Abstract
Phospholipase A2 enzymes are ubiquitously distributed throughout the prokaryotic and eukaryotic kingdoms and are utilized in a wide array of cellular processes and physiological and immunological responses. Several patatin-like phospholipase homologs of ExoU from Pseudomonas aeruginosa were selected on the premise that ubiquitin activation of this class of bacterial enzymes was a conserved process. We found that ubiquitin activated all phospholipases tested in both in vitro and in vivo assays via a conserved serine-aspartate catalytic dyad. Ubiquitin chains versus monomeric ubiquitin were superior in inducing catalysis, and ubiquitin-like proteins failed to activate phospholipase activity. Toxicity studies in a prokaryotic dual-expression system grouped the enzymes into high- and low-toxicity classes. Toxicity measured in eukaryotic cells also suggested a two-tiered classification but was not predictive of the severity of cellular damage, suggesting that each enzyme may correspond to unique properties perhaps based on its specific biological function. Additional studies on lipid binding preference suggest that some enzymes in this family may be differentially sensitive to phosphatidyl-4,5-bisphosphate in terms of catalytic activation enhancement and binding affinity. Further analysis of the function and amino acid sequences of this enzyme family may lead to a useful approach to formulating a unifying model of how these phospholipases behave after delivery into the cytoplasmic compartment.
INTRODUCTION
Phospholipase A2 (PLA2) enzymes represent a large class of proteins found throughout all phylogenetic kingdoms. They exert their activity on a number of phospholipid substrates, catalyzing the hydrolysis of the sn-2 ester bond to alter membrane structure and release a number of important biological mediators and precursor substrates (1). These enzymes are found in both intra- and extracellular environments. They are generally classified into six groups consisting of secreted forms (sPLA2), cytosolic forms (cPLA2), calcium-independent enzymes (iPLA2), platelet-activating factor acetylhydrolases (PAF-AH), lysosomal PLA2, and adipose-specific PLA2 (reviewed in references 2 and 3). Secreted PLA2 enzymes are typically ∼15 kDa and contain a histidine-asparatate catalytic dyad (4). In the presence of a calcium ion cofactor, they conduct lipid hydrolysis through interfacial activation, a process involving a substantial increase in catalytic activity upon binding to a phospholipid membrane surface. Previous studies suggest that this outcome mechanistically involves both the physical properties of membrane substrates and conformational changes within the PLA2 (5, 6). Many PLA2s display a binding preference for anionic membrane surfaces to promote activities including destruction of bacterial membranes in host defense (7), promotion of atherogenesis (reviewed in reference 8), processing of dietary phospholipids (reviewed in reference 9), recycling of apoptotic cells (10), potent myotoxicity after envenomation via snake bites and bee stings (reviewed in reference 11), sperm maturation, and skin homeostasis (reviewed in reference 12).
Cytosolic PLA2 enzymes differ from sPLA2 enzymes in both structure and function (reviewed in reference 13). Structurally, cPLA2 enzymes are much larger (∼85 kDa) and contain a catalytic domain linked to a smaller C2 domain. The catalytic domain is generally composed of a canonical α/β hydrolase fold with an active-site catalytic serine-aspartate catalytic dyad, along with a conserved arginine. Several phosphoserine residues are also required for maximal activity (14; reviewed in reference 15). Structural studies of the cPLA2 catalytic domain revealed that its dyad rests amid a funnel of hydrophobic residues covered by a lid, which is displaced upon substrate binding, leading to interfacial activation of the enzyme (16). The C2 domain contains a series of membrane-binding loops requiring calcium ions to neutralize their net anionic charges, allowing productive membrane binding. Further mechanisms for either electrostatic or hydrophobic membrane binding interactions have been described, with important implications for substrate specificity and biological function (reviewed in reference 17). Several mouse cPLAα knockout studies have shown that this enzyme is important in a number of physiological processes due to the release of arachidonic acid upon lipid hydrolysis (reviewed in reference 17). Released molecules are subsequently oxidized into one of a variety of eicosanoids, which generally have effects on inflammatory responses but also serve as regulators of tumorigenesis, atherosclerosis, thrombosis, and rhinitis (reviewed in references 17 and 18).
Phospholipases with homology to potato tuber patatin consitute another large family of enzymes (19). Patatin domains are prevalent among both prokaryotes and eukaryotes. Structural and bioinformatic studies show that this class of enzymes most closely resembles the intracellular iPLA2 (20). Patatin domains similarly harbor α/β hydrolase folds and serine-aspartate dyads, with the serine located in a conserved Gly-X-Ser-X-Gly hydrolase motif. Another motif, Gly-Gly-X-Arg (or Gly-Gly-X-Lys in the case of patatin B2), is usually present, and it is thought to function in stabilizing the transition state during the cleavage reaction of the sn-2 ester bond (16, 19, 21). Patatin differs from phospholipases such as cPLA2 in that the former is not subject to interfacial activation and does not contain a lid-like structure partially occluding access of the active site to substrate (16, 19, 22). Cytosolic PLA2s generally show increased substrate specificity for arachidonic acid-containing lipids, while patatin is considered more promiscuous in substrate preference (1, 22).
A growing number of reports demonstrate that bacterial pathogens, particularly Gram-negative species, use patatin-like PLA2 enzymes as effector molecules to target host cellular membranes (23–27). Previous work has shown that exoenzyme U (ExoU), a patatin-like phospholipase encoded by the opportunistic pathogen Pseudomonas aeruginosa, is injected by a type III secretion system (T3SS) into host cells as an effector (27–29). ExoU is potently activated by a proteinaceous cofactor present in mammalian and yeast cells that was recently identified as ubiquitin (28, 30). We reasoned that ExoU has evolved to utilize ubiquitin as its activation partner due in part to the ubiquitous presence of this protein in all eukaryotic cells and, more importantly, its absence from prokaryotic cells. This hypothesis is supported by previous work demonstrating that coexpression of ExoU with monoubiquitin in Escherichia coli induced rapid bacterial cell death (30).
The combination of a membrane-destructive hydrolase activated by a highly conserved, eukaryotic-specific protein may be widespread in T3SS+ Gram-negative bacteria considering that bioinformatic analyses revealed several close orthologs to ExoU (23–27). We queried three additional enzymes from bacterial species representing different ecological niches and pathogenic potentials to determine if ubiquitin activation was a common property of these proteins (24, 27, 31, 32). Functional characterization of each enzyme's enzymatic properties and substrate specificity in comparison to those of ExoU from P. aeruginosa should shed light on a biological role that each enzyme may play for the bacterium expressing it. Additionally, comparative information can be obtained from amino acid sequence alignments relative to the observed activity or activation potential. A universal description of the mechanism of activation of this family of phospholipases may emerge, which will ultimately be critical for the rational development of specific inhibitors or cell-targeting therapeutics.
MATERIALS AND METHODS
Reagents.
The antibodies used for detection by Western blotting were as follows: mouse antiubiquitin (Santa Cruz; sc-271289), mouse anti-His (GE Healthcare; 27-4701-01), anti-green fluorescent protein (anti-GFP) (Covance; MMS-118R), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Santa Cruz; SC-32233), anti-DnaK (Enzo; 8E2/2), and goat anti-mouse antibody–horseradish peroxidase (HRP) (Invitrogen; F-21453). All lipids purchased were from Avanti Polar Lipids, Inc. Recombinant monoubiquitin (U-100H), K63-linked diubiquitin (UC-300), NEDD8 (UL-812), SUMO-1 (UL-712), ISG15 (UL-601), and FAT10 (UL-900) were purchased from Boston Biochem, Inc.
Enzyme purification.
Pseudomonas aeruginosa ExoU (PAU), Burkholderia thailandensis ExoU (BTU), and Pseudomonas fluorescens ExoU (PFU) were expressed as hexahistidine-tagged fusion proteins from pET15b in Escherichia coli BL21(DE3) pLysS. Cultures were grown in Luria-Bertani (LB) broth with 30 μg/ml of chloramphenicol and 100 μg/ml of ampicillin to an optical density at 600 nm (OD600) of 0.5 at 37°C and induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h at 30°C. Cells were harvested and lysed by passage through a French pressure cell, and recombinant proteins were purified by cobalt metal affinity chromatography (Clontech) as described previously (30). Elution fractions were pooled and concentrated in 30-kDa molecular-mass-cutoff centrifugal concentrators (Millipore) before application to a Superose 6 size exclusion column (GE Healthcare) equilibrated in 10 mM Tris (pH 7.0), 150 mM NaCl, and 20% glycerol with an ÄKTA fast-performance liquid chromatography (FPLC) system (GE Healthcare). Peak fractions were concentrated and flash frozen in a dry ice-ethanol bath for storage at −80°C. The ExoU homolog encoded by the Photorhabdus asymbiotica genome was amplified from a pET15b vector to include the pET15b ribosome-binding site and hexahistidine tag and ligated into pJN105 as a SpeI-SacI fragment after removal of the endogenous SpeI sites via site-specific mutation (Change-IT; Affymetrix). This plasmid was introduced into strain BL21(DE3) pG-KJE8 and grown in LB broth with 30 μg/ml of chloramphenicol, 10 μg/ml of gentamicin, and 10 ng/ml of tetracycline to an OD600 of 0.5 at 37°C before a 2-h, 30°C induction with 0.5% arabinose. Purification of the P. asymbiotica enzyme (PYU) was identical to the above-described procedure except that the cells were lysed in buffer containing 6 M urea. Purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analyses, and protein concentrations were determined by A280 in a quartz cuvette at their respective extinction coefficients.
In vitro activity assay.
In vitro activity was assessed using the phospholipid mimetic N-((6-(2,4-dinitrophenyl)amino)hexanoyl)-2-(4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-pentanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (PED6) as described by Anderson et al. (30). Briefly, each well in a 96-well black-bottom plate (Costar) contained a 50-μl solution of 50 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.3), 750 mM monosodium glutamate (pH 6.3), 10 nM enzyme, 100 μM PED6, and various amounts of either monoubiquitin or K63-linked diubiquitin. Initial rates were calculated from the raw data acquired at 1-min intervals over 30 min using excitation and emission wavelengths of 488 and 515 nm, respectively, with a cutoff value of 495 nm (SpectraMax M5 plate reader; Molecular Devices). To evaluate the enhancement of PLA2 activity by phosphatidyl-4,5-bisphosphate (PIP2), either PIP2 or palmitoyl-oleoyl phosphatidylserine (POPS) was reconstituted to 1 mM in phosphate-buffered saline (PBS) and used in assays conducted with 10 μM enzyme, 30 μM PED6 substrate, and 5 μM monoubiquitin.
