Abstract
The genome of Bacillus subtilis 168 encodes eight rap-phr quorum-sensing pairs. Rap proteins of all characterized Rap-Phr pairs inhibit the function of one or several important response regulators: ComA, Spo0F, or DegU. This inhibition is relieved upon binding of the peptide encoded by the cognate phr gene. Bacillus subtilis strain NCIB3610, the biofilm-proficient ancestor of strain 168, encodes, in addition, the rapP-phrP pair on the plasmid pBS32. RapP was shown to dephosphorylate Spo0F and to regulate biofilm formation, but unlike other Rap-Phr pairs, RapP does not interact with PhrP. In this work we extend the analysis of the RapP pathway by reexamining its transcriptional regulation, its effect on downstream targets, and its interaction with PhrP. At the transcriptional level, we show that rapP and phrP regulation is similar to that of other rap-phr pairs. We further find that RapP has an Spo0F-independent negative effect on biofilm-related genes, which is mediated by the response regulator ComA. Finally, we find that the insensitivity of RapP to PhrP is due to a substitution of a highly conserved residue in the peptide binding domain of the rapP allele of strain NCIB3610. Reversing this substitution to the consensus amino acid restores the PhrP dependence of RapP activity and eliminates the effects of the rapP-phrP locus on ComA activity and biofilm formation. Taken together, our results suggest that RapP strongly represses biofilm formation through multiple targets and that PhrP does not counteract RapP due to a rare mutation in rapP.
INTRODUCTION
The behavior of bacterial communities is often regulated by quorum-sensing (QS) signaling pathways. In these pathways, a secreted molecular signal accumulates in the environment and activates a cognate receptor at sufficiently high concentrations. Gram-positive bacteria frequently utilize peptides as QS signals (1–3), thereby eliciting a variety of behaviors, including the secretion of various molecules (e.g., enzymes, antibiotics, surfactants, and exopolysaccharides [4]), the initiation of developmental processes (such as sporulation and biofilm formation), and horizontal gene transfer through transformation or conjugation (5–7).
Members of the Rap-Phr family of QS systems in the Gram-positive model Bacillus subtilis are involved in the regulation of competence, sporulation, and biofilm formation. The chromosome of strain B. subtilis 168 encodes eight full receptor signal systems and three orphan receptors from this family (6, 8). In each of the full systems, the phr gene encodes a prepeptide, which is secreted through the major secretory system and then further processed outside the cell, resulting in the formation of a mature penta- or hexapeptide signal (9). The mature peptide is transported into the cell by the oligopeptide-permease (Opp) complex, where it interacts with its Rap targets within the cytoplasm (10). All characterized Rap proteins contain two domains (10–12), a tricopeptide repeat (TPR) domain that interacts with the signaling peptide and a second domain that functions either as a phosphatase or as a contact-dependent inhibitor of a response regulator. Rap proteins exert their activity only in the peptide-free form as binding of the signaling peptide leads to the inactivation of the Rap protein and to subsequent derepression of the response regulator.
Three response regulators have been characterized as targets of Rap proteins in the so-called laboratory or domesticated strain 168 and include Spo0F, ComA, and DegU. Spo0F, a component of the spo0A activation phosphorelay, is repressed by RapA, RapB, RapE, RapH, RapI, and RapJ. The spo0A pathway regulates multiple processes in the cell; most notably, sporulation is activated at high Spo0A phosphorylation levels, while biofilm formation requires lower levels of phosphorylation (13, 14). Spo0A, through regulation of the transcriptional regulator AbrB and the sigma factor σH, also controls the transcription of many of the phr genes from an internal promoter within the rap-phr operon (15). The QS response regulator ComA was shown to be repressed by RapC, RapD, RapF, RapH, and RapK (16, 17). ComA is an activator of many of the Rap proteins, thereby forming a complex regulatory network between the various QS pathways (18). Finally, RapG represses phosphorylated DegU (DegU∼P) (19), whose main functions are to promote the secretion of multiple enzymes and repress social motility (20). Recently, it was hypothesized that a plasmid-borne Rap (specifically, Rap carried by plasmid pLS20 [RappLS20]) may also interact directly with an Xre-type plasmid-borne repressor (21).
The laboratory strain, B. subtilis 168, and its derivatives (including strain PY79 used in this work) have undergone multiple genetic changes during domestication from their ancestral strain, B. subtilis NCIB3610 (denoted 3610 in this work). These changes led to the loss of multiple phenotypes, such as the ability to produce an extracellular matrix and the surfactant surfactin, both necessary for biofilm formation (22). The lab strain also shows an increased frequency of genetic competence compared to its ancestor, a feature that facilitates its genetic manipulation in the laboratory (23). Currently, research dealing with biofilm formation in Bacillus subtilis has therefore shifted away from the domesticated strains such as 168 and its derivatives and is primarily focused on strain 3610, which is considered a more suitable representative of wild strains that display social phenotypes (24). It should be noted that in terms of their biofilm-forming properties, there is a relatively small number of mutations that distinguish the undomesticated strain 3610 and the lab strains (25). These include mutations in enzymes necessary for the production of exopolysaccharides and surfactin, as well as mutations in various regulatory proteins.
An additional major difference between the lab strains and strain 3610 is that strain 3610 possess an 80-kb plasmid, termed pBS32, which is absent from the lab strains (26). This plasmid is not found in other isolates of B. subtilis subsp. subtilis (27), but a highly homologous plasmid, pLS32, which is found in a more distant strain of B. subtilis subsp. natto, has been analyzed and sequenced (28). Notably, pBS32 has been shown to carry an additional rap-phr cassette, termed rapP-phrP, and RapP was specifically shown to contribute to the difference in biofilm-forming capabilities between strains 3610 and 168 (25) (Fig. 1). Recently, it was shown that RapP functions as a phosphatase of spo0F in vitro. Furthermore, it was demonstrated in vivo that a rapP deletion affects the expression of direct targets of Spo0A. RapP was also shown to inhibit the transcription of the eps and srfA operons, whose gene products are responsible for the production of exopolysaccharides (eps) (29, 30) and the surfactant surfactin (srfA). Both of these operons are regulated by numerous other regulators, and the experiments did not rule out the possibility that RapP affects these operons through other regulators. Finally, it was found that, unlike all other characterized Rap-Phr pairs, the effect of RapP on gene expression is not suppressed by the expression of its adjoining phrP signaling gene or by external addition of the presumed PhrP signaling peptides to the medium. It was observed, however, that weak suppression of RapP was achieved with the addition of the PhrH signaling peptide to the medium (29). RapP is therefore unique among all Rap-Phr pairs in that it appears not to interact with its cognate PhrP peptide. It is therefore unclear what the function of phrP is and what features of the RapP system can explain this difference between rapP-phrP and other rap-phr pairs.
FIG 1.
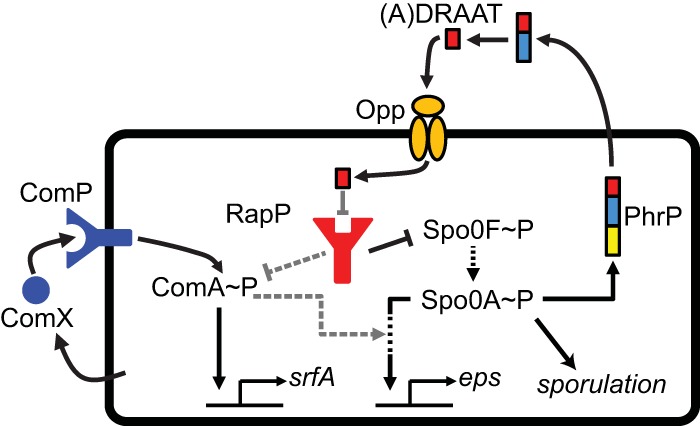
The proposed RapP interaction network. The scheme illustrates the RapP network in B. subtilis 3610 based on previous results (black arrows), including the additional interactions (gray arrows) which are proposed on the basis of results reported in this work. RapP (red) is a repressor of Spo0F activity and, directly or indirectly, represses ComA in an Spo0F-independent manner. RapP is repressed by the mature PhrP peptide (A)DRAAT, produced by secretion and subsequent cleavage of PhrP (the three-colored rectangle represents a signal peptide, an extracellularly cleaved prepeptide, and a mature peptide). ComA is activated by the ComX~P QS system (blue) and directly controls the transcription of several operons, including the srfA operon. The exopolysaccharide (eps) operon is indirectly controlled by ComA. Spo0A, the master regulator of sporulation and biofilm formation, indirectly regulates the expression of the eps operon and of the phrP gene. Spo0F is part of the phosphorelay that activates Spo0A and is a known target of many Rap proteins. rapP of strain NCIB3610 encodes an atypical substitution that prevents its repression by the PhrP signaling peptide.
