. The next morning, each strain was subcultured (1:40) in 5 ml of fresh LB medium with the appropriate antibiotic. After 2 h, 200 μl of each strain was transferred into a black-bottom 96-well plate (96-well black flat-bottom polystyrene; Corning). The optical density at 600 nm (OD600) and the luminescence (counts per second [cps]) were measured at late log phase (OD600 of 0.6 to 0.8) by using a Wallac Victor 2 multilabel plate reader (PerkinElmer Life Sciences). In the case of plasmid pCA-nlpE, NlpE overexpression was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) 2 h after subculturing of strains MBC4, MBC9, and MBC14. Each strain was assayed in quintuplicate. The average cps/OD600 values, standard deviations, and statistical significance were calculated by using GraphPad Prism 6.0.
Protein purification.
The maltose binding protein (MBP)-CpxR fusion protein was purified from JM109(pMCR) as previously described, with some variations (22). A single colony of JM109(pMCR) was grown overnight in 10 ml of LB medium with 150 μg/ml of ampicillin. The next morning, the entire overnight culture was used to inoculate 1 liter of fresh LB medium with 0.2% glucose. Cells were grown until they reached an OD600 of 0.3, and MBP-CpxR overproduction was induced with 0.1 M IPTG for 4 h. Next, cells were pelleted by centrifugation at 4,000 rpm for 20 min. The bacterial culture was disrupted by passage through a French press at 20,000 lb/in2. The crude extract was then treated overnight with 10 ml of amylose resin (New England Biolabs) for batch purification. This preparation was poured into a column and treated, as previously described, to purify the MBP-CpxR fusion protein (22).
Mobility shift assay.
Binding reactions were carried out with buffer (10 mM MgSO4, 20 mM potassium glutamate, 1 mM dithiothreitol [DTT], 50 μg/ml bovine serum albumin [BSA], 1 mM EDTA, 50 mM KCl, 10 mM Tris [pH 7.4]) in the absence or presence of a low-molecular-weight phosphodonor, 20 mM acetyl phosphate. Fifteen-microliter reaction mixtures containing various concentrations of MBP-CpxR were incubated at 37°C for 30 min. Next, 1.5 pmol of purified PCR product containing the promoter of interest was added, and the mixture was incubated at 37°C for another 30 min. Next, 6× DNA loading dye (0.25% xylene cyanol, 30% glycerol in water, 0.25% bromophenol blue) was added, and reaction mixtures were loaded onto a 5% nondenaturing Tris-borate-EDTA (TBE) polyacrylamide gel (Bio-Rad), electrophoresed in 1× TBE running buffer, and strained with ethidium bromide for visualization.
β-Galactosidase assays.
Single colonies of bacterial strains were inoculated in quintuplicate in 2 ml of LB medium with the indicated antibiotic and grown overnight at 30°C with shaking. The next morning, each strain was subcultured (1:40) in 2 ml of fresh LB medium and grown until late log phase (OD600 = 0.6). Cultures were centrifuged at 4,000 rpm for 10 min and resuspended in the same volume of Z buffer, according to a previously described protocol (23). β-Galactosidase activity was measured by using a Wallac Victor 2 multilabel plate reader (PerkinElmer Life Sciences) in a 96-well plate (96-well clear flat-bottom polystyrene; Corning). Average Miller units, standard deviations, and statistical significance were calculated by using WorkOut 2.0 and GraphPad Prism 6.0.
Preparation of peptidoglycan.
Peptidoglycan was prepared, as previously described, with small changes (24), from cultures of E. coli grown in LB medium at 37°C with aeration, except for MC4100 and cpxA* (cpxA24, cpxA102, and cpxA711) derivatives, which were grown at 30°C. In control experiments, we determined that muropeptide quantities were not appreciably altered by growth at 30°C compared to growth at 37°C (data not shown). Cells were grown overnight in 100 ml of LB medium with the appropriate antibiotics and then subcultured 1:20 into 100 ml of LB medium for several hours until an OD600 of 1.0 was reached. The cells were then incubated on ice until the temperature of the culture was 4°C. Samples were centrifuged at 4,000 rpm for 10 min at 4°C (5810R; Eppendorf), resuspended in 3 ml of distilled H2O, and slowly mixed drop by drop with an equal volume of 6% (wt/vol) boiling SDS with vigorous stirring until the mixture was homogeneous. The cells were boiled for 5 min, transferred into 10-ml glass tubes, and heated at 90°C for 72 h in a Baxter heat block. Sacculi were concentrated by centrifugation at 65,000 rpm for 15 min at 20°C in an ultracentrifuge (Beckman-Optima TLX with a TLA110 rotor). The pellet was washed with water until no SDS was detected, according to the method of Hayashi (25). The last pellet from the washing procedure was suspended in 1 ml of 10 mM Tris-HCl (pH 7.2) and digested first with 100 μg/ml α-amylase (EC 3.2.1.1; Sigma-Aldrich, St. Louis, MO) for 1 h at 37°C and then with 100 μg/ml preactivated pronase E (EC 3.4.24.4; Merck, Darmstadt, Germany) at 60°C for 90 min. The enzymes were inactivated by boiling for 20 min in 1% (final concentration) SDS. The cell walls were collected by centrifugation as described above and washed three times with water. Peptidoglycan was stored in water at 4°C.
Preparation and separation of muropeptides.
Peptidoglycan was digested in 50 mM phosphate buffer (pH 4.9) with Cellosyl (Hoechst AG, Frankfurt, Germany), at a 100-μg/ml final concentration, at 37°C overnight. The enzyme was inactivated by boiling of the sample for 2 min in a water bath and centrifugation (Eppendorf centrifuge at maximum speed for 10 min) to remove insoluble debris. The supernatant was mixed with a 1/3 volume of 0.5 M sodium borate buffer (pH 9.0) and reduced with excess sodium borohydride (NaBH4) for 30 min at room temperature. The pH was tested with pH indicator strips (Acilit; Merck) and adjusted to pH 3 with orthophosphoric acid. All samples were filtered (Millex-GV filters with a 0.22-μm pore size and a 2.5-mm diameter; Millipore, Cork, Ireland) and stored at −20°C. Separation of the reduced muropeptides was performed essentially as described previously by Glauner et al. (26, 27). Muropeptides were analyzed by using a binary-pump Waters high-performance liquid chromatography (HPLC) system (Waters Corporation) fitted with a reverse-phase RP18 Aeris peptide column (250 by 4.6 mm with a 3.6-μm particle size; Phenomenex, USA) and a dual-wavelength absorbance detector (Waters UV-1570 spectrophotometer). The eluted muropeptides were monitored by measuring the absorbance at 204 nm. When required, the individual peaks were collected, vacuum dried, and stored at −20°C.
Quantification of muropeptides.
Individual muropeptides were quantified from their integrated areas (Waters Breeze) by using samples of known concentrations as standards. Concentrations of the standard muropeptides were determined as described previously by Work (28). The average PG chain length was calculated by dividing the molar amount of anhydro-muropeptide (chain termini) by the total molar amount of muropeptides in muramidase-digested PG, and the degree of cross-linking was calculated by calculating the molar ratio of dimers and trimers to total muropeptide, as described previously by Glauner et al. (26, 27). In each experiment, muropeptides were quantified from three biological replicates, and the average molar ratio (mol%) values are presented (see Tables 3 to 5; see also Table S1 in the supplemental material). Experimental variation resulted in changes for each muropeptide measurement of ≤5%.
TABLE 3.
