Abstract
The DtxR family consists of metal-dependent transcription factors (DtxR-TFs) that regulate the expression of genes involved in metal homeostasis in the cell. The majority of characterized DtxR-TFs belong to Bacteria. In the current work, we applied a comparative genomics approach to predict DNA-binding sites and reconstruct regulons for DtxR-TFs in Archaea. As a result, we inferred 575 candidate binding sites for 139 DtxR-TFs in 77 genomes from 15 taxonomic orders. Novel DNA motifs of archaeal DtxR-TFs that have a common palindromic structure were classified into 10 distinct groups. By combining functional regulon reconstructions with phylogenetic analysis, we selected 28 DtxR-TF clades and assigned them metal specificities and regulator names. The reconstructed FetR (ferrous iron), MntR (manganese), and ZntR (zinc) regulons largely contain known or putative metal uptake transporters from the FeoAB, NRAMP, ZIP, and TroA families. A novel family of putative iron transporters (named Irt), including multiple FetR-regulated paralogs, was identified in iron-oxidizing Archaea from the Sulfolobales order. The reconstructed DtxR-TF regulons were reconciled with available transcriptomics data in Archaeoglobus, Halobacterium, and Thermococcus spp.
INTRODUCTION
Transition metals, including iron, manganese, and zinc, are responsible for a diverse array of biochemical reactions and other biological functions in prokaryotes. The particular properties of ferrous iron (Fe2+) and ferric iron (Fe3+) ions make them widely used redox-sensing elements in many metalloenzymes, from heme-containing cytochromes to Fe-S proteins. Manganese is used in free radical-detoxifying enzymes, including superoxide dismutase and catalase and some other enzymes. Zinc was found in many proteins, including enzymes of nucleic acid metabolism and ribosomal proteins, where it plays a role in both catalysis and protein structure. Many transition metals are taken up by specific transport systems, including FeoAB family transporters for ferrous iron (1, 2), Fbp-type ATP-binding cassette (ABC) transporters for ferric iron (3), Znu family ABC transporters for zinc (4), and NRAMP family MntR transporters for manganese (5). The precise maintenance of metal ion homeostasis, including metal uptake transporters, is vital for cells that have to balance between supplying enzymes with metal ion cofactors and decreasing the harmful effects of heavy metals (6, 7).
The regulation of gene expression by specific binding of transcription factors (TFs) to their DNA sites in response to a cellular signal (e.g., specific metal ions) is a common regulatory mechanism in microorganisms. Iron, manganese, and zinc homeostasis genes in prokaryotes are controlled by TFs from two major families of metalloregulators, Fur and DtxR (8). The ferric uptake regulator Fur and zinc uptake regulator Zur constitute the majority of the Fur family metalloregulators, being widely distributed in diverse lineages of Bacteria but not Archaea, whereas the manganese- and nickel-specific regulators Mur and Nur were found in only some bacterial lineages (9, 10). Manganese-responsive DtxR family regulators were studied in many bacteria, including MntR in Escherichia coli (11), Bacillus subtilis (12), Staphylococcus aureus (13), Corynebacterium diphtheria (14), and C. glutamicum (15, 16), ScaR in Streptococcus gordonii (17), and TroR in Treponema pallidum (18), where they mostly control genes for manganese uptake transporters. In contrast, iron-responsive TFs from the DtxR family were found only in Actinobacteria and include two experimentally studied TFs, the diphtheria toxin repressor DtxR in Corynebacterium diphtheriae (19) and the iron-dependent regulator IdeR in Mycobacterium tuberculosis (20, 21), which control large networks of genes involved in iron homeostasis and other cellular functions, such as the toxin gene in C. diphtheriae. The natural ligand for IdeR/DtxR is ferrous iron, but divalent ions of nickel, cobalt, manganese, and zinc also bind to and activate the regulators in vitro (22). Direct sensing of cytoplasmic ferrous iron or manganese ions by DtxR family TFs (DtxR-TFs) represses target genes by increasing affinities of TFs to their DNA sites. The metalloregulators from the DtxR family also were implicated in the control of iron homeostasis in several archaeal species, including Halobacterium sp. strain NRC-1 and H. salinarium (23, 24), Pyrococcus furiosus (25), and Thermococcus kodakaraensis (26).
Previously, we applied the comparative genomics approach to predict TF-binding sites (TFBSs) and sets of TF-regulated genes (regulons) and finally reconstruct metal-responsive transcriptional regulatory networks controlled by the Fur family regulators in Bacteria (27–33). Here, we extended this analysis toward transcriptional regulons for the DtxR family in Archaea. As a result, we inferred novel TF-binding DNA motifs and reconstructed regulons for the majority of DtxR-TFs in available genomes of Archaea from three phyla, Euryarchaeota, Crenarchaeota, and Korarchaeota, representing at least 15 taxonomic orders. The comparative analysis of reconstructed regulons revealed considerable variability in gene content of the iron homeostasis FetR regulons, whereas the manganese (MntR) and zinc (ZntR) uptake regulons are mostly conserved but less widely distributed among archaeal lineages. This comprehensive reconstruction of transcription regulation by the DtxR family can serve as a basis for understanding the evolution of metal-responsive transcriptional networks in Archaea.