Here, we report the results of genetic analysis that examined three layers of regulation of the RapP pathway: (i) the transcriptional regulation of pathway, (ii) the effect on downstream targets, and (iii) RapP interaction with PhrP. We find that the transcriptional regulation of rapP-phrP is reminiscent of the transcriptional organization of other rap-phr pairs (31) and that rapP has an additional target beside Spo0F, which regulates the repression of srfA and eps in a ComA-dependent manner. Finally, we find that a highly conserved amino acid of the Rap TPR domain has been replaced in the rapP allele of strain 3610 (rapP3610). Reconstitution of the consensus amino acid in RapP restored the interaction between PhrP and RapP and eliminated the effects of RapP on biofilm formation.
MATERIALS AND METHODS
Growth media.
Routine growth experiments were performed in Luria-Bertani (LB) broth containing 1% tryptone (Difco), 0.5% yeast extract (Difco), and 0.5% NaCl. Most gene expression measurements were done using Spizizen minimal medium (SMM) containing 2 g liter−1 (NH4)2SO4, 14 g liter−1 K2HPO4, 6 g liter−1 KH2PO4, 1 g liter−1 disodium citrate, and 0.2 g liter−1 MgSO4·7H2O supplemented with 125 mg liter−1 MgCl2·6H2O, 5.5 mg liter−1 CaCl2, 13.5 mg liter−1 FeCl2·6H2O, 1 mg liter−1 MnCl2·4H2O, 1.7 mg liter−1 ZnCl2, 0.43 mg liter−1 CuCl2·4H2O, 0.6 mg liter−1 CoCl2·6H2O, 0.6 mg liter−1 Na2MoO4·2H2O, and 0.5% glucose. Biofilm growth was done on MSgg medium plates; MSgg medium contains 100 mM morpholinepropanesulfonic acid (MOPS) (pH 7), 0.5% glycerol, 0.5% glutamate, 5 mM potassium phosphate (pH 7), 50 μg ml−1 tryptophan, 50 μg ml−1 phenylalanine, 2 mM MgCl2, 700 μM CaCl2, 50 μM FeCl3, 50 μM MnCl2, 2 μM thiamine, and 1 μM ZnCl2 (22). Media were solidified using 1.5% agar (Difco). The following antibiotics were used (concentrations): macrolides-lincosamides-streptogramin B (mls; 1 μg ml−1 erythromycin, 25 μg ml−1 lincomycin), spectinomycin (Sp; 100 μg ml−1), chloramphenicol (Cm; 5 μg ml−1), and kanamycin (Kan; 15 μg ml−1). Isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma) was added to the medium when appropriate, at the concentrations indicated in the text and figure legends.
Strain and mutant construction.
All of the strains used in this study are listed in Table 1. Deletion of comA from the PY79 chromosome and its replacement with a chloramphenicol resistance cassette were performed through the long flanking homology PCR method (32) using the primers comA-P1, comA-P2, comA-P3, and comA-P4.
TABLE 1.
Strain list
| Genetic backgrounda | Strain name | Genotype or description | Source, reference, or construction methodb |
|---|---|---|---|
| 3610 | AES1109 | B. subtilis NCIB3610 wild type | Bacillus genetic stock center |
| PY79 | AES101 | B. subtilis PY79 wild type | Bacillus genetic stock center |
| PY79 | AES1419 | amyE::(PrapA-3×YFP Sp) | This study |
| PY79 | AES1502 | amyE::(PrapA-3×YFP Sp) ΔcomA::Cm | AES1403→AES1419 |
| PY79 | AES1403 | ΔcomA::Cm | This study |
| PY79 | AES1444 | amyE::(PrapA-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) | AES1370→AES1419 |
| PY79 | AES1503 | amyE::(PrapA-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) ΔcomA::Cm | AES1370→AES1502 |
| PY79 | AES1334 | amyE::(Psrf-3×YFP Sp) | This study |
| PY79 | AES1379 | amyE::(Psrf-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) | AES1370→AES1334 |
| PY79 | BS225 | Δspo0A::Km | Kind gift of the Ben-Yehuda lab |
| PY79 | AES1472 | amyE::(Psrf-3×YFP Sp) Δspo0A::Km | BS225→AES1334 |
| PY79 | AES1874 | amyE::(Psrf-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) Δspo0A::Km | AES1370→AES1472 |
| PY79 | DS1993 | ΔdegU::mls | Kind gift of the Kearns lab |
| PY79 | AES1875 | amyE::(Psrf-3×YFP Sp) ΔdegU::mls | AES1334→DS1993 |
| PY79 | AES1606 | amyE::(Psrf-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) ΔdegU::mls | AES1370→1875 |
| PY79 | AES1876 | amyE::(Psrf-3×YFP Sp) ΔdegU::mls Δspo0A::Km | BS225→AES1875 |
| PY79 | AES1877 | amyE::(Psrf-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) ΔdegU::mls Δspo0A::Km | AES1370→AES1876 |
| 3610 | AES1401 | amyE::(PrapP-3×YFP Sp) | This study |
| PY79 | AES1377 | amyE::(PrapP-3×YFP Sp) | This study |
| 3610 | AES1411 | amyE::(PrapP-3×YFP Sp) | This study |
| 3610 | AES1452 | amyE::(PrapP-3×YFP Sp) ΔcomA::Cm | AES1403→AES1401 |
| 3610 | AES1714 | amyE::(PphrP-3×YFP Sp) Δspo0A::Km | BS225→AES1411 |
| PY79 | RL620 | ΔabrB::mls | 50 |
| 3610 | AES1668 | amyE::(PphrP-3×YFP Sp) ΔabrB::mls | RL60→AES1411 |
| PY79 | RL2201 | Δspo0H::Km | 50 |
| 3610 | AES1413 | amyE::(PphrP-3×YFP Sp) Δspo0H::Km | RL2201→AES1411 |
| PY79 | AES1819 | amyE::(Peps-3×YFP Sp) | This study |
| PY79 | AES1820 | sacA::(PrapP-rapP-phrP Cm) amyE::(Peps-3×YFP Sp) | AES913→AES1819 |
| PY79 | AES1827 | amyE::(Peps-3×YFP Sp) ΔcomA::Cm | AES1403→AES1819 |
| PY79 | AES1828 | amyE::(Peps-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) ΔcomA::Cm | AES1370→AES1827 |
| 3610 | DS2569 | pBS32 plasmid cured | Kind gift of the Kearns lab |
| 3610 | AES1821 | pBS32 plasmid cured; amyE::(Psrf-3×YFP Sp) | AES1334→DS2569 |
| 3610 | pBS32 plasmid cured; amyE::(Psrf-3×YFP Sp) zba88::(Phs-rapP Sp Cm Km) | AES1370→ AES1821 | |
| 3610 | AES1836 | pBS32 plasmid cured; amyE::(Psrf-3×YFP Sp) Δspo0A::Km | BS225→AES1821 |
| 3610 | AES1837 | pBS32 plasmid cured; amyE::(Psrf-3×YFP Sp) Δspo0A::Km zba88::(Phs-rapP Sp Cm Km) | AES1370→AES1836 |
| 3610 | AES1605 | amyE::(Psrf-3×YFP Sp); plasmid cured | AES1334→DS2569 |
| 3610 | AES1707 | ΔrapP-phrP sacA::(PrapP-rapPN236T-phrP Cm) | AES1656→DS8796 |
| PY79 | AES1709 | amyE::(Psrf-3×YFP Sp) zba88::(Phs-phrP Sp Cm Km) sacA::(PrapP-rapPN236T Cm) | AEC735→AES1477 |
| PY79 | AES1478 | amyE::(Psrf-3×YFP Sp) zba88::(Phs-phrP Sp Cm Km) sacA::(PrapP-rapP-phrP Cm) | AES913→AES1477 |
| PY79 | AES1477 | amyE::(Psrf-3×YFP Sp) zba88::(Phs-phrP Sp Cm Km) | This study |
| PY79 | AES1678 | amyE::(Psrf-3×YFP Sp) sacA::(PrapP-rapPN236T Cm) | AES1656→ AES1337 |
| PY79 | AES1380 | amyE::(Psrf-3×YFP Sp) sacA::(PrapP-rapP-phrP Cm) | AES913→AES1337 |
| 3610 | AES1873 | ΔrapP-phrP amyE::(Psrf-3×YFP Sp) sacA::(PrapP-rapPN236T Cm) | AES1337→AES1707 |
| 3610 | DS8796 | ΔrapP-phrP | 29 |
| 3610 | AES1336 | amyE::(Psrf-3×YFP Sp) | This study |
| PY79 | AES1418 | amyE::(Ppel-3×YFP Sp) | This study |
Description of strains and their construction methods. Further details on the construction of relevant plasmids is given in Materials and Methods.