Peptidoglycan composition of cpx mutant strains
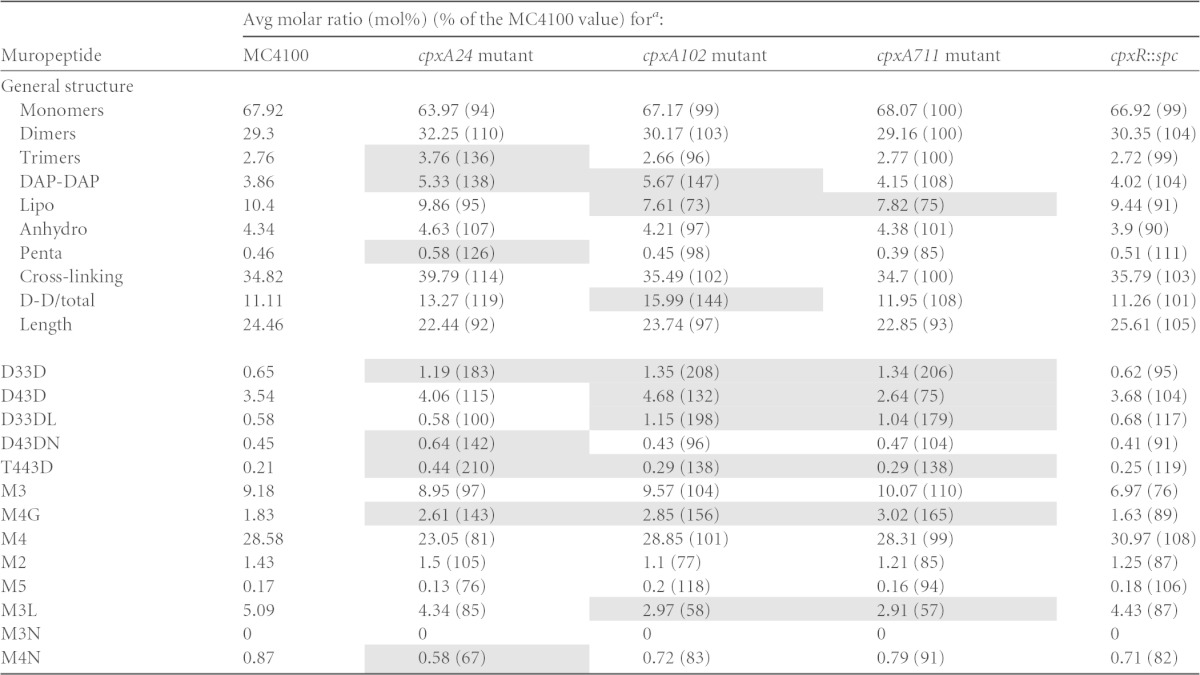
The molar ratios of the different muropeptide structures were calculated for MC4100 or isogenic derivatives carrying a cpxR::spc, cpxA24, cpxA102, or cpxA711 allele, as previously described (26). The different muropeptide structures are abbreviated according to length, where M indicates a monomer, D indicates a cross-linked dimer, and T indicates a cross-linked trimer. The numbers indicate the length of the stem peptide, and the last letter indicates whether the muropeptide contains a DAP-DAP cross-link (D) or is anchored to Braun's lipoprotein (L) or if there is a 1,6-anhydro-muramic acid residue (N) or a glycine (G) at the end of the stem peptide. D-D/total is the ratio of DAP-DAP cross-links to the total cross-links. The averages of data from three measurements from biological replicates are shown. Shading indicates measurements that were different from the MC4100 value by 25%.
TABLE 5.
Peptidoglycan composition of cpxA24 and isogenic ygaU, ldtD, and ygaU ldtD strains
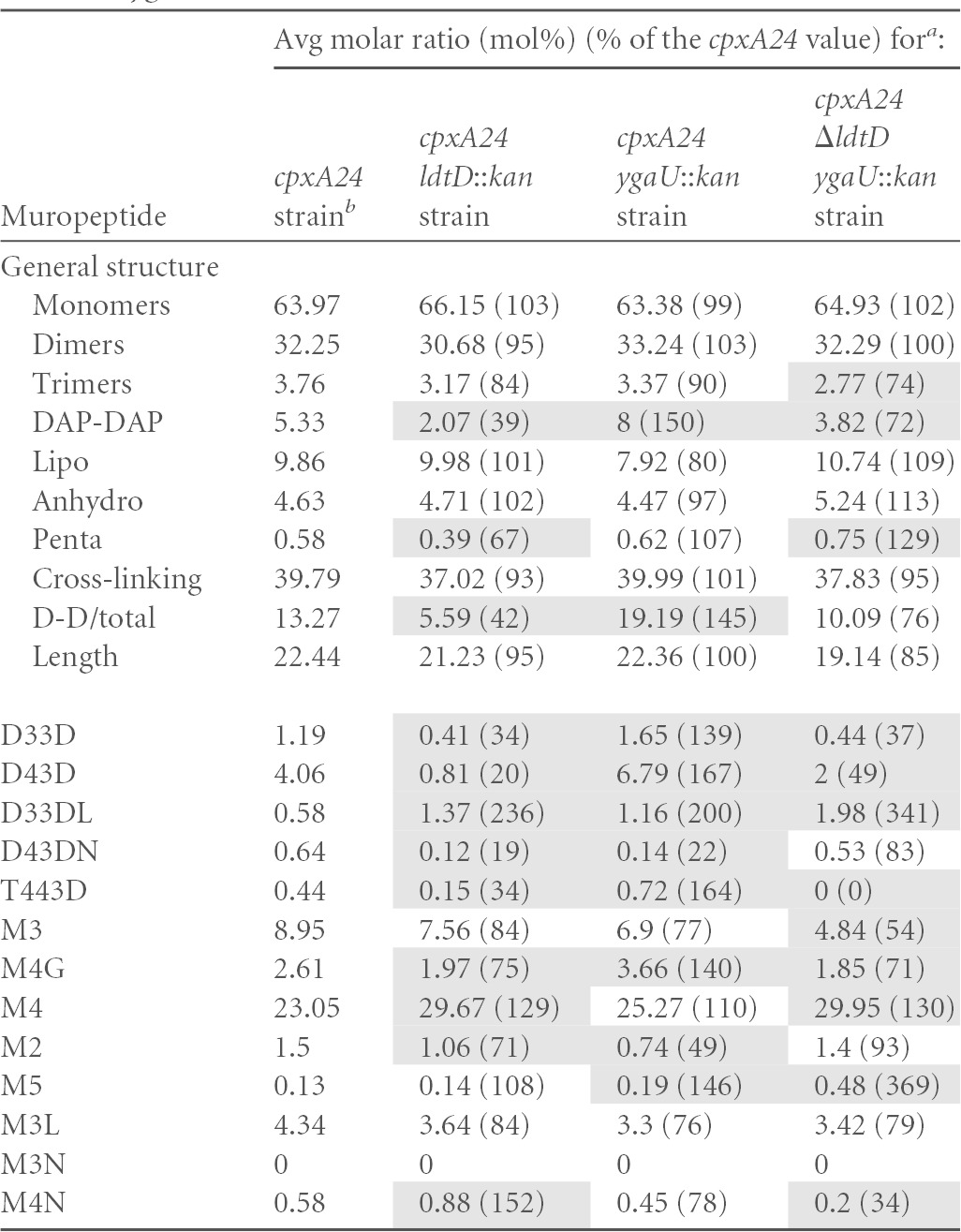
The molar ratios of the different muropeptide structures were calculated for a cpxA24 mutant or isogenic derivatives carrying ygaU::kan, ldtD::kan, or ΔldtD and ygaU::kan alleles, as previously described (26). All strains were grown at 30°C. The different muropeptide structures are abbreviated according to length, where M indicates a monomer, D indicates a cross-linked dimer, and T indicates a cross-linked trimer. The numbers indicate the length of the stem peptide, and the last letter indicates whether the muropeptide contains a DAP-DAP cross-link (D) or is anchored to Braun's lipoprotein (L) or if there is a 1,6-anhydro-muramic acid residue (N) or a glycine (G) at the end of the stem peptide. D-D/total is the ratio of DAP-DAP cross-links to the total cross-links. The averages of data from three measurements from biological replicates are shown. Shading indicates measurements that were different from the cpxA24 value by 25%.
The average muropeptide composition for the cpxA24 mutant was determined once and is indicated here and in Table 3 (see also Table S1 in the supplemental material).
RESULTS
The Cpx pathway regulates expression of ygaU, ldtD, and slt.