MATERIALS AND METHODS
Archaeal genomes were downloaded from GenBank (34). We excluded closely related strains and selected 83 nonredundant archaeal genomes for comparative analysis (see Table S1 in the supplemental material). Repertoires of TFs from the DtxR family were identified by similarity searches and domain predictions in the Pfam database (35) (see Table S2). DtxR-TFs consist of two characteristic domains, an N-terminal helix-turn-helix (HTH) DNA-binding domain (PF01325) and a C-terminal metal-binding and dimerization domain (PF02742). Near one-third of all found DtxR-TFs in Archaea (50 out of 164) contain an additional C-terminal domain (PF04023; also known as the SH3 domain), which is shared with the FeoA component of ferrous iron transporters. Mbur_0783 and Tneu_0335 are truncated proteins with only the HTH domain retained; however, their orthologs in closely related genomes are full-length DtxR-TFs, suggesting that these truncations are caused by sequencing errors. Multiple alignment of DtxR-TFs was constructed using MUSCLE (36) and based on the protein sequences of only the HTH and metal-binding domains and excluding the optional SH3 domains. The phylogenetic tree of DtxR-TFs (see Fig. S1) was built using a maximum-likelihood algorithm implemented in PhyML 3.0 (37), using bootstrapping with 100 replicates, and visualized by Dendroscope (38). The MntR regulator from Bacillus subtilis (BSU24520) was added to the tree as an outgroup. Individual clades of TF orthologs were selected on the phylogenetic tree using a minimum bootstrap value of 50 for the most ancestral node in each clade.
For the identification of TF-binding site (TFBS) motifs and regulon reconstruction, we used a previously established comparative genomics approach (39) implemented on the RegPredict Web server (40) and the GenomeExplorer software (41). Orthologs were defined by the bidirectional best-hit criterion in GenomeExplorer and validated by phylogenetic trees from the MicrobesOnline database (42). We started regulon reconstruction from genome context analysis of DtxR-TF gene neighborhoods using MicrobesOnline (42). As a result, for each clade of DtxR-TFs, we collected an initial training set of potentially coregulated genes that mostly encode metal ion transporters and other metal homeostasis genes. For de novo identification of a candidate TFBS motif in the training set of potential upstream regions, we used the Discover Profile tool in RegPredict (40). A search for palindromic DNA motifs of 16 to 18 bp in length was carried out within putative promoter regions from −300 to +25 bp relative to the translational gene start. Motifs were further validated by the construction of multiple alignments of orthologous DNA fragments using MUSCLE (36). Conservative palindromic sites were selected as potential binding sites for DtxR-TFs and included in final training sets used for the construction of positional weight matrices (PWMs). Sequence logos for the derived clade-specific DNA motifs were drawn using WebLogo (43) and compared to each other. Highly similar motifs were unified, resulting in the final selection of 10 distinct motifs. The constructed final motif PWMs were further used to search for additional regulon members using the Run profile tool in RegPredict and GenomeExplorer. The lowest score in the final TFBS training set was used as the threshold for a site search. False-positive TFBS predictions were eliminated using the consistency check approach (31, 39). For reconstructed regulons, we selected genes that were preceded by a candidate TFBS in three or more genomes. New sites were selectively added to training sets to improve PWMs. For clades consisting of a single DtxR-TF, we looked for known metal-associated genes in its close gene neighborhood (less than 10 genes upstream and downstream) and searched for putative TFBSs within upstream regions of these genes using all known DtxR-TF motifs. We assigned TFBSs to a specific DtxR-TF using (i) their mutual colocalization on the chromosome and (ii) the co-occurrence of TFBSs for orthologous candidate target genes with DtxR-TF orthologs for the studied clade on the tree. Potential TFBSs of paralogous DtxR-TFs were combined into one regulon. In the Halobacteriales, predicted targets of FetR TFs from clades 23 and 24 also were assigned to the combined FetR regulon, as their TFBS motifs were indistinguishable.
Biological functions of predicted target genes were assigned by combination of similarity searches against the Swiss-Prot/UniProt database (44) and domain architecture analysis in Pfam (35). Genome context analysis of regulators and candidate target genes was performed using MicrobesOnline (42). Transmembrane segments in metal transporters were predicted using TMPred (45).
RESULTS AND DISCUSSION
Repertoire of DtxR family transcription factors in Archaea.
To analyze variances in regulons controlled by DtxR-TFs in the domain Archaea, we collected a set of 164 DtxR family proteins in 83 archaeal genomes (see Table S1 in the supplemental material). These genomes belong to four phyla: Euryarchaeota (56 genomes), Crenarchaeota (25 genomes), Korarchaeota (1 genome), and Thaumarchaeota (1 genome). The number of DtxR family proteins varies significantly with taxonomy. The Halobacteriales, Archaeoglobales, Methanomicrobiales, Methanosarcinales, and Nitrosopumilales orders have from three to five DtxR-TF genes per genome, whereas the other studied taxa have only one or two regulators.
To identify orthologous groups of archaeal DtxR-TFs, we built a phylogenetic tree of these proteins and divided it into 39 clades (see Fig. S1 in the supplemental material). Nine of these clades were singletons, whereas the remaining 30 clades contain 2 to 13 proteins per group (see Table S2). In a few cases, a phylogenetic clade contains regulators from different archaeal lineages. For instance, clade 2 includes seven proteins from the Methanomicrobiales order and two other proteins from the Methanosarcinales and Thermoproteales. Clades 17, 18, and 32 are composed of proteins from various orders that all belong to the Methanomicrobia class. Interestingly, DtxR-TFs from the Desulfurococcales order (8 genomes, each containing a single regulator) are not monophyletic but were distributed between two singleton clades and three clades that each contained a pair of orthologous regulators.
Reconstruction of DtxR family regulons in archaeal genomes.