An arrow indicates transformation of genomic DNA. The recipient strain is on the right. Plasmid construction is described in Materials and Methods.
To generate inducible zba88::(Phs-rapP) and zba88::(Phs-phrP) constructs (where Phs is the hyperspank promoter), a PCR product containing the relevant open reading frame (ORF) was amplified using the hsRapP-F/hsRapP-R and hsPhrP-F/hsPhrP-R primer pairs. The PCR products were digested with NheI and SphI and ligated downstream of the hyperspank promoter of the pDR111 vector containing spectinomycin resistance (33).
The construction of transcriptional fusions to yfp was performed by PCR amplification of the relevant promoter using 3610 genomic DNA as a template. To generate the promoter fusions, PCR fragments were amplified using the following primer pairs: Psrf, Psrf-F/Psrf-R; Peps, Peps-F/Peps-R; PrapP, PrapP-F/PrapP-R; PphrP, PphrP-F/PphrP-R; PrapA, PrapA-F/PrapA-R; PslrA, PslrA-F/PslrA-R; PslrR, PslrR-F/PslrR; PsinI, PsinI-F/PsinI-R (Table 2). After fragments were digested with the appropriate enzymes (purchased from New England BioLabs), they were ligated to the plasmid pDL30-3×YFP containing three copies of the yellow fluorescent protein (YFP) (34). The resulting constructs were integrated into the amyE site on the chromosome using spectinomycin resistance for selection.
TABLE 2.
Primer list
| Namea | Sequenceb | Enzyme |
|---|---|---|
| PrapP-F | AAGTGAATTCTTCATCCGGAGACTATTTAT | EcoRI |
| PrapP-R | GTGGGGATCCTCAAATACCTCCTTTTCTTT | BamHI |
| PphrP-F | AAGTGAATTCCGCTTTAGAAACAGCTGAAA | EcoRI |
| PphrP-R | GTGGGGATCCATGAAGCGCTAGATACTCCA | BamHI |
| PrapA-F | CGACCGAATTCCAAAACTTACAGAAGGCTT | EcoRI |
| PrapA-R | GCAGAGCTAGCTGAATTACCCGAGATATGTC | NheI |
| Peps-F | GTGCGGAATTCGTCGTTATTTCGTTCATTAT | EcoRI |
| Peps-R | AGTTGGCTAGCTCATGTATTCATAGCCTTCA | NheI |
| Psrf-F | ATGGGGAATTCCGTTGTAAGACGCTC | EcoRI |
| Psrf-R | AGGTGGCTAGCTTTATAAGCAGTGAACAT | NheI |
| rapP-phrP-F | AGGAGGATATCTTCATCCGGAGACTATTTATGAACAA | EcoRV |
| rapP-phrP-R | CTCCTGCATGCTTAGGTGGTAGCACCATTCTTGCA | SphI |
| rapP-R | ATGAAGCATGCTTACATTTTTTCATTTAAATG | SphI |
| hsRapP-F | ATCCAGCTAGCAAAGAAAAGGAGGTATTTGATTG | NheI |
| hsRapP-R | ATCCGGCATGCCAAGAGCGCTAAACAAATTG | SphI |
| hsPhrP-F | ATCCAGCTAGCAAATTCAAAGGGGGAAACATTTAAATG | NheI |
| hsPhrP-R | ATCCGGCATGCTTAAGTTGCTGCTCTATCTG | SphI |
| rapP-phrP-NtoT-F | [phos]AGCTCATTTTAACGTGGGATTA | |
| rapP-phrP-NtoT-R | [phos]GAACCAATTAAATGTTGCTCCC | |
| comA-P1 | AAGTTGGACCGGACTGGAAT | |
| comA-P2 | TTTTCTAATGTCACTAACCTGCCAAACTGTTCGCTCGGTTCAG | |
| comA-P3 | AGTAATCCGCCCGACGGTATAGCGGTCCATTGAATACAGC | |
| comA-P4 | GGTGAGCCGGTGATGTTTAC |
See Materials and Methods for descriptions. Primer rapP-phrP-NtoT-F contains the mutated base necessary for creating the T236N substitution by performing a C-to-A mutation. This base is shown in boldface in the sequence. F, forward; R, reverse.
The restriction enzyme recognition sequences are underlined. [phos] indicates the 5′ phosphorylated primer.
Construction of sacA::(PrapP rapP-phrP Cm) and sacA::(PrapP rapP Cm) was performed by PCR amplification of the annotated rapP and phrP genes or of rapP together with 400 bp upstream of rapP using the primer pair rapP-phrP-F/rapP-phrP-R or rapP-phrP-F/rapP-R, respectively. The PCR products were digested with EcoRV and SphI and ligated into the sacA locus of pSac-cm.
A rapP gene encoding the T-to-N amino acid change at position 236 (rapPT236N) was constructed by site-directed mutagenesis. The 5′ phosphorylated primer pair rapP-phrP-TtoN-F/rapP-phrP-TtoN-R, in which the primers are adjoining but with reversed alignments, was designed to amplify the entire sacA::(PrapP rapP Cm)-containing vector and change the codon ACC (threonine) to AAC (asparagine). The PCR product was used for self-ligation and transformed to PY79 cells.
All of the mutations and constructs were transferred to PY79 cells by transformation (35) and to 3610 by phage SPP1-mediated generalized transduction (36). Integration of amyE integration plasmids into the zba88::amyE Ω Cm Kan strain was done in two steps. First, the plasmid was integrated into a PY79 strain carrying the zba88 construct and screened for an Amy+ phenotype. A genomic prep of the resulting strain was then inserted into other strains with selection for either Kan or Cm, depending on the genetic background of the integrated genome.
Biofilm formation assay.
Bacteria were grown overnight in MSgg broth, diluted 1:100, and grown to an optical density at 600 nm (OD600) of ∼1.0. Five microliters of the culture was plated on MSgg agar, and the plates were incubated at 30°C for 72 h.
Peptide synthesis.