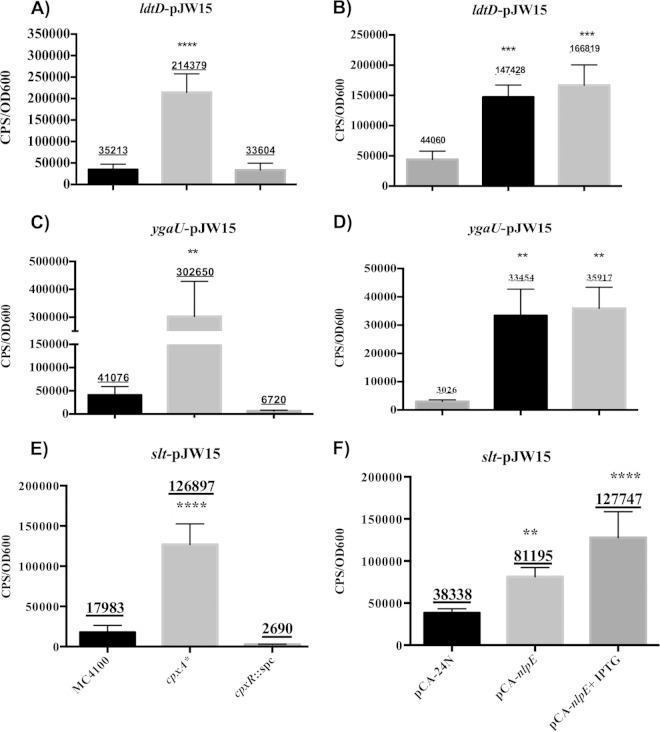
Microarray analysis showed that ygaU, ldtD, and slt transcription increases >2-fold in the presence of NlpE overproduction, suggesting that the expression of these genes is Cpx regulated (11). In order to test this hypothesis, we performed luciferase assays with luminescent reporter genes to determine whether the activation of the Cpx pathway resulted in increased expression levels of ygaU, ldtD, and slt. The promoters of these genes were cloned into the luciferase vector pJW15 (4). The expression of each promoter was measured at late log phase (OD600 of ∼0.6) in different genetic backgrounds: MC4100, cpxA* (TR10, which contains a cpxA24 gain-of-function allele that results in the constitutive activation of the Cpx response independently of any inducing cues) (22), and a cpxR mutant (TR51), where the induction of transcription of Cpx-regulated genes in response to envelope stress is abolished (29) (Fig. 1A, C, and E). We observed significant differences (P < 0.05 by analysis of variance [ANOVA]) in the transcriptional activity of the ygaU, ldtD, and slt reporters in the cpxA24 background compared to the wild type. Specifically, the ldtD-lux, ygaU-lux, and slt-lux reporters were upregulated 5-, 6-, and 7-fold, respectively (Fig. 1A, C, and E) in the presence of constitutive activation of the Cpx pathway. The luminescence levels in the cpxR background for the ygaU-lux and the slt-lux reporters were lower than those in the wild-type (Fig. 1C and E), indicating that CpxR might be necessary for the basal transcription of ygaU and slt. In contrast, the transcription of the ldtD-lux reporter in the cpxR mutant background was close to that in the wild type, suggesting that CpxR is not necessary for the basal transcription of ldtD (Fig. 1A).
FIG 1.
The ldtD, ygaU, and slt promoters are Cpx regulated. All the strains were grown at 30°C. Expression of light from the luminescent reporter genes of ldtD (A and B), ygaU (C and D), and slt (E and F) was measured in the following strains: MC4100, TR10 (cpxA24), TR51 (cpxR::spc), MC4100(pCA-nlpE), and the vector control for NlpE overexpression, MC4100(pCA24N). NlpE overexpression was induced with 0.1 mM IPTG after cultures grown overnight were subcultured (1:40) and grown for 6 h at 30°C. Light was measured at mid- to late log phase (OD600 = 0.6 to 0.8) and quantified by dividing the counts per second (CPS) by the OD600 of each strain. Each bar represents the mean for three replicates, shown above each bar. Error bars represent the standard deviations. Asterisks indicate that the value is significantly different from that of MC4100 or MC4100(pCA24N) (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). Each experiment was repeated at least three times.
We further tested the Cpx regulation of ldtD, ygaU, and slt by measuring the expression of the luminescent reporter genes in the presence of the Cpx inducer NlpE (12). We induced NlpE overexpression from the ASKA library pCA-nlpE clone with 0.1 mM IPTG (21) (Fig. 1B, D, and F). We observed increased transcription of the ldtD, ygaU, and slt reporter genes in the presence of NlpE overexpression compared to that observed with the vector control pCA24N (Fig. 1B, D, and F). Interestingly, the expression levels of all reporters were elevated by the leaky expression of NlpE from plasmid pCA in the absence of the inducer IPTG (Fig. 1B, D, and F). This suggests that the expression of ldtD, ygaU, and slt is sensitive to small increases in Cpx pathway activation. The transcription levels of the ldtD and slt reporter genes were further elevated when nlpE expression was induced (Fig. 1B, D, and F). In agreement with previously reported observations, we observed that the expression levels of the ldtD, ygaU, and slt promoters were markedly higher in the cpxA24 background than in the presence of NlpE overexpression (19).
CpxR binds to the promoters of the ldtD, ygaU, and slt genes.
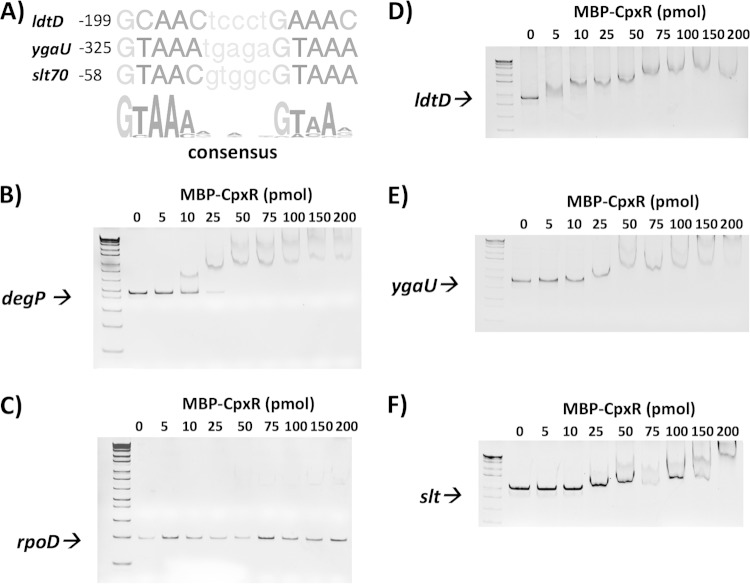
Our results showed that the Cpx pathway regulates ldtD, ygaU, and slt transcription. Bioinformatics analysis revealed the presence of CpxR consensus binding sites upstream of the transcription start site of each of these genes (Fig. 2A). We investigated the interaction between CpxR and the ldtD, ygaU, and slt promoters by performing electrophoretic mobility shift assays (EMSAs). EMSAs were performed by using different concentrations of purified MBP-CpxR (0 to 200 pmol) and 1.5 pmol of the PCR product containing the promoters of degP, rpoD, ldtD, ygaU, and slt (Fig. 2B to F). As expected, phosphorylated CpxR (CpxR∼P) bound strongly to the positive-control degP promoter, causing shifting starting at 25 pmol of protein (10). CpxR∼P did not bind appreciably to the negative-control rpoD promoter, even at high concentrations of CpxR∼P (>100 pmol). In contrast, for ldtD, some shifting was evident at 5 pmol of MBP-CpxR∼P, while the ygaU and slt promoters exhibited slower mobilities in the presence of 10 and 25 pmol of CpxR∼P, respectively (Fig. 2). For all three genes, all the promoter DNA was shifted at concentrations of 25 pmol and above (Fig. 2). Furthermore, we also observed a higher-molecular-weight complex for the promoters of degP, ldtD, ygaU, and slt at concentrations of between 75 and 200 pmol. These data may suggest the existence of an additional, weaker CpxR binding site(s) upstream of these promoters. No unbound promoter DNA was observed at CpxR∼P concentrations of >25 pmol in the presence of either the degP, ldtD, ygaU, or slt promoter (Fig. 2B, D, and F). These observations show that CpxR binds to the ldtD, ygaU, and slt promoters with at least an affinity similar to that of the degP promoter. CpxR∼P appears to have the highest affinity for the ldtD promoter, followed by those of ygaU and slt (Fig. 2). Altogether, our data suggest that CpxR directly regulates the transcription of these genes.
FIG 2.
CpxR binds to the promoters of the ldtD, ygaU, and slt genes. (A) Alignments of putative CpxR binding sites in the promoter regions of ldtD, ygaU, and slt. Shown at bottom is the CpxR consensus binding site (http://www.prodoric.de/vfp/index2.php). The numbers indicate the positions of the putative CpxR binding site relative to the transcription start site. (B to F) Various concentrations of purified MBP-CpxR (0 to 200 pmol) were added to 1.5 pmol of the PCR-amplified promoters of degP (positive control) (B), rpoD (negative control) (C), ldtD (D), ygaU (E), and slt (F). Reaction mixtures were loaded onto a 5% nondenaturing TBE polyacrylamide gel (Bio-Rad), electrophoresed in 1× TBE running buffer, and stained with ethidium bromide for visualization.
