Abstract
Glycolipids are found mainly in photosynthetic organisms (plants, algae, and cyanobacteria), Gram-positive bacteria, and a few other bacterial phyla. They serve as membrane lipids and play a role under phosphate deprivation as surrogates for phospholipids. Mesorhizobium loti accumulates different di- and triglycosyl diacylglycerols, synthesized by the processive glycosyltransferase Pgt-Ml, and two so far unknown glycolipids, which were identified in this study by mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy as O-methyl-digalactosyl diacylglycerol (Me-DGD) and glucuronosyl diacylglycerol (GlcAD). Me-DGD is a novel glycolipid, whose synthesis depends on Pgt-Ml activity and the involvement of an unknown methyltransferase, while GlcAD is formed by a novel glycosyltransferase encoded by the open reading frame (ORF) mlr2668, using UDP-glucuronic acid as a sugar donor. Deletion mutants lacking GlcAD are not impaired in growth. Our data suggest that the different glycolipids in Mesorhizobium can mutually replace each other. This may be an adaptation mechanism to enhance the competitiveness in natural environments. A further nonphospholipid in Mesorhizobium was identified as a hydroxylated form of an ornithine lipid with the additional hydroxy group linked to the amide-bound fatty acid, introduced by the hydroxylase OlsD. The presence of this lipid has not been reported for rhizobia yet. The hydroxy group is placed on the C-2 position of the acyl chain as determined by NMR spectroscopy. Furthermore, the isolated ornithine lipids contained up to 80 to 90% d-configured ornithine, a stereoform so far undescribed in bacteria.
INTRODUCTION
Glycerolipids represent the main building blocks for biological membranes. Based on their head groups, they can be classified into different groups, with the phospho- and glycolipids as the predominant representatives. While phospholipids are widespread and found in almost all organisms, glycolipids are restricted mainly to photosynthetic organisms (plants, algae, and cyanobacteria), Gram-positive bacteria, and a few members of other bacterial phyla, including some representatives of Gram-negative bacteria (Proteobacteria) (1). An important group of bacteria with high ecological and economical significance are the Rhizobiales (Alphaproteobacteria), with many members being capable of fixing atmospheric nitrogen in symbiosis with legumes. These bacteria often contain the charged glycolipid sulfoquinovosyl diacylglycerol (SQD), as shown for Sinorhizobium and Rhizobium (2, 3). A further important bacterium is the plant pathogen and model organism Agrobacterium tumefaciens (Rhizobiaceae), which accumulates four different glycolipids under phosphate deprivation (4). These lipids were identified as digalactosyl diacylglycerol (DGD) and glucosylgalactosyl diacylglycerol (GGD), with all sugars in these two lipids bound in β-anomeric configuration, and as monoglucosyl diacylglycerol (MGlcD) and glucuronosyl diacylglycerol (GlcAD), with α-anomeric configuration of the sugars. GGD and DGD in Agrobacterium are synthesized by the processive glycosyltransferase Pgt-Atum (4, 5), while MGlcD and GlcAD are formed by a recently described single α-glycosyltransferase (6). These glycolipids in Agrobacterium serve as surrogates for phospholipids under phosphate deprivation, as shown for other organisms (plants, cyanobacteria, and bacteria) (1). The nodule bacterium Mesorhizobium loti (Phyllobacteriaceae, Rhizobiales), a symbiont of Lotus japonicus, shows a similar lipid composition and response to phosphate deprivation as observed for Agrobacterium. Mesorhizobium contains a processive glycosyltransferase, Pgt-Ml, with homology to Pgt-Atum. Pgt-Ml synthesizes DGD, GGD, and different triglycosyl diacylglycerols (TGDs) with galactose and/or glucose in their head groups (5, 7). Mesorhizobium further accumulates two unknown glycolipids, U1 and U2 (7), with U1 depending on Pgt-Ml activity. U2 seems to be similar to GlcAD detected in Agrobacterium, as concluded from the similar chromatographical behavior of the two lipids (4, 6).
Despite the high diversity of bacterial glycolipids, the number of glycosyltransferases isolated so far is much lower (1). Two of the best-studied glycosyltransferases are the MGlcD and diglucosyl diacylglycerol (DGlcD) synthase from the cell wall-lacking bacterium Acholeplasma laidlawii (Mollicutes) (8, 9). The two enzymes are classified in glycosyltransferase family 4 (GT4) in the Carbohydrate Active Enzymes database (http://www.cazy.org/), a comprehensive collection of glycosyltransferases (10, 11). The sugars in MGlcD and DGlcD are bound in α-anomeric configuration. Additional well-characterized glycosyltransferases are Pgt-Atum and Pgt-Ml and the processive glycosyltransferases from different Gram-positive bacteria (4, 5, 7, 12, 13). Pgt-Atum and Pgt-Ml are members of GT21, while the processive glycosyltransferases from Gram-positive bacteria belong to GT28. Enzymes in both GT21 and GT28 are β-glycosyltransferases.
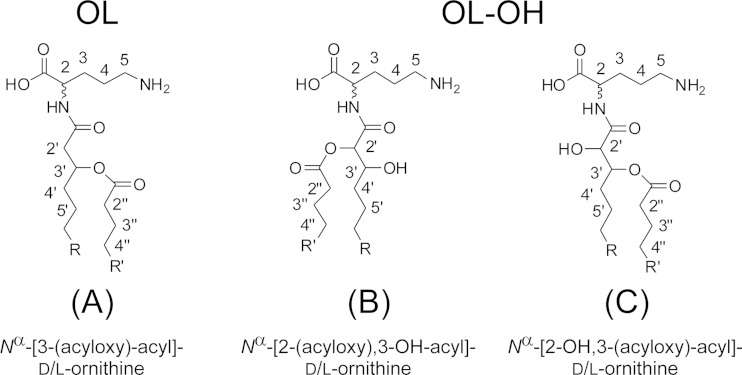
Besides glycolipids, different organisms (bacteria and algae) accumulate additional phosphate-free membrane lipids (14), such as diacylglyceryl trimethylhomoserine (DGTS) and ornithine lipid (OL). The ornithine head group of OL is directly linked via an amide bond to a hydroxy fatty acid. A second fatty acid is esterified to the hydroxy group of this amide-bound fatty acid. There are several modifications of OL with respect to hydroxylation of different positions in the molecule. The inserted hydroxy group can be found at one of the two fatty acids and/or the ornithine head group (4, 15, 16). The position of the hydroxy group of the ester-linked fatty acid was resolved with gas chromatography-mass spectrometry (GC-MS) at C-2 (16), while there are no structural details available on the positions of the hydroxy groups inserted at the amide-bound fatty acid or at the head group. Like glycolipids, DGTS and OL serve as surrogates for phospholipids, as shown for Sinorhizobium, Agrobacterium, and Mesorhizobium (4, 7, 14).
The elucidation of the head group structures of the unknown glycolipids U1 and U2, the identification of the glycosyltransferase synthesizing U2, and the structural analysis of ornithine lipids from M. loti are the subjects of this study.
(Part of the research concerning O-methyl-digalactosyl diacylglycerol [Me-DGD] from Mesorhizobium was presented at the FEBS Workshop on Microbial Lipids: Diversity in Structure and Formation, Bern, Switzerland, 16 to 19 May, 2013).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
If not otherwise indicated, M. loti strain R7a was used in this study. Mesorhizobium was grown as described previously (7) with slight changes. M. loti Δagt was grown in the presence of gentamicin at a final concentration of 20 mg liter−1. For growth curve experiments, M. loti wild type and the Δagt mutant were cultivated first in yeast extract-tryptone (YT) medium. After 3 days the cells were harvested by centrifugation, including a washing step of the cells with sterile distilled water. The cells were transferred to AB minimal medium [12.5 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES) buffer] with high (1.25 mM) or low (0.5 μM) phosphate (with most of the phosphate replaced with 10 mM KCl and 5 mM NaCl). The growth was started at an A600 of 0.05 and stopped after about 60 h. A simplified protocol was used to harvest cell material for lipid analyses. For this purpose, cells grown on YT medium were harvested by centrifugation and transferred to a 5-fold volume of AB minimal medium lacking phosphate (phosphate was replaced with 10 mM KCl and 5 mM NaCl). To grow cells under phosphate-replete conditions, AB minimal medium containing 1.25 mM phosphate was inoculated with 1 ml liter−1 of a Mesorhizobium culture grown in YT medium. All cultures in AB minimal medium were incubated for 6 days. Escherichia coli strain BL21(DE3) (Novagen) was used as an expression host for the Agt open reading frame (ORF) mlr2668. Expression cultures were induced with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) at an A600 of 0.6, incubated for 4 h at 37°C, and used for lipid analyses or enzyme assays. Rhizobium tropici was grown in YT medium. Agrobacterium tumefaciens C58 was grown in yeast extract-peptone (YEP) as described previously (4). Agrobacterium expression cultures were grown in the presence of kanamycin (50 mg/liter).
Cloning of Agt and OlsD expression vectors.
Complete genome sequence data are available only for M. loti MAFF303099 and not for R7a (17). Therefore, all of the DNA sequences used in this study were retrieved from strain MAFF303099. DNA used as the template for PCR was extracted from strain R7a. The ORF mlr2668 was amplified with the primers BN900 (CCTAGGTATGCGCATATTGATGGTCACC) and BN901 (GGATCCTCAGGCCGCCAGCTTGCGCG) containing XmaJI and BamHI restriction sites (underlined) and cloned in pGEM-T Easy (Promega). The fragment mlr2668 was released and inserted into the bacterial expression vector pTnVagro (5) after excision of the former ORF by digestion with XmaJI/BamHI, resulting in the new expression vector pTnV-mlr2668. E. coli BL21(DE3) (Novagen) was used as the expression host. The ORF mll1937 was amplified with the primers BN1535 (CCTAGGTATGTCCCGCACCTACGATCTTG) and BN1231 (GGATCCTCAGCCGCTGAACGACACGC). The cloning strategy was similar to that described for ORF mlr2668, with pJet1.2 (Thermo Scientific) as the cloning vector. The resulting expression vector was pTnV-mll1937 used for expression in A. tumefaciens.