A synthetic PhrP 5-mer peptide (NH2-DRAAT-COOH), PhrP 6-mer peptide (NH2-ADRAAT-COOH), and PhrH 6-mer peptide (NH2-TDRNTT-COOH) were purchased from GL Biochem (Shanghai, China) at >98% purity. Aliquots (10 mM) were prepared by resuspension of the lyophilized peptides in H2O and stored at −20°C.
Gene expression analysis.
Flow cytometry was used to quantify gene expression at the single-cell level using a Beckman-Coulter Gallios system with a 488-nm laser. The cells were grown in SMM to an OD600 of ∼0.1, diluted by a factor of 106 in fresh SMM, and grown for about 20 h to the beginning of the exponential phase. Samples were taken at several time points, and the OD and YFP levels were measured. A minimum of 20,000 cells were analyzed for each sample. The results are presented as the mean YFP level of the population for a specific OD (calculated by interpolation from actual OD measurements) and the standard error of the mean. The results represent the average of at least three independent experiments. YFP levels are measured as a ratio between the measured strain and the autofluorescence of a wild-type strain. Autofluorescence of the wild type did not change by more than 5% at different stages of growth in SMM.
RESULTS
The transcriptional regulation of rapP-phrP in 3610 is similar to the regulation of other rap-phr pairs.
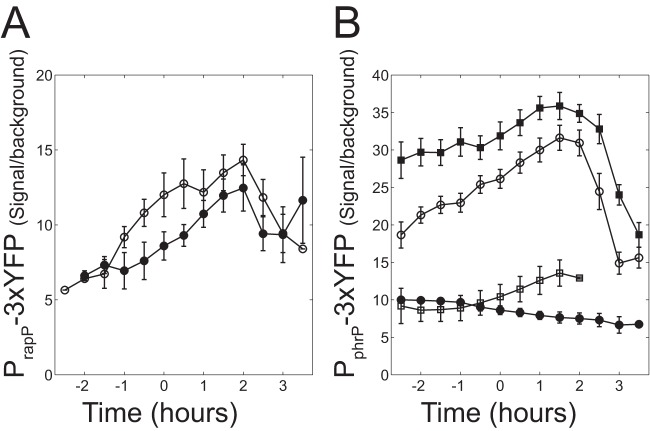
We first examined whether the transcriptional regulation of the rapP-phrP operon reflects the unique independence of RapP from PhrP or whether it is similar to other rap-phr pairs. To this end, we cloned the regions immediately upstream of the rapP and phrP ORFs into the 3×YFP reporter plasmid and integrated each of the reporters separately into the amyE locus of strain 3610. The expression of each promoter was monitored in several genetic backgrounds (Fig. 2). Expression of PrapP-3×YFP showed only a moderate increase as the cells approached the stationary phase (Fig. 2A). PrapP-3×YFP expression was slightly lower in a comA deletion background, suggesting a possible weak regulation by ComA. The phrP internal promoter reporter (PphrP-3×YFP) doubled its activity during transition from early log to stationary phase (Fig. 2B). Promoter activity was markedly reduced in a Δspo0A background (Fig. 2B). In order to gain further insight into the activation of PphrP-3×YFP by Spo0A, the expression of the reporter was monitored in strains carrying null mutations in sigH or abrB. As shown in Fig. 2B, the strain carrying a sigH deletion exhibited reduced expression while the strain carrying an abrB deletion showed an increase in expression levels.
FIG 2.
Transcriptional regulation of rapP and phrP promoters. Cells were grown in SMM, and their YFP fluorescence was measured as a function of time as described in Materials and Methods. Time zero is defined as the time when the OD equals 1.5. (A) YFP levels of wild-type 3610 (AES1401, open circle) and the ΔcomA 3610 (AES1452, filled circle) strains carrying a PrapP-3×YFP reporter. (B) YFP levels in the wild-type strain 3610 background carrying a PphrP-3×YFP reporter with the following genetic modifications: wild type (AES1411, open circles), Δspo0A (AES1714, filled circles), ΔabrB (AES1668, filled squares), and ΔsigH (AES1413, open squares). Expression levels in this and the following figures are given as the ratio between the mean expression levels of the designated genotype to the autofluorescence of the wild-type PY79. We find autofluorescence to be very stable when cells are grown in minimal medium. Error bars represent the standard error of the mean for at least three independent experiments.
The results shown in Fig. 2 indicate that the transcriptional regulation of the rapP-phrP locus resembles the transcriptional regulation of other rap-phr loci even though its activity does not depend on an interaction with PhrP.
RapP regulates the expression of comA-dependent genes in an spo0F-independent pathway.
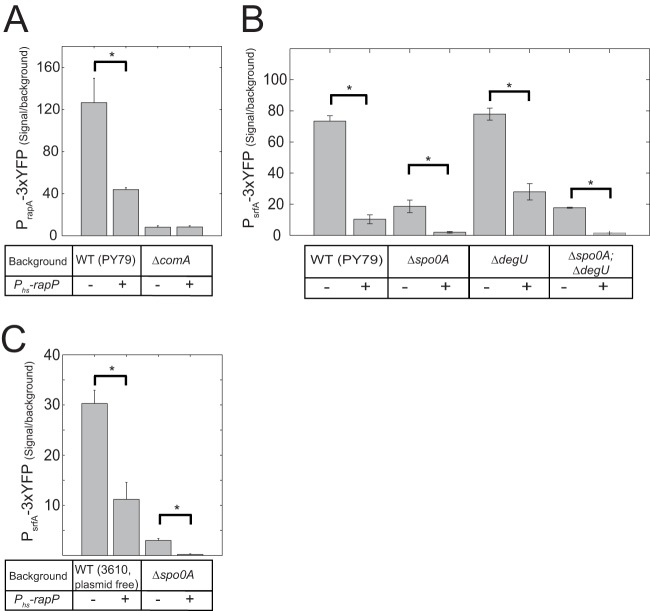
It was previously shown that RapP strongly represses the expression of the srfA promoter in strain 3610 (29). Since srfA expression is modulated by multiple QS systems whose functions are not well characterized in strain 3610, we examined whether this repression is conserved when RapP is expressed in the genetic background of the lab strain PY79 in which QS is well characterized. The use of strain PY79 also allowed us to ignore interactions of RapP with plasmid-borne genes (as described with other Rap proteins [21, 37]). In addition, the activity could be assessed without the indirect effects that surfactin may have on gene expression (38). To this end, rapP together with its promoter was cloned into the sacA locus of PY79. The rapP gene was also cloned together with its native ribosomal binding site downstream of the IPTG-inducible hyperspank promoter (Phs) and subsequently integrated into an ectopic amyE locus positioned at 33° on the bacterial chromosome, as previously described (34, 39, 40). To monitor srfA expression, we cloned the previously characterized (41) 560-bp promoter of the srfA operon in front of the 3×YFP reporter, which was integrated into the native amyE locus. The resulting PsrfA-3×YFP reporter was then introduced into three strains, parent PY79 and the Phs-rapP and PrapP-rapP strain backgrounds as described in Materials and Methods. Expression of rapP in strain PY79, either from its native promoter or from the hyperspank promoter supplemented with 10 μM IPTG, led to an ∼10-fold reduction in YFP levels (Fig. 3B; see also Fig. 5B).
FIG 3.
RapP effect on ComA reporters in various genetic backgrounds. Promoter activity was measured in each genotype with (+) or without (−) a Phs-rapP construct. (A) Shown are the YFP expression levels of four strains carrying the PrapA-3×YFP reporter in the following backgrounds: wild-type PY79 (AES1419), Phs-rapP (AES1444), ΔcomA (AES1502), and Phs-rapP ΔcomA (AES1503). (B) YFP levels were measured in a PY79 PsrfA-3×YFP background without (−) and with (+) an IPTG-inducible rapP construct. The following genetic backgrounds are used: wild-type PY79 (AES1334), Phs-rapP (AES1379), Δspo0A (AES1472), Δspo0A Phs-rapP (AES1874), ΔdegU (AES1875), ΔdegU Phs-rapP (AES1606), ΔdegU Δspo0A (AES1876), and ΔdegU Δspo0A Phs-rapP (AES1877). (C) The experiment is the same as that described for panel B but in a background of a plasmid-free derivative of strain 3610. The following genotypes were used: the wild-type plasmid-free strain (strain AES1605), Phs-rapP strain (AES2472), Δspo0A strain (AES2436), and Δspo0A Phs-rapP strain (AES2437). All measurements were performed at an OD600 of 1.5. All genetic backgrounds with the Phs-rapP construct were induced with 10 μM IPTG. In this and the following figures, the difference between all pairs marked with an asterisk are statistically significant (t test, P < 0.05).