For the reconstruction of regulons controlled by these DtxR-TFs, we started from the comparative genomic analysis of regulators from clades containing two or more proteins and then proceeded with regulon inference for the remaining singleton DtxR-TFs. The genome context analysis revealed that most of the DtxR family genes are neighbors with genes encoding uptake transporters for metal ions or iron siderophores. Alternatively, some DtxR-TF genes are colocalized with other iron homeostasis genes, e.g., the FeS assembly suf genes.
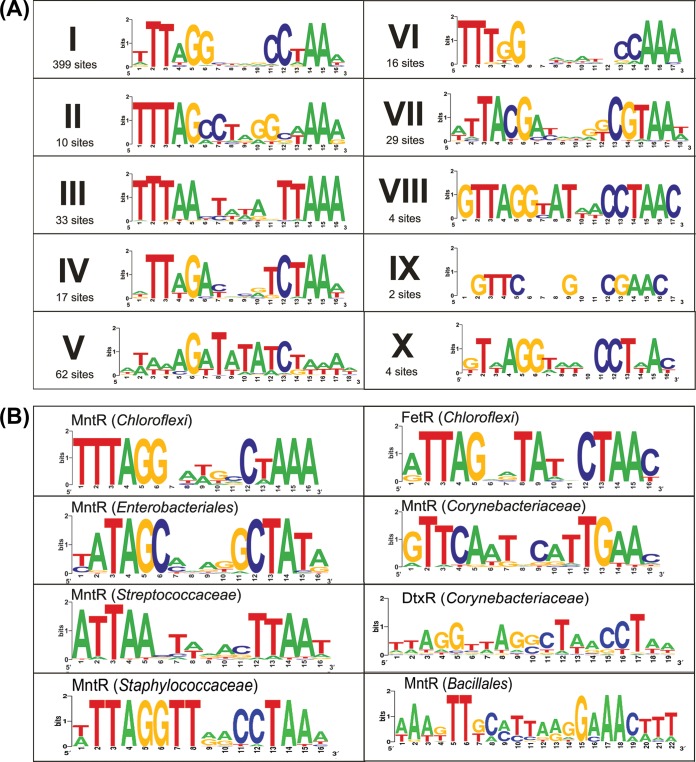
To find putative TFBS motifs, we searched upstream regions of these potentially coregulated genes for conserved palindromic DNA motifs across related genomes that contain DtxR-TFs from the same clade. By utilizing the de novo regulon reconstruction procedure, as described in Materials and Methods, we identified 575 putative TFBSs for 139 studied regulators unevenly distributed across 77 genomes from 15 taxonomic orders of Archaea (see Table S3 in the supplemental material). We grouped these putative TFBSs into 10 distinct DNA motifs that were designated motifs I to X (Fig. 1A). All binding motifs of DtxR-TFs in Archaea have a common palindromic structure, which is consistent with previous studies of DtxR-TFs in Bacteria (12, 14, 15, 31, 46). The overwhelming majority of putative DtxR-TF binding sites in Archaea are even palindromes (16 or 18 nucleotides [nt] long), whereas noncanonical palindromes (17 nt long) were found for only 8% of regulators (motifs VI, VII, and IX). Based on the predicted TFBS motifs, we searched for additional sites in archaeal genomes and applied the comparative genomics approach for regulon reconstruction (see Table S3).
FIG 1.
Predicted DNA-binding motifs for DtxR family regulators in Archaea (A) and Bacteria (B). Ten major groups of archaeal DNA motifs are shown by roman numerals. The number of candidate binding sites used to build a motif logo is shown below the roman numeral. Known DNA motifs of DtxR-TFs from different taxonomic groups of Bacteria were collected from the RegPrecise database (65).
The reconstructed regulons demonstrate significant differences in the number of predicted target genes and operons. The majority of archaeal DtxR-TF regulons include one to three target operons that contain 10 or fewer genes. Large- and midsize regulons involved in iron homeostasis were identified in the Halobacteriales, Thermococcales, Sulfolobales, and Methanobacteriales orders. By assessing the functional content of the reconstructed regulons, we tentatively predicted biological functions and effectors of DtxR-TFs (Table 1). These include 93 FetR regulons potentially involved in iron uptake and homeostasis (from 22 DtxR-TF clades), 36 MntR regulons for manganese uptake transporters (representing six clades), and 10 ZntR regulons containing potential zinc uptake transporters that correspond to a single clade on the DtxR-TF phylogenetic tree (see Table S1 in the supplemental material). Below we present the detailed functional analysis of the reconstructed metal uptake and homeostasis regulons in archaeal genomes.
TABLE 1.