Construction of the M. loti Δagt and ΔolsD deletion mutants by disruption of the mlr2668 and mll1937 loci, respectively, using a Gmr cassette.
With respect to the Δagt mutant, the 5′ and 3′ sequences (flanking the gentamicin resistance [Gmr] cassette) used for homologous recombination were amplified with the primer pair BN946 (TGATCAAGGTTCCAGCCTAGG) and BN947 (CCATGGGAAAGGTCATCGGCGAGCGA) and the pair BN948 (ACGCGTAAGCTCAAGTCCCGCTACCC) and BN949 (GCAAGCACCGTCAGACAGAG), respectively. NcoI and MluI restriction sites are underlined. The two PCR products were cloned in pGEM-T Easy. For further cloning, the 5′ sequence was released by digestion with NcoI and NsiI, which is a restriction site of the cloning vector. The vector containing the 3′ sequence was linearized with MluI and NsiI. The Gmr cassette (4) was released by digestion with MluI/NcoI. All three restricted fragments were ligated in one step with the Gmr cassette cloned in antisense orientation to the flanking sequences. The resulting vector was used to transform M. loti (R7a). The correct disruption of the mlr2668 locus was confirmed by PCR with the following primer combinations: P1 (GGAAGTCAGCGCCGATCATG) and P2 (AGTGGCTCTCTATACAAAGTTG); P3 (TTTGCCATACGGTCGAGAACG) and P4 (TTCGGTCAAGGTTCTGGACC); and P1 and P4. The strategy for deletion of mll1937 was similar to that described for the Δagt mutant. However, the 3′ sequence was released by digestion with MluI and PsiI, which is a restriction site of the cloning vector. The vector containing the 5′ sequence was linearized with NcoI and PsiI. The 5′ and 3′ sequences were amplified before cloning with the primer pair BN1236 (TGAATGCATCGAAGCAACGTCC) and BN1237 (CCATGGACGAGATCGCGGCCGATTTG) and the pair BN1238 (ACGCGTGAAACGCGCATAGCCCTGTTG) and BN1239 (GTCAGGCTTTCGTGCAGGAAC), respectively (restriction sites are underlined). The correct disruption of the mll1937 locus was confirmed by PCR with the following primer combinations: P5 (AATGCCGATGTCGAATGCAACC) and P2; P6 (AGGCAAACCTCAGCGTCAAG) and P4; and P5 and P6.
Lipid isolation, separation, and analysis by GC-MS and Q-TOF MS.
Lipids were isolated, separated, and analyzed as described by Geske et al. (4). Glycolipids from Mesorhizobium and E. coli (expressing mlr2668) were separated by preparative one-dimensional thin-layer chromatography (TLC). Purification of GlcAD from Mesorhizobium required two steps due to comigrating lipids. In the first step total lipid extracts from Mesorhizobium were separated on TLC plates pretreated with ammonium sulfate (0.15 M) and activated by heat (120°C for 2.5 h) prior to use. In this system U2 is protonated and migrates clearly above monogalactosyl diacylglycerol (MGD) (4). U2 and comigrating lipids were scraped off and extracted from the silica material with chloroform-methanol (2:1) in an ultrasonic bath for 30 min. The extracted lipid mixture was subjected to a second TLC step using nontreated plates to obtain pure U2 lipid, which migrates similarly to DGD in this TLC system (this study). The hydroxylated ornithine lipid (OL-OH) from Mesorhizobium (R7a) as well as OL from Mesorhizobium (R7a and MAFF303099), R. tropici, and Agrobacterium were separated by preparative one-dimensional TLC in the solvent chloroform-methanol-H2O (65:25:4). In the event that OL-OH and OL were not sufficiently separated from other lipids, a second purification step was necessary using the solvent chloroform-methanol-acetic acid-H2O (90:15:10:3.5). For quadrupole time of flight mass spectrometry (Q-TOF MS), the fragmentation energy was 30 V for OL-OH, 20 V for GlcAD/U2, and 12 V for MGlcD and Me-DGD/U1.
NMR spectroscopy.
All nuclear magnetic resonance (NMR) spectroscopic measurements were performed at 27°C using 3-mm or 5-mm tubes on a Bruker Avance III 700 MHz NMR instrument (equipped with an inverse 5-mm quadruple-resonance Z-gradient cryoprobe). Deuterated solvents were purchased from Deutero GmbH (Kastellaun, Germany). Prior to NMR analysis, MGlcD was per-O-acetylated with acetic anhydride in pyridine (1:2) and further purified as described previously (16). Prior to the measurement in CDCl3, this purified per-O-acetylated glycolipid was evaporated twice with CDCl3 under a stream of nitrogen to reduce residual water. Chemical shifts were referenced to internal chloroform (δH 7.26; δC 77.16). Samples which were not per-O-acetylated (GlcAD, OL, and OL-OH) were exchanged twice from methanol (MeOH)-d4 under a stream of nitrogen and were measured in CDCl3–MeOH-d4–D2O (40:53:7). Chemical shifts were referenced to external tetramethylsilane (δH 0.0, δC 0.0) in the same solvent mixture. NMR spectroscopy of Me-DGD was performed on the O-deacylated sample after treatment with absolute hydrazine at 37°C for 30 min. The sample was measured in D2O, and chemical shifts were reported according to an external standard of acetone (δH 2.225, δC 31.45). All data were acquired and processed by using Bruker Topspin, version 3.0 and higher. 1H NMR assignments were confirmed by two-dimensional 1H,1H-COSY (where COSY is correlation spectroscopy) and 1H,1H-TOCSY (total correlation spectroscopy) experiments. 13C NMR assignments were indicated by two-dimensional 1H,13C-heteronuclear single quantum correlation (HSQC), based on the 1H NMR assignments. Interresidue connectivity and further evidence for 13C assignment were obtained normally from two-dimensional 1H,13C-heteronuclear multiple bond correlation (HMBC) and 1H,13C-HSQC-TOCSY.
Absolute configuration of ornithine in ornithine lipids.
The determination of the absolute configuration of ornithine was performed by acidic butanolysis with (R)-2-butanol as described previously (18). The acetylated butyl derivatives were measured by gas chromatography-mass spectrometry, and the configuration was determined by using l-ornithine as a reference. GC-MS analysis was performed on an Agilent 5975 inert XL mass-selective detector (MSD) instrument equipped with an HP-5ms capillary column (30 m by 0.25 mM; film thickness, 0.25 μM) and applying a temperature gradient of 150°C (kept for 3 min) to 320°C at 5°C/min.
Enzyme assays.
For enzyme assays E. coli BL21(DE3) cultures containing the expression vector pTnV-mlr2668 were harvested by centrifugation. The pellet was resuspended in 1 ml of buffer 1 (5, and cells were disrupted with glass beads using a Precellys homogenizer (Peqlab). Cell debris was removed by centrifugation at 70 × g for 1 min. The assays were performed in a final volume of 205 μl with 100 μl of buffer 2 (15 mM Tricine-KOH, pH 7.2, 30 mM MgCl2, 3 mM dithiothreitol [DTT]), 50 μl UDP-glucuronic acid or UDP-glucose (40 pmol/μl), 50 μl of E. coli protein extract, and 5 μl of 18:2/18:2-diacylglycerol [DAG(18:2/18:2)] (10 nmol/μl in ethanol). After incubation for 60 min at 28°C, the assays were terminated by the addition of 3 ml of chloroform-methanol (2:1) and 0.5 ml NaCl solution (0.9%). The lipids were extracted as described previously (4, 7) and separated by TLC. MGD and DGD were used as reference lipids to identify the positions of MGlcD and GlcAD on the TLC plate. Corresponding bands were scraped off the silica plate and extracted for 30 min with chloroform-methanol (2:1) in an ultrasonic bath. The extracted reaction products were analyzed with the Q-TOF mass spectrometer (Agilent) by direct nanospray infusion in the positive mode as described previously (4, 7) using fragmentation energies of 12 V for MGlcD and 20 V for GlcAD.
RESULTS
Identification of a new glycosyltransferase (ORF mlr2668) from M. loti synthesizing MGlcD and GlcAD after expression in E. coli.
As shown in an earlier study, Mesorhizobium accumulates two unknown glycolipids, U1 and U2, when grown under phosphate starvation (7). A glycolipid with the same chromatographical characteristics as the mesorhizobial U2 was also described for Agrobacterium (4, 6). The presence of similar glycolipids in the two organisms indicated the existence of glycosyltransferases with homologous sequences in Agrobacterium and Mesorhizobium. To identify the glycosyltransferase responsible for the synthesis of U2, we performed database searches and identified 14 candidate sequences present in the two organisms. The ORF mlr2668 showed high sequence similarity to atu2297 from Agrobacterium, an ORF investigated in a parallel study (6). Expression of mlr2668 led to the accumulation of two novel glycolipids in E. coli (Fig. 1). One of the lipids migrating slightly above MGD was expected to be α-MGlcD, deduced from its chromatographical behavior (6, 19). The anomeric configuration of this lipid can be further inferred from the sequence of mlr2668, which shows that it is a member of GT4 (20), coding for an α-glycosyltransferase. The second glycolipid migrating similarly to DGD showed chromatographic characteristics similar to those of U2 (4, 7). For further analyses, the two glycolipids were subjected to NMR spectroscopy. The identity of the first lipid could be confirmed as MGlcD, with the NMR data in accordance with previously reported values (19), and the second was identified as GlcAD (see Table S1 in supplemental material). The sugars in the two lipids are bound in α-anomeric configuration. Therefore, the ORF mlr2668 codes for an α-glycosyltransferase (named Agt) synthesizing MGlcD and GlcAD. However, the accumulation of MGlcD in Mesorhizobium has never been described (7). Possible explanations for the absence of MGlcD in Mesorhizobium may be (i) that this lipid is not synthesized in detectable amounts by Agt in Mesorhizobium, (ii) that MGlcD is an intermediate, which is below the detection limit, or (iii) that its synthesis depends on other conditions than phosphate starvation.
FIG 1.

TLC showing the accumulation of two new glycolipids in E. coli expressing Agt from Mesorhizobium. The two glycolipids were further analyzed by subsequent NMR spectroscopy and identified as MGlcD and GlcAD. No glycolipids were observed in the empty vector control. Glycolipids were stained with α-naphthol–H2SO4 at 137°C.