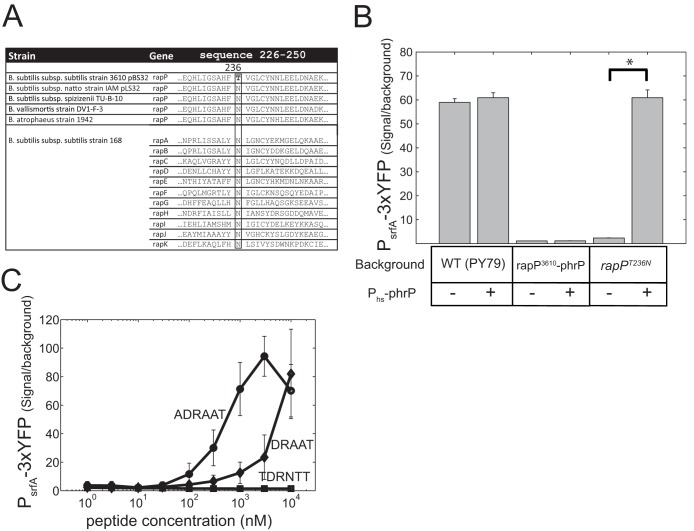
FIG 5.
rapP3610 codes for an N236T substitution mutation compared to the consensus, which renders it insensitive to the PhrP peptide. (A) Sequence alignment of amino acids 226 to 250 from RapP3610 with the sequences of other RapP homologues and with other Rap proteins found in strain 3610. Asparagine residue 236 is conserved in all Rap proteins except for the product of rapP3610, where it is replaced with threonine. Residue 236 of RapP is shown in boldface, and the aligned column is boxed for emphasis. (B) The addition of an IPTG-inducible phrP gene suppresses the effect of rapPT236N but not of rapP3610 on PsrfA-3×YFP expression. Shown are YFP expression levels of a PsrfA-3×YFP reporter in strain PY79 with different backgrounds, where each genotype was measured with (+) or without (−) a Phs-phrP construct. Genotypes are as follows: wild-type PY79 (AES1334), Phs-phrP (AES1477), PrapP-rapP3610 (AES1380), PrapP-rapP3610 Phs-phrP (AES1478), PrapP-rapPT236N (AES1678), and PrapP-rapPT236N Phs-phrP (AES1709). Expression was measured at an OD600 of 1.5, and IPTG was added at 10 μM when needed. (C) PsrfA-3×YFP expression was monitored in a PrapP-rapPT236N (AES1678) background as a function of the concentration of the externally added peptides. Three peptides were tested for their ability to repress rapPT236N activity: the putative signaling peptides of PhrP (DRAAT or ADRAAT) and the hexapeptide signal of PhrH (TDRNTT).
While srfA transcription is strongly activated by ComA, there are several other transcriptional regulators that directly regulate this operon, and an interaction between RapP and any of these regulators may lead to the same observed effect. If RapP exerts its effect on the srfA promoter through the repression of ComA activity, we would expect to see other genes in the ComA regulon respond in a similar fashion. To check this hypothesis, we cloned the promoters of pel and rapA, two known direct targets of ComA (4), into the 3×YFP vector. We compared the expression of the srfA, pel, and rapA reporters in both the parental PY79 background and the inducible Phs-rapP background. The expression of all three genes was markedly reduced when expression of rapP was induced compared to the wild-type level (Fig. 3A and B; see also Fig. S1 in the supplemental material). Unlike the srfA promoter, the promoter of rapA has a strong residual expression in a comA null mutant background. We found that RapP had no effect on this residual expression (Fig. 3A), demonstrating the epistatic interaction between RapP and ComA. These results suggest that RapP inhibits ComA activity, either directly or indirectly.
ComA activity is modulated in strain PY79 by Spo0A activity through Spo0A regulation of phrC and other phr genes (15). If RapP repression of ComA activity is dependent on RapP-catalyzed dephosphorylation of Spo0F, we would expect that RapP would have no effect on ComA activity in an spo0A or spo0F null mutant. To explore this hypothesis, the PsrfA-3×YFP reporter was introduced into strains carrying either the Δspo0A or Δspo0A Phs-rapP genetic background, and YFP levels were compared. We found that in a Δspo0A mutant, PsrfA-3×YFP expression was reduced 4-fold compared to that of the parental PY79 strain. YFP levels were further reduced 9-fold when RapP was induced in the spo0A null background (Fig. 3B). We further verified that RapP inhibition of the PsrfA-3×YFP reporter was independent of degU or of the combination of degU and spo0A (Fig. 3B). These results clearly indicated that RapP has additional targets beyond Spo0F which mediate its inhibition of ComA activity.
Since strain 3610 differs from strain PY79 in multiple loci, the question arose as to whether the spo0A-independent effect of RapP on srfA activity could also be observed when these other mutations are taken into account. We therefore examined the epistatic interaction between spo0A and rapP in a derivative of strain 3610 that was cured of the pBS32 plasmid (strain DS2569). The same pattern of interactions was observed; namely, srfA expression in an a Phs-rapP Δspo0A background was significantly lower than expression in an spo0A null background (Fig. 3C).
The eps operon is positively regulated by ComA, and this regulation is epistatic to rapP.
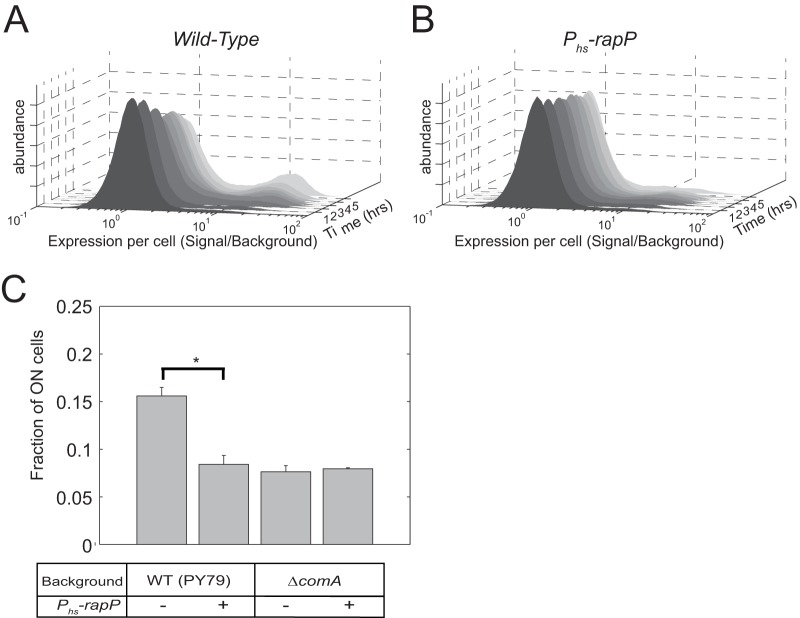
It was previously shown that RapP strongly affects the biofilm phenotype of B. subtilis 3610 (25, 29). In this regard it was reported that RapP regulates the expression of the eps operon, which encodes all structural genes needed for the production and secretion of the exopolysaccharide component of the biofilm matrix (29, 30). To examine this regulation further, we constructed a Peps-3×YFP transcriptional reporter containing the 224 bp immediately upstream of the epsA open reading frame and introduced it into strain PY79. Flow cytometry was used to quantify gene expression at the single-cell level. When the cells were grown on minimal medium, the eps reporter was expressed in a bimodal manner (Fig. 4A). In a small subpopulation of cells, YFP was expressed at levels approximately 15 times higher than the background level (the ON cell population), whereas the majority of the cells did not express significant levels of YFP (the OFF cell population). The fraction of ON cells increased over time, reaching about 20% of the total population several hours after the onset of the stationary phase (Fig. 4A). Integration of the Phs-rapP construct into PY79 and its induction in the presence of 10 μM IPTG did not significantly change the mean fluorescence levels of either the ON or OFF population. Instead, the fraction of the ON population was kept at a constant basal level of less than 10% of the population, without the significant increase in the fraction of ON cells observed in the stationary phase in strain PY79 (Fig. 4B).