Content of reconstructed DtxR family regulons in Archaea
| Clade | Archaeal phylum/order(s)a | Regulonb | TF no. | Motif | Target operon(s)c |
|---|---|---|---|---|---|
| 2 | E/Methanomicrobiales | FetR | 8 | I | feoAB, fetR |
| 3 | E/Methanosarcinales | FetR | 7 | I | feoAB, fetR |
| 4 | E/Methanomicrobiales | FetR | 2 | I | fepBCD, fetR, and 3 other targets |
| 5 | E/Archaeoglobales | FetR | 3 | I | feoAB, fetR |
| 6 | C/Desulfurococcales | FetR | 2 | I | feoAB, fetR, cl00388 |
| 7 | E/Thermococcales | FetR | 7 | I | feoABC, fetR, efeU, sufCB, COG0803, COG0428, COG1433, COG1149, and 12 other targets |
| 8 | E/unclassified | FetR | 2 | I | feoABC, COG1433, COG1149, and 3 other targets |
| 9 | C/Thermoproteales | MntR | 7 | II | mntH, mntR, COG0428 |
| 10 | C/Desulfurococcales | FetR | 1 | I | feoAB |
| 11 | E/Methanopyrales | FetR | 1 | I | feoAB |
| 12 | E/Methanomicrobiales | FetR | 1 | I | feoAB, fetR |
| 13 | K/unclassified | FetR | 1 | I | feoAB |
| 15 | E/Methanococcales | FetR | 8 | III | feoAB, fetR, hdrA, COG1142 |
| 16 | E/Archaeoglobales | FetR | 5 | I | cfd, COG1433, COG1149 |
| 17 | E/Methanobacteriales | FetR | 6 | I | feoAB, fetR, copA |
| 18 | E/Methanosarcinales | FetR | 6 | I | feoAB, fetR |
| 21 | E/Halobacteriales | MntR | 13 | IV | mtsABC, mntR, COG0428, UPF0016 |
| 23, 24 | E/Halobacteriales | FetR | 17 | I | fbpABC, fhuGCD, fetR, sufCB, dps, rhbA-ddc-rhbCDEF, COG0428, and 16 other targets |
| 26 | C/Sulfolobales | FetR | 8 | V | irt, doxDA, COG1131-COG0843, PF1925, and 6 other targets |
| 27 | C/Desulfurococcales | FetR | 2 | V | feoAB, fetR |
| 29 | E/Archaeoglobales | FetR | 3 | I | feoB, fetR |
| 30 | E/Methanosarcinales | FetR | 2 | I | feoB, fetR |
| 31 | E/Methanobacteriales | FetR | 1 | I | feoB |
| 32 | E/Methanosarcinales, Methanomicrobiales, Methanocellales | ZntR | 10 | VI | znuABC, nikQOM, zntR, COG4430 |
| 35 | C/Sulfolobales | MntR | 8 | VII | mntH, vit, COG0428 |
| 37 | E/unclassified | MntR | 2 | VIII | mtsABC, mntR |
| 38 | E/Thermoplasmatales | MntR | 4 | X | mntH |
| 39 | E/Archaeoglobales | MntR | 2 | IX | mtsABC, mntR |
Phyla are indicated by one-letter acronyms: E, Euryarchaeota; C, Crenarchaeota; K, Korarchaeota.
Functional roles of target genes were homeostasis of iron (FetR), zinc (ZntR), and manganese (MntR) ions.
Detailed information on studied genomes, regulators, and identified target operons can be found in Tables S1 and S2 in the supplemental material.
Iron regulons. (i) Iron uptake transporters.
In all reconstructed FetR regulons, we observed that the majority of target genes constitute metal ion transporters from different families. The most widely distributed transporter under the predicted FetR regulation is the ferrous iron transporter FeoAB, which was observed in 56 reconstructed iron regulons. The FeoAB transporter is predicted to be regulated by FetR in all studied archaeal lineages except the Halobacteriales and Sulfolobales (Table 1). The reconstructed FetR regulons in the Halobacteriales include the ferric iron ABC transporter fbpABC and multiple paralogs of the siderophore ABC transporter genes fhuCDG. In the Thermococcales and Halobacteriales, the FetR regulons include COG0428, encoding the ZIP family metal ion transporters that are known to take up ferrous iron, manganese, zinc, and cadmium ions (47). In the Sulfolobales order, the reconstructed FetR regulons contain up to six paralogs of novel putative iron transporters (named Irt, for iron-regulated transporters). The novel Irt transporters are most similar to the DHA2 family of drug-H+ antiporters from the major facilitator superfamily (MFS) according to the TCDB database (48). The exact function of Irt transporters remains to be elucidated.
Additional transporters were found in several FetR regulons. In the Thermococcales genomes, the regulons include proteins homologous to the ferrous iron transporter EfeU, characterized in bacteria (49), and proteins from the COG0803 family of metal-binding components of ABC transporters. Members of the latter family are known to transport manganese (12) and zinc (50). In the Sulfolobales, we found a conserved FetR-regulated operon encoding a hypothetical ABC-type exporter (COG1131-COG0842). The siderophore transporter FepBCD was observed in the Methanospirillum hungatei FetR regulon, whereas in the Methanobacteriales genomes it includes the copA genes, encoding the heavy-metal transport ATPase (∼70% similarity to CopA from Archaeoglobus fulgidus [51]).
(ii) Other iron homeostasis genes.
Besides iron and other metal transport systems, the reconstructed FetR regulons contain other genes potentially involved in iron homeostasis. In five Halobacteriales genomes, FetR presumably controls the rhbA-ddc-rhbCDEF operon encoding the rhizobactin siderophore biosynthesis pathway. In the Thermococcales, Archaeoglobales, and Aciduliprofundum spp., the FetR regulons contain several genes encoding potential Fe-S cluster assembly enzymes, including the SufB and SufC proteins (52), 4Fe-4S cluster binding proteins from the COG1149 family, the Fe/Mo-cofactor binding proteins from the COG1433 family, and homologs of eukaryotic CFD1, encoding an Fe-S cluster assembly protein (53). Moreover, the extended FetR regulons in the Thermococcales include genes encoding proteins with either metal-binding or Fe-S cluster sites: metal ion-binding endonuclease Nfo, metal-dependent hydrolase COG1237, Fe-S cluster tRNA wyosine derivative biosynthesis protein Taw1, Fe-S cluster sarcosine oxidase SoxAB, iron-binding methionine aminopeptidase Map, and Fe-S cluster oxidoreductase COG0731. In the Methanococcales, we found a conserved FetR regulon member, the hdrA gene, encoding a close homolog of the CoB-CoM heterodisulfide reductase subunit from Methanothermobacter marburgensis (∼75% similarity) that contains a 4Fe-4S cluster and catalyzes the final step in the methanogenic pathway (54). FetR regulons in eight Halobacteriales members contain dps genes encoding ferritin proteins that are capable of assembling into homopolymeric spheres and store ferrous iron (55). In two Metallosphaera and one Sulfolobales genome, the FetR regulons contain the doxDA operon, encoding a terminal quinol oxidase involved in electron transfer from sulfur oxidation, as well as genes encoding the putative sulfite exporter PF01925 (56).