Dual-expression bacterial assays.
Each respective E. coli strain was cultivated with antibiotics formulated to select for plasmid retention (30 μg/ml of chloramphenicol [pJY2], 10 μg/ml of gentamicin [pJN105], and 30 μg/ml of kanamycin [pCOLA-Duet]) on LB agar containing 0.5% glucose before each experiment. All toxin genes were amplified from a pET15b vector to include the pET15b ribosome-binding site and hexahistidine tag for ligation into the pJN105 plasmid. Isolated colonies were scraped from growth plates and suspended into LB broth (OD600 = 2.0) with antibiotics but lacking glucose and grown statically for 40 min at 30°C. Each culture was diluted to an OD600 of 0.25 and either serially diluted for the spot plate assay or grown in LB broth (200 rpm and 30°C) with either glucose (0.5%) or inducing reagents (0.5 mM IPTG and 0.5% arabinose) for the desired periods. Samples from specific time points were plated onto LB agar with glucose and antibiotics for the enumeration of surviving cells. SDS-PAGE and Western blotting were conducted directly on 15 μl of lysates at the desired time point from a starting inoculation of 1 × 108 CFU/ml with anti-His (1:8,000 dilution), antiubiquitin (1:10,000 dilution), or anti-DnaK (1:10,000 dilution) antibodies as probes for detection with HRP-conjugated anti-mouse IgG (1:10,000 dilution). SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) was used for signal detection.
Detection of ExoU and homolog proteins expressed in transfected HeLa cells by Western blotting.
HeLa cells (CCL-2 from ATCC) were seeded at a density of 1 × 105 in a 35-mm culture dish and grown overnight in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. Cells were washed with Hanks balanced salt solution (HBSS) 3 times and transfected with the FuGENE HD transfection reagent (Promega) mixed with 1.6 μg of DNA at the ratio of 2.2:1 according to the manufacturer's instructions. The enhanced GFP (eGFP) control plasmid was transfected using 400 ng of DNA at a 0.5:1 ratio of DNA to FuGENE (the total amount of FuGENE was identical in all experiments). Transfected cells were stained with propidium iodide (50 ng/ml; Sigma) for 5 min at 37°C. Images were acquired and processed under the same conditions for all samples, except that plasmid-eGFP and PFU-S96A-eGFP images were acquired with half the exposure time and processed at 50% of the intensity of the other samples due to increased relative signal intensity. Plasmid PFU-eGFP was processed at 50% of the intensity of the other samples as well. Fluorescence microscopy images were acquired by a Nikon Eclipse Ti-U inverted microscope equipped with a CoolSNAP ES2 charge-coupled-device (CCD) camera (Photometrics) and a multifluorescent Sedat Quad ET filter set (multichroic splitter; Chroma). NIS-Elements software (Nikon) was used for image acquisition, processing, and analysis. The objective lens used was 10× Plan Fluor (numerical aperture [NA], 0.3) from Nikon. In each experiment, 3 fields of images were acquired per sample and quantified. The data represent the means and standard deviations (SD) of values from 3 independent experiments.
Western blot detection of homolog-eGFP proteins was conducted using cells from 60-mm plates seeded with 7.2 × 105 cells. Cells were transfected with 400 μg of plasmid DNA at a 1:2.5 DNA/FuGENE ratio. After 24 h of incubation, cells were scraped in 900 μl of sucrose buffer (250 mM sucrose, 3 mM imidazole, and cOmplete mini-protease inhibitors [EDTA free; Roche] in PBS), transferred to a microcentrifuge tube, and collected by centrifugation at 5,000 × g and 4°C for 15 min. Floating cells in the culture medium were also collected by centrifugation at 680 × g for 10 min and combined with the harvested cells. Cell pellets were combined with 50 μl of 2× lysis buffer (6 mM imidazole, 2× cOmplete mini-protease inhibitors [EDTA free], 100 μg/ml of DNase, and 100 μg/ml of RNase in PBS), and 50 μl of 5× protein sample buffer was added. After 15 min of incubation at room temperature, samples were boiled for 5 min prior to analysis by SDS-PAGE and Western blot analysis. GFP-tagged proteins were probed with an anti-GFP monoclonal antibody (1:5,000 dilution) followed by an HRP-conjugated anti-mouse IgG (1:10,000 dilution). As a loading control, GAPDH was probed with an anti-GAPDH monoclonal antibody (1:2,000 dilution) and an HRP-conjugated anti-mouse IgG.
LDH release assay to measure cytotoxicity.
HeLa cells (6.4 × 104) were seeded in a 24-well tissue culture plate, grown overnight, and transfected using the FuGENE HD transfection reagent mixed with 400 ng of DNA at a 2.2:1 reagent-to-DNA ratio. After 24 h of incubation, aliquots of the tissue culture medium were subjected to centrifugation to exclude floating cells and debris. The supernatant (50 μl) was subjected to lactate dehydrogenase (LDH) activity measurements using the CytoTox 96 non-radioactive cytotoxicity assay (Promega) according to the manufacturer's instructions. Lactate dehydrogenase activity was detected by using a SpectraMax M5 microplate reader. Assays were performed at least 3 times as independent experiments; error bars in figures indicate SD from the means.
Liposome binding assays.
Liposomes were produced by subjecting the component mixtures of organically solubilized lipids to a nitrogen stream and subsequent overnight desiccation at room temperature. Dried lipid films were suspended in a 30% sucrose solution and subjected to multiple freeze-thaw cycles before extrusion through a 200-nm filter to generate uniform, unilamellar vesicles. These liposomes were then pelleted at 100,000 × g and 4°C before suspension in 10 mM 3-(N-morpholino)propanesulfonic acid (MOPS; pH 7.0) and 150 mM NaCl buffer (buffer A) to a final lipid concentration of 40 mM. Various concentrations of each liposome preparation (0 to 12 mM) were combined with 1 μM enzyme in a 100-μl solution of buffer A and incubated for 1 h at 4°C with constant rotation. The mixture was then subjected to centrifugation for 1 h at 4°C and 100,000 × g to pellet liposomes and bound enzyme. Immediately upon conclusion of the centrifugation, 25 μl of the supernatant was withdrawn and utilized for SDS-PAGE analysis. Densitometric analysis of enzyme remaining in the supernatant was performed after staining the gels with Coomassie brilliant blue and quantifying the loss of signal intensity using ImageJ 1.440 software (http://imagej.nih.gov/ij).
RESULTS
Bioinformatic interrogation of ExoU orthologs.
A previous report indicated that enzymes with patatin-like domains annotated in the current database of sequenced bacterial genomes total at least 4,400 proteins (33). To determine if enzymes similar to ExoU also utilize ubiquitin as a cofactor, we performed an initial bioinformatic survey. To reduce the number of candidate proteins, we narrowed the list to Gram-negative bacteria with T3S, putative T3S, or T4S systems and amino acid lengths exceeding 500 residues. Additional sequences beyond the catalytic domain are important for activation of ExoU and thus may also perform the same function among its orthologs (34–36). Altogether, our analysis yielded 17 proteins that may have phospholipase activity activated by ubiquitin (Table 1). Each protein contains a Gly-X-Ser-X-Gly hydrolase motif, a Gly-Gly-X-K/R glycine-rich motif, and an Asp-Gly-Gly motif likely to contain the catalytic aspartic acid residue. This group of enzymes was largely represented by species of Pseudomonas and Rickettsia (Table 1). Additional candidates were encoded in the genomes of facultative intracellular organisms such as Legionella, Burkholderia thailandensis, and certain strains of Photorhabdus asymbiotica (23, 31, 37). Aeromonas diversa and Vibrio vulnificus also contain orthologs to ExoU and differ from the other strains in that they are capable of colonizing anaerobic environments such as the gastrointestinal tract (38, 39). Analysis of the predicted isoelectric points of these proteins reveals a span of values, ranging from acidic in the V. vulnificus enzyme (pI = 4.5) to basic for the P. extremeaustralis enzyme (pI = 10.0). The average percent identity to ExoU from P. aeruginosa is 30%, ranging from the most identity observed for the P. asymbiotica enzyme (53%) to the least identity observed for the V. vulnificus protein (19%). The average size of the enzymes is slightly smaller than that of ExoU (687 amino acid residues), at 636 residues.
TABLE 1.
T3S or T4S orthologs of P. aeruginosa ExoU
| Organism | Lifestyle | No. of residues | % identity | pIa | Accession no. | Reference |
|---|---|---|---|---|---|---|
| Pseudomonas aeruginosa | Aerobic | 687 | 5.6 | YP_792285 | 27 | |
| Achromobacter arsenitoxydans | Aerobic | 591a | 23 | 5.7 | WP_008163363 | 58 |
| Aeromonas diversa | Facultative anaerobic | 560 | 42 | 5.4 | WP_005356718 | 38 |
| Burkholderia thailandensis | Facultative intracellular | 670 | 35 | 9.1 | WP_006028569 | 31 |
| Legionella longbeachae | Facultative intracellular | 652 | 21 | 6.3 | YP_003456657 | 59 |
| Legionella pneumophila subsp. pneumophila | Facultative intracellular | 621 | 21 | 5.8 | YP_005187154 | 23 |
| Legionella pneumophila subsp. pneumophila strain Philadelphia 1 | Facultative intracellular | 665a | 20 | 6.5 | AAU28471 | 60 |
| Photorhabdus asymbiotica | Facultative intracellular | 676 | 53 | 8.7 | YP_003039880 | 24 |
| Pseudomonas extremaustralis | Aerobic | 639 | 42 | 10.0 | ZP_10438440 | 61 |
| Pseudomonas fluorescens | Aerobic | 643 | 41 | 9.0 | YP_002875210 | 32 |
| Pseudomonas plecoglossicida | Aerobic | 613 | 40 | 4.9 | EPB94356 | 62 |
| Pseudomonas syringae | Aerobic | 629 | 43 | 5.0 | ACU65062 | 63 |
| Rickettsia bellii | Obligate intracellular | 587 | 21 | 5.6 | YP_538364 | 64 |
| Rickettsia massiliae | Obligate intracellular | 598 | 20 | 7.9 | YP_005302159b | 26 |
| Rickettsia montanensis | Obligate intracellular | 611 | 20 | 9.1 | YP_005391134b | 26 |
| Rickettsia prowazekii | Obligate intracellular | 598 | 20 | 9.0 | YP_00599896b | 25 |
| Vibrio vulnificus | Facultative anaerobic | 779 | 19 | 4.5 | AAO10069b | 39 |
Theoretical calculation based on amino acid sequence (http://workbench.sdsc.edu).