Activation of the Cpx pathway causes changes in peptidoglycan composition.
LdtD and Slt have been experimentally shown to be involved in peptidoglycan modification (13, 17). Furthermore, YgaU contains a predicted LysM peptidoglycan association domain (13, 17), and the expression of ldtD, ygaU, and slt is upregulated during activation of the Cpx pathway (11). These observations suggest that changes in peptidoglycan structure may occur when the Cpx response is activated. To test this idea, we purified and performed HPLC analysis on sacculi isolated from MC4100 and isogenic strains carrying either a cpxR::spc null allele or one of three different cpxA* alleles (cpxA24, cpxA102, and cpxA711). Each of the cpxA* alleles contains a mutation that results in the constitutive activation of the Cpx response (22). The cpxA24, cpxA102, and cpxA711 alleles lead to strong, medium, and weak induction, respectively, of the Cpx response when a Cpx-regulated lacZ reporter gene is used to measure Cpx pathway activation (22). Our measurements of the muropeptide composition of the wild type routinely varied by 5% or less between experimental replicates. Thus, we defined substantial changes in cell wall composition as values for the mutant strains that differed from the corresponding values obtained for the wild-type strain by at least 25% (Table 3, shading). Throughout this study, we retained this criterion (i.e., a change in muropeptide composition of 25% or more relative to the control) when comparing cell wall components of different strains. Using this measure, we found that no muropeptide species were appreciably altered in a strain lacking cpxR (Table 3). Indeed, most species were within 10% of the value for wild-type strain MC4100. The largest change observed was a 24% decrease in the level of monomers with tripeptide stems (M3) (Table 3). These data might suggest that basal Cpx pathway activity is needed to maintain the expression of one or more peptidases involved in modifying peptidoglycan.
Several changes in the muropeptide composition of >25% relative to value for MC4100 were observed for the strains carrying a cpxA24, cpxA102, or cpxA711 allele. Most notably, two or more of these strains contained elevated levels of muropeptide species with DAP-DAP cross-links (total DAP-DAP, D33D, D43D, D33DL, and T443D) (Table 3). Additionally, levels of lipoprotein-linked muropeptides were diminished by 25% or more, including the M3L species, in at least two strains carrying cpxA* alleles. Finally, the amounts of the glycine-modified monomer M4G in strains carrying a cpxA24, cpxA102, or cpxA711 allele were increased by 43%, 56%, and 65%, respectively, relative to the amount in wild-type strain MC4100 (Table 3). Overall, these results indicate that DAP-DAP cross-links become more frequent when the Cpx response is activated, suggesting that this type of cross-link plays an important role in the stability of the envelope of E. coli in the presence of Cpx-inducing cues. This change is accompanied by decreased levels of those muropeptides associated with lipoprotein (Table 3). In contrast, the absence of CpxR does not seem to greatly affect cell wall composition in the absence of pathway activation, suggesting that it is not centrally involved in the regular synthesis and composition of peptidoglycan (at least under the conditions examined here).
LdtD is necessary for increased DAP-DAP cross-linking when the Cpx response is activated.
We wondered if slt, ygaU, or ldtD could be involved in the altered DAP-DAP cross-linking observed for strains carrying activated cpxA* alleles (cpxA24, cpxA102, and cpxA711). Since slt has been demonstrated to encode a lytic transglycosylase, and we observed only small changes in peptidoglycan length in the cpx mutants (Table 3), we focused on ldtD, a demonstrated l,d-transpeptidase, and ygaU, an uncharacterized gene with a LysM peptidoglycan binding domain. To determine if LdtD and/or YgaU was involved in the observed changes in DAP-DAP cross-linking observed for the cpxA* (cpxA24, cpxA102, and cpxA711) mutants, we used HPLC to compare muropeptides derived from sacculi isolated from wild-type MC4100, a cpxA24 mutant, and isogenic strains lacking ldtD and/or ygaU (Tables 4 and 5).
TABLE 4.
Peptidoglycan composition of ygaU, ldtD, and ygaU ldtD mutants
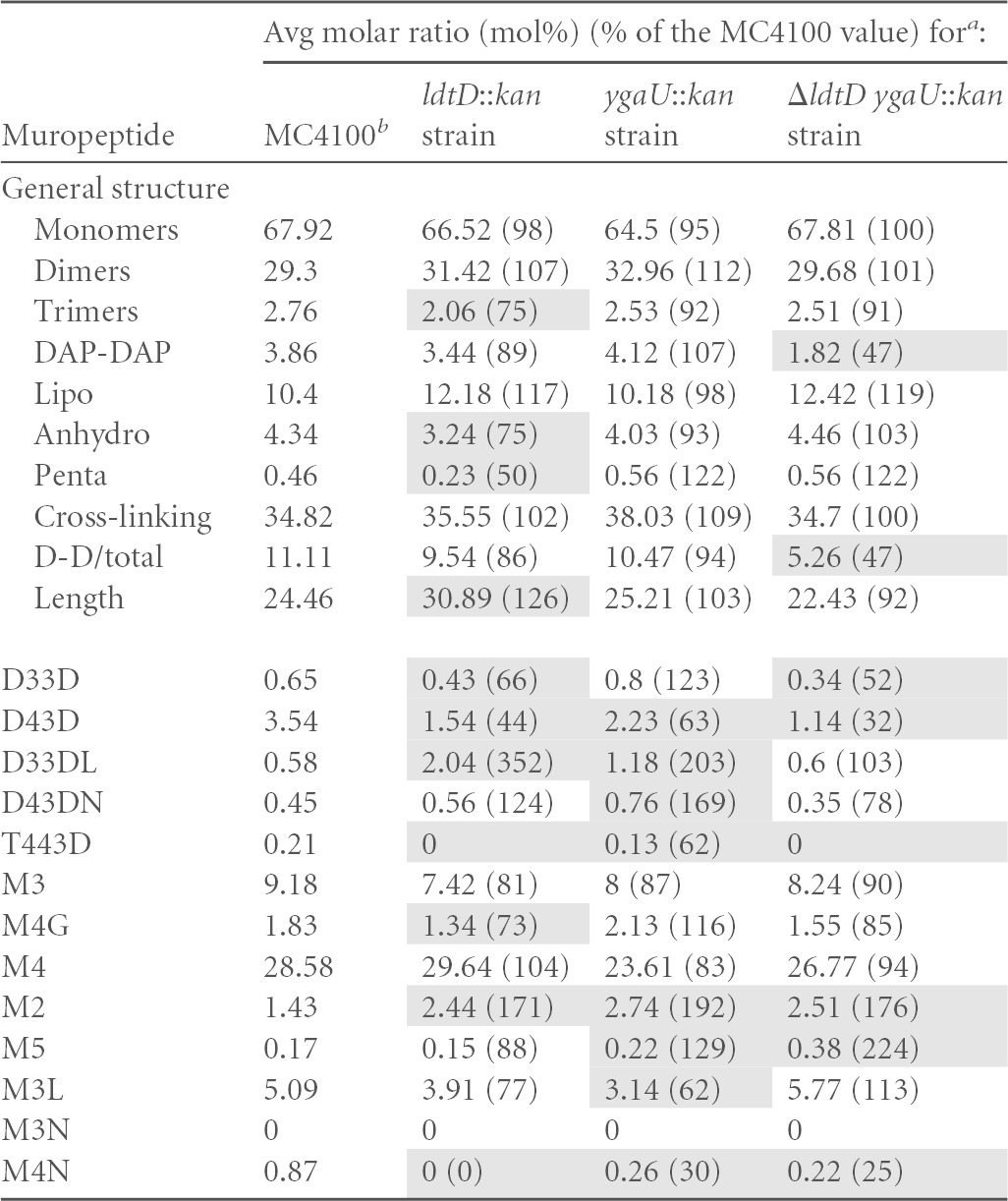
The molar ratios of the different muropeptide structures were calculated for MC4100 or isogenic derivatives carrying ygaU::kan, ldtD::kan, or ΔldtD and ygaU::kan alleles, as previously described (26). All strains were grown at 30°C. The different muropeptide structures are abbreviated according to length, where M indicates a monomer, D indicates a cross-linked dimer, and T indicates a cross-linked trimer. The numbers indicate the length of the stem peptide, and the last letter indicates whether the muropeptide contains a DAP-DAP cross-link (D) or is anchored to Braun's lipoprotein (L) or if there is a 1,6-anhydro-muramic acid residue (N) or a glycine (G) at the end of the stem peptide. D-D/total is the ratio of DAP-DAP cross-links to the total cross-links. The averages of data from three measurements from biological replicates are shown. Shading indicates measurements that were different from the MC4100 value by 25%.