Manganese and zinc uptake regulons. (i) TroA family manganese and zinc transporters.
The reconstructed MntR and ZntR regulons in most lineages contain metal uptake ABC transporters from the TroA family. In Bacteria, this family is known to include transporters with different metal ion specificities. Most of the characterized members are specific to either zinc (ZnuABC) (4, 50, 57–59) or manganese (MtsABC) (60). The predicted ZntR-regulated ABC transporters in the Methanosarcinales and Methanomicrobiales orders are most similar to the zinc-specific transporter P73085_SYNY3 from Cyanobacteria (57); thus, they were named ZnuABC. The TroA family proteins from the Archaeoglobales and Aciduliprofundum spp. belong to the MntR regulons; thus, they were named MtsABC. One of these MntR regulators, AF_1984 in A. fulgidus, previously was shown to bind manganese as an effector (61). The MntR-regulated transporter from the TroA family in the Halobacteriales previously was shown to be differentially expressed in Halobacterium sp. strain NRC-1 in response to manganese (23); thus, it was named MtsABC.
(ii) Predicted manganese transporters from other families.
The reconstructed MntR regulons in three archaeal lineages (Sulfolobales, Thermoproteales, and Thermoplasmatales) include the mntH gene, encoding an NRAMP family divalent metal transporter. The closest characterized homologs of these archaeal transporters are the MntH manganese transporters from B. subtilis and E. coli (5, 12, 62). In four Halobacteriales genomes, the inferred MntR regulons include the UPF0016 gene, encoding a hypothetical protein from an uncharacterized family (PF01169), which is predicted to contain seven transmembrane helixes, suggesting its localization within the cytoplasmic membrane. In Halorhabdus utahensis and Haloquadratum walsbyi, UPF0016 is the only predicted member of the MntR regulons; thus, it was predicted to function as a manganese transporter. The COG0428 ZIP family transporters were observed in the predicted MntR regulons in five Sulfolobales genomes, one Thermoproteus neutrophilus genome, and two Halobacteriales genomes. Finally, all reconstructed MntR regulons in the Sulfolobales genomes contain a second type of predicted manganese transporter from the VIT family (63).
Refinement of reconstructed regulons with experimental data.
To confirm our regulon reconstructions, we compared them with previously published experimental studies of DtxR-TFs in Archaea. In A. fulgidus, electromobility shift assay (EMSA) and DNase footprinting have confirmed the binding ability of AF_1984 (MntR, clade 39) to its own promoter region (61), which is consistent with our data. In P. furiosus, the quantitative PCR analysis of the PF0851 (FetR, clade 7) deletion mutant has detected the differential expression of genes PF0723 (efeU or ftr1) and PF0858 (feoA) under iron-limiting conditions compared to the wild type (25). Both of these genes are preceded by the predicted FetR binding sites (see Tables S3 in the supplemental material). Specific binding of the PF0851 protein to the ftr1 and feoA promoter regions was further confirmed using EMSA (25). In Thermococcus kodakaraensis, the global transcriptional analysis of the TK0107 (FetR, clade 7) mutant has revealed 5- to 27-fold changes for the TK0652 (COG0428), TK0714-16 (feoAAB), and TK0958-57 (feoAB) operons under iron-limited conditions (26), confirming the predicted FetR-binding sites in the upstream regions of these three operons (see Table S3). In Halobacterium sp. strain NRC-1, the analysis of VNG_0536G (MntR, clade 21; also named SirR) mutant has revealed the involvement of this regulator into manganese-dependent repression of the mtsABC (or zurAM-ycdH) operon (23). The c 2.0 database for systems-level models of gene regulation in prokaryotes contains groups of coregulated genes predicted by analysis of combined gene expression and conditional data (64). One of these groups, represented by the condition-dependent coregulated module hc38890 in Halobacterium sp. strain NRC-1, contains the following 10 operons from FetR regulon the reconstructed in this work: VNG_6210G (rhbA-ddc-rhbCDEF), VNG_0249G (PF00127-hyp4), VNG_0924G (fbpABC-hyp7), VNG_0925C (rgp), VNG_1036H (PF13618-COG2303), VNG_1720H (fhuD), VNG_2299H (X-COG0492), VNG_2442H (hyp6), VNG_2549C (fhuDGC), and VNG_2562H (fhuDGC). The regulatory motif inferred for the hc38890 module in EGRIN also is consistent with the FetR binding motif predicted in our work (Fig. 1A, motif I). In summary, several of the regulons and DNA-binding motifs predicted in this work are consistent with previous experimental results for DtxR-TFs, confirming that the comparative genomics techniques can be applied successfully for reconstruction of transcription regulation in Archaea.
Conclusions.
Transcriptional control of metal homeostasis in Archaea is mediated by regulators from the DtxR protein family. By applying the comparative genomics approach, we tentatively predicted DNA binding motifs and reconstructed DtxR-TF regulons in 77 archaeal genomes. Functional analysis of the reconstructed regulons allowed us to predict potential functions of TFs and coregulated genes. Three major functional groups of DtxR-TFs in Archaea are FetR, MntR, and ZntR, controlling the homeostasis of iron, manganese, and zinc, respectively.