Currently annotated as a hypothetical protein or putative esterase.
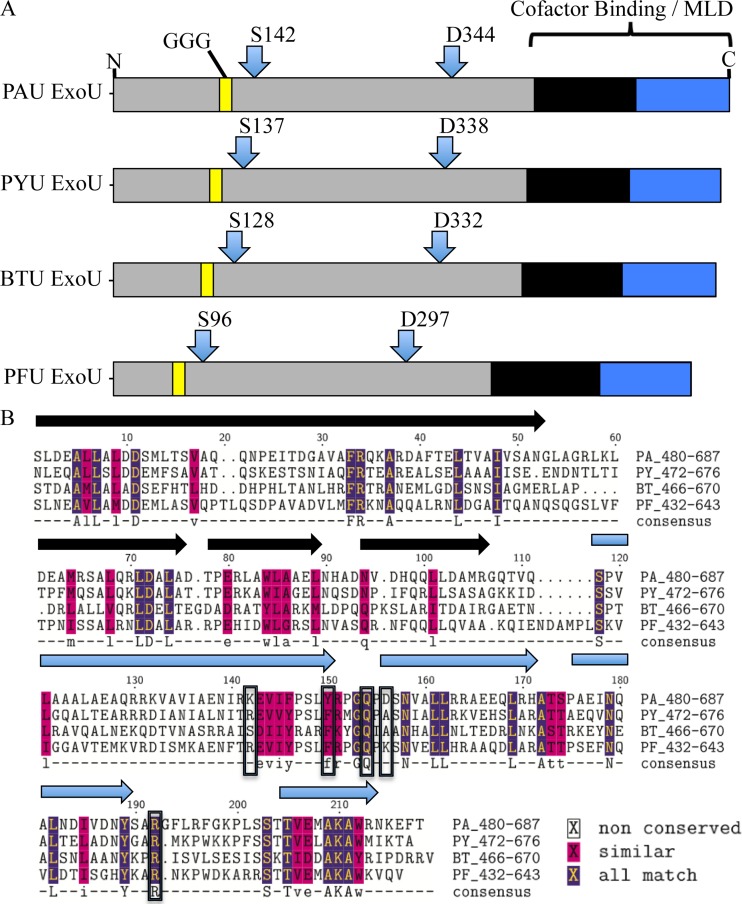
To begin to determine whether patatin orthologs of ExoU also require a ubiquitin cofactor, three enzymes from bacteria with differing ecological niches (see Discussion) were chosen from Table 1 for further analysis. As with ExoU, the potential requirement of ubiquitin may suggest that the functional activity and/or biological role depends on delivery into a eukaryotic cellular environment. A representative schematic depicting each selected protein at comparative scale is shown in Fig. 1A. The proteins are designated PAU, PYU, BTU, and PFU for enzymes encoded by P. aeruginosa, P. asymbiotica, B. thailandensis, and P. fluorescens, respectively. The proteins appear to retain similar domain structures, with the approximate locations of the catalytic residues and glycine-rich motifs conserved. The BTU and PYU orthologs are similar in total number of amino acids to ExoU, while the PFU enzyme, at 643 residues, is closer to the average enzyme listed in Table 1. All of the selected proteins carry a fairly basic isoelectric point (pI ∼ 9.0), in contrast to ExoU's predicted pI of 5.6.
FIG 1.
Comparison of three patatin-like homologs to ExoU from P. aeruginosa. (A) Diagram of the relative size and location of important residues in each of the four enzymes. GGG, glycine-rich motif postulated to participate in an oxyanion hole; S and D, catalytic residues serine and aspartate, which form a dyad; blue and black domains, regions characterized in ExoU to possess membrane and/or cofactor interaction activity. (B) Sequence alignment of the C-terminal domains corresponding to the black and blue C-terminal domains in panel A. The alpha-helical secondary structure of ExoU is shown as either black or blue arrows above the alignment for reference. In the alignment, sequence identity or similarity is shown as purple or pink highlights, respectively. Boxed amino acids were shown in previous studies to be important for ExoU activity when monoubiquitin serves as a cofactor. A consensus sequence is shown as calculated by the CLUSTALW algorithm.
ExoU contains a two-domain region downstream of the catalytic domain which has been shown in other studies to be important for both membrane and ubiquitin interactions (23, 34, 36, 40–42) (Fig. 1A). To determine if a common motif might be associated with ubiquitin binding or activation, a CLUSTALW alignment of this region was performed (43) (Fig. 1B). In previous studies, structural homology between ubiquitin binding proteins and ExoU was used to target structurally analogous residues for their functional contribution to phospholipase activity and ubiquitin interaction. Residues previously shown to significantly affect activity when monoubiquitin is used as a cofactor (36) are marked within black boxes in Fig. 1B. Overall, there are several conserved regions with high similarity or identity to residues that have been shown to be important for ExoU activity throughout the alignment. Notable clusters of identity are coincident with secondary structural predictions of helices, suggesting that an overall maintenance of structure is important. Significant conservation around the C-terminal helix is observed. The presence of this sequence has been previously shown to be vital for ExoU toxicity and ubiquitylation in host cells (44). This analysis was inconclusive toward predicting whether ubiquitin serves as a cofactor to activate BTU, PYU, or PFU. The lack of an identifiable motif, the large region involved (2 domains, according to the ExoU structure [41, 42]), the conservation of structure, and studies previously performed in our laboratory are, however, consistent with the hypothesis that a collective functional surface rather than the interaction with specific residues may be important for activation by ubiquitin (36).
Kinetic analysis of ExoU relative to the homologs.
To definitively test for both phospholipase activity and activation by ubiquitin, purified enzymes were analyzed for the ability to cleave a phospholipid mimetic molecule (PED6) in the absence and presence of either monoubiquitin or diubiquitin (45). The diubiquitin used in these assays is linked via lysine 63 to the distal ubiquitin's C-terminal glycine. Multiple preparations of purified proteins demonstrated the ability to cleave PED6 only in the presence of an ubiquitin cofactor (data not shown).
Initial optimization experiments revealed that the standard pH (6.3) and salt (750 mM monosodium glutamate) conditions of the assay resulted in the highest levels of activity for each enzyme (data not shown), thus allowing a more direct comparison of catalytic constants. The PYU enzyme was roughly 2- to 3-fold more efficiently activated (Kact) by monoubiquitin than both the PAU and BTU enzymes and 16-fold more efficiently activated than PFU proteins (Table 2). As activation of phospholipase activity for all the enzymes is dependent upon ubiquitin in a highly purified system, the Kact, though not a quantitative measure of binding, can serve as a binding surrogate. Additionally, specific interaction of each purified protein with ubiquitin was demonstrated by biolayer interferometry (see Fig. S1 in the supplemental material). PAU catalysis of the substrate (kcat) was about ∼3-fold faster than that of PFU and 5- to 10-fold faster than those of BTU and PYU under saturating levels of activator and substrate. All together, the results showed that the PAU enzyme appeared approximately 3-fold more catalytically efficient (kcat/Kact) over the next best enzyme, PYU, and nearly 20-fold better than the least productive enzyme, PFU, when monoubiquitin was used as the activator.
TABLE 2.
In vitro kinetic data for enzyme catalysis of PED6 substrate via ubiquitin activation
| Enzymea | Monoubiquitin activation |
K63-linked Ub2 activation |
||||
|---|---|---|---|---|---|---|
| Kact (M)b | kcat (s−1)c | kcat/Kact (M−1 s−1) | Kact (M) | kcat (s−1) | kcat/Kact (M−1 s−1) | |
| PAU | (2.6 ± 0.85) × 10−5 | 4.7 ± 0.46 | 1.8 × 105 | (3.9 ± 0.98) × 10−8 | 3.7 ± 0.18 | 9.5 × 107 |
| PYU | (9.2 ± 5.60) × 10−6 | 0.5 ± 0.03 | 5.4 × 104 | (4.8 ± 0.17) × 10−8 | 0.5 ± 0.03 | 1.0 × 107 |
| BTU | (2.1 ± 0.55) × 10−5 | 0.9 ± 0.06 | 4.2 × 104 | (4.6 ± 2.82) × 10−7 | 0.5 ± 0.08 | 1.1 × 106 |
| PFU | (1.5 ± 0.37) × 10−4 | 1.5 ± 0.16 | 1.0 × 104 | (5.2 ± 0.19) × 10−7 | 2.8 ± 0.28 | 4.8 × 106 |
No enzyme activity was detectable in the absence of ubiquitin.
Activation constant, or the concentration of ubiquitin activator required to reach half-maximal enzyme initial rate of PED6 cleavage, obtained from a one-site binding model nonlinear regression analysis of a theoretical curve generated using GraphPad Prism 5.0 (GraphPad Software Inc.) software (n = 3).
Catalytic constants obtained from nonlinear regression analysis of a theoretical curve generated by GraphPad Prism 5.0 using the conversion of 59,293 relative fluorescence units, equal to 1 nmol of PED6.