The glycolipids U1 and U2 from Mesorhizobium contain O-methyl-digalactose and glucuronic acid in their head groups, respectively.
The structural determination of the head groups of the unknown glycolipids U1 and U2 from Mesorhizobium may reveal details about the enzymes involved in the synthesis of these lipids. For this purpose the lipids were separated via TLC and analyzed with Q-TOF MS and NMR spectroscopy. The analysis with Q-TOF MS enables the identification of molecules by their masses and fragmentation patterns, but it does not distinguish between different epimeric and anomeric configurations of the sugars. Figure 2 shows the Q-TOF tandem MS (MS/MS) spectra and illustrates the head group structures of U1 and U2. The fragmentation of glycolipids with Q-TOF MS/MS leads to a neutral loss of the sugar head group and to the detection of fragments representing DAG and monoacylglycerol (MAG). The neutral loss of 373.1578 u (Fig. 2A) was consistent with the loss of a methylated dihexose as an ammonia adduct, while the neutral loss of 211.0760 u derived from hexuronic acid as ammonia adduct (Fig. 2B). Therefore, U1 could be identified as methylated dihexosyl diacylglycerol (Fig. 2A), and U2 was identified as hexuronosyl diacylglycerol (Fig. 2B). The head group structures of the two lipids were further analyzed by NMR. Thus, U1 was identified as β-d-Galp-(1→6)-(3-O-Me)-β-d-Galp-(1→3)-1,2-di-O-acyl-sn-Gro (NMR data of de-O-acylated Me-DGD are shown in Fig. S1 in the supplemental material; see also Table S2 in the supplemental material), and U2 was identified as α-GlcAD. NMR shifts of U2 were congruent with the ones reported for GlcAD isolated from Agt expressing E. coli (see Table S1 in the supplemental material). These results suggest that Me-DGD is synthesized by a β-glycosyltransferase, probably Pgt-Ml, since the presence of this lipid depends on Pgt-Ml activity (7), with involvement of an unknown O-methyltransferase. The formation of GlcAD in Mesorhizobium depends on an α-glycosyltransferase because of the α-anomeric configuration of the sugar.
FIG 2.

Q-TOF MS/MS spectra of U1 identified as O-methyl-dihexosyl diacylglycerol (A) and U2 identified as monohexuronosyl diacylglycerol (B). Each spectrum shows one abundant representative molecular species. The ions were selected as ammonium adducts with the calculated masses 1,004.7240 u (U1) and 842.6353 u (U2) and fragmented. The fragments with m/z 631.5640 or 631.5630 represent DAG with 18:1-Me and/or 19:0c fatty acids as a protonated form after loss of H2O. 18:1-Me and 19:0c fatty acids are isobaric and thus not distinguishable with respect to their masses. The neutral loss of 373.1578 u (1,004.7218 u minus 631.5640 u) is derived from dihexose with an O-methyl group (as an ammonia adduct); the neutral loss of 211.0760 u (842.6393 u minus 631.5630 u) is derived from hexuronic acid (as an ammonia adduct). The ions m/z 353.3041 and 353.3017 represent MAG-18:1-Me and/or MAG-19:0c (each as a protonated form after loss of H2O). COO-R1 and COO-R2 represent 18:1-Me and/or 19:0c fatty acyl residues bound to the glycerol backbone. The head group structures of U1 and U2 were further determined by NMR, with U1 identified as a methylated form of DGD (Me-DGD) with the O-methyl group linked to the C-3 position of the innermost galactose and U2 as α-GlcAD, as illustrated in the figure.
Deletion of mlr2668 leads to the loss of GlcAD in Mesorhizobium.
The characterization of the deletion mutant of Agt (Δagt strain) may reveal a possible function of Agt in glycolipid biosynthesis in Mesorhizobium. Therefore, we disrupted the mlr2668 locus by insertional mutagenesis (confirmed by PCR [data not shown]) and analyzed the lipids of the wild type and the Δagt strain grown under phosphate starvation. Separation of the lipid extracts via two-dimensional TLC showed the lack of GlcAD in the mutant (Fig. 3B), while this lipid was present in the wild type (Fig. 3A). The absence of GlcAD in the Δagt strain suggests the involvement of Agt in the biosynthesis of this lipid, as shown for the homologous enzyme encoded by ORF atu2297 synthesizing GlcAD in Agrobacterium (6). All of the other glycolipids in Mesorhizobium (DGD, GGD, TGD, and Me-DGD) were unaffected by the disruption of mlr2668.
FIG 3.
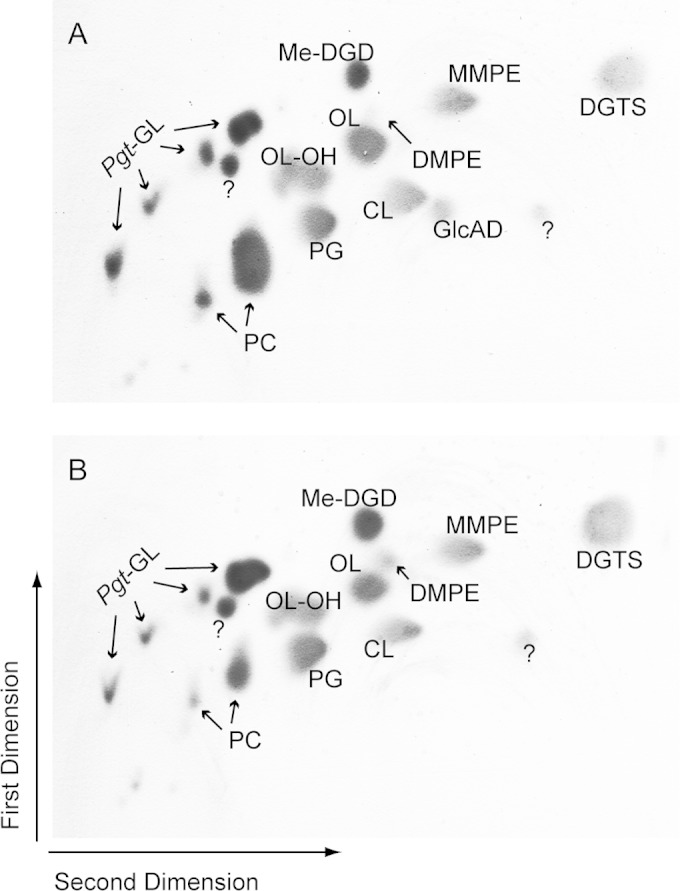
Two-dimensional TLC of lipid extracts from Mesorhizobium wild type (A) or the Δagt strain (B) grown under phosphate deprivation, with GlcAD absent in the mutant. MMPE, monomethyl-phosphatidylethanolamine; DMPE, dimethyl-phosphatidylethanolamine; PG, phosphatidylglycerol; CL, cardiolipin. Pgt-GL, glycolipids synthesized by Pgt-Ml; OL-OH (this study); question mark, unknown lipid.
Heterologous expression of Agt in Agrobacterium leads to the synthesis of GlcAD but not of MGlcD in detectable amounts.
To reveal further details about the role of Agt in glycolipid biosynthesis in Mesorhizobium, expression experiments were performed with Agt from Mesorhizobium and the MGlcD synthase from Acholeplasma (alMGS), which was shown to synthesize only MGlcD after expression in different hosts (9). Agrobacterium was chosen as an appropriate expression host for these two glycosyltransferases because this organism provides the metabolic equipment to synthesize both GlcAD and MGlcD (6). The enzymes were expressed in a glycolipid-free Agrobacterium double-knockout mutant with disruption of the Pgt-Atum locus and of atu2297 (6). The cells were grown under phosphate starvation. These conditions are necessary to enhance the accumulation of glycolipids in Agrobacterium (4). The lipids were analyzed via TLC and Q-TOF MS/MS. The cells expressing Agt from Mesorhizobium accumulate GlcAD when MGlcD is absent, as shown in Fig. 4 (see also Fig. S2 in the supplemental material). The line expressing alMGS from Acholeplasma accumulates MGlcD but lacks GlcAD. These experiments show that the metabolic equipment allows the synthesis of the two lipids in Agrobacterium. Therefore, the lack of MGlcD in Agrobacterium cells expressing Agt from Mesorhizobium may indicate a higher activity of Agt with respect to the synthesis of GlcAD toward MGlcD under the tested conditions.
FIG 4.
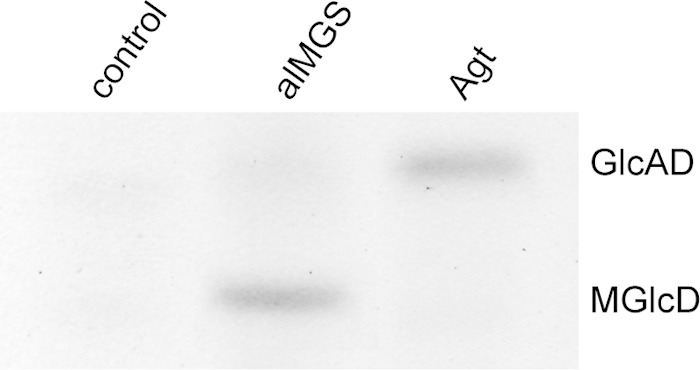
TLC showing the accumulation of MGlcD and GlcAD in a glycolipid-free Agrobacterium strain (6) as an expression host transformed with the empty vector, the MGlcD synthase alMGS from Acholeplasma, or Agt from Mesorhizobium. Expression of alMGS leads to the synthesis of MGlcD and that of Agt leads to the synthesis of GlcAD. The lipid extracts were separated on TLC plates pretreated with ammonium sulfate. This acidification leads to the protonation of GlcAD. Due to decreased polarity, GlcAD migrates above MGlcD. Glycolipids were stained with α-naphthol–H2SO4 at 137°C.
Enzyme assays with recombinant Agt protein confirm the synthesis of MGlcD and GlcAD by Agt with UDP-glucose and UDP-glucuronic acid as sugar donors.