The average muropeptide composition for MC4100 was determined once and is indicated here and in Table 3 (see also Table S1 in the supplemental material).
Our results confirmed the previously reported finding that LdtD functions as an l,d-transpeptidase (13). In particular, deletion of ldtD in wild-type strain MC4100 led to reduced levels of species containing DAP-DAP cross-links, including D33D, D43D, and T443D (Table 4). Additionally, ldtD deletion in this strain background caused an increase in the levels of lipoprotein-linked DAP-DAP-cross-linked dimers (D33DL); changes in the abundance of the monomeric species M4G, M2, and M4N; and an increase in the length of glycan strands (Table 4), suggesting that the activities of other cell wall modification enzymes are impacted in the absence of LdtD (Table 4). In the cpxA24 mutant, the loss of ldtD caused similar, although more dramatic, changes in DAP-DAP cross-linking. In this strain, nearly all DAP-DAP-cross-linked species were altered, including decreases in the amounts of D33D, D43D, D43DN, T443D, and M4G species (19 to 75% of the amount in the cpxA24 parent) (Table 5). Strikingly, the overall level of DAP-DAP cross-linking was reduced in the absence of LdtD to levels below that of even the wild-type strain (compare Tables 4 and 5). Similarly to MC4100, the abundance of lipoprotein-linked DAP-DAP dimers was also increased when ldtD was removed from the cpxA24 strain, and several monomeric species of muropeptide exhibited altered levels (M4, M4N, and M2) (Table 5). Cumulatively, these results are consistent with the known role of LdtD in l,d-transpeptidation and suggest that it is necessary for the elevated DAP-DAP cross-linking observed when the Cpx response is induced, consistent with its strong Cpx-mediated upregulation (Fig. 1A and B).
The uncharacterized gene ygaU impacts l,d-transpeptidase activity.
Interestingly, deletion of the uncharacterized ygaU gene also impacted the abundance of muropeptide species indicative of changes in l,d-transpeptidase activity in both strains (Tables 4 and 5). When ygaU was deleted from wild-type strain MC4100, levels of the DAP-DAP-cross-linked D33D, D33DL, and D43DN species were 123%, 203%, and 169% of those of the wild type (Table 4). Conversely, levels of the D43D and T443D muropeptides decreased by >25% compared to the MC4100 value (Table 4). The monomeric muropeptides M2, M5, M3L, and M4N were also present at altered levels in the absence of ygaU (Table 4). In the cpxA24 ygaU::kan double mutant, the overall level of DAP-DAP cross-linking was 50% higher than that observed in the cpxA24 single mutant, and this reflected increases in the abundances of the D33D, D43D, D33DL, and T443D muropeptide species (Table 5). Levels of the monomeric glycine-substituted tetrapeptide species M4G and the M2 and M5 monomeric muropeptides were also altered by >25% compared to the cpxA24 value (Table 5). Deletion of ygaU differentially affected some peptidoglycan species in the MC4100 versus the cpxA24 strain background. Namely, while the levels of the cross-linked D43D and T443D species were diminished upon ygaU removal in MC4100, these muropeptides were present at increased levels in a cpxA24 ygaU double mutant (Tables 4 and 5). Similarly, in the MC4100 ygaU mutant, the levels of D43DN and M2 muropeptides were elevated, while they were diminished in the cpxA24 ygaU strain (Tables 4 and 5). Altogether, these data suggest that ygaU could be involved in l,d-transpeptidation and indicate that it affects cell wall modification differently in a Cpx-activated strain background.
The alteration of 3-3 cross-linking observed upon the deletion of ygaU suggested that this previously uncharacterized gene might be involved in this cell wall modification (Tables 4 and 5). In an effort to complement this phenotype, we examined the composition of sacculi derived from ygaU mutant strains expressing ygaU from a leaky IPTG-inducible promoter in the absence of IPTG (see Table S1 in the supplemental material). Under these conditions, the altered levels of the DAP-DAP-cross-linked D43D and D43DN species, as well as those of the M2 monomer, were partially or fully complemented in both the MC4100 ygaU and cpxA24 ygaU mutants (see Table S1 in the supplemental material). Surprisingly, however, we were unable to complement the majority of the effects that the mutation of ygaU had on both MC4100 and the cpxA24 mutant. Rather, the overexpression of YgaU for the most part exacerbated the peptidoglycan modifications observed upon the deletion of ygaU (see Table S1 in the supplemental material). Additionally, ygaU overexpression led to the appearance of changes in other muropeptide species that were not appreciably altered in the ygaU mutant strains. In fact, virtually all of the muropeptide species measured were altered by >25% compared to the value found for the MC4100 or cpxA24 strain (see Table S1 in the supplemental material). As observed when ygaU was deleted singly from either MC4100 or the cpxA24 mutant, the overexpression of ygaU had strain-dependent effects. In general, ygaU overexpression had more dramatic effects in the MC4100 background (more muropeptide species were altered, and the changes relative to MC4100 were of a greater magnitude) (see Table S1 in the supplemental material). Cumulatively, these data show that YgaU overexpression has pleiotropic effects on cell wall composition and that some of these effects are strain dependent.
Some ygaU mutant phenotypes are dependent on ldtD.
The absence of ygaU resulted in changes in DAP-DAP cross-linking, and yet this gene lacks any of the active-site residues or domains associated with l,d-transpeptidases. Thus, we wondered if the similarly Cpx-regulated LdtD enzyme could be involved with this phenotype. To test this hypothesis, we eliminated both ygaU and ldtD in the MC4100 and cpxA24 strain backgrounds and compared the muropeptide profiles of these strains to those of the single mutants (Tables 4 and 5). In some cases, in both strain backgrounds, the loss of ldtD reversed the changes in DAP-DAP cross-linking observed in the ygaU mutant. For example, in MC4100, the elevated levels of D33D, D33DL, and D43DN observed in a ygaU mutant were negated by the simultaneous deletion of ldtD (Table 4). Similarly, in the cpxA24 strain background, the increased amounts of overall DAP-DAP, D33D, D43D, T443D, and M4G muropeptide species seen in the absence of ygaU were not observed in the cpxA24 ygaU ldtD triple mutant (Table 5). The effect of the removal of both ygaU and ldtD on other muropeptide components, however, appeared to be additive or even synergistic. For example, levels of the M5 monomer were greatly elevated in the double mutant (224% of the MC4100 level), while they were less elevated (129% of the MC4100 level) in the ygaU mutant and slightly diminished (88% of the MC4100 level) in the ldtD mutant (Table 4). Similarly, the levels of the lipoprotein-linked DAP-DAP D33DL species and the M5 monomer were elevated in both the cpxA24 ygaU and cpxA24 ldtD strains and were even further increased in the cpxA24 ygaU ldtD triple mutant (Table 5). These observations suggest that in some cases, the effect of YgaU on cell wall modification is dependent on LdtD, while in others, YgaU and LdtD act independently, or even synergistically, to affect peptidoglycan alterations.
The absence of YgaU and LdtD leads to the activation of the Cpx pathway.
The activation of the Cpx pathway occurs upon the simultaneous absence of PBPs 4, 5, and 7 and AmpH, which function as d,d-carboxypeptidases and d,d-endopeptidases to trim pentapeptide stems and cleave 4-3 cross-links during peptidoglycan biosynthesis, respectively. Thus, the Cpx pathway is sensitive to some changes in peptidoglycan structure (31). Our results indicate that YgaU and LdtD are involved in Cpx-mediated changes in cross-linking and, therefore, presumably in mediating adaptation to envelope stresses sensed by the Cpx response (Fig. 1 and Tables 3 and 5). Accordingly, we wondered if the removal of ygaU and/or ldtD would generate a Cpx-sensed envelope stress signal. To test this hypothesis, we examined Cpx-mediated gene expression using a chromosomal cpxP-lacZ reporter gene in wild-type strains and those in which ldtD and ygaU had been deleted individually or in combination (Fig. 3). We observed significant differences in cpxP-lacZ activity in all the mutants tested (P < 0.05 by ANOVA). The single and double mutants had an ∼2-fold increase in Cpx pathway activity compared to the wild type, indicating that a Cpx-inducing signal is generated under these conditions.