Previous studies have suggested that PAU possesses a higher affinity and is more efficiently activated by polyubiquitin chains than monomers (30, 36). We used K63-linked diubiquitin in these experiments as a model of polyubiquitin chains, as it has been shown that ExoU is modified by a K63-linked diubiquitin in host cells and because linear and K48-linked chains appear to activate ExoU to roughly similar levels under the conditions of our in vitro assay (30, 44). The use of diubiquitin as an activator resulted in a 100- to 1,000-fold increase in activation efficiency (Kact) for all of the enzymes compared to monoubiquitin-induced activation. PAU and PYU reached their activation constants at 40 to 50 nM diubiquitin, while BTU and PFU enzymes required the presence of roughly a 500 nM concentration of an activator. The trends in substrate turnover (kcat) mirrored those with monoubiquitin activation, indicating that cofactor saturation was achieved in both cases. In summary, the PAU enzyme was roughly 10-fold catalytically superior (kcat/Kact) to PYU in vitro following diubiquitin activation. PFU and BTU enzymes exhibited catalytic efficiencies (kcat/Kact) approximately 50- to 100-fold less than that of PAU and 5- to 10-fold less than that of PYU.
Ubiquitin-like proteins SUMO-1 (≤80 μM tested), FAT10 (≤6.6 μM tested), and ISG15 (≤13 μM tested) did not induce activation above controls without an activator under standard assay conditions (data not shown). NEDD8 (≤100 μM tested) appeared to induce cleavage above the baseline in all four homologs at high concentrations; however, this level of activation was at least 20- to 30-fold lower in stimulating phospholipase activity than that of monoubiquitin (data not shown). It is not surprising that small amounts of activity due to the presence a high concentration of NEDD8 were detected, as this molecule is nearly structurally identical to ubiquitin, with 58% sequence identity and 80% sequence similarity (46). Several studies aimed at quantifying intracellular levels of ubiquitin have suggested that nucleated cells contain somewhere between 8.0 × 107 and 1.5 × 108 ubiquitin molecules per cell (∼85 to 150 μM depending on the assumption of cellular volume) (47–49). Data on the intracellular concentration of NEDD8 are sparse; however, one report suggested that the total free amounts of this protein and ubiquitin were similar (46). Given that slightly more than half of the total cell ubiquitin pool is typically in a conjugated form, and each of the homologs displays more than a 100- to 1,000-fold decrease in the Kact for diubiquitin compared to monoubiquitin, we speculate that the most physiologically relevant activators of ExoU and its orthologs are likely polyubiquitin chains.
ExoU orthologs recognize bacterial membranes as substrates via a conserved catalytic dyad.
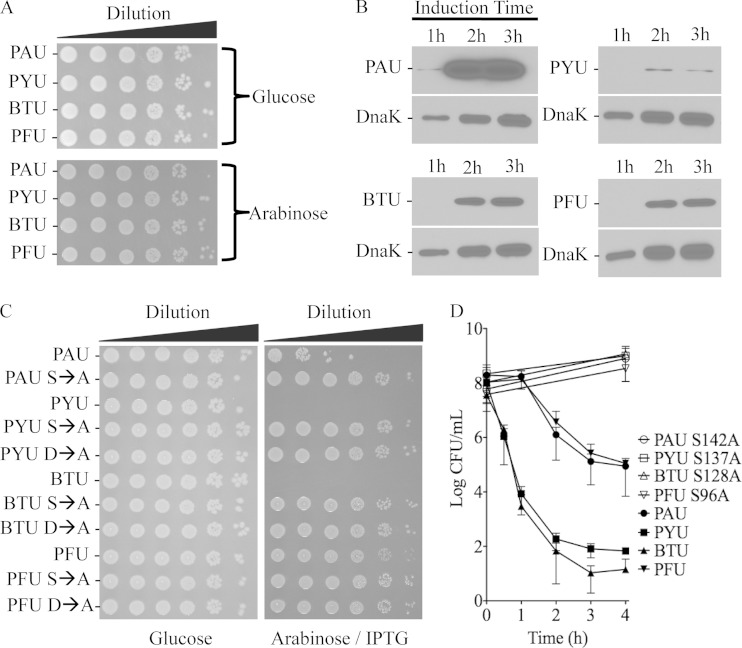
To test enzyme toxicity in a more physiological environment, we used a variation of the E. coli dual-expression model in which both enzyme and ubiquitin are expressed from separate inducible plasmids (30). This system allows for a simplified, semiquantitative, and controlled method for studying the outcomes of intoxication in living cells lacking the ubiquitylation/deubiquitylation machinery found in eukaryotes. Each enzyme was cloned with an N-terminal hexahistidine tag into the arabinose-inducible, gentamicin-selectable plasmid pJN105 (50). Ubiquitin was cloned as a nontagged construct into the kanamycin resistance vector, pCOLADuet, held under repression by the pJY2 plasmid. Initial experiments tested the toxicity of each enzyme alone in E. coli when induced. Cell death was not evident either in liquid cultures (data not shown) or on agar medium (Fig. 2A and C, glucose) when ubiquitin was totally absent from the system. Serial spot plate dilutions of each strain onto medium containing arabinose showed growth titers identical to that of the glucose control (Fig. 2A). Western blot analysis detecting the histidine tag for each enzyme over a 3-h time course revealed that PAU was much more highly expressed than the other toxins relative to the DnaK chaperone loading control. BTU and PFU enzymes had similar, more moderate expression profiles, while the PYU construct expressed the smallest amount of protein (Fig. 2B). Western blot detection of any protein was not achieved at any time point prior to 1 h postinduction (data not shown).
FIG 2.
Recombinant enzyme-ubiquitin coexpression is differentially toxic to E. coli. (A) Spot plates of 10-fold serially diluted strains containing plasmids pJY2 and pJN105 encoding His-tagged enzyme on agar medium containing either glucose (noninducing conditions) or l-arabinose (inducing conditions for production of the cloned enzyme). (B) Western blot of total bacterial lysates from the same strains as in panel A (lacking an expression construct containing ubiquitin) under inducing conditions. Total lysates were probed for the histidine tag of each enzyme after harvesting a constant volume at 1, 2, or 3 h of growth. The overall replication of bacteria was followed by blotting for DnaK, which increased at each time point. (C) Spot plates of 10-fold serially diluted bacteria containing an inducible monoubiquitin construct with an inducible construct expressing each parental enzyme or catalytic point mutant derivative spotted onto medium containing either glucose or l-arabinose. (D) Graph of cell viability of E. coli strains used in panel C when induced in liquid medium for the time indicated before plating on LB agar with glucose and antibiotics (n = 3 or 4).
Wild-type or single catalytic serine-to-alanine point mutant derivative enzymes were next expressed with monoubiquitin in the dual-expression system to test if these enzymes were active against the bacterial inner membrane and if the proposed catalytic residues were indeed necessary for substrate cleavage (Fig. 2C and D). We found that all enzymes were activated by monoubiquitin when bacterial membranes served as substrates; however, the degrees of toxicity appeared to differ between the proteins (Fig. 2C). Strains expressing PFU suffered about a log inhibition in growth under inducing conditions on agar medium compared to plates containing glucose as a negative control. The PAU enzyme was more intermediate in growth inhibition (∼3 logs) on solid medium, while PYU and BTU decreased apparent growth by at least 6 logs under dual inducing conditions (arabinose and IPTG). To confirm that this growth inhibition was cell death, we carried out cell viability time course assays for each strain. These data recapitulated the spot plate assays in that the PYU and BTU enzymes were extremely toxic to E. coli when induced with monoubiquitin (Fig. 2D). Cell death was apparent after 30 min of induction and continued until nearly the entire population of cells was nonviable at approximately 2 h postinduction. Attempts to detect either PYU or BTU protein in the presence of the ubiquitin-encoding plasmid by immunoblotting were unsuccessful even in the absence of induction, further supporting a high degree of toxicity (see Fig. S2 in the supplemental material). Strains producing PAU or PFU enzymes showed approximately equal levels of cell death, but the levels were more moderate than those of strains expressing monoubiquitin and BTU or PYU, respectively. Cell death mirrored the timing of Western blot detection, occurring after 1 h of induction and culminating in an approximately 3-log decrease in cell viability by 2 h of induction. Control serine-to-alanine catalytic point mutant derivatives showed a steady rate of cell growth throughout the assay, consistent with the abrogation of phospholipase activity (Fig. 2D). When considering the amount of enzyme expression and cell death, the PYU protein appears to be the most toxic enzyme of this group in targeting the bacterial membrane. The BTU enzyme could be considered the second most toxic, while the PFU enzyme may be more lethal than PAU in bacteria given the similar amount of cell death in liquid medium and PAU's higher relative expression level.
Relative toxicity of each enzyme in a transfection assay of HeLa cells.