Enzyme assays were performed with protein extracts prepared from E. coli cells expressing Agt or the empty vector as a control. UDP-glucose or UDP-glucuronic acid was used as a sugar donor, and DAG(18:2/18:2) was used as a sugar acceptor. Such DAG species are not detectable in E. coli. The formation of MGlcD and GlcAD, each with 18:2 fatty acids, was analyzed via Q-TOF MS/MS. We could detect MGlcD(18:2/18:2) and GlcAD(18:2/18:2) in the assays supplemented with the respective sugar donors (Fig. 5). In contrast to bacterial glycolipids with low desaturation degrees, these in vitro-synthesized glycolipids with higher desaturation degrees showed a slightly deviant fragmentation pattern, with two different fragment ions, both representing DAG. Ions with m/z 617.4981 (Fig. 5) are derived from protonated DAG(18:2/18:2), while ions with m/z 599.4812 represent protonated DAG(18:2/18:2) characterized by the loss of H2O. The higher desaturation degree in glycolipids obviously supports the enhanced generation of the higher mass fragment ions with m/z 617.4981 at the expense of ions with m/z 599.4812 (Fig. 5). The empty vector controls did not show any formation of the expected glycolipids. These results demonstrate that Agt is able to form MGlcD and GlcAD. Furthermore, the lack of MGlcD in Agt-expressing Agrobacterium cells or in Mesorhizobium indicates different substrate specificities of Agt for UDP-glucuronic acid and UDP-glucose.
FIG 5.
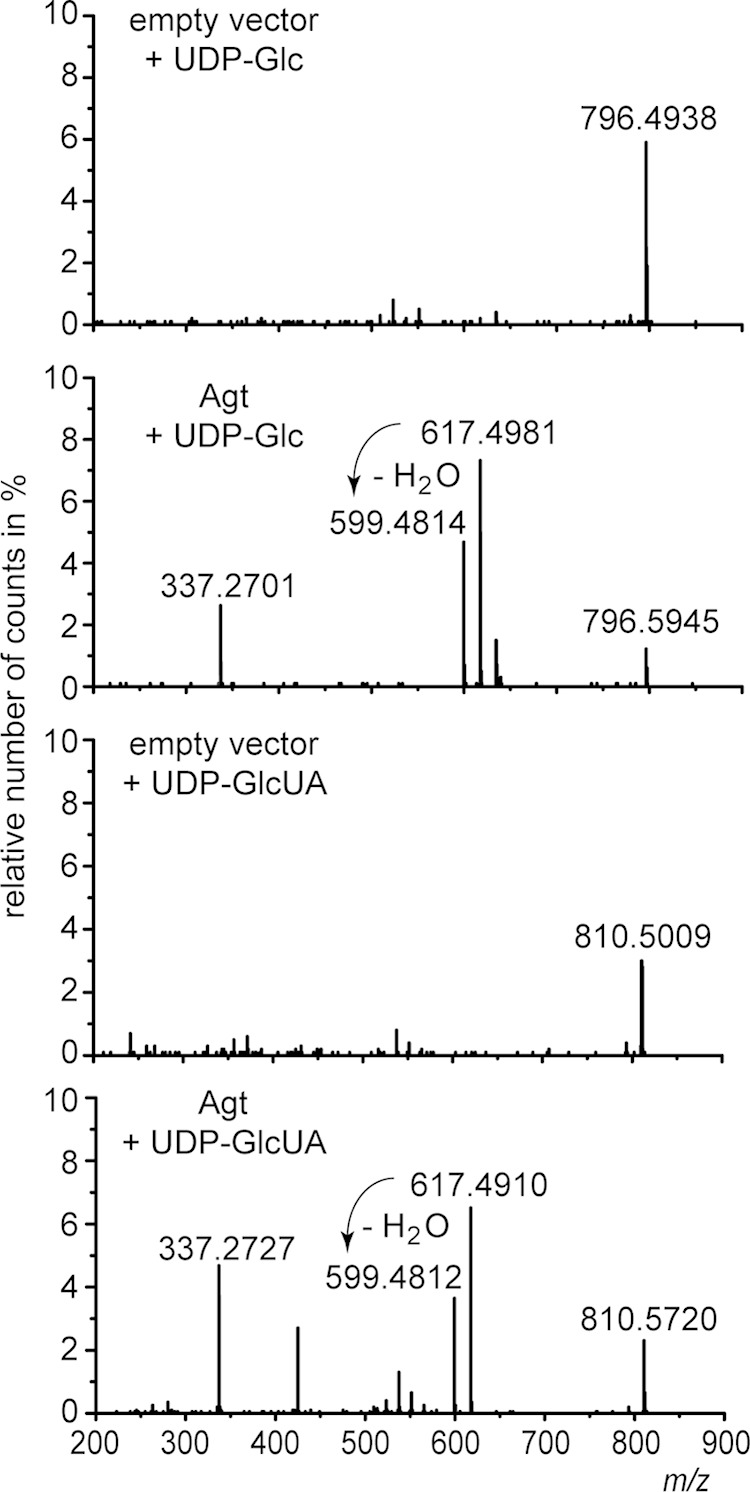
Q-TOF MS/MS spectra of MGlcD and GlcAD synthesized in enzyme assays with protein extracts from E. coli cells expressing Agt from Mesorhizobium or harboring the empty vector as a control. The assays were supplemented with DAG(18:2/18:2) as a sugar acceptor and UDP-glucose (UDP-Glc) or UDP-glucuronic acid (UDP-GlcUA) as sugar donors. Parental ions with m/z 796.5934 or 810.5727 representing ammonium adducts of MGlcD or GlcAD, respectively, were selected in the positive-ion mode. The fragment ions with m/z 617.4981 (or 617.4910) represent the protonated form DAG(18:2/18:2), while fragment ions with m/z 599.4812 (or 599.4814) represent the protonated form of DAG(18:2/18:2) after loss of H2O; fragment ions with m/z 337.2701 (or 337.2727) represent the protonated form of MAG(18:2) after loss of H2O. The respective fragments were absent in the spectra of the control assays. The neutral losses of 197.1131 u (796.5945 u minus 599.4814 u) or 211.0908 u (810.572 u minus 599.4812 u) represent the ammonia adducts derived from glucose or glucuronic acid, respectively. These spectra prove the formation of both MGlcD and GlcAD (by supplementation of UDP-Glc or UDP-GlcUA, respectively) by Agt.
Identification of novel ornithine lipids in Mesorhizobium.
Mesorhizobium is characterized by a very complex lipid composition when grown under phosphate starvation. Many of the lipids accumulating under these conditions were analyzed recently (7), but there are still some unidentified spots visible on TLC plates, as shown in Fig. 3. Therefore, we investigated an unknown lipid, which migrated slightly more slowly than OL; thus, this lipid was of a higher polarity. Analyses by Q-TOF MS/MS revealed a new hydroxylated form of OL (OL-OH) with the hydroxy group bound to the amide-linked fatty acid, as shown in Fig. 6.
FIG 6.
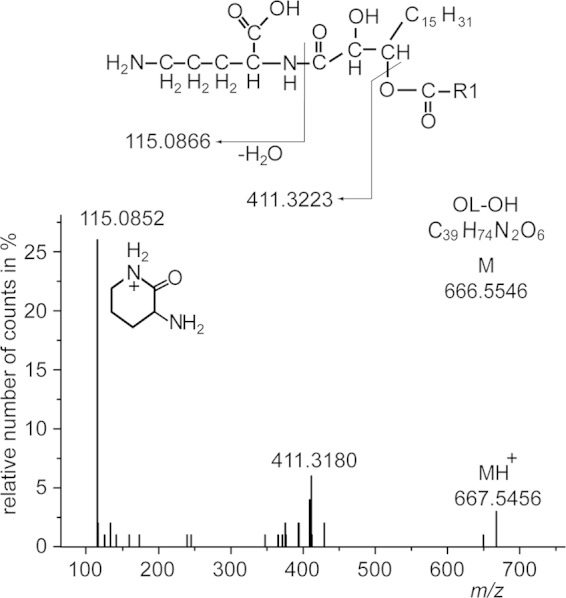
Q-TOF MS/MS spectrum of OL-OH analyzed in the positive-ion mode. The fragment pattern represents a main molecular OL-OH species with a 16:0 fatty acid esterified to the primary hydroxy group of an amide-bound dihydroxy-18:1 fatty acid. The exact position of the secondary hydroxy group cannot be determined by Q-TOF MS/MS. The protonated form of this species (parental ion) yields a mass of m/z 667.5456 (calculated mass is 667.5618 u). The neutral loss of 256.2276 u (667.5456 u minus 411.3180 u) is derived from 16:0 as an ester-linked fatty acid. The fragment ion m/z 411.3180 represents the lyso form of this ornithine lipid containing the additional hydroxy group linked to the amide-bound 18:1-hydroxy fatty acid. The low-mass fragment ion at m/z 115.0852 is consistent with the presence of ornithine in this lipid (45), forming a ring structure with loss of a water molecule (COO-R1 represents a 16:0 acyl chain).