FIG 3.
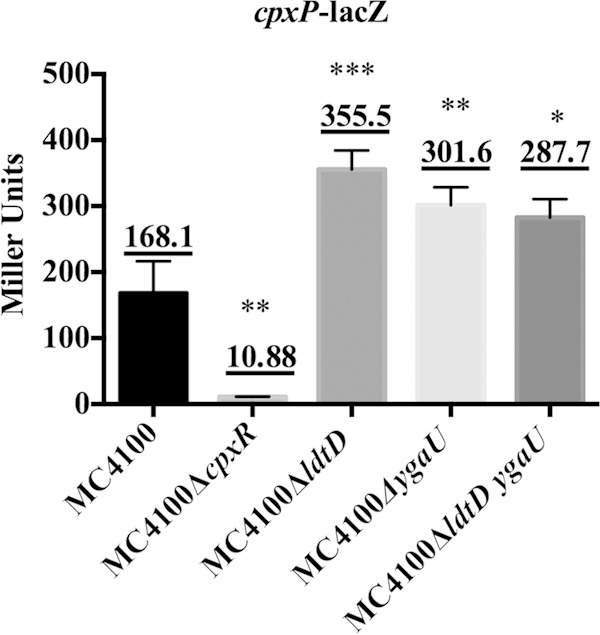
Absence of ldtD and/or ygaU induces the Cpx pathway. A β-galactosidase assay was carried out on the MC4100 (TR50), MC4100 ΔcpxR (TR70), MC4100 ΔygaU (MBC24), MC4100 ΔldtD (MBC23), and MC4100 ΔldtD ygaU (MBC25) strains carrying a single chromosomal copy of the cpxP-lacZ reporter gene. Cultures grown overnight were subcultured (1:40) and grown until the cells reached late log phase (final OD600 = ∼0.7). Each bar represents the mean for three replicates, shown above each bar. Error bars represent the standard deviations. Asterisks represent changes in β-galactosidase levels that are significantly different from those in MC4100 (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001). This assay was repeated at least two times, and a representative data set is shown.
DISCUSSION
The Cpx pathway regulates the expression of the l,d-transpeptidase LdtD, the predicted cell wall-associated protein YgaU, and the soluble lytic transglycosylase Slt.
Recent transcriptomic studies revealed that the Cpx pathway regulates a complex response that has more functions than initially thought (11, 32, 33), including genes known or predicted to be involved in peptidoglycan modification, such as ygaU, ldtD, and slt (34). Our data further support these findings. Interestingly, the transcription of all three of these genes has previously been associated with stressful conditions. The expression level of ldtD was elevated in the presence of acidic pH (35) and DNA-damaging agents (36). ygaU is a member of the general stress response regulated by σS (37), and its transcription increases in response to osmotic stress (30). Finally, slt is predicted to be regulated by the heat shock sigma factor σ32, and its expression is induced by the stringent response (38). Our work demonstrates that envelope stress also results in elevated transcription levels of these genes (Fig. 1). Cumulatively, these findings suggest that ldtD, ygaU, and slt play important roles in cell wall modifications that enable adaptation to a variety of stresses expected to directly or indirectly affect the envelope and/or growth.
Bioinformatics analyses and EMSAs indicate that the Cpx regulation of ldtD, ygaU, and slt may be direct (Fig. 2). CpxR∼P binds to the DNA upstream of these genes with affinities that appear to be similar to, or greater than, that of the moderately Cpx-regulated gene degP (10, 39). The putative CpxR binding site upstream of slt occurs very close to the predicted σ32 promoter (Fig. 2A), as is the case for other strongly Cpx-regulated genes (19), providing compelling evidence for direct transcriptional activation of this gene by CpxR∼P. Interestingly, ygaU is found just 84 bp downstream of a Cpx-regulated gene of unknown function, yqaE, and the perfect CpxR binding site that we identified upstream of ygaU is actually found upstream of yqaE (Fig. 2). Although a σS promoter is predicted to exist directly upstream of ygaU, in between yqaE and ygaU, when we assayed a luminescent reporter carrying only this promoter, we found that it was not activated by the Cpx response (data not shown). This finding suggests that these genes are transcribed as a single unit under Cpx-inducing conditions. In support of this conclusion, the expression levels of yqaE and ygaU are elevated to a similar degree when NlpE is overexpressed (11).
Increased DAP-DAP cross-linking upon Cpx response activation involves the l,d-transpeptidase LdtD.
HPLC analysis of sacculi derived from strains carrying a constitutively activated cpxA* mutation (cpxA24, cpxA102, or cpxA711) showed that the cell wall is altered during the activation of the Cpx pathway. These alterations include an increase in the abundance of DAP-DAP cross-links, which may play a role in stabilizing the envelope in the presence of Cpx-inducing cues such as alkaline pH (9), high osmolarity (40), SDS (41), and high temperature (42). Our data indicate that increased DAP-DAP cross-linking during Cpx response activation is mostly mediated by the l,d-transpeptidase LdtD, as the absence of this enzyme under these conditions led to obvious decreases in the levels of these cross-links (Table 5). Notably, the levels of DAP-DAP cross-linking dropped below levels observed in the uninduced, wild-type MC4100 strain background when ldtD was deleted from the cpxA24 strain (Table 5), indicating the necessity of LdtD for this modification during Cpx response induction. In support of this, no changes in the expression of the gene encoding the only other characterized l,d-transpeptidase, ynhG, in E. coli (13) were observed upon NlpE overexpression (11).
Other muropeptide changes indicative of alterations in l,d-transpeptidase activity were also evident in Cpx-activated strains, including alterations in lipoprotein-linked subunits as well as monomeric species bearing a glycine-modified tetrapeptide. Of these, LdtD also appears to be involved in the changes to the M4G muropeptide, since the removal of ldtD in the cpxA24 strain background negated the increased abundance of this species caused by constitutive Cpx response activation (Table 5). The lipoprotein-linked muropeptides, however, were unaffected when ldtD was absent from the cpxA24 strain, suggesting that some other transpeptidase activity is altered upon Cpx response activation. The conclusion that other cell wall modification enzymes (besides LdtD) may be affected upon Cpx response induction is supported by the observation that the deletion of ygaU and/or ldtD as well as the overexpression of ygaU have differential effects in the cpxA24 and MC4100 strain backgrounds (Tables 4 and 5; see also Table S1 in the supplemental material). Whether or not these differences are due to direct, Cpx-regulated changes in the expression of genes encoding additional peptidoglycan modification enzymes or are a result of Cpx-mediated changes to the envelope that impact the formation and/or activity of complexes containing such enzymes remains to be determined.
Does YgaU regulate l,d-transpeptidase activity?
Although the deletion or overexpression of ygaU leads to elevated levels of 3-3 cross-links produced by l,d-transpeptidases (Tables 4 and 5; see also Table S1 in the supplemental material), we think that it is unlikely that YgaU itself functions as an l,d-transpeptidase, since it lacks the highly conserved Cys residues that define the catalytic active site of these enzymes (43) as well as a signal sequence or transmembrane domains that would localize it to the periplasm. Instead, we favor the idea that YgaU may act to regulate l,d-transpeptidase activity. When ygaU was deleted, we observed an increase in the level of DAP-DAP cross-links in the cpxA24 strain background relative to the level in wild-type strain MC4100 (Tables 4 and 5). Interestingly, this phenotype is similar to that observed upon LdtD overexpression in the absence of other l,d-transpeptidases (13) and was mostly abrogated when ldtD and ygaU were simultaneously deleted (Tables 4 and 5). These results suggest that the absence of ygaU may upregulate LdtD activity. The HPLC results obtained upon YgaU overexpression also indicated cell wall changes characteristic of elevated l,d-transpeptidase activity, such as increased levels of DAP-DAP cross-links, D33D and D443D muropeptides, and M3 monomers and decreased levels of M4 monomers (see Table S1 in the supplemental material). While it is paradoxical that both the deletion and overexpression of YgaU seem to result in a similar upregulation of l,d-transpeptidase activity, both results argue that YgaU regulates this process. We suggest that the relative amounts of YgaU and LdtD, and possibly other peptidoglycan modification enzymes, may be important for determining the overall effect of YgaU on DAP-DAP cross-linking. This speculation is supported by the fact that the deletion or overexpression of ygaU exerts some effects on peptidoglycan composition that are dependent on the strain background (Tables 4 and 5; see also Table S1 in the supplemental material). In this regard, it will be interesting to determine what other cell wall modification genes may be Cpx regulated.