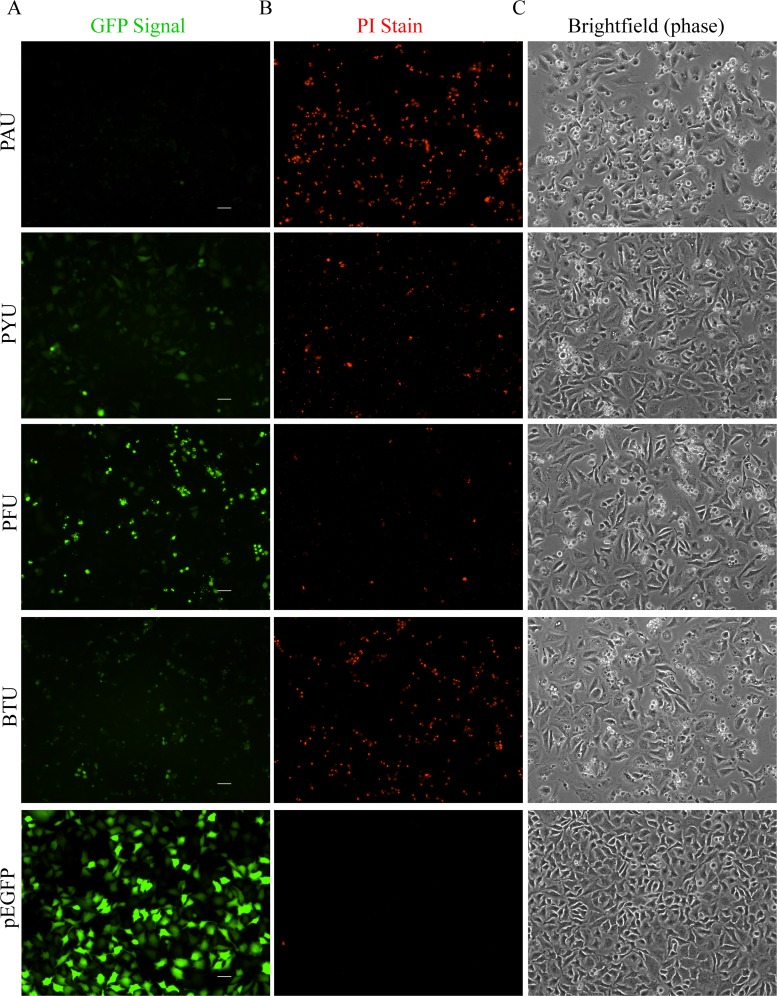
We next examined if the toxicity phenotype patterns observed in prokaryotes correlated with toxicity in a eukaryotic environment by transfecting HeLa cells with plasmids expressing each homolog from identically designed constructs. This initial assessment aimed to broadly observe the levels of enzyme expression, activity, and cellular response. To facilitate Western blotting and directly visualize the expressed enzymes, all clones were constructed as N-terminal fusions to enhanced GFP (eGFP); prior studies indicated that ExoU retains biologically relevant levels of toxicity and trafficking when fused to eGFP (35, 44, 51). Fluorescence microscopy 24 h after transfection showed no detectable eGFP signal in cells expressing the PAU enzyme, recapitulating earlier studies demonstrating the apparently high toxicity of ExoU in this environment (Fig. 3A) (51). Surprisingly, eGFP signal was visible from cells transfected with constructs encoding the PYU-eGFP and BTU-eGFP enzymes. The cells appeared to tolerate more expression of PFU-eGFP than of the other homologs, suggestive of a lower relative toxicity (Fig. 3A). Control cultures expressing only eGFP displayed a strong signal from a confluent monolayer. Compared to the parental version of the clones, the PAU catalytic null mutant was now detectable, and a moderate increase in expression of the other mutant toxins was observed (see Fig. S3 in the supplemental material).
FIG 3.
Representative microscopic analysis of HeLa cell cultures transfected with enzyme-eGFP fusions expressed from the cytomegalovirus (CMV) promoter. (A) Fluorescence signal from eGFP fused to wild-type sequences of each enzyme or an eGFP control at 24 h posttransfection. (B) Propidium iodide staining of the corresponding fluorescence images in panel A. (C) Bright-field (phase-contrast) images of the corresponding fluorescence images in panel A. Scale bars represent 50 μm.
Propidium iodide (PI) staining of each cell culture inversely correlated with the intensity of eGFP signal from the native sequence toxin fusions. The intercalation of PI into endogenous nucleic acids is an indicator of plasma membrane damage. Compromised plasma membranes were clearly detected in the PAU and BTU transfections, as seen by the intensity of the red signal in the corresponding micrographs (Fig. 3B). The PYU transfections resulted in PI stain levels visibly lower than those obtained with PAU or BTU transfections; PFU expression resulted in minimal PI staining comparatively. The eGFP-only and catalytic mutant enzyme controls did not result in PI staining beyond background staining levels in vehicle controls (Fig. 3B; see also Fig. S2B in the supplemental material). Lastly, bright-field contrast images of the same microscopic fields in Fig. 3A and B show cell-rounding phenotypes in each of the transfection experiments utilizing catalytically active toxin genes (Fig. 3C). The PAU toxin appears to be the most potent in disrupting cell integrity via this general assessment. The BTU enzyme appears to be slightly more potent in a rounding phenotype and monolayer perturbation than PYU or PFU; however, it is clear that cell rounding is evident in those transfections at levels above the GFP control well. Given that a minimal amount of PI staining was detected under expression of PYU and PFU, it is possible that those enzymes disrupt cell morphology at a location other than the plasma membrane or that their activity is sufficiently low to avoid membrane rupture.
Quantitative analysis of homolog expression in HeLa cells.
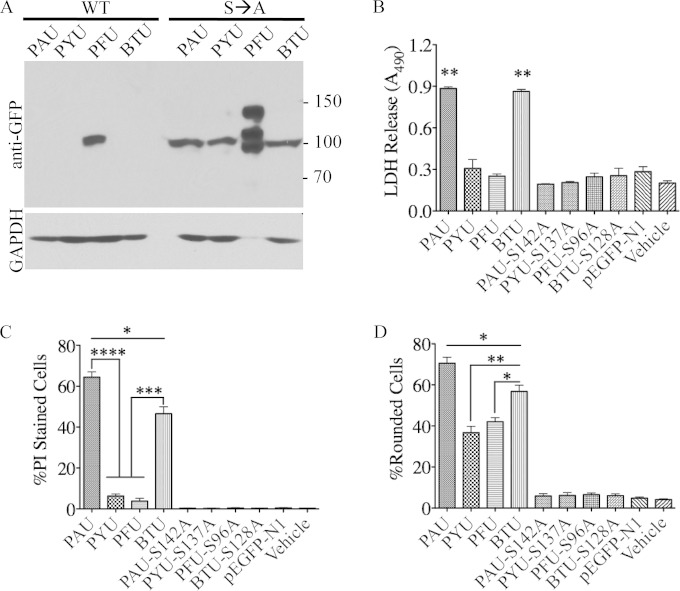
Microscopic examination of transfected cells was useful, but the lack of apparent toxicity for PTU and PFU enzymes (GFP+ and PI−) may be due to many factors that impact overall protein expression, including codon usage and rates of protein degradation. Even in the bacterial dual-expression system there were clear differences in protein detection between the different toxin genes cloned in an identical fashion. To address potential differences in toxicity related to expression, we measured relative protein expression by Western blotting and toxicity by LDH release and quantified multiple fields of PI-stained and rounded cells.
Immunoblot detection of the homologs correlated well with microscopically detected GFP fluorescence. PFU expression was higher than expression of the other proteins for both parental and catalytic serine point mutant derivatives (Fig. 4A). It is notable that PFU is the only parental protein detected, indicative of its lower inherent toxicity to HeLa cells. All noncatalytic serine-to-alanine derivatives were detectable in Western blot analyses of transfected cultures. Additionally, higher-molecular-mass bands were present in the S96A PFU-GFP lane in Fig. 4A. Presumably these bands correspond to posttranslationally modified versions of noncatalytic PFU (such as ubiquitylated PFU) that are detectable due to the high levels of expression. During transfection, noncatalytic derivatives of PAU and BTU enzymes are produced to similar levels. In contrast to the relatively poor prokaryotic expression levels, we observed only a slight decrease in S137A PYU enzyme levels compared to the levels of other mutant enzymes in HeLa cells (Fig. 4A).
FIG 4.
Quantitative analysis of HeLa cell cultures transfected with enzyme-eGFP fusions expressed from the CMV promoter. (A) Western blot signal of each enzyme-eGFP fusion from an equivalent amount of HeLa cell lysate at 24 h posttransfection. The S96A PFU-eGFP lysate was diluted 1:5 compared to the other samples due to the higher expression levels. Numbers are molecular masses in kilodaltons. (B) LDH release from HeLa cells transfected with equivalent amounts of plasmid DNA at 24 h posttransfection. Results are means from 4 independent experiments. (C) Average percentage of PI-positive cells per field in 3 independent fields from 3 replicate experiments. (D) Average percentage of rounded cell phenotypes in 3 independent microscopic fields from 3 replicate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.0005; ****, P < 0.0001.
A variety of assays were performed to measure different stages of cell death. The end stages of cell death were measured by LDH release, which requires membrane permeability to a large molecule. PI staining was used to measure the initial stages of plasma membrane compromise, and cell rounding was used as an indicator of cytoskeletal collapse. LDH release correlated well with PI signal from our microscopy study (Fig. 3B) in that only PAU and BTU intracellular enzyme expression resulted in levels of LDH release that were significantly different from those in the vehicle, vector, and transfection assays using genes encoding noncatalytic enzymes (Fig. 4B). Initial stages of toxicity were easily detectable with PI staining of cells transfected with the PAU and BTU constructs (Fig. 4C) (64% ± 5% and 47% ± 6% of cells are permeable to PI, respectively). In contrast, the number of cells permeable to PI in PYU transfections was 6% ± 2%, and that in PFU transfections was 4% ± 2%. Unpaired t tests indicated that the values measured for PYU transfections were statistically significant (P < 0.005) compared to values obtained from transfection assays performed with eGFP-only controls or genes encoding noncatalytic forms of each enzyme. A t test between PYU and PFU PI-stained cells, however, indicated no statistical difference between the two (P = 0.22). We interpret these combined LDH and PI staining results to suggest that PAU and BTU are more active against the eukaryotic cell plasma membrane than PYU and PFU.
Enumeration of cells rounded per microscopic field indicated a marked difference in the number of perturbed cells among PYU- and PFU-expressing HeLa cells (37% ± 5% and 42% ± 3% rounded for PYU and PFU, respectively) over their counterpart controls (<7% rounded). This analysis suggests that PYU and PFU express a biological activity that appears not to result in a significant compromise of the plasma membrane (Fig. 4D). To ensure that the GFP fusions had minimal impact on toxic activity, all clones were reconstructed without fusion to eGFP and tested again for LDH release, for PI staining, and by bright-field microscopy of rounded cells. These results recapitulated the fusion construct data, suggesting that the fusion itself likely does not explain the apparent lack of activity against the plasma membrane (see Fig. S4 in the supplemental material).
Differential binding of phospholipase A2 bacterial enzymes to plasma membrane mimetic liposomes.