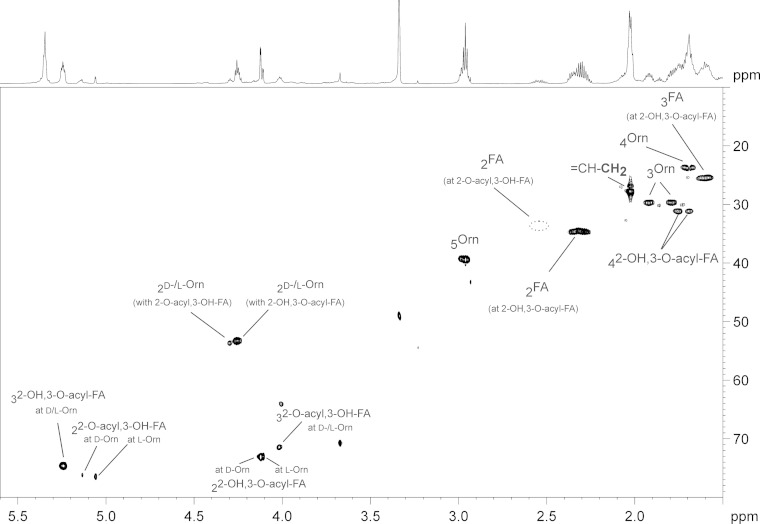
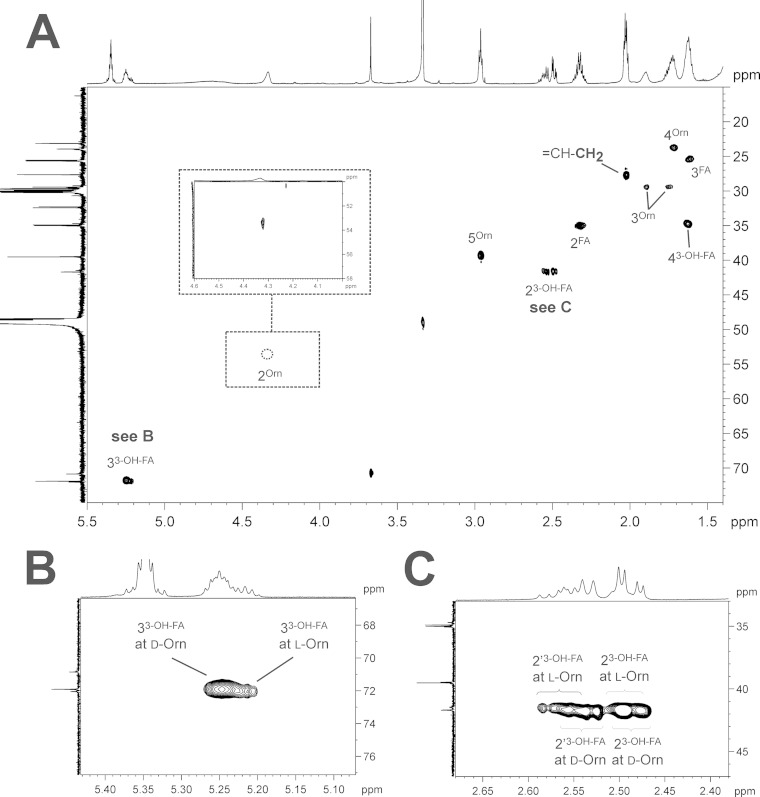
For positional determination of the hydroxy group, the mesorhizobial OL-OH lipid was subjected to NMR spectroscopy. The NMR analysis clearly revealed the presence of the additional hydroxy function at the C-2 position of the amide-bound fatty acid, thus indicating the presence of 2,3-dihydroxy fatty acids in mesorhizobial OL-OH. The presence of such types of fatty acids in bacteria has rarely been reported, with examples so far in bacterial galactosylceramides or sulfonolipids (21) or in lipopolysaccharides isolated from some Legionella strains (22, 23). Interestingly, the NMR spectra of OL-OH (Fig. 7; see also Fig. S3 and S4 in the supplemental material) contained four different sets of signals for this OL-OH. The complete 1H and 13C NMR chemical shift data are given in Table S3 in the supplemental material. The NMR analysis showed that the heterogeneity is caused in part by two different acylation variants. The major compounds were found to be OL-OH with an amide-linked 2-OH,3-O-acyl-dihydroxy fatty acid (Fig. 8C); the minor compounds are OL-OH containing an amide-linked 2-O-acyl,3-OH-dihydroxy fatty acid (Fig. 8B). Additional experiments indicated that the 2-O-acyl,3-OH species are most likely artifacts of acid-catalyzed acyl migration and do not result from a targeted biological transfer (see the supplemental material). Acyl migration is also described for glycerolipids (24). To determine whether the occurrence of isoforms is a common phenomenon in mesorhizobial ornithine lipids, we investigated the mesorhizobial OL (the most likely biosynthetic precursor of OL-OH) by NMR as well (its structure is shown in Fig. 8A). The respective 1H,13C-HSQC NMR is depicted in Fig. 9A; the complete 1H and 13C NMR chemical shift data are given in Table S4 in the supplemental material. As previously analyzed by mass spectrometry (data not shown), this OL consists of ornithine with an amide-bound 3-OH-fatty acid which is, in turn, esterified. Interestingly, in the HSQC spectrum two sets of signals for α- and β-protons (2-H′/2′-H′ and 3-H′) of the 3-OH-fatty acid could be observed as well (Fig. 9B and C), indicating that two isoforms of the OL are present. A detailed discussion of the NMR spectra of mesorhizobial OL and OL-OH can be found in the supplemental material.
FIG 7.
Section (δH 5.6 to 1.5; δC 80 to 10) of the 1H,13C-HSQC NMR spectrum (700 MHz) obtained from mesorhizobial OL-OH (acidic isolation procedure) including assignment of signals. Four different sets of signals were obtained due to the mixed presence of d- and l-ornithine as well as two different acylation species (2-O-acyl,3-OH and 2-OH,3-O-acyl) combined with each ornithine configuration. Orn, ornithine; FA, fatty acid.
FIG 8.
Chemical structures of the ornithine lipids isolated from M. loti, as indicated.
FIG 9.
(A) Section (δH 5.5 to 1.4; δC 75 to 15) of the 1H,13C-HSQC NMR spectrum (700 MHz) obtained from mesorhizobial OL including assignment of signals. The magnified regions for positions 3 (B) and 2 (C) of the 3-OH-fatty acid clearly indicate the presence of two sets of signals, caused by the mixed presence of d- and l-ornithine. Orn, ornithine; FA, fatty acid.
To clarify the reason for the two sets of signals in mesorhizobial OL as well as the two different sets of signals within one acylation variant of OL-OH, we determined the absolute configuration of ornithine by acidic butanolysis with (R)-2-butanol and subsequent GC-MS analysis. As a standard, we used commercially available l-ornithine (see Fig. S5 in the supplemental material); as a biological control we used the OL of R. tropici, isolated under conditions analogous to those used for the mesorhizobial OL. This analysis revealed that the two isoforms, indeed, are caused by the presence of both stereoconfigurations of ornithine. In OL and OL-OH of M. loti R7a we could detect 80 to 85% d-ornithine and 15 to 20% l-ornithine, respectively (see Fig. S6B and C in the supplemental material), whereas the OL from R. tropici contained only l-ornithine (see Fig. S6A). To the best of our knowledge, this is the first description of naturally occurring ornithine lipids containing d-ornithine. Integration of selected signals in 1H NMR spectra further corroborated the GC-MS results (see the supplemental material).
The first step in OL formation in different bacteria (such as Sinorhizobium meliloti) is catalyzed by the acyltransferase OlsB condensing ornithine with a fatty acid to form lyso-OL (25). OlsB from these organisms may be selective for l-ornithine, as suggested by the presence of l-ornithine-containing OL in R. tropici (see Fig. S6A in the supplemental material). We included OL from A. tumefaciens C58 in our studies to get more insights in the distribution of d-/l-ornithine lipids in different bacteria, with the result that A. tumefaciens was also found to synthesize exclusively l-ornithine-containing OL (see Fig. S7A in the supplemental material), catalyzed by the involvement of an agrobacterial OlsB homolog (26). As there are no genome sequence data available for the Mesorhizobium strain R7a used in this study, it is not clear whether this organism contains OlsB, which might be responsible for the synthesis of the d-ornithine-containing OL. Therefore, we analyzed OL from the strain M. loti MAFF303099, which contains the ORF mlr3216 coding for a putative OlsB. The analyses revealed, that similar to R7a, OL from the MAFF303099 strain contains mainly d-ornithine (∼93%) and, to a minor, extent l-ornithine (∼7%) (see Fig. S7C). These results may indicate that d-ornithine-containing OL is synthesized by the mesorhizobial OlsB homolog (mlr3216), exhibiting a regioselectivity for the synthesis of d-ornithine-containing OL. In addition, sequence alignments of different OlsB homologs (including A. tumefaciens, R. tropici, M. loti MAFF303099, S. meliloti, Rhodobacter sphaeroides, and Burkholderia cenocepacia) were performed to get any insights that might allow us to infer conclusions about the regioselectivity of the different homologs for d- or l-ornithine. However, these alignments revealed no obvious differences (see Fig. S8 in the supplemental material). Further detailed sequence analyses and investigations of the role of OlsB from Mesorhizobium in the formation of d-ornithine-containing OL will be part of a future study.
The hydroxylated OL in Mesorhizobium is synthesized by the hydroxylase OlsD.
Mesorhizobium contains the ORF mll1937 with homology of its deduced amino acid sequence to the hydroxylase OlsD from Burkholderia cenocepacia that introduces a hydroxy group to the amide-bound fatty acid of OL (15). To confirm that mll1937 codes for a functional OlsD, responsible for the synthesis of OL-OH in Mesorhizobium under phosphate deprivation, we generated a ΔolsD deletion mutant, and we further expressed mll1937 in A. tumefaciens C58 as host, providing OL as the substrate (4). The Mesorhizobium ΔolsD mutant and wild type (as positive control) were grown under phosphate deprivation, the condition required for OL-OH accumulation. The lipid extracts of the two cultures were analyzed via two-dimensional TLC. In contrast to the wild type (Fig. 3A), OL-OH was not detectable in the ΔolsD mutant (Fig. 10A), showing that mll1937 is required for OL-OH formation in Mesorhizobium. The Agrobacterium strain expressing mll1937 was grown under phosphate-replete conditions. The gene was expressed under the control of an inducible promoter; therefore, it does not require phosphate deprivation. Furthermore, the native agrobacterial OlsE activity, which is responsible for the hydroxylation of the ornithine head group of OL, is low under these conditions (4), avoiding a competitive situation for the substrate OL. Expression of mll1937 in Agrobacterium resulted in the formation of two novel lipid compounds separated via two-dimensional TLC (Fig. 10B). One of these lipids was identified with Q-TOF MS/MS as OL-OH (see Fig. S9A in the supplemental material) with the hydroxy group linked to the amide-bound fatty acid. The ion with m/z 681.5762 represents the main molecular OL-OH species (protonated form) with 19:0c as the ester-linked fatty acid, indicated by the neutral loss of 296.2717 u. The fragment with m/z 385.3045 corresponds to the lyso-form of OL-OH, where the amide-bound 16:0-hydroxy fatty acid carries the additional hydroxyl group. The fragment m/z 115.0856 represents the ornithine head group. The second lipid was identified as an ornithine lipid with two hydroxy groups [OL-(OH)2] with the ion m/z 697.5728 as the main molecular species (see Fig. S9B). The lyso form of this lipid contains two additional hydroxy groups as derived from the fragment ion m/z 401.2985. One of the hydroxy groups is linked to the amide-bound fatty acid (16:0 derivative), and the second one is linked to the ornithine head group, indicated by the fragment ion m/z 131.0806. The neutral loss of 296.2743 u represents 19:0c as an ester-linked fatty acid. These results confirm that the ORF mll1937 coding for a functional OlsD enzyme in Mesorhizobium is active under phosphate deprivation.