How YgaU might regulate LdtD and/or other PG modification enzymes is an open question. YgaU is localized to the inner membrane (our unpublished data), likely by virtue of its BON domain (16), while its LysM domain is predicted to bind the NAG moiety of PG (44, 45). How, then, could YgaU bind to the NAG moiety of PG and regulate 3-3 cross-linking imposed by LdtD from the cytoplasm? The simplest possibility invokes an interaction between YgaU and the predicted transmembrane or cytoplasmic domains of LdtD. The cytoplasmic membrane location of YgaU and its LysM domain suggests that interaction with the NAG moieties of precursor PG fragments that have not yet been translocated across the inner membrane might act to regulate the interaction of YgaU with LdtD (and possibly other cell wall modification enzymes) and/or the effect of YgaU on LdtD. This model is particularly attractive in light of the fact that recent studies indicate that the Cpx response functions to mediate envelope stress adaptation largely at the inner membrane (11, 33). Perhaps YgaU responds to a buildup of PG precursors arising as a result of inner membrane malfunction that precludes or slows down PG precursor translocation into the periplasm. In this scenario, perhaps YgaU-regulated increases in the abundances of cross-links provide some adaptive advantage, maybe by stabilizing the existing peptidoglycan. The regulation of inner membrane-localized cell wall modification enzymes by proteins in both the cytoplasm and the outer membrane was demonstrated previously (46). It is known that outer membrane lipoproteins function to regulate the activity of PG synthesis enzymes at the inner membrane, through an as-yet-uncertain mechanism (47, 48). Similarly, cytoskeletal proteins in the cytoplasm also guide PG assembly to coordinate cell growth and division (49, 50). Further studies of interactions between YgaU, LdtD, and PG will shed light on the activity of this so-far-uncharacterized protein.
The Cpx response is sensitive to changes in peptidoglycan composition.
We observed an increase in the activity of the Cpx pathway in single and double mutants carrying ygaU and/or ldtD mutations, which suggests that the composition of the cell wall affects the regulation of the Cpx pathway. These results support previously reported findings (31) that showed that the Cpx pathway is also activated in the absence of PBPs 4, 5, and 7 and AmpH. Nonetheless, it is somewhat surprising that these single mutations affected Cpx response activation, since mutations affecting single peptidoglycan modification genes generally have very small or no effects on the tested phenotypes, including resistance to a variety of stresses. Thus, our observation may suggest a specific role for the ygaU and ldtD genes that is relevant to the generation of Cpx-inducing signals. Evans et al. proposed that the missing carboxypeptidase and endopeptidase activities of PBPs 4, 5, and 7 and AmpH could lead to the accumulation of an activating muropeptide species or to the removal of an inhibitory molecule (31). Another possibility is that alterations in the cell wall impact inner membrane functions. The induction of the Cpx response changes the expression of a set of genes that are enriched for inner membrane proteins, including a large group of transporter proteins, and also downregulates the production of envelope-spanning protein complexes involved in motility, secretion, and transport (11, 32, 51). In this light, it is interesting that a number of these complexes depend upon direct interactions with the peptidoglycan for assembly and function (52–55). These observations may suggest that cell wall alterations could impact the activity and stability of secretion and transport complexes, potentially generating a Cpx-inducing signal. Regardless of the mechanism, it is clear that changes in peptidoglycan composition can result in a Cpx-activating signal. Although connections between the outer membrane and the cell wall are well documented, those involving the inner membrane are less well understood. Hopefully, characterization of how the Cpx inner membrane stress response reacts to cell wall changes will be informative on this point.
Why does the Cpx response stimulate changes in the cell wall?
Although it is clear that the Cpx response can be induced by alterations in cell wall composition, we do not yet understand why the Cpx response regulates genes involved in peptidoglycan remodeling. Weatherspoon-Griffin et al. (56) have connected the Cpx-upregulated production of peptidoglycan amidases to antimicrobial peptide resistance. Furthermore, as previously mentioned, changes in 3-3 cross-linking, similar to those observed here upon Cpx response activation, have also been implicated in responses to a variety of stress conditions (27, 57–62). Thus, YgaU and LdtD may help stabilize the bacterial envelope in the presence of Cpx-inducing conditions, perhaps by reinforcing the structure of the cell wall (58). Increased cell wall stability may be important while the cell recovers from stresses that destabilize the cytoplasmic membrane. Another possibility is that the adaptations mediated at the inner membrane by the Cpx response impact peptidoglycan synthesis and maturation at this cellular location, resulting in altered or impaired cell wall biogenesis. The Cpx-mediated upregulation of LdtD and YgaU could act to counter such problems associated with adaptation, by modifying the existing cell wall until homeostasis returns.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Canadian Institute of Health Research operating grant 97819 and Natural Science and Engineering Research Council grant RGPIN 238422-2013. T.L.R. was supported by a senior scholar award from Alberta Innovates Health Solutions. We acknowledge financial support (project number BFU2009-09200) from the Spanish Ministry of Economy and Competitiveness to J.A.A.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02449-14.
REFERENCES
- 1.Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. Protein quality control in the bacterial periplasm. Annu Rev Microbiol 65:149–168. doi: 10.1146/annurev-micro-090110-102925. [DOI] [PubMed] [Google Scholar]
- 2.van Alphen W, Selm N, Lugtenberg B. 1978. Pores in the outer membrane of Escherichia coli K12. Mol Gen Genet 159:75–83. doi: 10.1007/BF00401750. [DOI] [PubMed] [Google Scholar]
- 3.Jones CH, Danese PN, Pinkner JS, Silhavy TJ, Hultgren SJ. 1997. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J 16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacRitchie DM, Ward JD, Nevesinjac AZ, Raivio TL. 2008. Activation of the Cpx envelope stress response down-regulates expression of several locus of enterocyte effacement-encoded genes in enteropathogenic Escherichia coli. Infect Immun 76:1465–1475. doi: 10.1128/IAI.01265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nevesinjac AZ, Raivio TL. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J Bacteriol 187:672–686. doi: 10.1128/JB.187.2.672-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleerebezem M, Crielaard W, Tommassen J. 1996. Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions. EMBO J 15:162–171. [PMC free article] [PubMed] [Google Scholar]
- 7.Leblanc SK, Oates CW, Raivio TL. 2011. Characterization of the induction and cellular role of the BaeSR two-component envelope stress response of Escherichia coli. J Bacteriol 193:3367–3375. doi: 10.1128/JB.01534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- 9.Danese N, Silhavy J. 1997. The sigma(E) and the Cpx signal transduction systems control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev 11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 10.Pogliano J, Lynch S, Belin D, Lin C, Beckwith J. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev 11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 11.Raivio TL, Leblanc SK, Price NL. 2013. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol 195:2755–2767. doi: 10.1128/JB.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol 177:4216–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magnet S, Dubost L, Marie A, Arthur M, Gutmann L. 2008. Identification of the L,D-transpeptidases for peptidoglycan cross-linking in Escherichia coli. J Bacteriol 190:4782–4785. doi: 10.1128/JB.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bateman A, Bycroft M. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J Mol Biol 299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- 15.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 16.Yeats C, Bateman A. 2003. The BON domain: a putative membrane-binding domain. Trends Biochem Sci 28:352–355. doi: 10.1016/S0968-0004(03)00115-4. [DOI] [PubMed] [Google Scholar]
- 17.Höltje JV, Mirelman D, Sharon N, Schwarz U. 1975. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol 124:1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong JL, Vogt SL, Raivio TL. 2013. Using reporter genes and the Escherichia coli ASKA overexpression library in screens for regulators of the Gram negative envelope stress response. Methods Mol Biol 966:337–357. doi: 10.1007/978-1-62703-245-2_21. [DOI] [PubMed] [Google Scholar]