The high degree of toxicity of PYU in the dual-expression system but apparent low toxicity in eukaryotic cells led us to reason that a preference in membrane substrate composition may account for this observed pattern. The inner membranes of prokaryotes differ from the inner leaflet plasma membranes of eukaryotes in several ways (52). Most notably, E. coli harbors a large, negatively charged lipid species called cardiolipin (∼5% molar composition) in its membrane, with the remaining composition heavily consisting of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE; 70% molar composition) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG; 25% molar composition). Eukaryotic plasma membrane inner leaflets are more complex than those of prokaryotes. In addition to POPE, these membranes typically consist of equal amounts of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC; 30% molar composition), with another roughly 40% of the makeup being cholesterol and 1-palmitoyl-2-oleoyl-sn-glycero-3-(phospho-l-serine) (POPS). Importantly, another component found only in eukaryotic membranes is phosphatidyl-4,5-bisphosphate (PIP2), usually constituting ∼0.5% of the molar composition of the plasma membrane (53). This lipid species was previously shown to have a stimulating effect on ExoU phospholipase activity in vitro and in vivo (40, 54). Additional studies suggest that PIP2 tightly associates with ExoU and is an important substrate (54). The interaction with and cleavage of PIP2 by ExoU play a biologically significant role in cell death by disruption of focal adhesion-membrane complexes, leading to a loss of cytoskeletal integrity and eventual rupture of the plasma membrane in temporally separable steps (54).
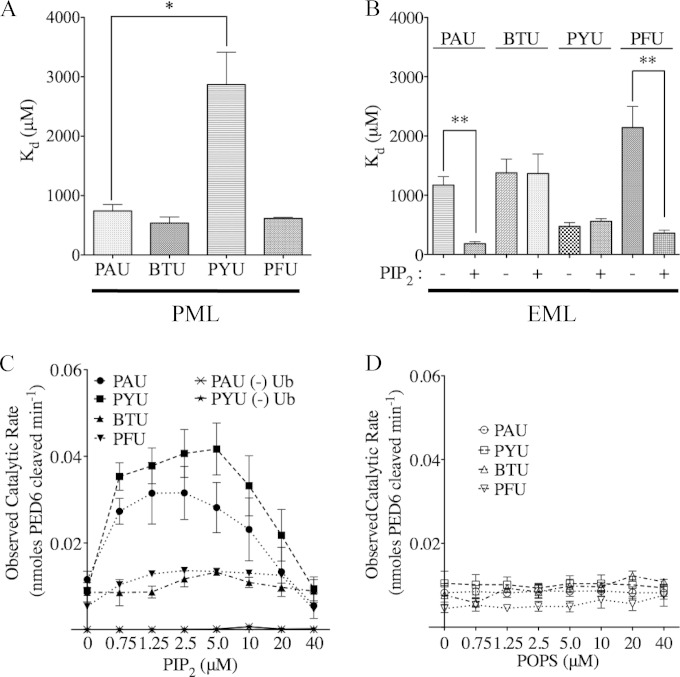
To compare membrane-binding characteristics of these enzymes for prokaryotic (PML) or eukaryotic (EML) model liposomes, we performed lipid binding assays with each type of model liposome and determined the relative dissociation constants. Serine-to-alanine point mutant enzymes were purified for this analysis to avoid possible confounding effects due to catalysis. Binding isotherms were constructed from SDS-PAGE densitometric analyses of enzyme remaining in the supernatant after binding and centrifugation steps. Analysis of each protein binding to prokaryotic-like membrane liposomes revealed dissociation constants of 742, 537, and 613 μM for PAU, BTU, and PFU enzymes, respectively. The PYU enzyme displayed a roughly 4-fold-lower binding affinity, with a dissociation constant (Kd) of 2.8 mM (Fig. 5A). Affinities for eukaryote-like membranes were next tested using compositions that either lacked or included 0.5% PIP2 (Fig. 5B). Both Pseudomonas enzymes displayed a significant increase in affinity (decrease in Kd) for the PIP2-containing version of liposomes over liposomes lacking the molecule (182 μM to 1.1 mM for PAU and 360 μM to 2.1 mM for PFU). Conversely, neither the BTU or PYU enzyme displayed sensitivity to PIP2 in terms of an increase in affinity for this liposome model using this assay. The BTU molecule appears to have a nearly 3-fold-higher affinity for the prokaryotic membrane mimics than either eukaryotic model, while the PYU toxin displayed a Kd of 500 μM regardless of the presence of PIP2 (a 5.6-fold increase over that of the prokaryotic model).
FIG 5.
Liposome binding and the effect of PIP2 on enzyme activity. (A) Apparent affinity constants for each enzyme binding to the PML model liposomes determined by a supernatant depletion assay. (B) Apparent affinity constants of each enzyme binding to the EML model liposomes, plus or minus PIP2, as determined by a supernatant depletion assay. (C) Observed rate of PED6 hydrolysis by each enzyme under subsaturating enzymatic conditions in the presence of increasing amounts of PIP2. As a negative control, PAU and PYU enzymes were assayed for PED6 hydrolysis in the absence of ubiquitin. (D) Observed rate of PED6 hydrolysis by each enzyme under subsaturating enzymatic conditions in the presence of POPS. All experimental data are from at least 3 independent experiments. *, P < 0.05; **, P < 0.01.
Enzyme assays were next conducted to address whether PIP2 had a dose-dependent effect in our assay system. Subsaturating conditions of enzyme, cofactor, and PED6 substrate were present in all wells, as either PIP2 or POPS was added up to an approximately 1:1 concentration with PED6. Both the PAU and PYU enzyme activities were boosted 3- to 4-fold upon the addition of PIP2 at a 1:6 ratio to PED6 substrate (5 μM PIP2 to 30 μM PED6) (Fig. 5C). The addition of PIP2 alone did not result in detectable PED6 cleavage by PAU or PYU under the conditions of the assay (Fig. 5C). No enhancement in activity was observed with POPS (Fig. 5D). The reason for attenuation of the activity enhancement with larger amounts of PIP2 is unclear at this point but may be related to product interference with either an enzyme-PED6 interaction or fluorescence signal. Conversely, neither the BTU nor PFU enzymes seemed sensitive to the addition of either lipid in terms of catalytic activity. Together, these data suggest the possibility that there is more than a single type of PIP2-binding site in this family of enzymes, given that the PFU enzyme displayed an increase in liposome binding affinity but not catalysis and that the PYU enzyme did not show in increase in binding affinity, but substrate catalysis was enhanced.
DISCUSSION
This study was designed to test the hypothesis that ubiquitin activation of injected phospholipases is a conserved mechanism found throughout multiple species of Gram-negative bacteria. Several homologs of ExoU were selected on the basis that a differing lifestyle of the parental strain might have affected specific properties of each phospholipase while maintaining the general activation mechanism. This premise would be consistent with the numerous physiological processes regulated by various subsets of PLA2 enzymes depending on their structure, location, and expression level.
Pseudomonas aeruginosa is an extremely adaptable opportunistic pathogen of both plants and animals found ubiquitously through soil, freshwater, and marine environments (55). In contrast, P. fluorescens is not considered to be pathogenic and usually exists in a symbiotic relationship within the plant rhizosphere (56). Photorhabdus asymbiotica typically exists in an entomopathogenic life cycle with the Heterorhabditis nematode genus. Monoxenically colonized nematode juveniles regurgitate these bacteria into an infected insect's hemolymph to cause host death in less than 48 h. Several cases of human infection have been reported recently, and from these cases, it appears as though P. asymbiotica may be capable of acting as a primary pathogen (37). Isolates of this strain were shown to be facultative intracellular organisms capable of replicating in and inducing apoptosis of human macrophage-like cells (57). Burkholderia thailandensis is a member of the betaproteobacteria class and a close relative of the category B select agent Burkholderia pseudomallei. Like some strains of Photorhabdus, B. thailandensis appears to be a facultative intracellular organism; however, it is generally considered nonpathogenic to humans. It has also been shown to require a functional Bsa T3S system for successful escape from endocytic vacuoles of HeLa cells (31). This study focused on enzymes secreted from two pathogenic and two nonpathogenic strains, with one pathogenic and one nonpathogenic facultative intracellular organism species represented in each category.
The selection of phospholipase orthologs from bacterial genera with different lifestyles, hosts, and/or native environments may have impacted the variation of toxin properties that we observed. The lack of a distinct pattern in enzyme properties (ubiquitin activation, binding constants, toxicities, and PIP2 enhancement) prohibited a simplistic correlation between enzyme activities and pathogenic versus nonpathogenic or intracellular versus extracellular lifestyles. Our overall findings are consolidated into a global summary (Table 3). Table 3 reports toxicity in both eukaryotes and prokaryotes as inversely correlated to protein expression levels. For example, the lower the relative protein expression and higher LDH release or lower recoverable CFU were associated with highly toxic enzymes. It is apparent that although each enzyme is activated by ubiquitin and utilizes a conserved serine-aspartate catalytic dyad, toxicity profiles between different enzymes in different hosts vary. In prokaryotes, the enzymes partition into two categories whereby the PYU and BTU enzymes were highly toxic and the Pseudomonas enzymes were not as potent. We postulated that both PYU and BTU would be equally toxic in HeLa cells via the transfection method and were surprised to find the attenuated toxicity profile of PYU in comparison to its lethality to E. coli. Lactate dehydrogenase release and propidium iodide staining, an indication of plasma membrane permeability, were comparatively lower in experiments utilizing expression constructs PYU and PFU versus PAU and BTU; however, a significant degree of cell rounding compared to that in controls was observed with each construct, indicating that a biological effect was still occurring. As a result, PAU and BTU toxins appear to partition to a more cytotoxic subset of bacterial phospholipases than PYU and PFU enzymes when expressed inside HeLa cells.
TABLE 3.
Summary of ExoU homolog properties
| Propertya | PAU | PYU | BTU | PFU |
|---|---|---|---|---|
| Host niche | Extracellular | Facultative intracellular | Facultative intracellular | Extracellular |
| Prokaryotic expression | ***** | * | *** | *** |
| Prokaryotic toxicity (broth culture) | ** | ***** | ***** | ** |
| Eukaryotic expressionb | ** | * | ** | ***** |
| Plasma membrane damagec | ***** | ** | **** | * |
| PIP2 affinity enhancement | Yes | No | No | Yes |
| PIP2 activity enhancement | Yes | Yes | No | No |
The number of asterisks for each category represents an estimate of the relative levels of expression, toxicity, and plasma membrane damage from the sum of replicate Western blot analyses, dual-expression experiments in broth inductions, and transfection analyses.
Observed expression from noncatalytic sequence.