FIG 10.

Characterization of OlsD as the responsible enzyme for OL-OH formation in Mesorhizobium. (A) Two-dimensional TLC of a lipid extract from Mesorhizobium ΔolsD grown under phosphate deprivation lacking OL-OH. (B) Separated lipid extract from Agrobacterium as a host for heterologous expression of OlsD grown under phosphate-replete conditions with accumulation of two novel hydroxylated ornithine lipids, OL-OH and OL-(OH)2.
The loss of GlcAD in the Δagt mutant is compensated for by a strong accumulation of DGD/GGD.
The loss of GlcAD may affect the composition of the remaining lipids in the Δagt mutant. Therefore, we quantified the lipids of the Mesorhizobium wild type and the Δagt strain to reveal details about the relations between the different membrane lipids. The two lines were grown under phosphate-replete and -depleted conditions, and the lipids were quantified by measuring their fatty acid methyl (Me) esters via GC-MS. As expected, we observed no differences in the lipid patterns between the wild type and mutant grown under full nutrition (Fig. 11). Under phosphate starvation, the wild type and Δagt strain responded with the accumulation of different glycolipids, DGTS and OL-OH, accompanied by the reduction of the amounts of phospholipids (Fig. 11). OL-OH accumulated to about 12% both in the wild type and the mutant, while it was not detectable under conditions of full nutrition. Apart from OL-OH, the two lines show distinct differences in their responses. The loss of GlcAD in the Δagt strain affects different lipids, especially GGD/DGD and phosphatidylcholine (PC). The proportion of GGD/DGD is strongly increased from 6% in the wild type to 15% in the Δagt mutant. This strong increase in neutral glycolipids considerably exceeded the loss of GlcAD, which was about 2% in the wild type. Me-DGD and DGTS were only slightly increased in the mutant. On the other hand, the phospholipid PC was strongly decreased from 20% in the wild type to 8% in the Δagt strain. Phosphatidylglycerol (PG) was the only phospholipid which was increased, from 11% (wild type) to 14% (Δagt strain). The remaining membrane lipids were hardly affected by the loss of GlcAD. These data suggest an interconnection of GlcAD, DGD/GGD, PC, and possibly PG metabolism and a redistribution of the membrane lipids to compensate for the loss and a possible function of the charged glycolipid GlcAD.
FIG 11.
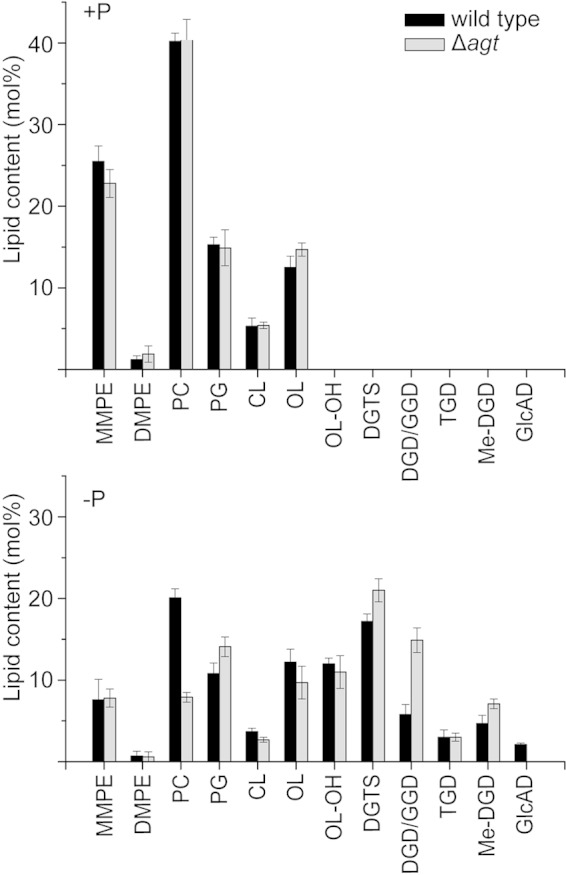
Lipid composition of Mesorhizobium wild type and the Δagt strain grown under phosphate-replete (+P) or -depleted (−P) conditions. The bars represent mean values ± standard deviations of three independent measurements. MMPE, monomethyl phosphatidyl ethanolamine; DMPE, dimethyl phosphatidyl ethanolamine; PG, phosphatidyl glycerol; CL, cardiolipin.
Agt is not required for growth under phosphate deprivation.
As shown in a previous study (7), the Pgt-Ml-dependent glycolipids (DGD, GGD, TGD, and Me-DGD) are not important for growth of Mesorhizobium under phosphate starvation. In this study, we analyzed the growth of the Δagt strain to reveal effects originating from the lack of GlcAD. Therefore, the wild-type and the mutant lines were grown in minimal medium with high (25 mM) or low (15 μM) phosphate. As expected, growth of the two lines was reduced under phosphate deprivation, but there was no difference between the wild type and Δagt mutant (Fig. 12). Therefore, these data suggest that GlcAD is not required for growth of Mesorhizobium under normal and phosphate-deficient conditions.
FIG 12.

Growth curves of Mesorhizobium wild type (wt) and the Δagt strain grown with 1.25 mM (+P) or 0.5 μM (−P) phosphate show no differences under the respective conditions. Mean values ± standard deviations were calculated from three independent measurements.
DISCUSSION
In a previous study Mesorhizobium was shown to accumulate a series of different glycolipids under phosphate starvation, with GGD, DGD, TGD, and U1 depending on Pgt-Ml activity, while the enzyme synthesizing U2 remained unknown (7). In this study, we could determine the head group structures of U1 (Me-DGD) and U2 (GlcAD), and we identified the responsible glycosyltransferase (encoded by the ORF mlr2668) synthesizing GlcAD in Mesorhizobium. Further, we describe for the first time novel ornithine lipids in Mesorhizobium with d-ornithine and hydroxylation at C-2 of the amide-bound fatty acid.
Me-DGD is a novel glycolipid which has never been described before for any organism. The synthesis of this lipid depends, on the one hand, on Pgt-Ml activity because this lipid is absent in the Pgt-Ml deletion mutant (7). On the other hand, the methyl group of this lipid is probably introduced by an unknown O-methyltransferase acting on the C-3 of galactose with (S)-adenosylmethionine as a likely methyl donor. Methyl groups on sugars are found mainly in lipopolysaccharides or in some other glycans of bacteria, but they have never been observed in bacterial glycolipids (27). Organisms of medical relevance, like mycobacteria or rhizobia, have been well investigated, but little information is available about the function of the methyl group in the glycans of these organisms or about methyltransferases acting on sugars. It is not even completely clear whether the methylation occurs on the glycan or on the sugar donor (27). The most probable scenario for Me-DGD synthesis in Mesorhizobium would be the methylation on the level of DGD. The presence of the methyl group leads to a reduced polarity of Me-DGD compared to DGD (7). Furthermore, methylation may have a strong effect on the head group hydration and the physicochemical properties of Me-DGD, as shown for a synthetic methyl-MGlcD (28). Therefore, reduced head group hydration of Me-DGD may lead to a slightly decreased ability to form bilayer structures in comparison to DGD, which is a bilayer-stabilizing glycolipid. Depending on the growth conditions, Acholeplasma can regulate its lipid composition by modification of the glycolipid head group. This organism contains MGlcD and DGlcD as main membrane lipids, with MGlcD as a non-bilayer- and DGD as bilayer-forming lipid. To regulate the ratio of bilayer-forming to non-bilayer-forming lipids, DGlcD can be converted into a non-bilayer-forming lipid by incorporation of a fatty acid into its head group in ester linkage (29). It is not known whether Mesorhizobium shows a similar response (i.e., by methylation of DGD), but it is possible that membrane properties are adjusted by changing the ratios of DGD and Me-DGD.
GlcAD is a rare glycolipid found in only a few bacterial species, such as in the cauliform bacteria (Caulobacterales) and relatives belonging to the Alphaproteobacteria (1). In addition to GlcAD, these organisms contain different glycolipids, such as MGlcD and SQD (30–33). These glycolipids represent mainly bulk membrane lipids, while a function as surrogate for phospholipids was reported for only a few species (34, 35). With respect to rhizobia, the anaerobic phototrophic bacterium Blastochloris viridis (Rhizobiales) was the only member accumulating GlcAD, which is synthesized independently of the phosphate supply (36). Therefore, Mesorhizobium is, besides Agrobacterium (4, 6), one of the first members of nonphototrophic Rhizobiales synthesizing GlcAD exclusively as a response to phosphate starvation. The accumulation of GlcAD was recently described for Arabidopsis thaliana, in which this lipid also plays a role under phosphate deprivation. The plant GlcAD is not synthesized by a novel glycosyltransferase, but its formation partly shares the pathway of SQD synthesis by involvement of the SQD synthase SQD2 probably transferring glucuronic acid onto DAG (37).
Although Agt is able to synthesize MGlcD (this study), this lipid has never been detected in Mesorhizobium. One explanation may be that its substrate specificity is much lower for UDP-glucose than for UDP-glucuronic acid under physiological conditions in Mesorhizobium or in Agrobacterium (used as expression host). The lack of MGlcD may also be explained by a greater accessibility of UDP-glucuronic acid to Agt or a higher concentration of UDP-glucuronic acid than UDP-glucose in Mesorhizobium cells. A further reason may be that the synthesis of MGlcD depends on conditions not tested in this study.