- 19.Price NL, Raivio TL. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J Bacteriol 191:1798–1815. doi: 10.1128/JB.00798-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. 2005. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 22.Raivio TL, Silhavy TJ. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol 179:7724–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Slauch JM, Russo FD, Silhavy TJ. 1991. Suppressor mutations in rpoA suggest that OmpR controls transcription by direct interaction with the alpha subunit of RNA polymerase. J Bacteriol 173:7501–7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.González-Leiza SM, de Pedro MA, Ayala JA. 2011. AmpH, a bifunctional DD-endopeptidase and DD-carboxypeptidase of Escherichia coli. J Bacteriol 193:6887–6894. doi: 10.1128/JB.05764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayashi K. 1975. A rapid determination of sodium dodecyl sulfate with methylene blue. Anal Biochem 67:503–506. doi: 10.1016/0003-2697(75)90324-3. [DOI] [PubMed] [Google Scholar]
- 26.Glauner B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem 172:451–464. doi: 10.1016/0003-2697(88)90468-X. [DOI] [PubMed] [Google Scholar]
- 27.Glauner B, Höltje JV, Schwarz U. 1988. The composition of the murein of Escherichia coli. J Biol Chem 263:10088–10095. [PubMed] [Google Scholar]
- 28.Work E. 1957. Reaction of ninhydrin in acid solution with straight-chain amino acids containing two amino groups and its application to the estimation of α-diaminopimelic acid. Biochem J 67:416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raivio TL, Popkin DL, Silhavy TJ. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol 181:5263–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber A, Kögl SA, Jung K. 2006. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J Bacteriol 188:7165–7175. doi: 10.1128/JB.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans KL, Kannan S, Li G, de Pedro M, Young KD. 2013. Eliminating a set of four penicillin binding proteins triggers the Rcs phosphorelay and Cpx stress responses in Escherichia coli. J Bacteriol 195:4415–4424. doi: 10.1128/JB.00596-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gangaiah D, Zhang X, Fortney KR, Baker B, Liu Y, Munson RS, Spinola SM. 2013. Activation of CpxRA in Haemophilus ducreyi primarily inhibits the expression of its targets, including major virulence determinants. J Bacteriol 195:3486–3502. doi: 10.1128/JB.00372-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol Lett 326:2–11. doi: 10.1111/j.1574-6968.2011.02406.x. [DOI] [PubMed] [Google Scholar]
- 34.Park JT. 1995. Why does Escherichia coli recycle its cell wall peptides? Mol Microbiol 17:421–426. doi: 10.1111/j.1365-2958.1995.mmi_17030421.x. [DOI] [PubMed] [Google Scholar]
- 35.Kannan G, Wilks JC, Fitzgerald DM, Jones BD, Bondurant SS, Slonczewski JL. 2008. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol 8:37. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khil PP, Camerini-Otero RD. 2002. Over 1000 genes are involved in the DNA damage response of Escherichia coli. Mol Microbiol 44:89–105. doi: 10.1046/j.1365-2958.2002.02878.x. [DOI] [PubMed] [Google Scholar]
- 37.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Betzner AS, Ferreira LCS, Höltje J-V, Keck W. 1990. Control of the activity of the soluble lytic transglycosylase by the stringent response in Escherichia coli. FEMS Microbiol Lett 67:161–164. doi: 10.1111/j.1574-6968.1990.tb13855.x. [DOI] [PubMed] [Google Scholar]
- 39.Danese PN, Snyder WB, Cosma CL, Davis LJ, Silhavy TJ. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev 9:387–398. doi: 10.1101/gad.9.4.387. [DOI] [PubMed] [Google Scholar]
- 40.Jubelin G, Vianney A, Beloin C, Ghigo JM, Lazzaroni JC, Lejeune P, Dorel C. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J Bacteriol 187:2038–2049. doi: 10.1128/JB.187.6.2038-2049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Danese PN, Silhavy TJ. 1998. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol 180:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mileykovskaya E, Dowhan W. 1997. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol 179:1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders AN, Pavelka MS. 2013. Phenotypic analysis of E. coli mutants lacking L,D-transpeptidases. Microbiology 159:1842–1852. doi: 10.1099/mic.0.069211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohnuma T, Onaga S, Murata K, Taira T, Katoh E. 2008. LysM domains from Pteris ryukyuensis chitinase-A: a stability study and characterization of the chitin-binding site. J Biol Chem 283:5178–5187. doi: 10.1074/jbc.M707156200. [DOI] [PubMed] [Google Scholar]
- 45.Petutschnig EK, Jones AM, Serazetdinova L, Lipka U, Lipka V. 2010. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem 285:28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Typas A, Banzhaf M, Gross CA, Vollmer W. 2012. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Typas A, Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ, Zietek M, Beilharz K, Kannenberg K, von Rechenberg M, Breukink E, den Blaauwen T, Gross CA, Vollmer W. 2010. Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143:1097–1109. doi: 10.1016/j.cell.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 50.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776. doi: 10.1016/S0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 51.Labandeira-Rey M, Brautigam CA, Hansen EJ. 2010. Characterization of the CpxRA regulon in Haemophilus ducreyi. Infect Immun 78:4779–4791. doi: 10.1128/IAI.00678-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aschtgen MS, Gavioli M, Dessen A, Lloubès R, Cascales E. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol Microbiol 75:886–899. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 53.Hausner J, Hartmann N, Lorenz C, Büttner D. 2013. The periplasmic HrpB1 protein from Xanthomonas spp. binds to peptidoglycan and to components of the type III secretion system. Appl Environ Microbiol 79:6312–6324. doi: 10.1128/AEM.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hizukuri Y, Kojima S, Yakushi T, Kawagishi I, Homma M. 2008. Systematic Cys mutagenesis of FlgI, the flagellar P-ring component of Escherichia coli. Microbiology 154:810–817. doi: 10.1099/mic.0.2007/013854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martynowski D, Grochulski P, Howard PS. 2013. Structure of a periplasmic domain of the EpsAB fusion protein of the Vibrio vulnificus type II secretion system. Acta Crystallogr D Biol Crystallogr 69:142–149. doi: 10.1107/S0907444912042710. [DOI] [PubMed] [Google Scholar]
- 56.Weatherspoon-Griffin N, Zhao G, Kong W, Kong Y, Morigen Andrews-Polymenis H, McClelland M, Shi Y. 2011. The CpxR/CpxA two-component system up-regulates two Tat-dependent peptidoglycan amidases to confer bacterial resistance to antimicrobial peptide. J Biol Chem 286:5529–5539. doi: 10.1074/jbc.M110.200352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burman LG, Park JT. 1983. Changes in the composition of Escherichia coli murein as it ages during exponential growth. J Bacteriol 155:447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goffin C, Ghuysen JM. 2002. Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol Mol Biol Rev 66:702–738. doi: 10.1128/MMBR.66.4.702-738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavollay M, Fourgeaud M, Herrmann JL, Dubost L, Marie A, Gutmann L, Arthur M, Mainardi JL. 2011. The peptidoglycan of Mycobacterium abscessus is predominantly cross-linked by L,D-transpeptidases. J Bacteriol 193:778–782. doi: 10.1128/JB.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mainardi JL, Legrand R, Arthur M, Schoot B, van Heijenoort J, Gutmann L. 2000. Novel mechanism of beta-lactam resistance due to bypass of DD-transpeptidation in Enterococcus faecium. J Biol Chem 275:16490–16496. doi: 10.1074/jbc.M909877199. [DOI] [PubMed] [Google Scholar]
- 61.Quintela JC, de Pedro MA, Zöllner P, Allmaier G, Garcia-del Portillo F. 1997. Peptidoglycan structure of Salmonella typhimurium growing within cultured mammalian cells. Mol Microbiol 23:693–704. doi: 10.1046/j.1365-2958.1997.2561621.x. [DOI] [PubMed] [Google Scholar]
- 62.Signoretto C, Lleo MDM, Canepari P. 2002. Modification of the peptidoglycan of Escherichia coli in the viable but nonculturable state. Curr Microbiol 44:125–131. doi: 10.1007/s00284-001-0062-0. [DOI] [PubMed] [Google Scholar]
- 63.Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86. doi: 10.1016/S0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 64.Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol 104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 65.Raivio TL, Laird MW, Joly JC, Silhavy TJ. 2000. Tethering of CpxP to the inner membrane prevents spheroplast induction of the cpx envelope stress response. Mol Microbiol 37:1186–1197. doi: 10.1046/j.1365-2958.2000.02074.x. [DOI] [PubMed] [Google Scholar]
- 66. Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. 1993. The activity of σE, an Escherichia coli heat-inducible σ-factor, is modulated by expression of outer membrane proteins. Genes Dev 7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 67.Bernal-Cabas M. 2014. The Cpx pathway causes changes in the peptidoglycan structure, turnover, and recycling. M.Sc. thesis University of Alberta, Edmonton, Canada [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.