Relative plasma membrane damage as determined by the LDH release assay, PI staining, and eGFP expression for parental phospholipases.
Control experiments testing the toxicity of the non-eGFP fusions recapitulated our results with GFP fusion proteins, suggesting that an alternative explanation for the observed toxicity, such as substrate specificity, may play a role in these observations. Experiments querying sensitivity to PIP2 support previous work in that this phospholipid is able to enhance ExoU catalysis of a substrate in the presence of ubiquitin under subsaturating enzymatic conditions (40, 54). This observation is also evident with the PYU enzyme but not BTU or PFU, suggesting a possible correlation between PIP2 enhancement and pathogenicity of the enzyme's host strain. Liposome binding experiments showed that both of the Pseudomonas enzymes seemed to have a higher affinity for PIP2-containing liposomes over those that lack the lipid, while the PYU and BTU proteins did not show this same sensitivity regarding affinity. Given that the PFU enzyme did not show a PIP2-stimulated increase in activity, we hypothesize that within this family of enzymes there may be multiple distinct sites of PIP2-protein interaction. Previous work has suggested that in addition to the catalytic site of ExoU, the C-terminal region contains a lipid binding sequence that facilitates ExoU's interaction with membrane substrates (35). A PIP2-sensitive peptide sequence may exist in this domain in addition to the catalytic domain. Work in our laboratory suggested that ExoU residue R661 may be important for substrate binding due to decreased catalytic rates but not ubiquitin activation constants in vitro (30). Additional work has suggested a link between R661 and PIP2 coactivation; however, no direct evidence of binding between the two has been shown to date (40). It should also be noted that PIP2 is a viable substrate of ExoU catalysis; thus, it is possible that the activity-boosting effect at certain concentrations is a result of PIP2 cleavage products (54). Binding assays performed in this study utilized serine-to-alanine point mutants for affinity measurements, which suggested that at least for the Pseudomonas enzymes, a by-product is likely not a required high-affinity binding target. It is also possible that other or additional residues are involved in this process, as this arginine is conserved in amino acid alignments.
In our prokaryotic toxicity model, we noted some discrepancies between assays done on spot plates and assays performed in liquid culture. For example, PFU was as toxic as PAU in broth but not as toxic in the spot test. We speculate that the dynamics of gene expression may be different in these two environments, perhaps depending on the surface exposure of bacteria to the inducers. In addition, using live-cell microscopy, we often observed leakage of DNA from the septum of dividing bacteria (30), suggesting that there may be transient weakness in newly synthesized membrane or cell wall structures. In broth cultures, hypertonic or hypotonic stresses may exacerbate these stresses, leading to more rapid death. Finally, perhaps growth on agar allows some recovery from the toxic effects of the phospholipases (reduction in toxin/ubiquitin transcription), masking actual death measured after broth growth. We find that the agar spot test is a useful complement to the broth assay in gaining a better understanding of the relative toxicity profiles of each enzyme.
In summary, we have shown that ubiquitin activation of bacterial phospholipase A2 enzymes is a conserved mechanism shared by many species of organisms. The absence of this activator from the prokaryotic environment allows for the safe regulation of such enzymes with relatively promiscuous activities against their membrane substrates. Even though this general mechanism of activation seems to be preserved, this work revealed different properties among the enzymes tested. Future work examining the trafficking and modification state(s) of the homologs in host cells may lend insights into possible explanations of the differential toxicities. It is possible that localized internal membrane disruption, rather than plasma membrane disruption, is the intended outcome of a subset of ubiquitin-activated bacterial phospholipases. A more complete understanding of the function of each of these enzymes will likely require study of them as injected molecules to recapitulate biologically relevant delivery. Lastly, although sequence alignments between enzymes from this class of proteins are useful, structural homology will likely be required to fully understand the mechanisms of activation and provide leads for future specific targets regarding therapeutic application and drug design.
Supplementary Material
ACKNOWLEDGMENTS
We thank Monika S. Casey for technical assistance.
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases grant 1R01 AI104922 to D.W.F.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02402-14.
REFERENCES
- 1.Leslie CC, Voelker DR, Channon JY, Wall MM, Zelarney PT. 1988. Properties and purification of an arachidonoyl-hydrolyzing phospholipase A2 from a macrophage cell line, RAW 2647. Biochim Biophys Acta 963:476–492. [DOI] [PubMed] [Google Scholar]
- 2.Burke JE, Dennis EA. 2009. Phospholipase A2 structure/function, mechanism, and signaling. J Lipid Res 50(Suppl):S237–S242. doi: 10.1194/jlr.R800033-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aloulou A, Ali YB, Bezzine S, Gargouri Y, Gelb MH. 2012. Phospholipases: an overview. Methods Mol Biol 861:63–85. doi: 10.1007/978-1-61779-600-5_4. [DOI] [PubMed] [Google Scholar]
- 4.Edwards SH, Thompson D, Baker SF, Wood SP, Wilton DC. 2002. The crystal structure of the H48Q active site mutant of human group IIA secreted phospholipase A2 at 1.5 A resolution provides an insight into the catalytic mechanism. Biochemistry 41:15468–15476. doi: 10.1021/bi020485z. [DOI] [PubMed] [Google Scholar]
- 5.Tatulian SA. 2001. Toward understanding interfacial activation of secretory phospholipase A2 (PLA2): membrane surface properties and membrane-induced structural changes in the enzyme contribute synergistically to PLA2 activation. Biophys J 80:789–800. doi: 10.1016/S0006-3495(01)76058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghomashchi F, Lin Y, Hixon MS, Yu BZ, Annand R, Jain MK, Gelb MH. 1998. Interfacial recognition by bee venom phospholipase A2: insights into nonelectrostatic molecular determinants by charge reversal mutagenesis. Biochemistry 37:6697–6710. doi: 10.1021/bi972525i. [DOI] [PubMed] [Google Scholar]
- 7.Koduri RS, Gronroos JO, Laine VJ, Le Calvez C, Lambeau G, Nevalainen TJ, Gelb MH. 2002. Bactericidal properties of human and murine groups I, II, V, X, and XII secreted phospholipases A(2). J Biol Chem 277:5849–5857. doi: 10.1074/jbc.M109699200. [DOI] [PubMed] [Google Scholar]
- 8.Webb NR. 2005. Secretory phospholipase A2 enzymes in atherogenesis. Curr Opin Lipidol 16:341–344. doi: 10.1097/01.mol.0000169355.20395.55. [DOI] [PubMed] [Google Scholar]
- 9.Kudo I, Murakami M, Hara S, Inoue K. 1993. Mammalian non-pancreatic phospholipases A2. Biochim Biophys Acta 1170:217–231. doi: 10.1016/0005-2760(93)90003-R. [DOI] [PubMed] [Google Scholar]
- 10.Atsumi G, Murakami M, Tajima M, Shimbara S, Hara N, Kudo I. 1997. The perturbed membrane of cells undergoing apoptosis is susceptible to type II secretory phospholipase A2 to liberate arachidonic acid. Biochim Biophys Acta 1349:43–54. doi: 10.1016/S0005-2760(97)00082-9. [DOI] [PubMed] [Google Scholar]
- 11.Gutiérrez JM, Lomonte B. 2013. Phospholipases A2: unveiling the secrets of a functionally versatile group of snake venom toxins. Toxicon 62:27–39. doi: 10.1016/j.toxicon.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Murakami M, Lambeau G. 2013. Emerging roles of secreted phospholipase A(2) enzymes: an update. Biochimie 95:43–50. doi: 10.1016/j.biochi.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Clark JD, Schievella AR, Nalefski EA, Lin LL. 1995. Cytosolic phospholipase A2. J Lipid Mediat Cell Signal 12:83–117. doi: 10.1016/0929-7855(95)00012-F. [DOI] [PubMed] [Google Scholar]
- 14.Pickard RT, Chiou XG, Strifler BA, DeFelippis MR, Hyslop PA, Tebbe AL, Yee YK, Reynolds LJ, Dennis EA, Kramer RM, Sharp JD. 1996. Identification of essential residues for the catalytic function of 85-kDa cytosolic phospholipase A2. Probing the role of histidine, aspartic acid, cysteine, and arginine. J Biol Chem 271:19225–19231. [DOI] [PubMed] [Google Scholar]
- 15.Leslie CC. 1997. Properties and regulation of cytosolic phospholipase A2. J Biol Chem 272:16709–16712. doi: 10.1074/jbc.272.27.16709. [DOI] [PubMed] [Google Scholar]
- 16.Dessen A, Tang J, Schmidt H, Stahl M, Clark JD, Seehra J, Somers WS. 1999. Crystal structure of human cytosolic phospholipase A2 reveals a novel topology and catalytic mechanism. Cell 97:349–360. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh M, Tucker DE, Burchett SA, Leslie CC. 2006. Properties of the group IV phospholipase A2 family. Prog Lipid Res 45:487–510. doi: 10.1016/j.plipres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Haeggström JZ, Rinaldo-Matthis A, Wheelock CE, Wetterholm A. 2010. Advances in eicosanoid research, novel therapeutic implications. Biochem Biophys Res Commun 396:135–139. doi: 10.1016/j.bbrc.2010.03.140. [DOI] [PubMed] [Google Scholar]