Glycolipids with one sugar in their head groups, like MGlcD, are non-bilayer-forming lipids (38), but due to its carboxy group, GlcAD is suggested to be a bilayer-forming lipid, as shown for the synthetic glucuronosyl dialkylglcerol (39). With a pK of about 5.5 determined for this synthetic glycolipid, GlcAD may become deprotonated with increasing pH and thus become a charged lipid at neutral pH. For a more detailed discussion about bilayer-forming properties of membrane lipids and a possible role of GlcAD, see Semeniuk et al. 6). Another important charged glycolipid is SQD, found in many phototrophic organisms (1). It also occurs in some nonphotosynthetic bacteria, especially in Gram-negative bacteria (1, 30, 31). With respect to rhizobia, different members contain either SQD (Rhizobium and Sinorhizobium) or GlcAD (Blastochloris and Agrobacterium) (1, 4, 6). Therefore, GlcAD in Mesorhizobium may play a role similar to that of SQD in other rhizobia.
A main function of SQD is the replacement of phospholipids, especially of PG, during phosphate deprivation (1). Because of its charged nature, GlcAD may be a counterpart of SQD, suggesting that the two lipids may have similar functions. The loss of GlcAD in the Δagt mutant (which is 2% in wild type) led to an increase in PG (11% in wild type; 14% in Δagt) (Fig. 11). This increase in the charged phospholipid PG during phosphate deprivation suggests that PG is required to compensate for loss of GlcAD and, furthermore, that GlcAD can serve as a surrogate for PG, as shown for SQD.
The deletion of mlr2668 has a strong effect on the lipid composition in the Δagt strain. The loss of small amounts of GlcAD seems to be overcompensated by accumulation of large amounts of GGD/DGD, accompanied by a strong reduction of PC. The loss of GlcAD may be further compensated for by increased amounts of the charged phospholipid PG. These data suggest an interconnection of GlcAD, DGD/GGD, PC, and PG and a compensation for the loss of a probable unknown function of the charged glycolipid GlcAD. A similar response was observed for Agrobacterium with deletion of atu2297, which is the homolog of mlr2668 (6). Moreover, the complete loss of all glycolipids in the glycolipid-free Agrobacterium double-knockout mutant (deletion of Pgt-Atum and atu2297) (6) led to a strong accumulation of DGTS. A similar response may be expected for a Mesorhizobium double-knockout mutant (deletion of Pgt-Ml and Agt), with DGTS compensating for the loss of glycolipids without further effect on growth under different conditions. The formation of Me-DGD seems not to be coupled to the levels of DGD as its precursor, probably due to different regulation mechanisms of Pgt-Ml and the unknown methyltransferase. As also shown for Sinorhizobium, the loss of one or two of the nonphosphorous lipids SQD, DGTS, or OL can be compensated for by increased accumulation of the remaining ones (4, 7, 40). Only the loss of DGTS and OL or the complete loss of all nonphosphorous lipids affects the growth and the plant-microbe interactions of Sinorhizobium. A mutual replacement of the different glycolipids may explain why the growth of the Δagt mutant is not affected under phosphate starvation compared to growth of the wild type. This ability may reflect the adaptability of these organisms to compete and survive in changing natural environments.
A glycolipid-free double-knockout mutant of Mesorhizobium may have no effects on growth and plant-microbe interactions, as shown for Agrobacterium (6). Therefore, only a triple knockout leading to the loss of all glycolipids and of DGTS may have consequences for the growth of Mesorhizobium and its symbiotic interactions with Lotus japonicus, as shown for Sinorhizobium (40). DGTS deletion mutants do not yet exist for Mesorhizobium. DGTS is formed in different organisms in two steps by involvement of the enzymes BtaA and BtaB (41), with homologs (mlr1574 and mlr1575, respectively) also found in Mesorhizobium. The mesorhizobial BtaA and BtaB homologs are not yet characterized, but a deletion of mlr1574 should abolish the complete DGTS biosynthesis pathway in Mesorhizobium. Moreover, a mutant reduced in the amount of PG or an Agt overexpression line accumulating GlcAD may give more insights into a possible interconnection of the phospholipid PG and GlcAD. This would mean that reduction of PG in the mutant should lead to the accumulation of GlcAD or that increased amounts of GlcAD by overexpression should lead to a reduction in PG.
The interconnection of GlcAD with different membrane lipids and its specific functions for growth under different conditions, for photosynthesis, and in plant-microbe interactions will be investigated in a later study.
In addition to glycolipids and DGTS, Mesorhizobium accumulates a new hydroxylated OL when grown under phosphate deprivation. The detection of this lipid in this study may be the result of changed culture conditions. Mesorhizobium was grown for 6 days under phosphate deprivation instead of 3 days as described earlier (7). Three genes are known to be responsible for hydroxylation of OL. OlsC and OlsE are found in Rhizobium and Agrobacterium hydroxylating the C-2 position of the ester-linked fatty acid and the ornithine head group, respectively (4, 16). A recently discovered hydroxylase, OlsD, forms OL-OH with the OH-group linked to the amide-bound fatty acid (15). This lipid, described for the first time for the pathogen Burkholderia cenocepacia (Betaproteobacteria) (15), may be the equivalent to OL-OH from Mesorhizobium (Alphaproteobacteria). However, the position of the hydroxy group was not determined. Therefore, this is the first report proving the position of the additional hydroxy group to be linked to the C-2 of the amide-bound fatty acid. Furthermore, Mesorhizobium is the first member of rhizobia containing this form of OL-OH. In contrast to Burkholderia, in which the hydroxylated form of OL is hardly synthesized (<1%) under acidic stress conditions, OL-OH accumulated in Mesorhizobium in large amounts (12%) as a response to phosphate deprivation (Fig. 11). Finally, our data suggest that OL-OH in Mesorhizobium serves as a surrogate for phospholipids under these conditions. In addition, hydroxylated forms of OL play a role in the response to other stress conditions (low pH or heat stress) in Rhizobium, where these lipids are also important for symbiosis (16, 42). The function of hydroxylated OL as a stress lipid was also speculated for Agrobacterium (4). Therefore, further functions of OL-OH in Mesorhizobium cannot be excluded, especially in symbiosis with Lotus japonicus. Even more interesting, our structural analysis showed that the ornithine lipids of Mesorhizobium comprise a mixture of d- and l-ornithine in the approximate ratio of 4:1 (or 9:1 in MAFF303099). As far as we know, this is the first description of naturally occurring ornithine lipids containing d-ornithine, while different bacteria investigated so far synthesize OL only with l-ornithine (43, 44). The accumulation of d-ornithine-containing OL may be explained by a higher specificity of the mesorhizobial OlsB for d- than for l-ornithine. However, it may not be excluded that the synthesis of d- or l-ornithine-containing OL might be regulated also on a substrate availability level adjusted to a certain ratio of d- and l-ornithine in the different organisms. It is also possible that d-ornithine-containing OL is synthesized by an unknown acyltransferase.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Ho 3870/1-3 to G.H.).
We thank Katharina Jakob and Birte Buske (Research Center Borstel [RCB]) for excellent technical assistance, Heiko Käßner (RCB) for NMR recordings, and Otto Holst (RCB) for discussion and critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02004-14.
REFERENCES
- 1.Hölzl G, Dörmann P. 2007. Structure and function of glycoglycerolipids in plants and bacteria. Prog Lipid Res 46:225–243. doi: 10.1016/j.plipres.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Weissenmayer B, Geiger O, Benning C. 2000. Disruption of a gene essential for sulfoquinovosyldiacylglycerol biosynthesis in Sinorhizobium meliloti has no detectable effect on root nodule symbiosis. Mol Plant Microbe Interact 13:666–672. doi: 10.1094/MPMI.2000.13.6.666. [DOI] [PubMed] [Google Scholar]
- 3.Cedergren RA, Hollingsworth RI. 1994. Occurrence of sulfoquinovosyl diacylglycerol in some members of the family Rhizobiaceae. J Lipid Res 35:1452–1461. [PubMed] [Google Scholar]
- 4.Geske T, vom Dorp K, Dörmann P, Hölzl G. 2013. Accumulation of glycolipids and other non-phosphorous lipids in Agrobacterium tumefaciens grown under phosphate deprivation. Glycobiology 23:69–80. doi: 10.1093/glycob/cws124. [DOI] [PubMed] [Google Scholar]
- 5.Hölzl G, Leipelt M, Ott C, Zähringer U, Lindner B, Warnecke D, Heinz E. 2005. Processive lipid galactosyl/glucosyltransferases from Agrobacterium tumefaciens and Mesorhizobium loti display multiple specificities. Glycobiology 15:874–886. doi: 10.1093/glycob/cwi066. [DOI] [PubMed] [Google Scholar]
- 6.Semeniuk A, Sohlenkamp C, Duda K, Hölzl G. 2014. A bifunctional glycosyltransferase from Agrobacterium tumefaciens synthesizes monoglucosyl and glucuronosyl diacylglycerol under phosphate deprivation. J Biol Chem 289:10104–10114. doi: 10.1074/jbc.M113.519298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devers EA, Wewer V, Dombrink I, Dörmann P, Hölzl G. 2011. A processive glycosyltransferase involved in glycolipid synthesis during phosphate deprivation in Mesorhizobium loti. J Bacteriol 193:1377–1384. doi: 10.1128/JB.00768-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edman M, Berg S, Storm P, Wikström M, Vikström S, Öhman A, Wieslander Å. 2003. Structural features of glycosyltransferases synthesizing major bilayer and nonbilayer-prone membrane lipids in Acholeplasma laidlawii and Streptococcus pneumoniae. J Biol Chem 278:8420–8428. doi: 10.1074/jbc.M211492200. [DOI] [PubMed] [Google Scholar]