FIG 4.
Dependence of eps expression on comA and rapP mutations. (A and B) The distribution of expression levels of YFP driven by the eps promoter as a function of time in strain AES1819 (PY79 Peps-3×YFP) (A) and strain AES1820 (PrapP-rapP-phrP Peps-3×YFP) (B). Note the difference between strains in the fraction of high-YFP-expressing cells (ON population) and how expression changes over time. (C) Shown are the fractions of the ON cells for strains that express the Peps-3×YFP reporter in the following genetic backgrounds: AES1819 (PY79 wild-type), AES1820 (PrapP-rapP-phrP), AES1827 (ΔcomA), and AES1828 (ΔcomA Phs-rapP). Inducible constructs were induced with 10 μM IPTG.
Since other RapP functions seem to be dependent on ComA (Fig. 3), it is possible that ComA has a role in the observed dependence of the eps operon expression profile on RapP. Indeed, a comA deletion reduced the fraction of the eps ON population to that observed in the RapP-expressing strain (Fig. 4C). Induction of RapP using the Phs-rapP in the comA null mutant background did not further reduce the frequency of ON cells (Fig. 4C). These results demonstrate that bistability of the eps operon is modulated by ComA and that its interaction with RapP contributes to eps expression. These results do not rule out that RapP also affects eps expression through Spo0A.
The effect of RapP on sporulation is ComA independent.
RapP has also been shown to affect genes directly targeted by Spo0A∼P (29). We therefore examined the involvement of RapP and ComA in sporulation (Table 3). We found that ComA had little effect on sporulation levels of strain PY79. On the other hand, sporulation levels were markedly reduced when Phs-rapP was introduced into strain PY79 or the PY79 comA null mutant strain, clearly demonstrating a ComA-independent role for RapP, in accordance with the direct interaction observed between RapP and Spo0F (29). We note that in order for rapP to have an effect on sporulation, much higher levels of IPTG (1 mM) were required than those used to observe the effects of RapP on eps and srfA gene expression.
TABLE 3.
The effect of comA and rapP on sporulation
| Strain | Relevant genotype | No. of viable cellsa | No. of heat-resistant cellsa | Sporulation efficiency (%)a | Mean sporulation efficiency (%)b |
|---|---|---|---|---|---|
| AES101 | PY79 | 5 × 108 | 2 × 108 | 40 | 36 ±8 |
| AES1403 | PY79 ΔcomA | 2.6 × 108 | 2.03 × 108 | 78 | 51 ±12 |
| AES1444 | PY79 Phs-rapP | 1.61 × 108 | 6.2 × 105 | 0.39 | 0.39 ± 0.06 |
| AES1503 | PY79 ΔcomA Phs-rapP | 1.04 × 108 | 3.47 × 105 | 0.33 | 0.32 ± 0.05 |
A single colony was inoculate into 1 ml of Difco sporulation medium (with 1 mM IPTG, if needed) and grown for 24 h. Serial dilutions were plated onto nonselective LB agar plates before and after the cells were heating to 80°C for 20 min. The results of a single experiment are shown, and values are representative.
Mean and standard error of sporulation efficiency for each strain. Statistics are based on 3 to 6 biological replicates for each strain. The difference between strains AES101 and AES1403 is not significant (P = 0.36, two-sample t test). The difference between strains AES1444 and AES1503 is not significant (P = 0.44, two-sample t test). The difference between strains AES1403 and AES1503 is significant (P = 0.01, two-sample t test).
The rapP allele of strain 3610 carries a substitution in a highly conserved residue whose reversion reconstitutes interaction with phrP.
It has previously been reported that repression by RapP is not alleviated by expression of the phrP gene or by exogenous addition of the candidate pentapeptide or hexapeptide signals (29). This behavior is unique to the RapP-phrP system; all other characterized Rap proteins which have an adjoining phr gene in the same operon are affected by the corresponding signaling peptide (9). The PhrP independence of RapP was retained when rapP and phrP were introduced into strain PY79. Moreover, overexpression of phrP had no effect on the RapP-dependent repression of srfA in strain PY79 (Fig. 5B).
Highly similar rapP homologues are present in plasmid pLS32 and in the chromosomal genomes of B. subtilis subsp. spizizenii TU-B-10, B. vallismortis strain DV1-F-3, and B. atrophaeus strain 1942. By aligning these proteins, we noticed that the rapP allele of pBS32 in strain 3610 (here, rapP3610) has an asparagine-to-threonine substitution at position 236, which results from an A-to-C substitution in position 707 of the gene (Fig. 5A). Sequence alignment of all other Rap proteins of strain 3610 show that they contain asparagine in the homologous position (Fig. 5A). Further alignment of 107 different Rap proteins from B. subtilis isolates showed that asparagine is conserved in all of them (see Fig. S2 in the supplemental material). The threonine substitution is therefore a unique variant. Structurally, the homologous asparagines in RapF and RapJ have been shown to strongly interact with the backbone of the signaling peptide (11, 12). More generally, asparagine residues typically mediate interaction with peptide backbone in other TPR domains (42).
In order to determine whether the N236T substitution rendered the RapP3610 allele insensitive to PhrP, we used in vitro mutagenesis to introduce a threonine-to-asparagine substitution at position 236 of the PrapP-rapP3610 construct. This allele, which we designate rapPT236N, was introduced into strain PY79 containing a PsrfA-3×YFP reporter. This led to the same level of repression of PsrfA-3×YFP expression as did the introduction of the rapP3610 allele (Fig. 5B), indicating that this change did not affect the activity of RapP in the absence of the phrP gene product. To determine whether introduction of phrP could derepress the RapPT236N allele but not the RapP3610 allele, we introduced the phrP gene, driven by the Phs inducible promoter, into the different strains. The addition of this construct to the rapP3610-phrP strain had no effect on PsrfA-3×YFP expression, irrespective of the level of IPTG used. On the other hand, the addition of the Phs-phrP construct to a strain containing rapPT236N restored PsrfA-3×YFP expression to its wild-type levels, demonstrating full suppression of RapPT236N activity (Fig. 5B). This derepression was also observed without the addition of IPTG, indicating that the leakiness of the hyperspank promoter produces a sufficient amount of signal to inhibit RapPN236T activity.
The Phr signaling peptides are penta- or hexapeptides often derived from the C terminus of the Phr polypeptide (9). The candidate PhrP signaling peptides are therefore DRAAT and ADRAAT. To quantify the activity of these peptides, we grew PY79 strains containing the PsrfA-3×YFP reporter and either Phs-rapP3610 or Phs-rapPT236N in various concentrations of the two peptides (Fig. 5C). YFP expression was restored to wild-type levels upon addition of a high concentration of either peptide to the strain carrying the rapPT236N allele, but for any given concentration, the hexapeptide showed higher activation than the pentapeptide. A significant increase in YFP expression levels was observed at peptide concentrations of 300 nM or higher, which is similar to the concentration that was reported for PhrG (19) but significantly higher than that of the PhrC signal, competence- and sporulation-stimulating factor (CSF) (6). No effect was observed by addition of either peptide to the rapP3610 strain or the addition of the PhrH hexapeptide to either strain (Fig. 5C).