The identified DtxR-TF binding motifs in Archaea have a common palindromic structure but are characterized by different consensus sequences; thus, they were classified into 10 distinct motifs (Fig. 1). Some of these archaeal motifs are similar to known DtxR-TF motifs in Bacteria. For instance, the group I FetR motif is similar to the MntR motifs in Chloroflexi and Staphylococcaceae and the group III FetR motif resembles the MntR motif in Streptococcaceae, whereas the MntR motifs from groups II, IV, and VIII are partially similar to the FetR motif in Chloroflexi (Fig. 1). Conservation of the DtxR-TF motifs across large phylogenetic distances allowed us to suggest that DtxR is an ancient family of TFs common to Bacteria and Archaea. A large-scale phylogenetic analysis of the previously studied DtxR family proteins from Bacteria (as captured in the RegPrecise database of transcriptional regulons [65]) and the archaeal DtxR-TFs (data not shown) identifies multiple branches of bacterial regulators that mixed with numerous groups of archaeal regulators, suggesting that the DtxR family was a frequent subject of various evolutionary processes, including divergent evolution (diversification of DNA motifs and effector specificities after duplication) and convergent evolution (appearance of a similar DNA motif in distantly related branches).
The majority of predicted members of the DtxR-TF regulons are metal transporters from the FeoAB, NRAMP, ZIP, and TroA families and are involved in the uptake of iron, manganese, and zinc ions. The novel Irt family of iron-regulated transporters was identified in acidophilic iron-oxidizing microorganisms from the Sulfolobales order. The previous transcriptomics studies in Metallosphaera sedula revealed that three Irt transporter genes from the predicted FetR regulon (Msed_0907, Msed_1001, and Msed_1095) are upregulated in the presence of ferrous iron in growth medium (66). The Irt family transporters are similar to drug-proton antiporters from the MFS superfamily, suggesting they can be involved in proton-dependent efflux of an excess of ferrous iron accumulated in the cytoplasm due to its high concentration in the environment. An additional predicted exporter of ferrous iron, COG1131-COG0842, was identified in the reconstructed FetR regulons in Sulfolobales.
In summary, this work demonstrates the power of the comparative genomics approach in applying the reconstruction of transcriptional regulons in poorly studied archaeal genomes. The reconstructed DtxR-TF regulons are useful for the genome context-based prediction of novel functions of transporters in archaeal genomes. Although many of the inferred regulons are supported by available experimental data, other regulon predictions still require experimental validation.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Russian Science Foundation (grant 14-14-00289). Additional funding was provided by the Genomic Science Program (GSP), Office of Biological and Environmental Research (OBER), U.S. Department of Energy (DOE).
Footnotes
This work is a contribution of the Pacific Northwest National Laboratory (PNNL) Foundational Scientific Focus Area.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02386-14.
REFERENCES
- 1.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo–transport of ferrous iron into bacteria. Biometals 19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 2.Kim H, Lee H, Shin D. 2012. The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem Biophys Res Commun 423:733–738. doi: 10.1016/j.bbrc.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 3.Anderson DS, Adhikari P, Nowalk AJ, Chen CY, Mietzner TA. 2004. The hFbpABC transporter from Haemophilus influenzae functions as a binding-protein-dependent ABC transporter with high specificity and affinity for ferric iron. J Bacteriol 186:6220–6229. doi: 10.1128/JB.186.18.6220-6229.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabbianelli R, Scotti R, Ammendola S, Petrarca P, Nicolini L, Battistoni A. 2011. Role of ZnuABC and ZinT in Escherichia coli O157:H7 zinc acquisition and interaction with epithelial cells. BMC Microbiol 11:36. doi: 10.1186/1471-2180-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kehres DG, Zaharik ML, Finlay BB, Maguire ME. 2000. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol Microbiol 36:1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x. [DOI] [PubMed] [Google Scholar]
- 6.Helmann JD. 2014. Specificity of metal sensing: iron and manganese homeostasis in Bacillus subtilis. J Biol Chem 289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koster W. 2005. Cytoplasmic membrane iron permease systems in the bacterial cell envelope. Front Biosci 10:462–477. doi: 10.2741/1542. [DOI] [PubMed] [Google Scholar]
- 8.Hantke K. 2001. Iron and metal regulation in bacteria. Curr Opin Microbiol 4:172–177. doi: 10.1016/S1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 9.Fillat MF. 2014. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch Biochem Biophys 546:41–52. doi: 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 10.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. doi: 10.1007/s10534-006-9070-7. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto K, Ishihama A, Busby SJ, Grainger DC. 2011. The Escherichia coli K-12 MntR miniregulon includes dps, which encodes the major stationary-phase DNA-binding protein. J Bacteriol 193:1477–1480. doi: 10.1128/JB.01230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Que Q, Helmann JD. 2000. Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol Microbiol 35:1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x. [DOI] [PubMed] [Google Scholar]