- 9.Berg S, Edman M, Li L, Wikström M, Wieslander Å. 2001. Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J Biol Chem 276:22056–22063. doi: 10.1074/jbc.M102576200. [DOI] [PubMed] [Google Scholar]
- 10.Campbell JA, Davies GJ, Bulone V, Henrissat B. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J 326:929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coutinho PM, Deleury E, Davies GJ, Henrissat B. 2003. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol 328:307–317. doi: 10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- 12.Jorasch P, Warnecke DC, Lindner B, Zähringer U, Heinz E. 2000. Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycoglycerolipids, glycophospholipids, glycosphingolipids and glycosylsterols. Eur J Biochem 267:3770–3783. doi: 10.1046/j.1432-1327.2000.01414.x. [DOI] [PubMed] [Google Scholar]
- 13.Jorasch P, Wolter FP, Zähringer U, Heinz E. 1998. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol Microbiol 29:419–430. doi: 10.1046/j.1365-2958.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- 14.Geiger O, González-Silva N, López-Lara IM, Sohlenkamp C. 2010. Amino acid-containing membrane lipids in bacteria. Prog Lipid Res 49:46–60. doi: 10.1016/j.plipres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 15.González-Silva N, López-Lara IM, Reyes-Lamothe R, Taylor AM, Sumpton D, Thomas-Oates J, Geiger O. 2011. The dioxygenase-encoding olsD gene from Burkholderia cenocepacia causes the hydroxylation of the amide-linked fatty acyl moiety of ornithine-containing membrane lipids. Biochemistry 50:6396–6408. doi: 10.1021/bi200706v. [DOI] [PubMed] [Google Scholar]
- 16.Vences-Guzmán MÁ Guan Z, Ormeño-Orrillo E, González-Silva N, López-Lara IM, Martínez-Romero E, Geiger O, Sohlenkamp C. 2011. Hydroxylated ornithine lipids increase stress tolerance in Rhizobium tropici CIAT899. Mol Microbiol 79:1496–1514. doi: 10.1111/j.1365-2958.2011.07535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko T, Nakamura Y, Sato S, Asamizu E, Kato T, Sasamoto S, Watanabe A, Idesawa K, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Mochizuki Y, Nakayama S, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res 7:331–338. doi: 10.1093/dnares/7.6.331. [DOI] [PubMed] [Google Scholar]
- 18.Gerwig GJ, Kamerling JP, Vliegenthart JF. 1979. Determination of the absolute configuration of mono-saccharides in complex carbohydrates by capillary G.L.C. Carbohydr Res 77:10–17. [DOI] [PubMed] [Google Scholar]
- 19.Hölzl G, Zähringer U, Warnecke D, Heinz E. 2005. Glycoengineering of cyanobacterial thylakoid membranes for future studies on the role of glycolipids in photosynthesis. Plant Cell Physiol 46:1766–1778. doi: 10.1093/pcp/pci189. [DOI] [PubMed] [Google Scholar]
- 20.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayberry WR. 1981. Dihydroxy and monohydroxy fatty acids in Legionella pneumophila. J Bacteriol 147:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonesson A, Jantzen E, Bryn K, Tangen T, Eng Zähringer JU. 1994. Composition of 2,3-dihydroxy fatty acid-containing lipopolysaccharides from Legionella israelensis, Legionella maceachernii and Legionella micdadei. Microbiology 140:1261–1271. doi: 10.1099/00221287-140-6-1261. [DOI] [PubMed] [Google Scholar]
- 24.Kodali DR, Tercyak A, Fahey DA, Small DM. 1990. Acyl migration in 1,2-dipalmitoyl-sn-glycerol. Chem Phys Lipids 52:163–170. doi: 10.1016/0009-3084(90)90111-4. [DOI] [PubMed] [Google Scholar]
- 25.Gao JL, Weissenmayer B, Taylor AM, Thomas-Oates J, López-Lara IM, Geiger O. 2004. Identification of a gene required for the formation of lyso-ornithine lipid, an intermediate in the biosynthesis of ornithine-containing lipids. Mol Microbiol 53:1757–1770. doi: 10.1111/j.1365-2958.2004.04240.x. [DOI] [PubMed] [Google Scholar]
- 26.Vences-Guzmán MÁ Guan Z, Bermúdez-Barrientos JR, Geiger O, Sohlenkamp C. 2013. Agrobacteria lacking ornithine lipids induce more rapid tumour formation. Environ Microbiol 15:895–906. doi: 10.1111/j.1462-2920.2012.02867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Staudacher E. 2012. Methylation—an uncommon modification of glycans. Biol Chem 393:675–685. doi: 10.1515/hsz-2012-013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trouard TP, Mannock DA, Lindblom G, Rilfors L, Akiyama M, McElhaney RN. 1994. Thermotropic phase properties of 1,2-di-O-tetradecyl-3-O-(3-O-methyl-β-d-glucopyranosyl)-sn-glycerol. Biophys J 67:1090–1100. doi: 10.1016/S0006-3495(94)80574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindblom G, Hauksson JB, Rilfors L, Bergenståhl B, Wieslander Å Eriksson PO. 1993. Membrane lipid regulation in Acholeplasma laidlawii grown with saturated fatty acids. Biosynthesis of a triacylglucolipid forming reversed micelles. J Biol Chem 268:16198–16207. [PubMed] [Google Scholar]
- 30.Abraham WR, Strömpl C, Meyer H, Lindholst S, Moore ER, Christ R, Vancanneyt M, Tindall BJ, Bennasar A, Smit J, Tesar M. 1999. Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int J Syst Bacteriol 49:1053–1073. [DOI] [PubMed] [Google Scholar]
- 31.Abraham WR, Strömpl C, Vancanneyt M, Bennasar A, Swings J, Lünsdorf H, Smit J, Moore ER. 2004. Woodsholea maritima gen. nov., sp. nov., a marine bacterium with a low diversity of polar lipids. Int J Syst Evol Microbiol 54:1227–1234. doi: 10.1099/ijs.0.02943-0. [DOI] [PubMed] [Google Scholar]
- 32.Batrakov SG, Nikitin DI, Pitryuk IA. 1996. A novel glycolipid, 1,2-diacyl-3-alpha-d-glucuronopyranosyl-sn-glycerol taurineamide, from the budding seawater bacterium Hyphomonas jannaschiana. Biochim Biophys Acta 1302:167–176. doi: 10.1016/0005-2760(96)00060-4. [DOI] [PubMed] [Google Scholar]
- 33.Batrakov SG, Nikitin DI, Sheichenko VI, Ruzhitsky AO. 1997. Unusual lipid composition of the gram-negative, freshwater, stalked bacterium Caulobacter bacteroides NP-105. Biochim Biophys Acta 1347:127–139. doi: 10.1016/S0005-2760(97)00060-X. [DOI] [PubMed] [Google Scholar]
- 34.Segers P, Vancanneyt M, Pot B, Torck U, Hoste B, Dewettinck D, Falsen E, Kersters K, De Vos P. 1994. Classification of Pseudomonas diminuta Leifson and Hugh 1954 and Pseudomonas vesicularis Busing, Doll, and Freytag 1953 in Brevundimonas gen. nov. as Brevundimonas diminuta comb nov and Brevundimonas vesicularis comb nov, respectively. Int J Syst Bacteriol 44:499–510. doi: 10.1099/00207713-44-3-499. [DOI] [PubMed] [Google Scholar]
- 35.Minnikin DE, Abdolrahimzadeh H, Baddiley J. 1974. Replacement of acidic phospholipids by acidic glycolipids in Pseudomonas diminuta. Nature 249:268–269. doi: 10.1038/249268a0. [DOI] [PubMed] [Google Scholar]
- 36.Linscheid M, Diehl BW, Övermöhle M, Riedl I, Heinz E. 1997. Membrane lipids of Rhodopseudomonas viridis. Biochim Biophys Acta 1347:151–163. doi: 10.1016/S0005-2760(97)00065-9. [DOI] [PubMed] [Google Scholar]
- 37.Okazaki Y, Otsuki H, Narisawa T, Kobayashi M, Sawai S, Kamide Y, Kusano M, Aoki T, Hirai MY, Saito K. 2013. A new class of plant lipid is essential for protection against phosphorus depletion. Nat Commun 4:1510. doi: 10.1038/ncomms2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wikström M, Xie J, Bogdanov M, Mileykovskaya E, Heacock P, Wieslander Å Dowhan W. 2004. Monoglucosyldiacylglycerol, a foreign lipid, can substitute for phosphatidylethanolamine in essential membrane-associated functions in Escherichia coli. J Biol Chem 279:10484–10493. doi: 10.1074/jbc.M310183200. [DOI] [PubMed] [Google Scholar]
- 39.Koynova RD, Tenchov BG, Kuttenreich H, Hinz HJ. 1993. Structure and phase behavior of a charged glycolipid (1,2-O-dialkyl-3-O-β-d-glucuronosyl-sn-glycerol). Biochemistry 32:12437–12445. doi: 10.1021/bi00097a023. [DOI] [PubMed] [Google Scholar]
- 40.López-Lara IM, Gao JL, Soto MJ, Solares-Pérez A, Weissenmayer B, Sohlenkamp C, Verroios GP, Thomas-Oates J, Geiger O. 2005. Phosphorus-free membrane lipids of Sinorhizobium meliloti are not required for the symbiosis with alfalfa but contribute to increased cell yields under phosphorus-limiting conditions of growth. Mol Plant Microbe Interact 18:973–982. doi: 10.1094/MPMI-18-0973. [DOI] [PubMed] [Google Scholar]
- 41.Riekhof WR, Andre C, Benning C. 2005. Two enzymes, BtaA and BtaB, are sufficient for betaine lipid biosynthesis in bacteria. Arch Biochem Biophys 441:96–105. doi: 10.1016/j.abb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Rojas-Jiménez K, Sohlenkamp C, Geiger O, Martínez-Romero E, Werner D, Vinuesa P. 2005. A ClC chloride channel homolog and ornithine-containing membrane lipids of Rhizobium tropici CIAT899 are involved in symbiotic efficiency and acid tolerance. Mol Plant Microbe Interact 18:1175–1185. doi: 10.1094/MPMI-18-1175. [DOI] [PubMed] [Google Scholar]
- 43.Lanéelle MA, Promé D, Lanéelle G, Promé JC. 1990. Ornithine lipid of Mycobacterium tuberculosis: its distribution in some slow- and fast-growing mycobacteria. J Gen Microbiol 136:773–778. doi: 10.1099/00221287-136-4-773. [DOI] [PubMed] [Google Scholar]
- 44.Batrakov SG, Shub MM, Rozynov BV, Bergel'son LD. 1974. Ornithine-containing lipid from Actinomyces globisporus. Chem Nat Compd 10:1–7. doi: 10.1007/BF00568208. [DOI] [Google Scholar]
- 45.Zhang X, Ferguson-Miller SM, Reid GE. 2009. Characterization of ornithine and glutamine lipids extracted from cell membranes of Rhodobacter sphaeroides. J Am Soc Mass Spectrom 20:198–212. doi: 10.1016/j.jasms.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.