Biofilm and ComA phenotypes associated with the pBS32 plasmid are abolished in the rapPT236N background.
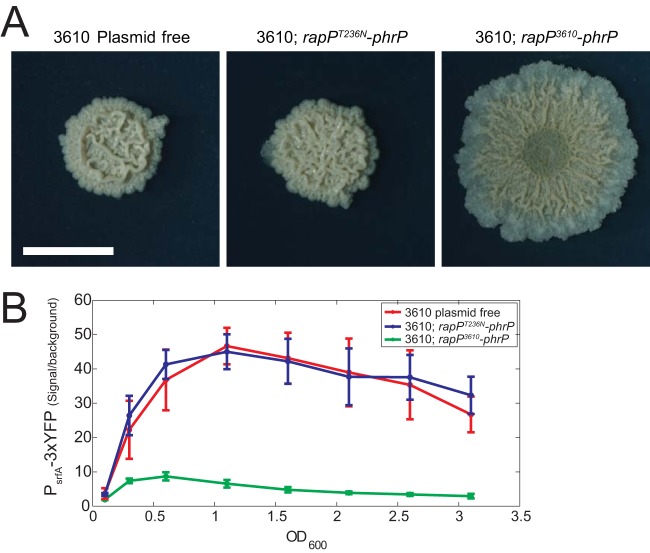
As PhrP inhibits RapP at a high peptide density, we wondered whether the replacement of rapP3610 with rapPT236N would render both biofilm formation and ComA activity independent of the plasmid pBS32. To this end, we compared the biofilm phenotypes of three strains:(i) strain DS2569, a plasmid-free version of strain 3610 (25, 26); (ii) strain AES1707 (3610 rapPT236N-phrP), where the rapP-phrP cassette was deleted from the pBS32 plasmid and the substituted rapPT236N-phrP cassette was inserted into the chromosomal sacA locus; and (iii) strain 3610. As shown in Fig. 6A, when the plasmid-free strain was plated on MSgg agar, its biofilm phenotype was highly similar to that of strain 3610 rapPT236N-phrP but markedly different from that of strain 3610.
FIG 6.
Replacement of rapP3610 by rapPT236N eliminates the phenotypic effects of the pBS32 plasmid on ComA activity and biofilm formation. (A) Biofilm formation by the 3610 plasmid-free strain (DS2569), 3610 rapPT236N-phrP strain (AES1707), and strain 3610 (rapP3610-phrP, AES1109). (B) YFP expression levels of PsrfA-3×YFP as a function of optical density during growth in SMM for the following backgrounds: 3610 plasmid-free strain (AES1605), 3610 rapPT236N-phrP strain (AES1873), and strain 3610 (rapP3610-phrP, AES1336).
We expect that the introduction of rapPT236N will also cancel the effect of RapP on ComA activity. To this end, a PsrfA-3×YFP reporter was introduced into the three strains described above in order to monitor ComA activity. We found that YFP expression levels were identical in the plasmid-free strain and the 3610 rapPT236N-phrP strain. Both strains showed expression levels which were 6 times higher than the level in strain 3610 (Fig. 6B). It should be noted that the PsrfA expression profile in the plasmid-free strain was still markedly different from PsrfA expression in the lab strain, PY79 (see Fig. S3 in the supplemental material), presumably due to additional mutations that distinguish the two strains (23, 25).
DISCUSSION
The Rap-Phr system is prevalent in the B. subtilis and Bacillus cereus groups of Bacillus species. All characterized Rap-Phr pairs form quorum-sensing systems, with the Phr peptide inhibiting the function of its cognate Rap. In this regard, RapP represents a unique case in which the association between Rap and Phr does not occur. Here, we show that this unique behavior is due to an atypical substitution in a highly conserved residue within the TPR domain of RapP. The full conservation of the asparagine residue may suggest that the atypical mutation arose during the early stages of domestication of the strain. Since strain 3610 is the earliest known ancestor of a domesticated strain such as B. subtilis 168, it is impossible to validate the hypothesis.
The typical organization of the Rap-Phr pairs implies that the effect of Rap proteins is most pronounced at low or intermediate cell densities. At higher cell densities, and especially under conditions where a sufficient amount of cells activate the Spo0A pathway, the effect of the Rap protein is suppressed by the Phr peptide. Indeed, deletion of Rap genes usually results in a modest effect on phenotype (16), especially during the beginning of the stationary phase (43, 44). The RapP3610 allele does not obey this rule since it is not counteracted by PhrP, and therefore its deletion has a strong effect on biofilm formation and ComA activity. We therefore propose that the biofilm phenotypes of the RapPT236N allele and of the plasmid-free strain are more typical than the biofilm phenotype of strain 3610.
Combining our genetic analysis with previous analysis of RapP (29) suggests that RapP acts on several targets. It was previously reported that RapP is involved in dephosphorylation of Spo0F (29). These biochemical results are in line with our finding that expression of RapP strongly affected sporulation efficiency (Table 3). On the other hand, our genetic analyses (Fig. 3 and 4) indicate that the control of ComA activity by RapP is partially independent of Spo0F. While it is tempting to speculate that RapP directly interacts with ComA, previous attempts to observe such interaction were unsuccessful (29). The identity of the second target of RapP, therefore, remains to be discovered.
We have also identified a positive regulatory effect of the QS master regulator, ComA, on the expression of the exopolysaccharide (eps) operon. This effect has been reported previously based on the analysis of microarray experiments (4). Using single-cell measurements, we find that ComA modulates the fraction of cells expressing the eps operon and not the level of expression. This indicates that ComA modulates the bistable positive feedback that governs the expression of biofilm-related genes (45). It was previously reported that ComA controls the Spo0A phosphorelay through the production of surfactin and the latter's effect on the phosphorelay kinase KinC (46). In addition, ComA has been shown to regulate biofilm formation through the activation of degQ (47). Strain PY79 does not produce surfactin and has a hypomorphic allele of the degQ promoter, implying that both of these pathways probably do not explain our results. Importantly, we find that ComA is a weak modulator of eps expression (the deletion leads only to a factor of 2 effect in the frequency of eps ON cells). We would therefore expect this effect to vary with environment and genotype, reconciling conflicting reports about the importance of this locus to biofilm formation (38, 48). It is important to note that RapP repression of the eps operon is probably mediated also by Spo0A.
Finally, many Rap-Phr systems have been shown to be encoded by mobile genetic elements (37), in a fashion similar to the RapP-PhrP system. In the mobile integrative and conjugative element ICEBs1, RapI-PhrI-mediated QS was shown to facilitate horizontal gene transfer and to dephosphorylate Spo0F. In addition, Rap60 encoded on the plasmid pTA1060 has been shown to regulate degradative enzyme production, possibly by dephosphorylation of Spo0F∼P (49). The sporadic occurence of plasmid pBS32 within the B. subtilis lineage implies that it is horizontally transferred, but the mechanism is unknown; therefore, it is yet unclear whether the RapP-PhrP system has an impact on the plasmid's horizontal transfer. The possible functions of Rap-Phr systems in mobile genetic elements and how they influence the spread and symbiosis of these elements with their host remain important open questions, the answers to which are likely to shed new light on the function of this broad and diverse family of QS systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel Kearns for strains and discussions and David Gutnick, Dave Dubnau, Matthew Neiditch, Gil Segal, Naama Geva-Zatorsky, and members of the Eldar lab for discussions.
This work was supported by funding from the Human Frontier Science Program grant CDA00015/2010-C (A.E.), Marie Curie International Reintegration Grant 268304 (S.O.B.), and European Research Council grant 281301 (A.E. and S.P).
We declare we have no conflicts of interest in the publication of this paper.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02382-14.