- 13.Ando M, Manabe YC, Converse PJ, Miyazaki E, Harrison R, Murphy JR, Bishai WR. 2003. Characterization of the role of the divalent metal ion-dependent transcriptional repressor MntR in the virulence of Staphylococcus aureus. Infect Immun 71:2584–2590. doi: 10.1128/IAI.71.5.2584-2590.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt MP. 2002. Analysis of a DtxR-like metalloregulatory protein, MntR, from Corynebacterium diphtheriae that controls expression of an ABC metal transporter by an Mn(2+)-dependent mechanism. J Bacteriol 184:6882–6892. doi: 10.1128/JB.184.24.6882-6892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brune I, Werner H, Huser AT, Kalinowski J, Puhler A, Tauch A. 2006. The DtxR protein acting as dual transcriptional regulator directs a global regulatory network involved in iron metabolism of Corynebacterium glutamicum. BMC Genomics 7:21. doi: 10.1186/1471-2164-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wennerhold J, Bott M. 2006. The DtxR regulon of Corynebacterium glutamicum. J Bacteriol 188:2907–2918. doi: 10.1128/JB.188.8.2907-2918.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoll KE, Draper WE, Kliegman JI, Golynskiy MV, Brew-Appiah RA, Phillips RK, Brown HK, Breyer WA, Jakubovics NS, Jenkinson HF, Brennan RG, Cohen SM, Glasfeld A. 2009. Characterization and structure of the manganese-responsive transcriptional regulator ScaR. Biochemistry 48:10308–10320. doi: 10.1021/bi900980g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Li W, Wei Y, Jiang Y, Tan X. 2013. Efficient preparation and metal specificity of the regulatory protein TroR from the human pathogen Treponema pallidum. Metallomics 5:1448–1457. doi: 10.1039/c3mt00163f. [DOI] [PubMed] [Google Scholar]
- 19.Boyd J, Oza MN, Murphy JR. 1990. Molecular cloning and DNA sequence analysis of a diphtheria tox iron-dependent regulatory element (dtxR) from Corynebacterium diphtheriae. Proc Natl Acad Sci U S A 87:5968–5972. doi: 10.1073/pnas.87.15.5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ranjan S, Yellaboina S, Ranjan A. 2006. IdeR in mycobacteria: from target recognition to physiological function. Crit Rev Microbiol 32:69–75. doi: 10.1080/10408410600709768. [DOI] [PubMed] [Google Scholar]
- 21.Schmitt MP, Predich M, Doukhan L, Smith I, Holmes RK. 1995. Characterization of an iron-dependent regulatory protein (IdeR) of Mycobacterium tuberculosis as a functional homolog of the diphtheria toxin repressor (DtxR) from Corynebacterium diphtheriae. Infect Immun 63:4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guedon E, Helmann JD. 2003. Origins of metal ion selectivity in the DtxR/MntR family of metalloregulators. Mol Microbiol 48:495–506. doi: 10.1046/j.1365-2958.2003.03445.x. [DOI] [PubMed] [Google Scholar]
- 23.Kaur A, Pan M, Meislin M, Facciotti MT, El-Gewely R, Baliga NS. 2006. A systems view of haloarchaeal strategies to withstand stress from transition metals. Genome Res 16:841–854. doi: 10.1101/gr.5189606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmid AK, Pan M, Sharma K, Baliga NS. 2011. Two transcription factors are necessary for iron homeostasis in a salt-dwelling archaeon. Nucleic Acids Res 39:2519–2533. doi: 10.1093/nar/gkq1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Y, Kumar S, Menon AL, Scott RA, Adams MW. 2013. Regulation of iron metabolism by Pyrococcus furiosus. J Bacteriol 195:2400–2407. doi: 10.1128/JB.02280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louvel H, Kanai T, Atomi H, Reeve JN. 2009. The Fur iron regulator-like protein is cryptic in the hyperthermophilic archaeon Thermococcus kodakaraensis. FEMS Microbiol Lett 295:117–128. doi: 10.1111/j.1574-6968.2009.01594.x. [DOI] [PubMed] [Google Scholar]
- 27.Blaby-Haas CE, Furman R, Rodionov DA, Artsimovitch I, de Crecy-Lagard V. 2011. Role of a Zn-independent DksA in Zn homeostasis and stringent response. Mol Microbiol 79:700–715. doi: 10.1111/j.1365-2958.2010.07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas CE, Rodionov DA, Kropat J, Malasarn D, Merchant SS, de Crecy-Lagard V. 2009. A subset of the diverse COG0523 family of putative metal chaperones is linked to zinc homeostasis in all kingdoms of life. BMC Genomics 10:470. doi: 10.1186/1471-2164-10-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panina EM, Mironov AA, Gelfand MS. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res 29:5195–5206. doi: 10.1093/nar/29.24.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panina EM, Mironov AA, Gelfand MS. 2003. Comparative genomics of bacterial zinc regulons: enhanced ion transport, pathogenesis, and rearrangement of ribosomal proteins. Proc Natl Acad Sci U S A 100:9912–9917. doi: 10.1073/pnas.1733691100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. 2006. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLoS Comp Biol 2:e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schroder J, Jochmann N, Rodionov DA, Tauch A. 2010. The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics 11:12. doi: 10.1186/1471-2164-11-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Todd JD, Sawers G, Rodionov DA, Johnston AW. 2006. The Rhizobium leguminosarum regulator IrrA affects the transcription of a wide range of genes in response to Fe availability. Mol Genet Genomics 275:564–577. doi: 10.1007/s00438-006-0115-y. [DOI] [PubMed] [Google Scholar]