- 19.Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, Pershing JC, Purcell JP, Alibhai MF. 2003. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 20.Wilson PA, Gardner SD, Lambie NM, Commans SA, Crowther DJ. 2006. Characterization of the human patatin-like phospholipase family. J Lipid Res 47:1940–1949. doi: 10.1194/jlr.M600185-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Hirschberg HJ, Simons JW, Dekker N, Egmond MR. 2001. Cloning, expression, purification and characterization of patatin, a novel phospholipase A. Eur J Biochem 268:5037–5044. doi: 10.1046/j.0014-2956.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 22.Strickland JA, Orr GL, Walsh TA. 1995. Inhibition of Diabrotica larval growth by patatin, the lipid acyl hydrolase from potato tubers. Plant Physiol 109:667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.VanRheenen SM, Luo ZQ, O'Connor T, Isberg RR. 2006. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun 74:3597–3606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brugirard-Ricaud K, Givaudan A, Parkhill J, Boemare N, Kunst F, Zumbihl R, Duchaud E. 2004. Variation in the effectors of the type III secretion system among Photorhabdus species as revealed by genomic analysis. J Bacteriol 186:4376–4381. doi: 10.1128/JB.186.13.4376-4381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Housley NA, Winkler HH, Audia JP. 2011. The Rickettsia prowazekii ExoU homologue possesses phospholipase A1 (PLA1), PLA2, and lyso-PLA2 activities and can function in the absence of any eukaryotic cofactors in vitro. J Bacteriol 193:4634–4642. doi: 10.1128/JB.00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman MS, Gillespie JJ, Kaur SJ, Sears KT, Ceraul SM, Beier-Sexton M, Azad AF. 2013. Rickettsia typhi possesses phospholipase A2 enzymes that are involved in infection of host cells. PLoS Pathog 9:e1003399. doi: 10.1371/journal.ppat.1003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish JP, Fleiszig SM, Wu C, Mende-Mueller L, Frank DW. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol 25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 28.Sato H, Frank DW, Hillard CJ, Feix JB, Pankhaniya RR, Moriyama K, Finck-Barbancon V, Buchaklian A, Lei M, Long RM, Wiener-Kronish J, Sawa T. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J 22:2959–2969. doi: 10.1093/emboj/cdg290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips RM, Six DA, Dennis EA, Ghosh P. 2003. In vivo phospholipase activity of the Pseudomonas aeruginosa cytotoxin ExoU and protection of mammalian cells with phospholipase A2 inhibitors. J Biol Chem 278:41326–41332. doi: 10.1074/jbc.M302472200. [DOI] [PubMed] [Google Scholar]
- 30.Anderson DM, Schmalzer KM, Sato H, Casey M, Terhune SS, Haas AL, Feix JB, Frank DW. 2011. Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU. Mol Microbiol 82:1454–1467. doi: 10.1111/j.1365-2958.2011.07904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haraga A, West TE, Brittnacher MJ, Skerrett SJ, Miller SI. 2008. Burkholderia thailandensis as a model system for the study of the virulence-associated type III secretion system of Burkholderia pseudomallei. Infect Immun 76:5402–5411. doi: 10.1128/IAI.00626-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loper JE, Hassan KA, Mavrodi DV, Davis EW II, Lim CK, Shaffer BT, Elbourne LD, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS III, Pierson EA, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT. 2012. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8:e1002784. doi: 10.1371/journal.pgen.1002784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang C, Flieger A. 2011. Characterisation of Legionella pneumophila phospholipases and their impact on host cells. Eur J Cell Biol 90:903–912. doi: 10.1016/j.ejcb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 34.Schmalzer KM, Benson MA, Frank DW. 2010. Activation of ExoU phospholipase activity requires specific C-terminal regions. J Bacteriol 192:1801–1812. doi: 10.1128/JB.00904-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veesenmeyer JL, Howell H, Halavaty AS, Ahrens S, Anderson WF, Hauser AR. 2010. Role of the membrane localization domain of the Pseudomonas aeruginosa effector protein ExoU in cytotoxicity. Infect Immun 78:3346–3357. doi: 10.1128/IAI.00223-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson DM, Feix JB, Monroe AL, Peterson FC, Volkman BF, Haas AL, Frank DW. 2013. Identification of the major ubiquitin-binding domain of the Pseudomonas aeruginosa ExoU A2 phospholipase. J Biol Chem 288:26741–26752. doi: 10.1074/jbc.M113.478529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa SC, Chavez CV, Jubelin G, Givaudan A, Escoubas JM, Brehelin M, Zumbihl R. 2010. Recent insight into the pathogenicity mechanisms of the emergent pathogen Photorhabdus asymbiotica. Microbes Infect 12:182–189. doi: 10.1016/j.micinf.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Farfán M, Spataro N, Sanglas A, Albarral V, Loren JG, Bosch E, Fuste MC. 2013. Draft genome sequence of the Aeromonas diversa type strain. Genome Announc 1(3):e00330-13. doi: 10.1128/genomeA.00330-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Testa J, Daniel LW, Kreger AS. 1984. Extracellular phospholipase A2 and lysophospholipase produced by Vibrio vulnificus. Infect Immun 45:458–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyson GH, Hauser AR. 2013. Phosphatidylinositol 4,5-bisphosphate is a novel coactivator of the Pseudomonas aeruginosa cytotoxin ExoU. Infect Immun 81:2873–2881. doi: 10.1128/IAI.00414-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gendrin C, Contreras-Martel C, Bouillot S, Elsen S, Lemaire D, Skoufias DA, Huber P, Attree I, Dessen A. 2012. Structural basis of cytotoxicity mediated by the type III secretion toxin ExoU from Pseudomonas aeruginosa. PLoS Pathog 8:e1002637. doi: 10.1371/journal.ppat.1002637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halavaty AS, Borek D, Tyson GH, Veesenmeyer JL, Shuvalova L, Minasov G, Otwinowski Z, Hauser AR, Anderson WF. 2012. Structure of the type III secretion effector protein ExoU in complex with its chaperone SpcU. PLoS One 7:e49388. doi: 10.1371/journal.pone.0049388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stirling FR, Cuzick A, Kelly SM, Oxley D, Evans TJ. 2006. Eukaryotic localization, activation and ubiquitinylation of a bacterial type III secreted toxin. Cell Microbiol 8:1294–1309. doi: 10.1111/j.1462-5822.2006.00710.x. [DOI] [PubMed] [Google Scholar]
- 45.Benson MA, Schmalzer KM, Frank DW. 2010. A sensitive fluorescence-based assay for the detection of ExoU-mediated PLA(2) activity. Clin Chim Acta 411:190–197. doi: 10.1016/j.cca.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hjerpe R, Thomas Y, Chen J, Zemla A, Curran S, Shpiro N, Dick LR, Kurz T. 2012. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem J 441:927–936. doi: 10.1042/BJ20111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carlson N, Rechsteiner M. 1987. Microinjection of ubiquitin: intracellular distribution and metabolism in HeLa cells maintained under normal physiological conditions. J Cell Biol 104:537–546. doi: 10.1083/jcb.104.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haas AL, Bright PM. 1985. The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem 260:12464–12473. [PubMed] [Google Scholar]
- 49.Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR. 2011. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods 8:691–696. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the L-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. [DOI] [PubMed] [Google Scholar]
- 51.Finck-Barbançon V, Frank DW. 2001. Multiple domains are required for the toxic activity of Pseudomonas aeruginosa ExoU. J Bacteriol 183:4330–4344. doi: 10.1128/JB.183.14.4330-4344.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeaman MR, Yount NY. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 53.Gennis RD. 1988. Biomembranes: molecular structure and function. Springer, New York, NY. [Google Scholar]
- 54.Sato H, Frank DW. 2014. Intoxication of host cells by the T3SS phospholipase ExoU: PI(4,5)P2-associated, cytoskeletal collapse and late phase membrane blebbing. PLoS One 9:e103127. doi: 10.1371/journal.pone.0103127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khan NH, Ahsan M, Taylor WD, Kogure K. 2010. Culturability and survival of marine, freshwater and clinical Pseudomonas aeruginosa. Microbes Environ 25:266–274. doi: 10.1264/jsme2.ME09178. [DOI] [PubMed] [Google Scholar]
- 56.Preston GM, Bertrand N, Rainey PB. 2001. Type III secretion in plant growth-promoting Pseudomonas fluorescens SBW25. Mol Microbiol 41:999–1014. doi: 10.1046/j.1365-2958.2001.02560.x. [DOI] [PubMed] [Google Scholar]
- 57.Costa SC, Girard PA, Brehelin M, Zumbihl R. 2009. The emerging human pathogen Photorhabdus asymbiotica is a facultative intracellular bacterium and induces apoptosis of macrophage-like cells. Infect Immun 77:1022–1030. doi: 10.1128/IAI.01064-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Hu Y, Gong J, Lin Y, Johnstone L, Rensing C, Wang G. 2012. Genome sequence of the highly efficient arsenite-oxidizing bacterium Achromobacter arsenitoxydans SY8. J Bacteriol 194:1243–1244. doi: 10.1128/JB.06667-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leggieri N, Gouriet F, Thuny F, Habib G, Raoult D, Casalta JP. 2012. Legionella longbeachae and endocarditis. Emerg Infect Dis 18:95–97. doi: 10.3201/eid1801.110579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chien M, Morozova I, Shi S, Sheng H, Chen J, Gomez SM, Asamani G, Hill K, Nuara J, Feder M, Rineer J, Greenberg JJ, Steshenko V, Park SH, Zhao B, Teplitskaya E, Edwards JR, Pampou S, Georghiou A, Chou IC, Iannuccilli W, Ulz ME, Kim DH, Geringer-Sameth A, Goldsberry C, Morozov P, Fischer SG, Segal G, Qu X, Rzhetsky A, Zhang P, Cayanis E, De Jong PJ, Ju J, Kalachikov S, Shuman HA, Russo JJ. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 61.Tribelli PM, Raiger Iustman LJ, Catone MV, Di Martino C, Revale S, Mendez BS, Lopez NI. 2012. Genome sequence of the polyhydroxybutyrate producer Pseudomonas extremaustralis, a highly stress-resistant Antarctic bacterium. J Bacteriol 194:2381–2382. doi: 10.1128/JB.00172-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mao Z, Li M, Chen J. 2013. Draft genome sequence of Pseudomonas plecoglossicida strain NB2011, the causative agent of white nodules in large yellow croaker (Larimichthys crocea). Genome Announc 1(4):e00586-13. doi: 10.1128/genomeA.00586-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarke CR, Cai R, Studholme DJ, Guttman DS, Vinatzer BA. 2010. Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc locus are common leaf colonizers equipped with an atypical type III secretion system. Mol Plant Microbe Interact 23:198–210. doi: 10.1094/MPMI-23-2-0198. [DOI] [PubMed] [Google Scholar]
- 64.Ogata H, La Scola B, Audic S, Renesto P, Blanc G, Robert C, Fournier PE, Claverie JM, Raoult D. 2006. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet 2:e76. doi: 10.1371/journal.pgen.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.