REFERENCES
- 1.Kleerebezem M, Quadri LE, Kuipers OP, De Vos WM. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol Microbiol 24:895–904. doi: 10.1046/j.1365-2958.1997.4251782.x. [DOI] [PubMed] [Google Scholar]
- 2.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 3.Lyon GJ, Novick RP. 2004. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides 25:1389–1403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 4.Comella N, Grossman AD. 2005. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol Microbiol 57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 5.Dunny GM. 2007. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc Lond B Biol Sci 362:1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazazzera BA, Solomon JM, Grossman AD. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in B. subtilis. Cell 89:917–925. doi: 10.1016/S0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 7.Li Y-H, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol 184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perego M, Hoch JA. 1996. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci U S A 93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lazazzera BA, Grossman AD. 1998. The ins and outs of peptide signaling. Trends Microbiol 6:288–294. doi: 10.1016/S0966-842X(98)01313-4. [DOI] [PubMed] [Google Scholar]
- 10.Perego M. 2013. Forty Years in the making: understanding the molecular mechanism of peptide regulation in bacterial development. PLoS Biol 11:e1001516. doi: 10.1371/journal.pbio.1001516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parashar V, Jeffrey PD, Neiditch MB. 2013. Conformational change-induced repeat domain expansion regulates Rap phosphatase quorum-sensing signal receptors. PLoS Biol 11:e1001512. doi: 10.1371/journal.pbio.1001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallego del Sol F, Marina A. 2013. Structural basis of Rap phosphatase inhibition by Phr peptides. PLoS Biol 11:e1001511. doi: 10.1371/journal.pbio.1001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molle V, Fujita M, Jensen ST, Eichenberger P, González-Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol Microbiol 50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- 15.McQuade RS, Comella N, Grossman AD. 2001. Control of a family of phosphatase regulatory genes (phr) by the alternate sigma factor sigma-H of Bacillus subtilis. J Bacteriol 183:4905–4909. doi: 10.1128/JB.183.16.4905-4909.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auchtung JM, Lee CA, Grossman AD. 2006. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol 188:5273–5285. doi: 10.1128/JB.00300-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura M, Fujita Y. 2007. Bacillus subtilis rapD, a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiol Lett 268:73–80. doi: 10.1111/j.1574-6968.2006.00559.x. [DOI] [PubMed] [Google Scholar]
- 18.Lazazzera BA, Kurtser IG, McQuade RS, Grossman AD. 1999. An autoregulatory circuit affecting peptide signaling in Bacillus subtilis. J Bacteriol 181:5193–5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogura M, Shimane K, Asai K, Ogasawara N, Tanaka T. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol Microbiol 49:1685–1697. doi: 10.1046/j.1365-2958.2003.03665.x. [DOI] [PubMed] [Google Scholar]
- 20.Murray EJ, Kiley TB, Stanley-Wall NR. 2009. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology 155:1–8. doi: 10.1099/mic.0.023903-0. [DOI] [PubMed] [Google Scholar]
- 21.Singh PK, Ramachandran G, Ramos-Ruiz R, Peiró-Pastor R, Abia D, Wu LJ, Meijer WJ. 2013. Mobility of the native Bacillus subtilis conjugative plasmid pLS20 is regulated by intercellular signaling. PLoS Genet 9:e1003892. doi: 10.1371/journal.pgen.1003892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeigler DR, Prágai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aguilar C, Vlamakis H, Losick R, Kolter R. 2007. Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol 10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konkol MA, Blair KM, Kearns DB. 2013. Plasmid-encoded ComI inhibits competence in the ancestral strain of Bacillus subtilis. J Bacteriol 195:4085–4093. doi: 10.1128/JB.00696-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durrett R, Miras M, Mirouze N, Narechania A, Mandic-Mulec I, Dubnau D. 2013. Genome sequence of the Bacillus subtilis biofilm-forming transformable strain PS216. Genome Announc 1(3):e00288–13. doi: 10.1128/genomeA.00288-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka T. 2010. Functional analysis of the stability determinant AlfB of pBET131, a miniplasmid derivative of Bacillus subtilis (natto) plasmid pLS32. J Bacteriol 192:1221–1230. doi: 10.1128/JB.01312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parashar V, Konkol MA, Kearns DB, Neiditch MB. 2013. A plasmid-encoded phosphatase regulates Bacillus subtilis biofilm architecture, sporulation, and genetic competence. J Bacteriol 195:2437–2448. doi: 10.1128/JB.02030-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu F, Kearns DB, McLoon A, Chai Y, Kolter R, Losick R. 2008. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol Microbiol 68:1117–1127. doi: 10.1111/j.1365-2958.2008.06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP, Perego M. 2007. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol 65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- 32.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265. doi:. [DOI] [PubMed] [Google Scholar]
- 33.Britton RA, Eichenberger P, Gonzalez-Pastor JE, Fawcett P, Monson R, Losick R, Grossman AD. 2002. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol 184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eldar A, Chary VK, Xenopoulos P, Fontes ME, Losón OC, Dworkin J, Piggot PJ, Elowitz MB. 2009. Partial penetrance facilitates developmental evolution in bacteria. Nature 460:510–514. doi: 10.1038/nature08150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harwood CR, Cutting SM (ed). 1990. Molecular biological methods for Bacillus. Wiley, New York, NY. [Google Scholar]
- 36.Yasbin RE, Young FE. 1974. Transduction in Bacillus subtilis by bacteriophage SPP1. J Virol 14:1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A 102:12554–12559. doi: 10.1073/pnas.0505835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.López D, Vlamakis H, Losick R, Kolter R. 2009. Paracrine signaling in a bacterium. Genes Dev 23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dworkin J, Losick R. 2001. Differential gene expression governed by chromosomal spatial asymmetry. Cell 107:339–346. doi: 10.1016/S0092-8674(01)00528-1. [DOI] [PubMed] [Google Scholar]
- 40.Frandsen N, Barák I, Karmazyn-Campelli C, Stragier P. 1999. Transient gene asymmetry during sporulation and establishment of cell specificity in Bacillus subtilis. Genes Dev 13:394–399. doi: 10.1101/gad.13.4.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn J, Dubnau D. 1991. Growth stage signal transduction and the requirements for srfA induction in development of competence. J Bacteriol 173:7275–7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 43.Veening JW, Hamoen LW, Kuipers OP. 2005. Phosphatases modulate the bistable sporulation gene expression pattern in Bacillus subtilis. Mol Microbiol 56:1481–1494. doi: 10.1111/j.1365-2958.2005.04659.x. [DOI] [PubMed] [Google Scholar]
- 44.Serra CR, Earl AM, Barbosa TM, Kolter R, Henriques AO. 15 September 2014. Sporulation during growth in a gut isolate of Bacillus subtilis. J Bacteriol doi: 10.1128/JB.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chai Y, Norman T, Kolter R, Losick R. 2010. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev 24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.López D, Fischbach MA, Chu F, Losick R, Kolter R. 2009. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106:280–285. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanley NR, Lazazzera BA. 2005. Defining the genetic differences between wild and domestic strains of Bacillus subtilis that affect poly-γ-dl-glutamic acid production and biofilm formation. Mol Microbiol 57:1143–1158. doi: 10.1111/j.1365-2958.2005.04746.x. [DOI] [PubMed] [Google Scholar]
- 48.Hamon MA, Lazazzera BA. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol Microbiol 42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- 49.Koetje EJ, Hajdo-Milasinovic A, Kiewiet R, Bron S, Tjalsma H. 2003. A plasmid-borne Rap-Phr system of Bacillus subtilis can mediate cell-density controlled production of extracellular proteases. Microbiology 149:19–28. doi: 10.1099/mic.0.25737-0. [DOI] [PubMed] [Google Scholar]
- 50.Silvaggi JM, Perkins JB, Losick R. 2006. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J Bacteriol 188:532–541. doi: 10.1128/JB.188.2.532-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.