- 34.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res 41:D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, Sonnhammer EL, Tate J, Punta M. 2014. Pfam: the protein families database. Nucleic Acids Res 42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 38.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. 2007. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinformatics 8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodionov DA. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem Rev 107:3467–3497. doi: 10.1021/cr068309+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res 38:W299–W307. doi: 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mironov AA, Vinokurova NP, Gelfand MS. 2000. Software for analysis of bacterial genomes. Mol Biol 34:222–231. doi: 10.1007/BF02759643. [DOI] [PubMed] [Google Scholar]
- 42.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res 38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.UniProt Consortium. 2014. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann K, Stoffel W. 1993. TMbase–a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 374:166. doi: 10.1515/bchm3.1993.374.1-6.143. [DOI] [Google Scholar]
- 46.Tao X, Murphy JR. 1992. Binding of the metalloregulatory protein DtxR to the diphtheria tox operator requires a divalent heavy metal ion and protects the palindromic sequence from DNase I digestion. J Biol Chem 267:21761–21764. [PubMed] [Google Scholar]
- 47.Guerinot ML. 2000. The ZIP family of metal transporters. Biochim Biophys Acta 1465:190–198. doi: 10.1016/S0005-2736(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 48.Saier MH Jr, Reddy VS, Tamang DG, Vastermark A. 2014. The transporter classification database. Nucleic Acids Res 42:D251–D258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grosse C, Scherer J, Koch D, Otto M, Taudte N, Grass G. 2006. A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol Microbiol 62:120–131. doi: 10.1111/j.1365-2958.2006.05326.x. [DOI] [PubMed] [Google Scholar]
- 50.Gaballa A, Helmann JD. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J Bacteriol 180:5815–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandal AK, Cheung WD, Arguello JM. 2002. Characterization of a thermophilic P-type Ag+/Cu+-ATPase from the extremophile Archaeoglobus fulgidus. J Biol Chem 277:7201–7208. doi: 10.1074/jbc.M109964200. [DOI] [PubMed] [Google Scholar]
- 52.Outten FW, Wood MJ, Munoz FM, Storz G. 2003. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J Biol Chem 278:45713–45719. doi: 10.1074/jbc.M308004200. [DOI] [PubMed] [Google Scholar]
- 53.Roy A, Solodovnikova N, Nicholson T, Antholine W, Walden WE. 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J 22:4826–4835. doi: 10.1093/emboj/cdg455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duin EC, Madadi-Kahkesh S, Hedderich R, Clay MD, Johnson MK. 2002. Heterodisulfide reductase from Methanothermobacter marburgensis contains an active-site [4Fe-4S] cluster that is directly involved in mediating heterodisulfide reduction. FEBS Lett 512:263–268. doi: 10.1016/S0014-5793(02)02281-0. [DOI] [PubMed] [Google Scholar]
- 55.Chiancone E, Ceci P, Ilari A, Ribacchi F, Stefanini S. 2004. Iron and proteins for iron storage and detoxification. Biometals 17:197–202. doi: 10.1023/B:BIOM.0000027692.24395.76. [DOI] [PubMed] [Google Scholar]
- 56.Weinitschke S, Denger K, Cook AM, Smits TH. 2007. The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology 153:3055–3060. doi: 10.1099/mic.0.2007/009845-0. [DOI] [PubMed] [Google Scholar]
- 57.Banerjee S, Wei B, Bhattacharyya-Pakrasi M, Pakrasi HB, Smith TJ. 2003. Structural determinants of metal specificity in the zinc transport protein ZnuA from Synechocystis 6803. J Mol Biol 333:1061–1069. doi: 10.1016/j.jmb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 58.Liu M, Yan M, Liu L, Chen S. 2013. Characterization of a novel zinc transporter ZnuA acquired by Vibrio parahaemolyticus through horizontal gene transfer. Front Cell Infect Microbiol 3:61. doi: 10.3389/fcimb.2013.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murphy TF, Brauer AL, Kirkham C, Johnson A, Koszelak-Rosenblum M, Malkowski MG. 2013. Role of the zinc uptake ABC transporter of Moraxella catarrhalis in persistence in the respiratory tract. Infect Immun 81:3406–3413. doi: 10.1128/IAI.00589-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun X, Ge R, Chiu JF, Sun H, He QY. 2008. Lipoprotein MtsA of MtsABC in Streptococcus pyogenes primarily binds ferrous ion with bicarbonate as a synergistic anion. FEBS Lett 582:1351–1354. doi: 10.1016/j.febslet.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 61.Bell SD, Cairns SS, Robson RL, Jackson SP. 1999. Transcriptional regulation of an archaeal operon in vivo and in vitro. Mol Cell 4:971–982. doi: 10.1016/S1097-2765(00)80226-9. [DOI] [PubMed] [Google Scholar]
- 62.Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MF. 2000. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol 35:1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x. [DOI] [PubMed] [Google Scholar]
- 63.Kim SA, Punshon T, Lanzirotti A, Li L, Alonso JM, Ecker JR, Kaplan J, Guerinot ML. 2006. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314:1295–1298. doi: 10.1126/science.1132563. [DOI] [PubMed] [Google Scholar]
- 64.Brooks AN, Reiss DJ, Allard A, Wu WJ, Salvanha DM, Plaisier CL, Chandrasekaran S, Pan M, Kaur A, Baliga NS. 2014. A system-level model for the microbial regulatory genome. Mol Syst Biol 10:740. doi: 10.15252/msb.20145160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novichkov PS, Kazakov AE, Ravcheev DA, Leyn SA, Kovaleva GY, Sutormin RA, Kazanov MD, Riehl W, Arkin AP, Dubchak I, Rodionov DA. 2013. RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics 14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Auernik KS, Maezato Y, Blum PH, Kelly RM. 2008. The genome sequence of the metal-mobilizing, extremely thermoacidophilic archaeon Metallosphaera sedula provides insights into bioleaching-associated metabolism. Appl Environ Microbiol 74:682–692. doi: 10.1128/AEM.02019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.